This work reports the functional analysis of a Bsister gene in monocots. Rice MADS29 is highly expressed in reproductive stages and affects early seed development and grain filling by inducing maternal tissue degradation.
Abstract
The MADS box transcription factors are critical regulators of rice (Oryza sativa) reproductive development. Here, we here report the functional characterization of a rice MADS box family member, MADS29, which is preferentially expressed in the nucellus and the nucellar projection. Suppressed expression of MADS29 resulted in abnormal seed development; the seeds were shrunken, displayed a low grain-filling rate and suppressed starch biosynthesis, and contained abnormal starch granules. Detailed analysis indicated that the abnormal seed development is due to defective programmed cell death (PCD) of the nucellus and nucellar projection, which was confirmed by a TUNEL assay and transcriptome analysis. Further studies showed that expression of MADS29 is induced by auxin and MADS29 protein binds directly to the putative promoter regions of genes that encode a Cys protease and nucleotide binding site–Leu-rich repeat proteins, thereby stimulating the PCD. This study identifies MADS29 as a key regulator of early rice seed development by regulating the PCD of maternal tissues. It provides informative clues to elucidate the regulatory mechanism of maternal tissue degradation after fertilization and to facilitate the studies of endosperm development and seed filling.
INTRODUCTION
The MADS box transcription factor (TF) family is characterized by the presence of a MADS box DNA binding domain in the N-terminal region (Shore and Sharrocks, 1995). The plant-specific MIKCc-type MADS box TFs contain three additional domains, the I region, K domain, and C-terminal region. The K domain is involved in protein–protein interaction, and the C-terminal region is predicted to be important for transcriptional activation (Cho et al., 1999; Egea-Cortines et al., 1999; Yang et al., 2003). Seventy-five MADS box genes were identified in the rice (Oryza sativa) genome and 38 of them are MIKCc-type TFs (Arora et al., 2007).
Studies showed that the MADS box TFs are the most important regulators in floral organ development and the main members of the ABC model (Lohmann and Weigel, 2002; Kater et al., 2006; Yamaguchi and Hirano, 2006). Bsister genes are the closest relatives of B genes and they may originate from an ancestral gene 400 to 300 million years ago according to database searches and phylogenetic analyses (Becker et al., 2002). Functional analyses showed that an Arabidopsis thaliana Bsister clade member TRANSPARENT TESTA (TT16) is required for proanthocyanidin accumulation in the endothelium (inner integument) of the seed coat (Nesi et al., 2002). FLORAL BINDING PROTEIN24 (FBP24), the ortholog of TT16 in petunia (Petunia hybrida), is highly expressed in the endothelium of ovule. Unlike the reduced-coloration seeds of tt16, the fbp24 knockdown line produces a few seeds, which also lack the endothelial layer, similar to those of tt16 (de Folter et al., 2006). Wheat (Triticum aestivum) Bsister is highly expressed in endothelium, but its function is still unknown (Yamada et al., 2009). Loss of function of another Arabidopsis Bsister clade member GORDITA (GOA/AGL63) results in larger fruits due to stimulated cell expansion, whereas overexpression of GOA results in the smaller flower organs and shorter fruits (Erdmann et al., 2010; Prasad et al., 2010). In rice, MADS29, MADS30, and MADS31 are three members of Bsister genes (Arora et al., 2007); however, the detailed expression pattern and functional data for them are scarce.
The flower morphology of rice is distinct from that of Arabidopsis. The rice flower consists of a lemma, palea, two lodicules, six stamens, and one carpel, and there is only a single ovule differentiated from the floral meristem (Yamaki et al., 2011). After flowering, the pollen tube conveys the sperm to the egg and the central cell in the embryo sac (known as double fertilization; Russell, 1993). When fertilization is completed, the nucellus cells undergo degenerative processes (Krishnan and Dayanandan, 2003) that are recognized as programmed cell death (PCD). In barley (Hordeum vulgare) grains, PCD also occurs in the nucellar projection cells (Dominguez et al., 2001).
The nucellar projection is a part of the nucellar tissue that faces the vascular tissue and has a morphology characteristic of transfer cells (Lombardi et al., 2007; Sreenivasulu et al., 2010). In the degenerating nucellus of Sechium edule, NO and H2O2 are produced, and exogenous NO and H2O2 are able to induce caspase-like activity in the nucellus (Lombardi et al., 2010). However, other signals that trigger the PCD process in nucellar projection cells are still unknown.
The regulatory mechanism of the PCD in the maternal tissue is still unclear. In barley, an analysis of the genes that are specifically expressed in the maternal tissues revealed that an aspartic protease-like protein is specifically expressed in the nucellus during nucellus degradation (Chen and Foolad, 1997), and JEKYLL is involved in the degradation process of the nucellar projection (Radchuk, 2006). In addition, the γ-VPE–type Cys proteases and a Ser protease are highly expressed in the nucellus (Sreenivasulu et al., 2006). However, only little is known about the upstream regulatory factors of PCD in the maternal tissue.
Caspases are essential for apoptosis and autophagy in animals (Rupinder et al., 2007). Although orthologs of caspases are absent in plants (Bonneau et al., 2008), some caspase-like proteases have been found using specific cleavage of the caspase substrates and typical caspase inhibitors (Grudkowska and Zagdanska, 2004; Bonneau et al., 2008). Cys proteases form a superfamily that is associated with PCD and display a caspase-like activity. The VPE proteins that belong to the legumain subfamily of Cys proteases display caspase-1–like activity and are involved in various PCD processes (Hatsugai et al., 2004; Lam, 2005). Cys proteases of the papain family are required for developmental PCD processes, including the senescence of leaves, flowers, and seeds (Tanaka et al., 1993; Hara-Nishimura et al., 2001; Eason et al., 2005). Two papain family members of rice, CP1 and Cys-EP, are necessary for the degeneration of tapetum and germinating endosperm (Schmid et al., 1999; Lee et al., 2004).
Previous studies indicated that most of the proteins involved in nucellus PCD processes are proteases; however, how they regulate the nucellus degradation, and, hence, the seed development remains unclear. Here, we report the functional characterization of a rice Bsister clade gene, MADS29 (Os02g07430), in the degradation of the nucellus and nucellar projection by regulating PCD. Suppression of MADS29 resulted in shrunken seeds due to the defective degradation of the nucellus and nucellar projection. Further studies showed that MADS29 regulates the degradation of the nucellus and nucellar projection after fertilization by promoting the expression of a Cys protease and PCD-related genes, which is achieved through direct binding to the promoter regions of these genes.
RESULTS
MADS29 Is Preferentially Expressed in Reproductive Tissues, Especially in the Nucellus and Nucellar Projection
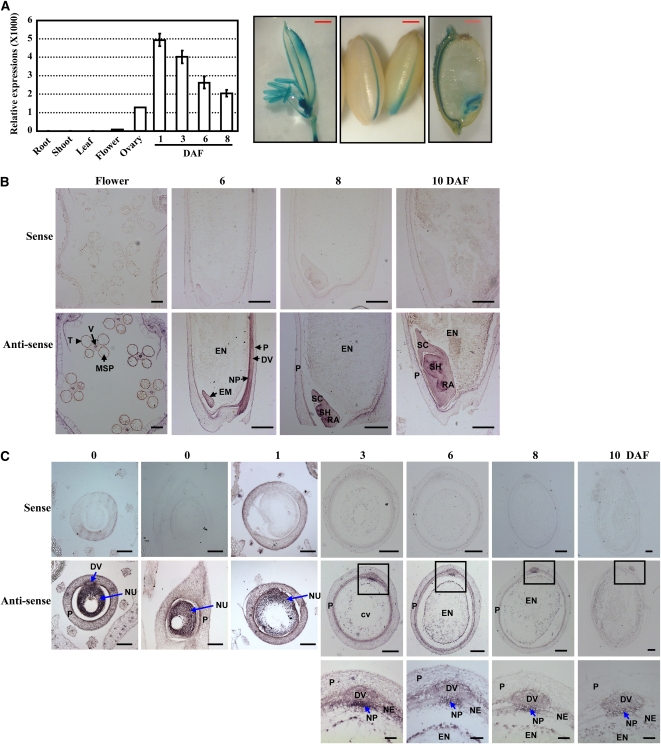
Our preliminary studies by microarray hybridization showed rice MADS29 is preferentially expressed in the ovaries and seeds but not in the vegetative tissues (Xue et al., 2009). Meanwhile, previous studies showed that MADS29 is highly expressed in the early stages of seed development (Lee et al., 2003; Arora et al., 2007). To investigate the expression profile of MADS29 further, quantitative RT-PCR (qRT-PCR) analyses were performed and results showed that MADS29 is highly expressed in the flower and developing seed, especially after fertilization, but is not detectable in the vegetative tissues, such as the roots, shoots, and leaves (Figure 1A, left panel). Promoter-β-glucuronidase (GUS) fusion studies analyzing at least five independent transgenic lines further revealed that MADS29 is expressed in the anther, ovary, seed, and embryo (Figure 1A, right panel). These results suggest that MADS29 may play a role in the early seed development.
Figure 1.
MADS29 Is Highly Expressed in the Nucellus and Nucellar Projection.
(A) qRT-PCR analysis of MADS29 expression in various tissues (left panel). Twenty-day-old plants were used to harvest shoots, leaves, and roots. The flowers and ovaries were harvested from plants after heading and before flowering. The data are presented as mean ± se (n = 3). Promoter-GUS fusion studies revealed the expression of MADS29 in flower, seed, and embryo at 20 DAF (right panel, bars = 1 mm).
(B) Spatial and temporal expression of MADS29 in flower and embryo. The transverse section of vacuolated pollen stage flower and the longitudinal sections of embryo at 6, 8, and 10 DAF were used for RNA in situ hybridization analysis. The expression of MADS29 in the tapetum, vascular bundle, and embryo is shown. Bars = 100 μm.
