The stabilization of a robust mitotic spindle is required for correct chromosome segregation. GIP proteins interact with microtubule nucleation complexes and localize on mitotic microtubule arrays. The analysis of knockdown mutants suggests that GIP proteins act in both the recruitment of these complexes at nucleation sites and the maintenance of spindle efficiency.
Abstract
Microtubules (MTs) are crucial for both the establishment of cellular polarity and the progression of all mitotic phases leading to karyokinesis and cytokinesis. MT organization and spindle formation rely on the activity of γ-tubulin and associated proteins throughout the cell cycle. To date, the molecular mechanisms modulating γ-tubulin complex location remain largely unknown. In this work, two Arabidopsis thaliana proteins interacting with GAMMA-TUBULIN COMPLEX PROTEIN3 (GCP3), GCP3-INTERACTING PROTEIN1 (GIP1) and GIP2, have been characterized. Both GIP genes are ubiquitously expressed in all tissues analyzed. Immunolocalization studies combined with the expression of GIP–green fluorescent protein fusions have shown that GIPs colocalize with γ-tubulin, GCP3, and/or GCP4 and reorganize from the nucleus to the prospindle and the preprophase band in late G2. After nuclear envelope breakdown, they localize on spindle and phragmoplast MTs and on the reforming nuclear envelope of daughter cells. The gip1 gip2 double mutants exhibit severe growth defects and sterility. At the cellular level, they are characterized by MT misorganization and abnormal spindle polarity, resulting in ploidy defects. Altogether, our data show that during mitosis GIPs play a role in γ-tubulin complex localization, spindle stability and chromosomal segregation.
INTRODUCTION
Microtubules (MTs) play essential roles at the cellular level by participating in cell shape, karyokinesis, cytokinesis, and a large variety of intracellular transports as well. At the organism level, MTs coordinate morphogenesis by defining cell division and expansion axes. They also act as targets and responding agents for incoming signals that regulate their assembly, organization, and dynamics. MTs polymerize in vivo from so-called nucleation complexes. In contrast with organisms with structured MT organizing centers (MTOCs), microtubular cytoskeleton assembly in higher plant cells occurs at various dispersed MT nucleation sites. Massive MT growth events take place in the spindle (Dhonukshe et al., 2006), the phragmoplast (Smertenko et al., 2011), and at both the nuclear envelope (NE) and the plasma membrane. Preexisting MTs are established MT nucleating sites in which γ-tubulin–containing complexes have been found (Stoppin et al., 1994; Binarová et al., 2006; Murata and Hasebe, 2007; Smertenko et al., 2011). These complexes are formed by the interaction of γ-tubulin with additional proteins named GAMMA-TUBULIN COMPLEX PROTEINs (GCPs). In animal cells, large complexes with a ring structure have been characterized and named γ-tubulin ring complexes (γ-TuRCs) (Zheng et al., 1995; Moritz et al., 2000). The assembly of the γ-TuRC requires the iterative association of a core structural subunit (γ-tubulin small complex [γ-TuSC]) composed of two molecules of γ-tubulin (also referred to as GCP1) and one of GCP2 and GCP3 (Kollman et al., 2010). Depending on the organism, the association of additional proteins, including GCP4-GCP6, NEDD1/GCP-WD, and MOZART2/GCP8, is necessary to obtain fully active MT nucleating complexes (Zhu et al., 2009; Guichard et al., 2010; Hutchins et al., 2010; Teixidó-Travesa et al., 2010; Guillet et al., 2011). γ-TuRCs associated with the nucleoporin complex Nup107-160 regulate MT polymerization at kinetochores in Xenopus laevis, participating in the formation of robust spindles (Mishra et al., 2010). Moreover, the Augmin protein complex binds the γ-TuRC and is critical for spindle MT-based MT generation, spindle integrity, and for both karyokinesis and cytokinesis (Lawo et al., 2009; Uehara et al., 2009).