(C) Spatial and temporal expression analyses of MADS29 during rice seed development. The transverse section and longitudinal section of the ovary and the transverse sections of seeds at 1, 3, 6, 8, and 10 DAF were used for the RNA in situ hybridization analysis, revealing the strong expression of MADS29 in the nucellus and nucellar projection. The high expression level of MADS29 in the nucellar projection (squared regions) is enlarged (bottom panel). Bars = 200 μm (top and middle panels, 3, 6, 8, and 10 DAF), 100 μm (top and middle panels, 0 and 1 DAF), or 50 μm (bottom panel).
For (B) and (C), DV, dorsal vascular bundle; EM, embryo; EN, endosperm; MSP, microspore; NE, nucellar epidermis; NP, nucellar projection; NU, nucellus; P, pericarp; RA, radicle; SC, scutellum; SH, shoot; T, tapetum; V, vascular bundle.
Considering that the putative promoter used for promoter-GUS fusion studies might miss some important cis-elements, RNA in situ hybridization analysis was performed with flower, ovary, and seed sections to determine more precisely the spatial and temporal patterns of MADS29 expression. The results revealed the specific expression of MADS29 in the nucellus and nucellar projection at the early stages of seed development. In the unfertilized flowers, the hybridization signals of MADS29 are detected in the vascular bundle and tapetum of anther, especially highly in the nucellus (Figures 1B and 1C). After fertilization, the nucellar cells begin to degrade and the endosperm cells start to accumulate; the hybridization signals are still strong in nucellar cells at 1 d after flowering (DAF). Following the degradation of the nucellar cells at 3 DAF, the MADS29 transcript is highly expressed in nucellar projection cells and vasculature especially in nucellar projection, while the expression in the epidermis, integument, and endosperm is very low. In accordance with the time frame of seed development, the MADS29 transcript is highly expressed in the nucellar projection cells at 6 and 8 DAF, while no detectable signal in the endosperm cells (Figure 1C).
The hybridization signal is weak in the nucellar projection at 8 DAF compared with 3 DAF, which is consistent with the qRT-PCR analysis. In addition, MADS29 is expressed throughout the embryo development (Figure 1B).
Suppressed Expression of MADS29 Results in the Shrunken Seeds and a Reduced Grain-Filling Rate
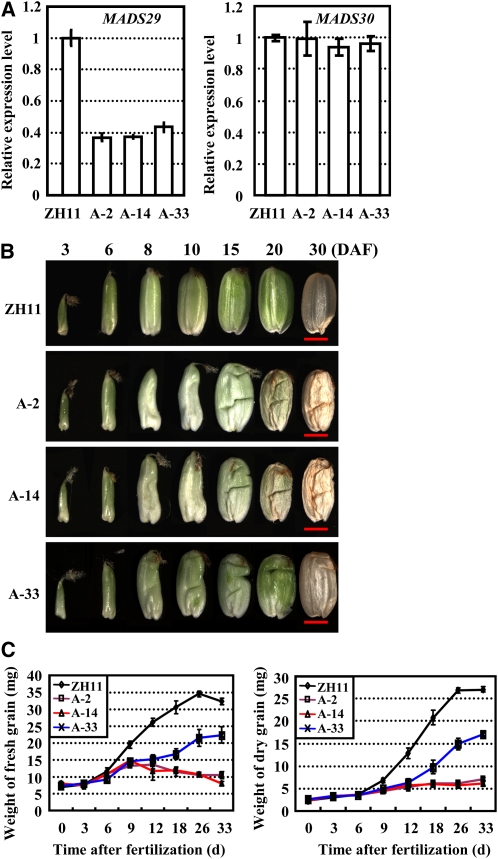
To study the physiological function of MADS29, we generated the Ubi:MADS29 binary antisense construct expressing a nonconserved region of MADS29 cDNA and transformed the construct into rice (Zhonghua 11 [ZH11]). More than 90 independent transgenic lines (A-MADS29) were obtained and most (>90%) of the transgenic plants with significant reduced levels of endogenous MADS29 expression (generally more than 60% reduction) developed aborted shrunken seeds (see Supplemental Figure 1 online). Three transgenic lines with a relatively weak suppression of MADS29 (A-2, A-14, and A-33; Figure 2A, left panel) and the corresponding confirmed lines after cultivation for two generations were used for further analysis (homozygous plants of A-33 were used). Determination of the copy number of transgenes in these A-MADS29 lines by DNA gel blot analysis revealed the presence of a single-copy transgene in A-2 and A-33 lines and two copies of transgene in the A-14 line (see Supplemental Figure 2 online), consistent with the suppressed expression of MADS29 (Figure 2A, left panel). MADS30 is the paralog of MADS29, but qRT-PCR analysis showed that the expression level of MADS30 is not affected in the transgenic lines (Figure 2A, right panel).
Figure 2.
Suppressed MADS29 Expression Results in Abnormal Seeds.
(A) qRT-PCR analysis confirmed the suppressed expression of MADS29 (left panel) and unaltered expression of MADS30 (right panel) in independent A-MADS29 transgenic lines (A-2, A-14, and A-33). Seeds at 6 DAF were analyzed. Data are presented as mean ± se (n = 3).
(B) Rice plants with suppressed MADS29 expression show abnormal seed development and phenotypic observation revealed the shrunken seeds of A-MADS29 lines. Seeds at 3, 6, 8, 10, 15, 20, and 30 DAF were observed. Bars = 2 mm.
(C) Characterization of the grain-filling rate in ZH11 and the A-MADS29 lines. The time course of grain fresh weight (left panel) and grain dry weight (right panel) are shown. The data are presented as mean ± sd (n > 10).
Because rice seeds develop rapidly following fertilization, the grain shape from fertilization to maturity was observed. A-MADS29 seeds of the same panicle are shrunken to various degrees (see Supplemental Figure 3 online) and show similar morphology with slightly differed thickness. Observation of the representative seeds of each line showed that during the first 6 DAF, there is no obvious difference between ZH11 and A-MADS29 seeds. After 8 DAF, the A-MADS29 seeds become shrunken, and this phenotype persists until 30 DAF because of a defect in dry matter accumulation, whereas the seeds are fully filled with dry matter by 30 DAF in ZH11 (Figure 2B).
Next, the grain-filling rate of the A-MADS29 lines was investigated. The results showed that there is no difference in either fresh weight or dry weight of the grains at 6 DAF (Figure 2C), whereas at 9 DAF, the grain fresh weight and dry weight of the A-MADS29 lines are significantly reduced compared with those of ZH11. At 9 DAF, both the fresh weight and dry weight of ZH11 seeds increase rapidly until DAF 26, while little increase occurs in the A-MADS29 seeds (A-2 and A-14). At later stages, seeds of the A-33 line accumulate dry matter rapidly from 12 DAF and the final weight reaches to ~63% of ZH11 seeds (Figure 2C), whereas A-2 and A-14 seeds have a more severe phenotype, which is consistent with the expression levels of MADS29 in these lines (Figure 2A).
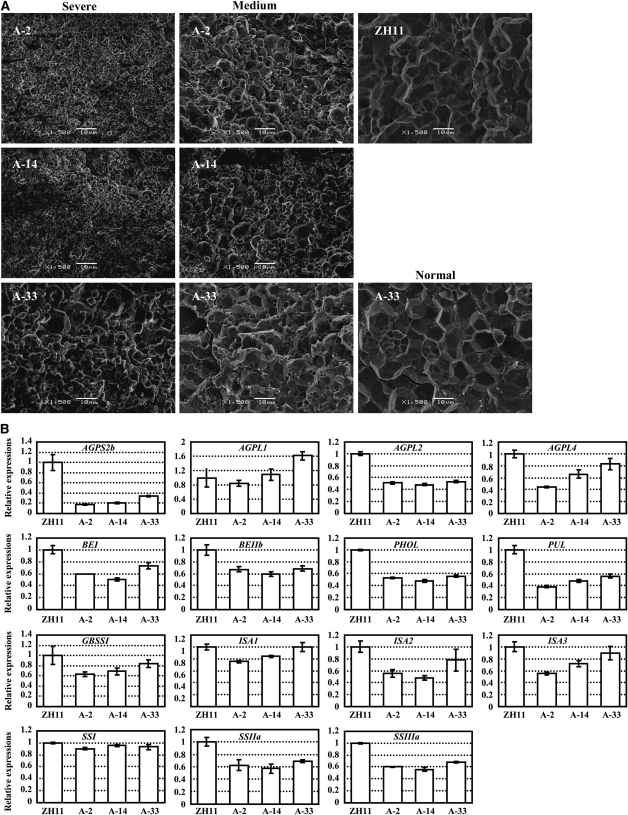
Previous studies showed that the grain-filling rate affects the morphology of the endosperm starch granules (Wang et al., 2008). Scanning electron microscopy observation was thus performed to examine whether the morphology of the endosperm starch granules was altered under suppressed MADS29 expression. A-MADS29 seeds of the same panicle present different degrees of shrunken phenotypes (see Supplemental Figure 3 online) and the seeds with the more severe phenotype show smaller and rounder starch granules when compared with those of ZH11 (Figure 3A, left panel). The seeds with a medium phenotype have rounder starch granules of intermediate size (some starch granules with smaller size; Figure 3A, middle panel) and fully filled A-33 seeds show normal-shaped starch granules (Figure 3A, right panel).
Figure 3.
Suppressed MADS29 Expression Results in Defective Starch Synthesis.
(A) Scanning electron microscope analysis of the starch granules of ZH11 and A-MADS29 seeds. Starch granules of plants with severe (left column) and medium (middle column) phenotypes and in ZH11 and a normal A-33 line (right column) are shown. Magnification, ×1500. Bars = 10 μm.
(B) qRT-PCR analysis confirmed the altered expressions of the starch biosynthesis genes in the A-MADS29 seeds. Seeds at 8 DAF were analyzed. The data are presented as mean ± se (n = 3).