In Arabidopsis thaliana, high order protein complexes composed of GCPs (GCP1 to GCP6) and NEDD1 are present (Seltzer et al., 2007; Zeng et al., 2009; Kong et al., 2010), and the Augmin complex colocalizes with GCPs throughout the cell cycle (Liu et al., 1994, 1993; Nakamura et al., 2010; Ho et al., 2011), indicating that the basic mechanisms of MT assembly are conserved in acentrosomal plant cells (Zeng et al., 2009; Murphy et al., 2001). Interconnections may take place between these complexes to ensure their proper functions.
The mechanisms controlling MT organization, spindle polarity, and stability require the regulation of MT assembly, including MT nucleation complex formation and recruitment as well as the regulation of polymer dynamics. Several studies have recently shown that γ-tubulin complexes are involved in regulating MT dynamics (Raynaud-Messina and Merdes, 2007; Bouissou et al., 2009) and are also critical for their organization. Depletion of γ-tubulin results in the disorganization of interphase MTs and the formation of altered spindles and phragmoplasts (Binarová et al., 2006; Pastuglia et al., 2006). GCP2 is required for the recruitment and/or positioning of γ-tubulin complexes on preexisting cortical MTs (Nakamura and Hashimoto, 2009; Nakamura et al., 2010), and GCP4 depletion causes MT disorganization (Kong et al., 2010). The coordinated activity of specific structural and motor mitogen-activated proteins (MAPs) associated with posttranslational modifications of spindle-associated proteins are also essential for robust spindle functions (Kawabe et al., 2005; Smertenko et al., 2006; Manning et al., 2007). All these data suggest that γ-tubulin and its associated proteins are not strictly involved in MT nucleation.
Beyond the identification of the core components of the γ-TuSC and γ-TuRC, very little information is available about the molecular mechanisms leading to the spatiotemporal regulated activity of the γ-tubulin complex (i.e., assembly, recruitment, and positioning at MTOCs, stabilization, and activation). Emerging findings have revealed the complexity of the γ-TuRC protein–protein interaction network (Choi et al., 2010; Teixidó-Travesa et al., 2010), suggesting that the efficiency of γ-TuRC functioning may be linked to transient interactions and posttranslational modifications (Johmura et al., 2011). Arabidopsis GCP2 and 3 contain nondirect NE targeting domains (Seltzer et al., 2007), but the diversity of MT nucleation sites in plants suggests that γ-tubulin complex localization requires additional factors for recruitment and anchoring at MTOCs.
A yeast two-hybrid screen using the Arabidopsis core subunit GCP3 as a bait led to the identification of a new GCP3-interacting protein named GCP3-INTERACTING PROTEIN1 (GIP1) in this study (Janski et al., 2008). Its human homolog MOZART1 (MZT1) is associated with the γ-TuRCs (Hutchins et al., 2010). Here, we describe the characterization of GIP1 and its homolog GIP2 in Arabidopsis through the analysis of T-DNA insertion mutants and GIP intracellular localization. Our data show that GIP proteins are present at the NE and colocalize with γ-tubulin, GCP3, or GCP4 in the spindle. GIP loss of function is linked with spindle defects, altered cellular patterning, and abnormal plant development.
RESULTS
The Arabidopsis Genome Contains Two GIP Genes That Are Constitutively Expressed
Searches in the Arabidopsis sequence databases revealed a GIP1 (At4g09550) homologous gene, which was named GIP2 (At1g73790). GIP2 encodes a 67–amino acid protein with a predicted molecular mass of 7.4 kD. The GIP1 and GIP2 proteins share 72% amino acid sequence identity (Figure 1, Table 1). GIP homologs are present and well conserved among various species. Interestingly, two members of the GIP family exist in the plant kingdom, while only one is present in the animal kingdom. GIPs are small proteins (67 to 99 amino acids in length) with a well-conserved central region. The percentages of identity among the GIP homologs presented range from 18% for Caenorhabditis elegans to 58% for rice (Oryza sativa) GIPs. The human homolog of GIP1 (MZT1) shares 35% of identity with the Arabidopsis protein. All these sequence analyses suggest that GIP proteins share important properties and functions that have been conserved throughout evolution. Quantitative RT-PCR analyses (see Supplemental Figure 1 online) show that GIP1 and GIP2 are ubiquitously expressed in Arabidopsis. However, GIP expression is higher in young tissues and meristematic cells, arguing in favor of a role in cycling cells.