Further analysis of starch characters showed a reduced content of apparent amylose (as it was difficult to obtain endosperm powder from seeds of A-2 and A-14 lines, A-33 seeds were analyzed; see Supplemental Figure 4A online) and the proportion of starch chains (degree of polymerization) in the range of 8-14 was significantly decreased, whereas the proportion of chains with a degree of polymerization in the range of 16-51 was slightly increased (see Supplemental Figure 4B online) under suppressed MADS29 expression.
Consistent with the altered starch synthesis, analysis of the expression levels of starch biosynthesis genes showed that among the 15 tested genes, most genes are downregulated under suppressed expression of MADS29, except AGPL1, ISAI, and SSI (Figure 3B). Interestingly, AGPS2b mediates the key step of starch biosynthesis in seeds and displays the most reduction under suppressed expression of MADS29 (Figure 3B), which might contribute to the shrunken phenotype as the agps2 mutant shows similar seeds (Lee et al., 2007).
MADS29 Affects Seed Development by Promoting the PCD of Maternal Tissues
To investigate whether the shrunken seed phenotype was due to a maternal defect, reciprocal crosses were performed. When A-MADS29 pollen was used to pollinate ZH11, normal seeds developed. However, when ZH11 pollen was pollinated to A-MADS29, shrunken seeds could be observed (see Supplemental Figure 5 online). Although it is unknown whether MADS29 is epigenetically regulated in diploid tissues, this result suggested that the abnormal seed development under suppressed MADS29 expression is possibly due to a maternal effect.
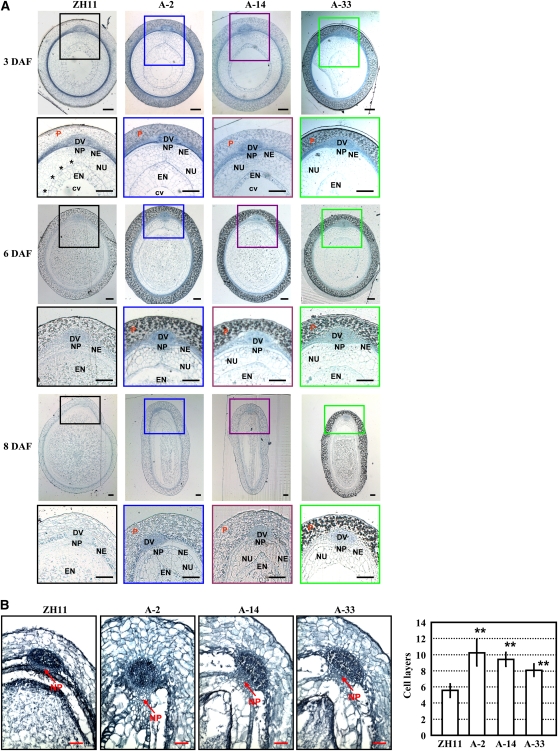
In a detailed examination of the defects in the A-MADS29 seeds, transverse section analysis of the seeds at 3, 6, and 8 DAF showed that at 3 DAF, the cell walls of the nucellus are collapsed in ZH11 seeds, while those of the A-MADS29 seeds still exist. At 6 DAF, the nucellus cells in ZH11 are completely degraded and the number of endosperm cells increase rapidly, whereas those of the A-MADS29 seeds are still present even at 8 DAF (Figure 4A). These results indicated that defects in nucellar degradation under suppressed MADS29 expression result in abnormal seed development.
Figure 4.
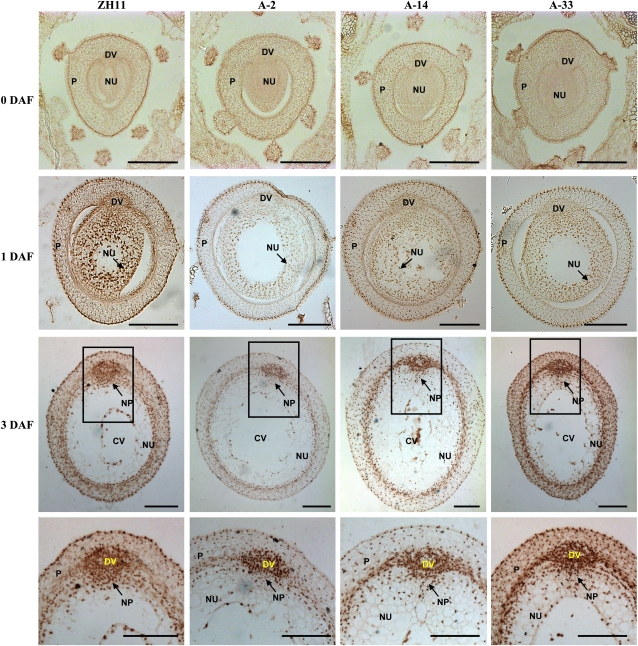
Suppressed MADS29 Expression Results in Nondegraded Nucellus and Nucellar Projection.
(A) Transverse section analysis of A-MADS29 and ZH11 seeds showed the nondegraded nucellus under suppressed MADS29 expression (bars = 100 μm). The squared regions are enlarged below (bars = 50 μm) and stars highlight the collapsed nucellus. Seeds at 3, 6, and 8 DAF were observed.
(B) Observation of transverse sections (left panel) and the quantification of cell layers (right panel) of seeds at 10 DAF revealed repressed nucellar projection degradation in the A-MADS29 lines. The data are presented as mean ± sd (n > 9), and a statistical analysis with a two-tailed Student’s t test indicates the significant differences from ZH11 (**P < 0.01). Bars = 50 μm.
CV, central vacuole; DV, dorsal vascular bundle; EN, endosperm; NE, nucellar epidermis; NP, nucellar projection; NU, nucellus; P, pericarp.
Because MADS29 is highly expressed in the nucellar projection cells, we then tested whether the degradation of the nucellar projection was defective in A-MADS29 seeds. Observation and quantification of the cell layers of the nucellar projection showed that the nucellar projection cells of the A-MADS29 seeds are dramatically thicker than those of ZH11 at 10 DAF (Figure 4B), suggesting that the degradation of the nucellar projection cells is suppressed and that MADS29 is required for the degradation of maternal tissue.
Previous studies indicated that the nucellus and the nucellar projection cells execute PCD following pollination (Dominguez et al., 2001; Sreenivasulu et al., 2010). Because the A-MADS29 lines contained undegraded nucellus cells and a thicker nucellar projection, it was proposed that the PCD was defective when MADS29 expression was suppressed. A terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) assay was then performed and results showed that prior to pollination there are no TUNEL-positive nuclei in the nucellar cells of ZH11 or A-MADS29 lines, which suggested that PCD is not initiated prior to pollination (Figure 5). However, 1 d after pollination (1 DAF), TUNEL-positive nuclei were detected in the nucellar cells of ZH11, whereas the signal was very weak in the A-MADS29 lines (Figure 5). At 3 DAF, the nucellar cells of ZH11 seeds were completely degraded, and TUNEL-positive nuclei were strongly detected in the nucellar projection cells and the dorsal vasculature. At this time point, only a weak TUNEL signal was detected in nucellar projection cells of A-MADS29 seeds; however, a strong TUNEL signal in dorsal vasculature was detected, which is similar to that in ZH11 seeds (Figure 5). These results are in agreement with the hypothesis that MADS29 promotes the degradation of the nucellus and nucellar projection by regulating PCD processes.
Figure 5.
Repressed PCD under Suppressed MADS29 Expression.
Analysis of the nuclear DNA fragmentation by the TUNEL assay revealed a decrease in TUNEL-positive nuclei under suppressed MADS29 expression. The ovary prior to pollination and seeds harvested at 1 and 3 DAF of ZH11 and A-MADS29 lines were used for analysis. The TUNEL-positive nuclei are stained dark brown. The squared regions are enlarged and shown in bottom panel. CV, central vacuole; DV, dorsal vascular bundle; NP, nucellar projection; NU, nucellus; P, pericarp. Bars = 200 μm.
Suppressed MADS29 Expression Results in the Alteration of Multiple Processes
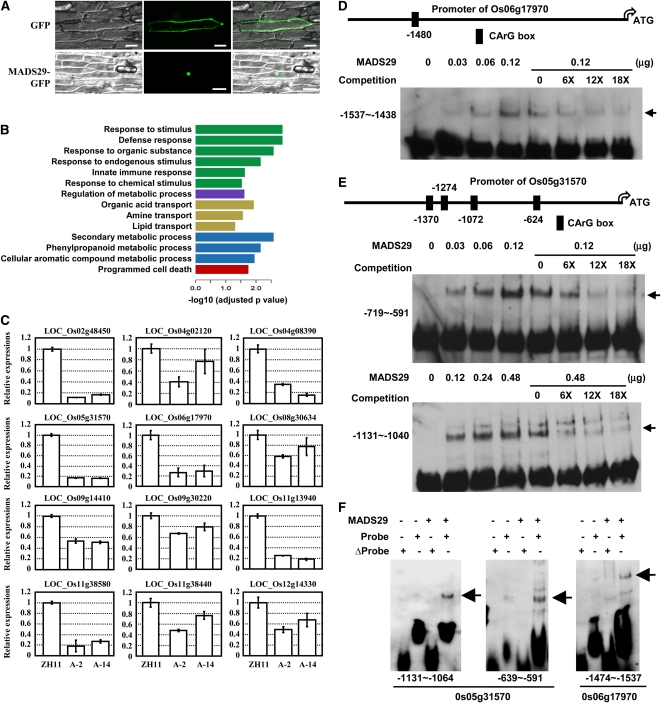
Because MADS29 is a MIKCc-type MADS box TF, how it regulates the downstream processes was then investigated. Analysis of the subcellular location of MADS29 via transient expression of the MADS29-green fluorescent protein (GFP) protein in onion epidermal cells showed that MADS29 was localized in the nuclei (Figure 6A), which is consistent with a role of MADS29 as a TF.
Figure 6.
MADS29 Is Localized in the Nucleus and Stimulates the Expression of PCD-Related Genes.
(A) Subcellular localization of the MADS29-GFP fusion protein. MADS29-GFP and GFP alone were transiently expressed in onion epidermal cells, and MADS29-GFP showed nuclear localization. Bars = 100 μm.
(B) Significantly overrepresented GO biological process terms for the 674 downregulated genes in the nucellus and nucellar projection of the A-MADS29 lines. The hypergeometric test was used to estimate the significance of the overrepresentation, and only GO terms with an adjusted P value <0.05 and at least 10 annotated genes were kept. Colors represent GO terms belonging to different categories: green bars, response to stimulus; purple bar, regulation of biological process; yellow bars, transport; blue bars, metabolic process; red bar, cell death. The negative logarithm (base 10) of the adjusted P value was used as the bar length.
(C) qRT-PCR analysis confirmed the downregulation of PCD-related genes in the nucellar projection of the A-MADS29 seeds. The data are presented as the mean ± se (n = 3).
(D) and (E) MADS29 directly binds to the putative promoter regions of Os06g17970 (D) and Os05g31570 (E). The positions of the CArG-box site in the promoter region are shown (top panel). EMSA assay showed that purified MADS29 protein directly binds the promoter region of Os06g17970 and Os05g31570 (bottom panel). Unlabeled DNA fragments were added (6-, 12-, and 18-fold of the labeled probes) and used as competitors. The shifted bands are indicated with arrows.
(F) EMSA assay confirmed that MADS29 directly binds to the CArG motif in the putative promoter of Os05g31570 and Os06g17970 in vitro. The original DNA region (normal probe) or a version lacking the CArG motif (Δprobe) was used for analysis. Purified MADS29 protein (0.1 μg) was used for the assay. The shifted bands are indicated with arrows.
To identify the downstream genes of MADS29, a genome-wide analysis of the gene expression profiles in the nucellar projection cells was performed. The maternal nucellar projection cells of ZH11 and A-MADS29 seeds (lines A-2 and A-14) at 3 DAF were harvested by laser capture dissection (see Supplemental Figure 6 online), and total RNA was extracted and amplified from two independent biological samples, which were used for hybridization with the Agilent 44k rice genome microarray. The coefficient of variation value of replicate probes (10 times) was <10% within each chip, indicating a reliable quality of hybridization. Detailed analysis showed that a total of 1096 genes display altered expression (P < 0.05 and more than twofold change), of which 422 and 674 are up- or downregulated, respectively, under suppressed MADS29 expression. Functional analysis by gene ontology (GO) annotation showed no significantly overrepresented processes/functions of upregulated genes, whereas many GO terms related to nucellar projection function, including the regulation of metabolic processes, transportation, and the response to stimuli, are significantly enriched among downregulated genes (Figure 6B). The enrichment of terms for organic acid, amine, and lipid transport is consistent with the decreased grain-filling rate, and, most importantly, the enrichment of PCD-related genes demonstrates the regulatory role of MADS29 in PCD (Figure 6B). Further qRT-PCR analyses confirmed the dramatically reduced expression levels of the PCD-related genes under suppressed MADS29 expression (Figure 6C, Table 1).
Table 1.
Valid Downregulated PCD-Related Genes under Suppressed MADS29 Expression
| Fold Change |
||||
| Locus | Annotation | Line A-2 | Line A-14 | P Value |
| Os02g48450 | Xylem Cys proteinase 2 precursor | −10.35 | −9.23 | 9.96E-04 |
| Os04g02120 | Expressed protein | −2.36 | −3.09 | 0.027 |
| Os04g08390 | LRR family protein | −3.15 | −3.16 | 0.13 |
| Os05g31570 | Disease resistance protein RGA4 | −2.94 | −10.79 | 0.012 |
| Os06g17970 | NBS-LRR disease resistance protein | −2.62 | −2.95 | 0.021 |
| Os08g30634 | DC1 domain containing protein | −2.95 | −3.66 | 0.024 |
| Os09g14410 | Expressed protein | −2.02 | −2.19 | 0.004 |
| Os09g30220 | Disease resistance protein RPM1 | −2.58 | −4.10 | 0.003 |
| Os11g13940 | NBS-LRR disease resistance protein | −3.14 | −2.32 | 0.023 |
| Os11g38440 | Expressed protein | −3.01 | −2.82 | 5.31E-05 |
| Os11g38580 | NBS-LRR–type disease resistance protein | −2.79 | −3.84 | 5.81E-04 |
| Os12g14330 | Disease resistance protein RPM1 | −4.10 | −4.92 | 0.002 |
Additionally, transcripts of 21 auxin signaling-related genes are downregulated under suppressed MADS29 expression (Table 2), including seven well-known indole-3-acetic acid (IAA) signaling early response genes (two AUX/IAA family proteins, four GH3 family proteins, and one ARF family member).
Table 2.
Auxin Signaling–Related Genes Were Downregulated under Suppressed MADS29 Expression
| Fold Change | ||||
| Locus | Annotation | Line A-2 | Line A-14 | P Value |
| Os01g09450 | AUX/IAA protein family protein | −3.36 | −2.26 | 0.042 |
| Os01g57610 | GH3 auxin-responsive promoter family protein | −9.59 | −6.11 | 5.49E-05 |
| Os01g66530 | Conserved hypothetical protein | −2.68 | −2.98 | 0.017 |
| Os01g68370 | Transcription activator VP1-rice | −15.68 | −28.06 | 0.001 |
| Os02g04810 | Auxin response factor 7 (ARF7) | −2.61 | −2.26 | 0.035 |
| Os02g07310 | Argonaute 4 protein | −4.59 | −2.9 | 1.31E-04 |
| Os02g10520 | Subtilisin-like protease | −10.71 | −3.02 | 0.005 |
| Os02g52990 | Auxin-responsive SAUR protein family protein | −3.63 | −5.47 | 0.027 |
| Os03g63650 | Brevis radix | −3.83 | −2.3 | 0.021 |
| Os04g51172 | Conserved hypothetical protein | −2.13 | −2.74 | 0.002 |
| Os05g05180 | GH3 auxin-responsive promoter family protein | −2.38 | −5.25 | 0.013 |
| Os05g10580 | Cullin family domain containing protein | −2.78 | −3.74 | 1.43E-04 |
| Os05g10690 | TF MYBS2 | −4.53 | −2.44 | 0.012 |
| Os05g42150 | Auxin-responsive-like protein (GH3) | −4.07 | −4.2 | 8.85E-04 |
| Os05g50610 | WRKY TF 34 | −8.37 | −8.52 | 6.00E-05 |
| Os06g07040 | AUX/IAA protein family protein | −17.47 | −3.9 | 0.015 |
| Os07g39320 | Homeodomain Leu zipper protein CPHB-4 | −2.97 | −2.24 | 0.025 |
| Os07g40290 | GH3 homolog | −5.42 | −3.14 | 0.010 |
| Os09g26590 | OsSAUR37 auxin-responsive SAUR gene | −2.56 | −2.41 | 7.25E-04 |
| Os12g42020 | Protein kinase domain containing protein | −2.61 | −3.46 | 0.003 |
MADS29 Directly Stimulates PCD-Related Genes
Previous studies showed that MADS box TFs can specifically bind to the CArG-box cis-element of target genes to regulate their expression (de Folter and Angenent, 2006). Indeed, an analysis of the putative promoters of the 12 examined PCD-related genes revealed the presence of a CArG-box [5′-CC(A/T)6GG-3′] in five and a variant CArG-box (5′-C(A/T)8G-3′) in 11 of these genes, which implies that MADS29 might stimulate PCD by directly regulating the expression of these PCD-related genes.
Interestingly, five of the 12 downregulated PCD-related genes encode putative nucleotide binding site–Leu-rich repeat (NBS-LRR) proteins, which have been characterized as signal transduction elements involved in the hypersensitive response (a pathway leading to PCD; Innes and DeYoung, 2006; Ting et al., 2008). To determine the possibly regulatory role of MADS29 on these NBS-LRRs, an electrophoretic mobility shift assay (EMSA) was performed, and the results showed that of the four tested NBS-LRR genes, MADS29 directly binds one fragment of the Os06g17970 putative promoter (−1537 to ~−1438; Figure 6D) and two fragments of the Os05g31570 putative promoter (−1131 to ~−1040 and −719 to ~−591; Figure 6E) in a dosage-dependent manner, and these interactions are completely inhibited by the addition of excess unlabeled fragments. No binding is observed when the CArG-boxes in these fragments are deleted (Figure 6F), indicating that the CArG-box is crucial for the binding and MADS29 regulates the expression of NBS-LRR genes through direct binding.
MADS29 Regulates the Expression of a Cys Protease through Direct Binding
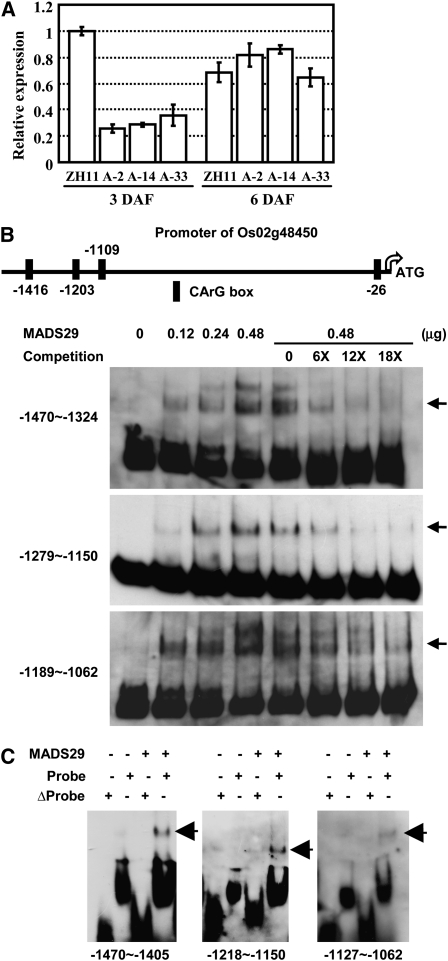
It is interesting to notice that among the PCD-related genes, a Cys protease (Os02g48450, belonging to the papain family of Cys proteases) is dramatically downregulated in the nucellar projection of the A-MADS29 seeds at 3 DAF (Figure 6C). There are 51 papain-type members in the rice genome (http://merops.sanger.ac.uk/), and Os02g48450 is the only of these whose expression is significantly downregulated under suppressed MADS29 expression. A detailed qRT-PCR analysis confirmed the reduced expression of Os02g48450 in the A-MADS29 seeds at 3 DAF and a similar expression to that of ZH11 seeds at 6 DAF (Figure 7A; the similar expression level at 6 DAF may due to the degradation of nucellus has finished in ZH11). The altered expression of Os02g48450 is consistent with the higher expression of MADS29 in the early stages of seed development (1 and 3 DAF; Figure 1A) and implies that this Cys protease is a possible target of MADS29 for the regulation of nucellar projection degradation and early seed development.