Figure 1.
Identification of GIP Homologs.
Amino acid sequence alignment of Arabidopsis GIP1 and GIP2 and putative homologs from various species. Conserved identical and similar amino acids are highlighted in gray. Two-letter species abbreviations are as follows: At, Arabidopsis; Ce, C. elegans; Dm, Drosophila; Hs, H. sapiens; Os, O. sativa; Pp, Physcomitrella patens; Sp, Schizosaccharomyces pombe; Xl, Xenopus laevis.
Table 1.
Percentage of Amino Acid Identity between Arabidopsis GIP1 and Some Eukaryotic Homologs
| Species Accession No. | Total No. of Amino Acids | Amino Acid No. Conserved Compared to At GIP1 | % of Identity |
| Arabidopsis GIP1 | 71 | 71 | 100.0 |
| At4g09550 | |||
| Arabidopsis GIP2 | 67 | 51 | 72.0 |
| At1g73790 | |||
| O. sativa GIP1 | 95 | 41 | 58.0 |
| O. sativa GIP2 | 99 | 41 | 58.0 |
| P. patens GIP1 | 68 | 44 | 62.0 |
| P. patens GIP2 | 68 | 41 | 58.0 |
| S. pombe | 85 | 20 | 28.0 |
| C. elegans GIP | 67 | 13 | 18.0 |
| Drosophila GIP | 82 | 16 | 22.5 |
| X. laevis GIP | 72 | 26 | 36.5 |
| H. sapiens GIP1/MOZART1 | 82 | 25 | 35.0 |
GIPs Interact in Vitro with the N-Terminal Domain of GCP3
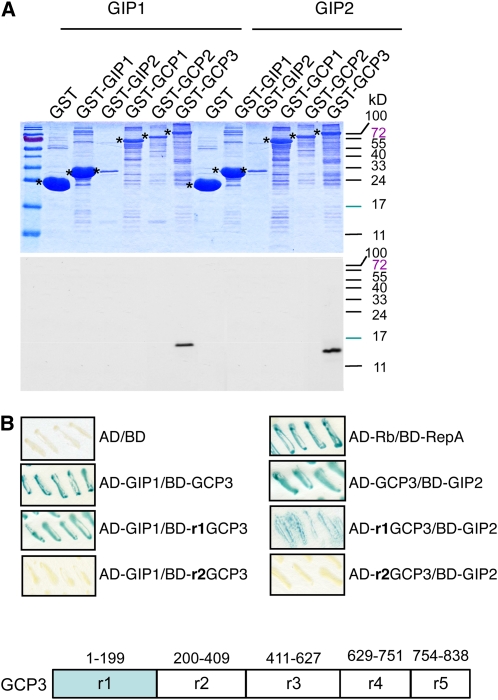
Arabidopsis GIP1 interacts with GCP3, both in a yeast two-hybrid system and in vitro. To confirm whether GIP2 behaves similarly, glutathione S-transferase (GST) pull-down assays were performed. Radiolabeled GIP2, such as GIP1 used here as a positive control, was detected in the GST-GCP3 fraction but not in the GST fraction (Figure 2A), indicating that GIP2 has the propensity to interact with GCP3. In parallel, the interaction of GIP2 with either GST-γ-tubulin, GST-GCP2, GST-GIP1, or GST-GIP2 was also tested. None of these fusion proteins interacted with GIP2.
Figure 2.
GIP1 and GIP2 Interact with GCP3.
(A) In vitro association of GIP1 and GIP2 with GCP3. Pull-down of [35S]Met-labeled GIP1 or GIP2 by GST and GST-GIP1, GST-GIP2, GST-GCP1, GST-GCP2, and GST-GCP3 immobilized on glutathione-Sepharose beads. Protein fractions were separated using SDS-PAGE and visualized through Coomassie blue staining (top panel) and by autoradiography (bottom panel). Asterisks indicate the positions of GST and GST fusion proteins on the left of the corresponding lane.