Figure 7.
MADS29 Directly Regulates Cys Protease to Affect PCD and Normal Endosperm Development.
(A) qRT-PCR analysis of Os02g48450 expression in seeds (3 and 6 DAF) of ZH11 and A-MADS29 lines. The data are presented as mean ± se (n = 3).
(B) MADS29 directly binds to the promoter regions of Cys protease (Os02g48450). The box shows the CArG-box site in the promoter region (top panel). EMSA assay showed that purified MADS29 protein can directly bind the promoter of Cys protease (bottom panel). Unlabeled DNA fragments were added as 6-, 12-, and 18-fold of the labeled probes and were used as competitors. The shifted bands are indicated with arrows.
(C) EMSA assay confirmed that MADS29 directly binds to the CArG motif in the putative promoter of Cys protease (Os02g48450) in vitro. The original DNA regions (normal probe) or versions lacking the CArG motif (Δprobe) for three DNA fragments were used for analysis. Purified MADS29 protein (0.1 μg) was used for the assay. The shifted bands are indicated with arrows.
An analysis of the Os02g48450 putative promoter region revealed the presence of three clustered variant CArG-box motifs [5′-C(A/T)8G-3′] in the upstream region between −1108 and −1425 bp (Figure 7B). Subsequent EMSA revealed the direct binding of purified MADS29 to these three regions (Figure 7B). The binding is enhanced with increased dosage of MADS29 protein and reduced with the addition of excess unlabeled fragments as competitors (Figure 7B). As expected, no binding is observed when the CArG-box in these fragments are deleted (Figure 7C), confirming that the Cys protease (Os02g48450) is the direct target of MADS29.
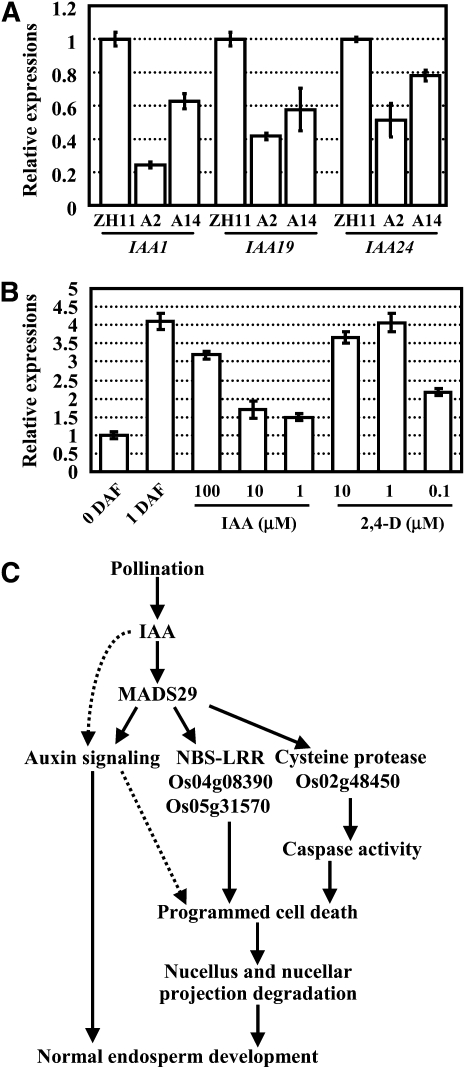
MADS29 Is Induced by Auxin
Microarray analysis revealed that auxin signaling-related genes are downregulated under suppressed MADS29 expression, which was further confirmed by qRT-PCR analyses (Figure 8A), implying that auxin may be involved in the PCD process to regulate seed development. To explore the possible involvement of MADS29 in the auxin effects, ovaries detached from pollinated and unpollinated ZH11 flowers were cultured in vitro and treated with IAA (1, 10, and 100 μM) or 2,4-D (0.1, 1, and 10 μM) for 24 h. qRT-PCR analysis indicated that the MADS29 expression is induced after pollination or by auxin treatment (Figure 8B).
Figure 8.
MADS29 Mediates Auxin Signaling and Hypothetical Model of MADS29 Effects in Regulation of Rice Seed Development.
(A) Validation of the microarray results by qRT-PCR analysis confirmed the downregulation of the IAA-responsive genes in nucellar projection of the A-MADS29 lines. The data are presented as mean ± se (n = 3).
(B) qRT-PCR analysis indicated that MADS29 expression is induced by IAA and 2,4-D. The spikelets were harvested before pollination and cultured in medium in the absence or presence of IAA (1, 10, and 100 μΜ) or 2,4-D (0.1, 1, and 10 μΜ) for 24 h. The spikelets harvested after pollination were cultured in the medium without hormone as a positive control. The data are presented as mean ± se (n = 3).
(C) Hypothetical model of MADS29 function in seed development. After pollination, the increased IAA content stimulates MADS29 expression, which activates the expression of the Cys protease and NBS-LRR proteins by directly binding to the promoter regions. The Cys protease and NBS-LRR proteins promote the degradation of the nucellus and nucellar projection by PCD processes, which are essential for normal endosperm development. Meanwhile, auxin regulates endosperm development through signal transduction or possibly by stimulating the PCD processes.
DISCUSSION
It has been hypothesized that Bsister clade MADS family members are more important for female organ development (Becker et al., 2002). MADS29 is highly expressed in female organs, similar to the previously reported Bsister genes, but expressed at low levels in the endothelium, which is different from the expression patterns of Arabidopsis FBP24 and wheat Bsister. The endothelium of the A-MADS29 lines develops normally (Figure 1C); however, considering the relatively lower suppression of MADS29 in analyzed A-MADS29 lines, further study using an mads29 knockout mutant will help to elucidate the role of MADS29 in rice endothelium formation. Interestingly, expression of MADS29 increases while that of FBP24 decreases after pollination, although expressions of both of them decrease during seed development (de Folter et al., 2006; Figure 1A). Besides seed development, MADS29 was reported as an important major quantitative trait loci candidate gene affecting germination rate (Li et al., 2011b), suggesting that in addition to playing roles in female organs like other reported Bsister clade genes, MADS29 also regulates other developmental processes.
Our studies showed that suppressed expression of MADS29 results in the reduced or delayed cell degradation of the nucellar projection and abnormal endosperm development and indicated that MADS29 is necessary for and a key regulator of the degradation of the nucellus and nucellar projection, which undergoing PCD during early rice seed development. Thus, we have identified a rice TF that regulates the degradation of the nucellus and nucellar projection. The barley homolog of MADS29 is also preferentially expressed in the nucellar projection (Thiel et al., 2008), suggesting that MADS29 and its homolog may play a critical and conserved function in the seed development of monocotyledons.
The nucellar cells differentiate during ovular development and begin to abort soon after fertilization. Previous studies indicated that the degradation of the nucellus after fertilization is required to facilitate the supply of nutrients to the young embryo and endosperm (Krishnan and Dayanandan, 2003; Sreenivasulu et al., 2010). Our studies indicated that the suppressed degradation of the nucellus and nucellar projection cells results in altered seed morphology, reduced accumulation of endosperm cells, and reduction of dry matter accumulation, further revealing that the nucellar degradation is also important for endosperm development at later stages and grain filling.
Nutrients are primarily supplied to the developing endosperm by the dorsal vascular bundle, through which nutrients flow into the nucellar projection and are transported to the starch endosperm by two pathways, either by the multiple aleuronic layers adjacent to the nucellar projection or by the nucellar epidermis and subsequently by the aleurone cells (Krishnan and Dayanandan, 2003). Because MADS29 is not expressed in the endosperm, the suppressed starch synthesis and abnormal starch granules may be a result of defective nutrient transportation, which is consistent with a previous nutrient transport study that used dye movement experiments to show that the dorsal vasculature affects grain filling (Krishnan and Dayanandan, 2003). Reduced or delayed degradation of the nucellus by suppressed MADS29 expression may prevent the transportation of the nutrients from nucellar tissues to endosperm, indicating that a direct connection between nucellar tissues and endosperm is necessary for nutrient transport.
Downregulated expressions of MADS6 and MADS17 and upregulated expression of MADS16 were detected under suppressed MADS29 expression. MADS16 is a B-class member and ectopic expression of MADS16 causes superman phenotypes (An et al., 2003). MADS6 is a key regulator of flower development via interaction with many homeotic genes (Li et al., 2011a). However, A-MADS29 plants display no obvious difference in floral organ identity, which is consistent with the high expression of MADS29 in seeds. In addition, MADS6 is expressed in endosperm and the mads6 mutant has less fully filled seeds with decreased starch content due to the decreased expressions of starch biosynthesis-related enzymes (Zhang et al., 2010). By contrast, the abnormal starch granules of A-MADS29 seeds are a result of a low grain-filling rate, revealing a unique role of MADS29, a preferentially expressed gene in the ovular and maternal tissues of developing seeds.
Consistent with the critical roles of hormones in plant growth and development, our studies indicated that auxin signaling is involved in the regulation of nucellar projection and endosperm development. IAA content is increased (approximately fivefold) in the rice ovary following pollination and the nucellus cells of unpollinated spikelets degenerate when cultured on medium containing auxin (Uchiumi and Okamoto, 2010), implying the role of IAA in nucellus degeneration. Considering that MADS29 expression is simultaneously upregulated (Figure 8B) and the transcripts of IAA-responsive genes are reduced under MADS29 suppression (Figure 8A, Table 2), it is supposed that MADS29 was induced by IAA after pollination to regulate the PCD of nucellus and nucellar projection and, hence, endosperm development (Figure 8C).
Among the 12 validated downregulated PCD-related genes, five of them belong to the NBS-LRR family, members of which are important receptors for pathogen effectors and are involved in the signal transduction of the resistance response (Innes and DeYoung, 2006; Ting et al., 2008). Plant NBS-LRR proteins have only been reported to be responsible for hypersensitive response, while their orthologs in mammals play roles in both developmental and pathogen-induced PCD (Aderem et al., 1999). Our results indicate that MADS29 stimulates NBS-LRR proteins to trigger PCD through directly binding their promoters, implying that plant NBS-LRR proteins are also associated with developmental PCD.
Interestingly, MADS29 directly binds and stimulates the expression of a Cys protease (Os02g48450), the only papain-type Cys protease that display reduced transcription under suppressed MADS29 expression. In animals, Cys proteases are involved in PCD by specifically cleaving target proteins (Thornberry and Lazebnik, 1998). Plant Cys protease members, including VPEs and metacaspases, possess caspase-like activities to trigger PCD (Hatsugai et al., 2004; Dangl et al., 2010). The papain family is the largest subfamily of Cys proteases and is associated with developmental PCD in plants (Grudkowska and Zagdanska, 2004), including of the tapetum and the germinating endosperm (Schmid et al., 1999; Lee et al., 2004), implying that Os02g48450 may also play a crucial role in reproductive tissues. The expression level of Os02g48450 is higher at 3 DAF than at 6 DAF (Figure 7A), which is consistent with the observation that the nucellus nearly collapses at 3 DAF and the seeds are full of endosperm cells at 6 DAF (Figure 4A), suggesting that this Cys protease is largely involved in the PCD of the nucellus following fertilization.
In conclusion, after fertilization, the increased IAA content induces the expression of MADS29, which then stimulates the degradation of the nucellus and the nucellar projection by directly stimulating a Cys protease and NBS-LRR proteins (Figure 8C). Acting through MADS29 or other pathways, auxin regulates the PCD processes required for normal endosperm development and grain filling.
METHODS
Plant Material and Growth Conditions
Wild-type rice (Oryza sativa cv Zhonghua 11 [ZH11]) was used for rice transformation. The rice seeds were germinated in sterilized water and then grown in pots in a phytotron with a 12-h-light (26°C)/12-h-dark (18°C) cycle. To measure the grain-filling rate, the plants were cultivated in an experimental field under natural growing conditions.
Promoter-GUS Fusion Studies and Histochemical Analysis of GUS activity
The ~2.8 kb putative promoter region of MADS29 (upstream of ATG) was amplified by PCR with primers (5′-GCCTGCTATACCTTCCTGATCGAG-3′ and 5′-CAGTTGCAGACAGTGGATGAGATG-3′) using ZH11 genomic DNA as template. Amplified DNA fragment was subcloned into SmaI site of pCAMBIA1300+pBI101.1 (Liu et al., 2003), and the resultant construct was introduced into the Agrobacterium tumefaciens strain EHA105 (Hood et al., 1993) by electroporation and transformed into ZH11 using immature embryos as materials. Independent lines of positive T2 homozygous transgenic progeny were used to detect the GUS activity. Photography was performed using a Nikon microscope (SMZ1500) with a digital camera (Nikon; control unit DS-U2).
In Situ Hybridization Analysis
A gene-specific region of the coding region of MADS29 was amplified by PCR from the cDNA KOME clone (AK109522; the primers are listed in Supplemental Table 1 online). The fragment was subcloned into a pGEM-T easy vector (Promega). The sense and antisense probes were transcribed in vitro under T7 promoter with RNA polymerase using a DIG RNA labeling kit. Wild-type ovaries and seeds from different developmental stages were fixed in a formaldehyde solution (4%), dehydrated through an ethanol series, embedded in paraffin (Sigma-Aldrich), and sectioned at 10 μm. In situ hybridization was performed according to the previous description (Coen et al., 1990).
DNA Gel Blot Analysis
The hygromycin gene was labeled by PCR with primers (5′-GCTTCTGCGGGCGATTTGTGT-3′ and 5′-GGTCGCGGAGGCTATGGATGC-3′) according to the manufacturer’s instructions (Roche). Forty micrograms of rice genomic DNA of ZH11 and A-MADS29 lines was digested with different restriction enzymes (BamHI, EcoRI, and SacI) and separated in 1% agarose gel. After transmembrane, cross-linking, hybridization, washing, blocking, and antibody incubation, the signal was detected with disodium 3-(4-methoxyspiro {1,2-dioxetane-3,2’-(5′-chloro)tricycle [3.3.1.13,7]decan}-4-yl)phenyl phosphate (CSPD; Roche).
Constructs and Observation of Seeds and Starch Granules
A 965-bp cDNA fragment of MADS29 was excised from the KOME clone by digestion with SacI and HindIII and subcloned into SmaI site of pUN1301 vector (Ge et al., 2004). The obtained construct was introduced into the Agrobacterium strain EHA105 and positive clones were used for rice transformation.
The rice seeds of ZH11 and A-MADS29 lines at different developmental stages were fixed with formalin–acetic acid–alcohol. The samples were embedded in Epon812 resin (Fluka) and polymerized at 60°C. Sections were cut, stained with 1% toluidine blue, microscopically examined (Leica DRM), and photographed. For the observation of endosperm starch granules, the completely dried rice seeds were cut in cross sections, and the surfaces of the cross sections were coated with gold and observed with a scanning electron microscope (JEOL JSM-6360LV).
Starch Content and Chain Length Distribution Analysis
The grains harvested in the field were dehulled and grounded to powder. To measure the total starch content, 50 mg powder was washed three times using 80% ethanol (v/v) and then extracted with 9.2 M and 4.6 M perchloric acid in order. The supernatant was diluted and analyzed by the anthrone method (Turner and Turner, 1960). The apparent amylose content was measured by the iodine colorimetric method as previously described (Juliano, 1971). To determine the chain length distributions of amylopectin, 5 mg of rice powder was digested with Pseudomonas amyloderamosa isoamylase (Sigma-Aldrich) and then analyzed by high-performance anion-exchange chromatography with pulsed amperometric detection (DX-500; Dionex) (Nagamine and Komae, 1996; Nishi et al., 2001).
TUNEL Assay
The ovaries and seeds of ZH11 and A-MADS29 transgenic lines were fixed in a formaldehyde solution (4%), dehydrated through an ethanol series, and embedded in paraffin (Sigma-Aldrich). Sections were obtained from the Paraplast Plus–embedded material. The Paraplast Plus was removed by a xylol treatment, and the sections were hydrated with an ethanol series and treated with proteinase K in PBS. The endogenous peroxidase activity was quenched by incubation with H2O2. The sections were incubated at 37°C for 60 min in the presence of TdT with a TUNEL apoptosis detection kit (DeadEnd colorimetric TUNEL system; Promega).
Subcellular Localization Studies of MADS29
The coding region of MADS29 was amplified using primers (5′-CATGCCAAGGTAGCAGGCATGGGGCGCGGC-3′, added NcoI site underlined; and 5′-CACTAGTACCCACAGCTGCAGGCCGTGGCC-3′, added SpeI site underlined). The amplified fragment was subcloned into pCAMBIA1302 (CAMBIA). GFP and MADS29-GFP fusion proteins were expressed transiently in onion epidermal cells with a PDS-1000/He Biolistic Particle Delivery System (Bio-Rad) as previously described (Scott et al., 1999). After incubation of the epidermal tissues, the green fluorescence was observed with confocal laser scanning microscopy (Olympus FV1000) using an argon laser excitation wavelength of 488 nm (GFP).
Laser Capture Microdissection, Microarrays, and GO Term Analysis
Nucellar projection cells of ZH11 and A-MADS29 seeds were isolated using a Veritas Microdissection System (Arcturus/Molecular Devices). Seeds harvested at 3 DAF were fixed in acetone and embedded in paraffin as previously described (Tang et al., 2006). A UV laser beam was used to cut the target cells, and an infrared laser beam was used to capture the target cells. The collected target cells were transferred to RNA extraction buffer for further analysis. RNA was extracted with the PicoPure RNA isolation kit (Arcturus/Molecular Devices) and linearly amplified using a TargetAmp 2-round Aminoallyl-aRNA amplification kit (Epicentre Biotechnologies).
The Agilent 4X44K rice microarrays (Agilent Technologies; containing 44,000 probes) were used to study the gene expression profiles in the nucellar projection cells of ZH11 and A-MADS29 seeds. Washing, staining, chip scanning, and data processing were performed according to the manufacturer’s instructions. The hybridization data have been deposited into the Gene Expression Omnibus database (GSE31893; http://www.ncbi.nlm.nih.gov/geo).
Only genes present in both of the two ZH11 samples were used for subsequent analysis. The linear statistical model in Limma package (Wettenhall and Smyth, 2004) from Bioconductor (http://www.bioconductor.org) was used to identify the significantly differentially expressed genes between ZH11 and A-MADS29 samples. The threshold for selection of significantly regulated genes was set as P value <0.05 and change ratio >2.0 in both of the A-MADS29 samples (lines A-2 and A-14).
To annotate the functions of significantly changed gene sets, BiNGO software was used for the analysis of the GO terms (http://www.psb.ugent.be/cbd/papers/BiNGO/). The 422 upregulated genes and 674 downregulated genes were compared with all the analyzed genes respectively. To determine the significance of the overrepresented GO terms, the hypergeometric test was performed and only the GO terms with at least 10 annotated genes and a Benjamini and Hochberg method (Benjamini and Hochberg, 1995) adjusted P value <0.05 and less than four terms away from the root term were retained.
qRT-PCR Analysis
Total RNAs were extracted from seeds at different developmental stages and various tissues of ZH11 and A-MADS29 transgenic plants using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. The isolated RNA was reverse transcribed and used as templates for qRT-PCR to analyze the gene expression levels quantitatively using the Rotor-Gene real-time thermocycler R3000 (Corbett Research) with real-time PCR Master Mix (Toyobo). A linear standard curve was generated using a series of dilutions for each PCR product. Rice ACTIN was amplified and used as an internal standard to normalize the expression of tested genes. The primers of the examined genes are listed in Supplemental Table 1 online, and primers for starch biosynthesis genes are previously described (Ohdan et al., 2005).
cis-Element Analysis and EMSA
The Plant Cis-acting Regulatory DNA Elements program (http://www.dna.affrc.go.jp/PLACE/) was used to analyze the 3000-bp upstream region of selected genes for the presence of a CArG-box.
To perform EMSA, the coding region of MADS29 was amplified using primers (5′-CGGGATCCATAGCAGGCATGGGGCGCGGC-3′, added BamHI site underlined; and 5′-CCCAAGCTTGCCACAGCTGCAGGCCGTGGCC-3′, added HindIII site underlined). The amplified fragment was subcloned into pET-32a(+) (Novagen) and was recombinantly expressed in the Escherichia coli BL21 strain. The His-tagged MADS29 protein was purified using Ni-NTA resin (Qiagen). DNA fragments of putative promoter regions (~100 bp) containing the CArG-box and altered versions of these fragments (lacking the CArG-box) were PCR amplified (primer sequences are listed in Supplemental Table 1 online) and used for EMSA.
The probes were labeled according to the manufacturer’s instructions (Roche). The labeled probes were incubated with different dosages of MADS29 proteins at room temperature in binding buffer (Beyotime) for 20 min. For the competition experiments, the purified MADS29 protein was incubated with unlabeled competitors for 10 min before adding the labeled probes. The gel electrophoresis was run on a 6% native polyacrylamide gel. After transfer to a membrane, cross-linking, blocking, and antibody incubation, the signal was detected with CSPD (Roche).
IAA and 2,4-D Treatment
Spikes of ZH11 were harvested before or 3 to ~6 h after pollination. The lemma, palea, lodicules, and filaments were removed and soaked in sterilized water for 2 min and then cultured in N6 medium containing IAA (Sigma-Aldrich) or 2,4-D (PhytoTechnology Laboratories) according to previous description (Uchiumi and Okamoto, 2010). The spikelets were cultured in greenhouse at 28°C for 24 h under a 12-h-light/12-h-dark cycle.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: MADS29 (Os02g07430), cDNA of MADS29 (AK109522), MADS30 (Os06g45650), Actin (Os03g50890), AGPS2b (Os08g25734), AGPL1 (Os05g50380), AGPL2 (Os01g44220), AGPL4 (Os07g13980), BEI (Os06g51084), BEII (Os02g32660), PHOL (Os03g55090), PUL (Os04g08270), GBSSI (Os06g04200), ISA1 (Os08g40930), ISA2 (Os05g32710), ISA3 (Os09g29404), SSI (Os06g06560), SSIIa (Os06g12450), SSIIIa (Os08g09230), tested PCD-related genes (Os02g48450, Os04g02120, Os04g08390, Os05g31570, Os06g17970, Os08g30634, Os09g14410, Os09g30220, Os11g13940, Os11g38440, Os11g38580, and Os12g14330), IAA1 (Os01g08320), IAA19 (Os05g48590), and IAA24 (Os07g08460).The hybridization data have been deposited into the Gene Expression Omnibus database (GSE31893; http://www.ncbi.nlm.nih.gov/geo).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Suppressed MADS29 Expression Results in Shrunken Seeds.
Supplemental Figure 2. DNA Gel Blot Analysis of the Transgene Copy Number in A-MADS29 Transgenic Lines.
Supplemental Figure 3. Suppressed Expression of MADS29 Results in the Decreased Grain Filling.
Supplemental Figure 4. Altered Starch Content and Amylopectin Structure under MADS29 Suppression.
Supplemental Figure 5. Seeds of Reciprocal Crosses between A-MADS29 Line (A-2) and Wild-Type ZH11.
Supplemental Figure 6. The Nucellar Projection Isolated Using Laser Capture Microdissection.
Supplemental Table 1. Primers Used for qRT-PCR and EMSA Analysis in This Study.
Acknowledgments
This study was supported by the State Key Project of Basic Research (2012CB944804 and 2011CB100202), the Ministry of Agriculture (2009ZX08009-135B), and the Chinese Academy of Sciences (SIBS2008004). We thank Shu-Ping Xu for assisting with the rice transformation and Wei-Hua Tang for assisting with the laser capture microdissection. We also thank Xiao-Shu Gao for the confocal microscopy observations and the transverse sections.
AUTHOR CONTRIBUTIONS
L.-L.Y. carried out the experiments, analyzed the data, and helped to write the article. H.-W.X. designed the experiments and wrote the article.
References
- Aderem A., Underhill D.M., Ozinsky A., Smith K.D. (1999). Toll-like receptor-2 mediates mycobacteria-induced proinflammatory signaling in macrophages. Proc. Natl. Acad. Sci. USA 96: 14459–14463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An G., Lee S., Jeon J.S., An K., Moon Y.H., Lee S., Chung Y.Y. (2003). Alteration of floral organ identity in rice through ectopic expression of OsMADS16. Planta 217: 904–911 [DOI] [PubMed] [Google Scholar]
- Arora R., Agarwal P., Ray S., Singh A., Singh V., Tyagi A.K., Kapoor S. (2007). MADS-box gene family in rice: Genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8: 242–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker A., Kaufmann K., Freialdenhoven A., Vincent C., Li M.A., Saedler H., Theissen G. (2002). A novel MADS-box gene subfamily with a sister-group relationship to class B floral homeotic genes. Mol. Genet. Genomics 266: 942–950 [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate - A practical and powerful approach to multiple testing. J. R. Statist. Soc. B. 57: 289–300 [Google Scholar]
- Bonneau L., Ge Y., Drury G.E., Gallois P. (2008). What happened to plant caspases? J. Exp. Bot. 59: 491–499 [DOI] [PubMed] [Google Scholar]
- Chen F.Q., Foolad M.R. (1997). Molecular organization of a gene in barley which encodes a protein similar to aspartic protease and its specific expression in nucellar cells during degeneration. Plant Mol. Biol. 35: 821–831 [DOI] [PubMed] [Google Scholar]
- Cho S.C., Jang S.H., Chae S.J., Chung K.M., Moon Y.H., An G.H., Jang S.K. (1999). Analysis of the C-terminal region of Arabidopsis thaliana APETALA1 as a transcription activation domain. Plant Mol. Biol. 40: 419–429 [DOI] [PubMed] [Google Scholar]
- Coen E.S., Romero J.M., Doyle S., Elliott R., Murphy G., Carpenter R. (1990). floricaula: A homeotic gene required for flower development in Antirrhinum majus. Cell 63: 1311–1322 [DOI] [PubMed] [Google Scholar]
- Dangl J.L., Coll N.S., Vercammen D., Smidler A., Clover C., Van Breusegem F., Epple P. (2010). Arabidopsis type I metacaspases control cell death. Science 330: 1393–1397 [DOI] [PubMed] [Google Scholar]
- de Folter S., Angenent G.C. (2006). Trans meets cis in MADS science. Trends Plant Sci. 11: 224–231 [DOI] [PubMed] [Google Scholar]
- de Folter S., Shchennikova A.V., Franken J., Busscher M., Baskar R., Grossniklaus U., Angenent G.C., Immink R.G. (2006). A Bsister MADS-box gene involved in ovule and seed development in petunia and Arabidopsis. Plant J. 47: 934–946 [DOI] [PubMed] [Google Scholar]
- Dominguez F., Moreno J., Cejudo F.J. (2001). The nucellus degenerates by a process of programmed cell death during the early stages of wheat grain development. Planta 213: 352–360 [DOI] [PubMed] [Google Scholar]
- Eason J.R., Ryan D.J., Watson L.M., Hedderley D., Christey M.C., Braun R.H., Coupe S.A. (2005). Suppression of the cysteine protease, aleurain, delays floret senescence in Brassica oleracea. Plant Mol. Biol. 57: 645–657 [DOI] [PubMed] [Google Scholar]
- Egea-Cortines M., Saedler H., Sommer H. (1999). Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 18: 5370–5379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann R., Gramzow L., Melzer R., Theissen G., Becker A. (2010). GORDITA (AGL63) is a young paralog of the Arabidopsis thaliana Bsister MADS box gene ABS (TT16) that has undergone neofunctionalization. Plant J. 63: 914–924 [DOI] [PubMed] [Google Scholar]
- Ge L., Chen H., Jiang J.F., Zhao Y., Xu M.L., Xu Y.Y., Tan K.H., Xu Z.H., Chong K. (2004). Overexpression of OsRAA1 causes pleiotropic phenotypes in transgenic rice plants, including altered leaf, flower, and root development and root response to gravity. Plant Physiol. 135: 1502–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grudkowska M., Zagdanska B. (2004). Multifunctional role of plant cysteine proteinases. Acta Biochim. Pol. 51: 609–624 [PubMed] [Google Scholar]
- Hara-Nishimura I., Yamada K., Matsushima R., Nishimura M. (2001). A slow maturation of a cysteine protease with a granulin domain in the vacuoles of senescing Arabidopsis leaves. Plant Physiol. 127: 1626–1634 [PMC free article] [PubMed] [Google Scholar]
- Hatsugai N., Kuroyanagi M., Yamada K., Meshi T., Tsuda S., Kondo M., Nishimura M., Hara-Nishimura I. (2004). A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science 305: 855–858 [DOI] [PubMed] [Google Scholar]
- Hood E.E., Gelvin S.B., Melchers L.S., Hoekema A. (1993). New Agrobacterium helper plasmids for gene-transfer to plants. Transgenic Res. 2: 208–218 [Google Scholar]
- Innes R.W., DeYoung B.J. (2006). Plant NBS-LRR proteins in pathogen sensing and host defense. Nat. Immunol. 7: 1243–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano B.O. (1971). A simplified assay for milled-rice amylose. Cereal Foods World 16: 334–340 [Google Scholar]
- Kater M.M., Dreni L., Colombo L. (2006). Functional conservation of MADS-box factors controlling floral organ identity in rice and Arabidopsis. J. Exp. Bot. 57: 3433–3444 [DOI] [PubMed] [Google Scholar]
- Krishnan S., Dayanandan P. (2003). Structural and histochemical studies on grain-filling in the caryopsis of rice (Oryza sativa L.). J. Biosci. 28: 455–469 [DOI] [PubMed] [Google Scholar]
- Lam E. (2005). Vacuolar proteases livening up programmed cell death. Trends Cell Biol. 15: 124–127 [DOI] [PubMed] [Google Scholar]
- Lee S., Jung K.H., An G., Chung Y.Y. (2004). Isolation and characterization of a rice cysteine protease gene, OsCP1, using T-DNA gene-trap system. Plant Mol. Biol. 54: 755–765 [DOI] [PubMed] [Google Scholar]
- Lee S., Kim J., Son J.S., Nam J., Jeong D.H., Lee K., Jang S., Yoo J., Lee J., Lee D.Y., Kang H.G., An G. (2003). Systematic reverse genetic screening of T-DNA tagged genes in rice for functional genomic analyses: MADS-box genes as a test case. Plant Cell Physiol. 44: 1403–1411 [DOI] [PubMed] [Google Scholar]
- Lee S.K., et al. (2007). Identification of the ADP-glucose pyrophosphorylase isoforms essential for starch synthesis in the leaf and seed endosperm of rice (Oryza sativa L.). Plant Mol. Biol. 65: 531–546 [DOI] [PubMed] [Google Scholar]
- Li H., Liang W., Hu Y., Zhu L., Yin C., Xu J., Dreni L., Kater M.M., Zhang D. (2011a). Rice MADS6 interacts with the floral homeotic genes SUPERWOMAN1, MADS3, MADS58, MADS13, and DROOPING LEAF in specifying floral organ identities and meristem fate. Plant Cell 23: 2536–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Sun P.L., Zhou H.J., Chen S., Yu S.B. (2011b). Identification of quantitative trait loci associated with germination using chromosome segment substitution lines of rice (Oryza sativa L.). Theor. Appl. Genet. 123: 411–420 [DOI] [PubMed] [Google Scholar]
- Liu W., Xu Z.H., Luo D., Xue H.W. (2003). Roles of OsCKI1, a rice casein kinase I, in root development and plant hormone sensitivity. Plant J. 36: 189–202 [DOI] [PubMed] [Google Scholar]
- Lohmann J.U., Weigel D. (2002). Building beauty: The genetic control of floral patterning. Dev. Cell 2: 135–142 [DOI] [PubMed] [Google Scholar]
- Lombardi L., Casani S., Ceccarelli N., Galleschi L., Picciarelli P., Lorenzi R. (2007). Programmed cell death of the nucellus during Sechium edule Sw. seed development is associated with activation of caspase-like proteases. J. Exp. Bot. 58: 2949–2958 [DOI] [PubMed] [Google Scholar]
- Lombardi L., Ceccarelli N., Picciarelli P., Sorce C., Lorenzi R. (2010). Nitric oxide and hydrogen peroxide involvement during programmed cell death of Sechium edule nucellus. Physiol. Plant. 140: 89–102 [DOI] [PubMed] [Google Scholar]
- Nagamine T., Komae K. (1996). Improvement of a method for chain-length distribution analysis of wheat amylopectin. J. Chromatogr. A 732: 255–259 [Google Scholar]
- Nesi N., Debeaujon I., Jond C., Stewart A.J., Jenkins G.I., Caboche M., Lepiniec L. (2002). The TRANSPARENT TESTA16 locus encodes the ARABIDOPSIS BSISTER MADS domain protein and is required for proper development and pigmentation of the seed coat. Plant Cell 14: 2463–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi A., Nakamura Y., Tanaka N., Satoh H. (2001). Biochemical and genetic analysis of the effects of amylose-extender mutation in rice endosperm. Plant Physiol. 127: 459–472 [PMC free article] [PubMed] [Google Scholar]
- Ohdan T., Francisco P.B., Jr, Sawada T., Hirose T., Terao T., Satoh H., Nakamura Y. (2005). Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J. Exp. Bot. 56: 3229–3244 [DOI] [PubMed] [Google Scholar]
- Prasad K., Zhang X., Tobon E., Ambrose B.A. (2010). The Arabidopsis B-sister MADS-box protein, GORDITA, represses fruit growth and contributes to integument development. Plant J. 62: 203–214 [DOI] [PubMed] [Google Scholar]
- Radchuk V. (2006). Jekyll encodes a novel protein involved in the sexual reproduction of barley. Plant Cell 18: 1652–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupinder S.K., Gurpreet A.K., Manjeet S. (2007). Cell suicide and caspases. Vascul. Pharmacol. 46: 383–393 [DOI] [PubMed] [Google Scholar]
- Russell S.D. (1993). The egg cell: Development and role in fertilization and early embryogenesis. Plant Cell 5: 1349–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., Simpson D., Gietl C. (1999). Programmed cell death in castor bean endosperm is associated with the accumulation and release of a cysteine endopeptidase from ricinosomes. Proc. Natl. Acad. Sci. USA 96: 14159–14164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott A., Wyatt S., Tsou P.L., Robertson D., Allen N.S. (1999). Model system for plant cell biology: GFP imaging in living onion epidermal cells. Biotechniques 26: 1125–, 1128–1132 [DOI] [PubMed] [Google Scholar]
- Shore P., Sharrocks A.D. (1995). The MADS-box family of transcription factors. Eur. J. Biochem. 229: 1–13 [DOI] [PubMed] [Google Scholar]
- Sreenivasulu N., Borisjuk L., Junker B.H., Mock H.-P., Rolletschek H., Seiffert U., Weschke W., Wobus U. (2010). Barley grain development toward an integrative view. Int. Rev. Cell Mol. Biol. 281: 49–89 [DOI] [PubMed] [Google Scholar]
- Sreenivasulu N., Radchuk V., Strickert M., Miersch O., Weschke W., Wobus U. (2006). Gene expression patterns reveal tissue-specific signaling networks controlling programmed cell death and ABA-regulated maturation in developing barley seeds. Plant J. 47: 310–327 [DOI] [PubMed] [Google Scholar]
- Tanaka T., Minamikawa T., Yamauchi D., Ogushi Y. (1993). Expression of an endopeptidase (EP-C1) in Phaseolus vulgaris plants. Plant Physiol. 101: 421–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W.H., Coughlan S., Crane E., Beatty M., Duvick J. (2006). The application of laser microdissection to in planta gene expression profiling of the maize anthracnose stalk rot fungus Colletotrichum graminicola. Mol. Plant Microbe Interact. 19: 1240–1250 [DOI] [PubMed] [Google Scholar]
- Thiel J., Weier D., Sreenivasulu N., Strickert M., Weichert N., Melzer M., Czauderna T., Wobus U., Weber H., Weschke W. (2008). Different hormonal regulation of cellular differentiation and function in nucellar projection and endosperm transfer cells: A microdissection-based transcriptome study of young barley grains. Plant Physiol. 148: 1436–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornberry N.A., Lazebnik Y. (1998). Caspases: Enemies within. Science 281: 1312–1316 [DOI] [PubMed] [Google Scholar]
- Ting J.P.Y., Willingham S.B., Bergstralh D.T. (2008). NLRs at the intersection of cell death and immunity. Nat. Rev. Immunol. 8: 372–379 [DOI] [PubMed] [Google Scholar]
- Turner D.H., Turner J.F. (1960). The hydrolysis of glucose monophosphates by a phosphatase preparation from pea seeds. Biochem. J. 74: 486–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiumi T., Okamoto T. (2010). Rice fruit development is associated with an increased IAA content in pollinated ovaries. Planta 232: 579–592 [DOI] [PubMed] [Google Scholar]
- Wang E., et al. (2008). Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat. Genet. 40: 1370–1374 [DOI] [PubMed] [Google Scholar]
- Wettenhall J.M., Smyth G.K. (2004). limmaGUI: A graphical user interface for linear modeling of microarray data. Bioinformatics 20: 3705–3706 [DOI] [PubMed] [Google Scholar]
- Xue L.J., Zhang J.J., Xue H.W. (2009). Characterization and expression profiles of miRNAs in rice seeds. Nucleic Acids Res. 37: 916–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Saraike T., Shitsukawa N., Hirabayashi C., Takumi S., Murai K. (2009). Class D and Bsister MADS-box genes are associated with ectopic ovule formation in the pistil-like stamens of alloplasmic wheat (Triticum aestivum L.). Plant Mol. Biol. 71: 1–14 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T., Hirano H.-Y. (2006). Function and dversification of MADS-box genes in rice. ScientificWorldJournal 6: 1923–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaki S., Nagato Y., Kurata N., Nonomura K.I. (2011). Ovule is a lateral organ finally differentiated from the terminating floral meristem in rice. Dev. Biol. 351: 208–216 [DOI] [PubMed] [Google Scholar]
- Yang Y.Z., Fanning L., Jack T. (2003). The K domain mediates heterodimerization of the Arabidopsis floral organ identity proteins, APETALA3 and PISTILLATA. Plant J. 33: 47–59 [DOI] [PubMed] [Google Scholar]
- Zhang J., Nallamilli B.R., Mujahid H., Peng Z. (2010). OsMADS6 plays an essential role in endosperm nutrient accumulation and is subject to epigenetic regulation in rice (Oryza sativa). Plant J. 64: 604–617 [DOI] [PubMed] [Google Scholar]