(B) Mapping of the GIP-interacting region of GCP3 with the yeast two-hybrid system. Interaction between GCP3 or the GCP3 region r1, r2, r3, r4, or r5 fused to the Gal4 binding domain (BD) or the Gal4 activation domain (AD) and GIP1 or GIP2 fused to binding domain or activation domain was assessed in a β-galactosidase filter assay. The appearance of a blue color indicates interaction between the proteins tested. The retinoblastoma protein (Rb1) of maize (Zea mays) and the RepA protein of wheat (Triticum aestivum) dwarf geminivirus (geminivirus of the genus Mastrevirus that infects monocotyledonous plants) were used as positive controls (Xie et al., 1996). Both GIPs interact with the r1 region only of GCP3. A schematic diagram of GCP3 regions (r1 to r5) is shown.
To identify the GCP3 domain(s) that could mediate the binding to GIPs, the interaction of the full-length sequence and of five GCP3 truncated regions corresponding to AA1-199 (r1), AA 200-409 (r2), AA 411-627 (r3), AA 629-751 (r4), and AA754-838 (r5) was investigated (Figure 2B). The 199–amino acid N-terminal region of GCP3 retained the interaction with both GIP1 and GIP2.
GIP1 Is Associated with γ-Tubulin Complexes
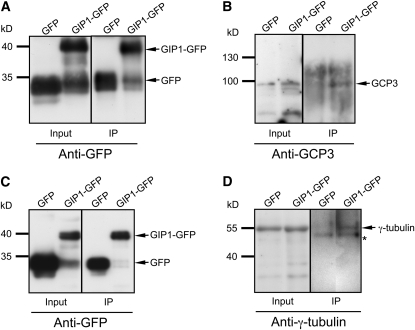
To determine whether GIPs were present in γ-tubulin complexes, Arabidopsis [gip1gip1 GIP2gip2] sesquimutants transformed by a P35S:GIP1:Etag:GFP (for green fluorescent protein) DNA construct were used for coimmunoprecipitation experiments. This construct was able to rescue the gip1 gip2 double mutant, validating its functionality (see details below). The GIP1-Etag-GFP fusion protein (GIP1-GFP) was immunodetected in total protein extracts by an anti-GFP antibody (Figures 3A and 3C). GFP and GIP1-GFP immunopurified fractions were prepared and separately subjected to immunoblotting using anti-GCP3 and anti-γ-tubulin antibodies. Both endogenous GCP3 and γ-tubulin could be detected in enriched GIP1-GFP fractions (Figures 3B and 3D), indicating that GIP1, GCP3, and γ-tubulin are part of the same protein complex in vivo.
Figure 3.
Coimmunoprecipitation of Endogenous GCP3 and γ-Tubulin with GIP1.
Total protein extracts were prepared from Arabidopsis seedlings expressing GFP or from [gip1gip1 GIP2gip2] sesquimutants transformed with the P35S:GIP1:Etag:GFP DNA construct (GIP1-GFP). Protein complexes were immunoprecipitated (IP) with anti-GFP antibodies. The inputs (0.5% of the total protein extracts used for immunoprecipitation) and immunoprecipitated proteins were analyzed by immunoblotting with anti-GFP ([A] and [C]), anti-GCP3 (B), or anti-γ-tubulin (D) antibodies. GFP and GIP1-GFP were detected in total and immunoprecipitated fractions. The positions of coimmunoprecipitated GFP (27 kD), GIP1-GFP (40 kD), GCP3 (95 kD), and γ-tubulin (54 kD) are indicated on the right by arrows. Each experiment was repeated four times independently and showed similar results. The molecular mass markers are shown on the left. In (D), the asterisk indicates the position of a nonspecific protein that is likely to correspond to rabbit IgG heavy chains.
GIPs Localize at the Nuclear Periphery and on Mitotic MT Arrays
The expression and localization of GIP1-GFP or GIP2-GFP was followed in the root apical meristem of 1-week-old Arabidopsis seedlings transformed with either the P35S:GIP1:Etag:GFP or the P35S:GIP2:Etag:GFP DNA construct. The expression of the Etag-GFP fusion proteins was checked using confocal microscopy and through immunoblotting analyses using anti-Etag antibodies (see Supplemental Figure 2A online). It appeared that the expression of GIP1-GFP or GIP2-GFP did not affect the plant phenotype throughout development.
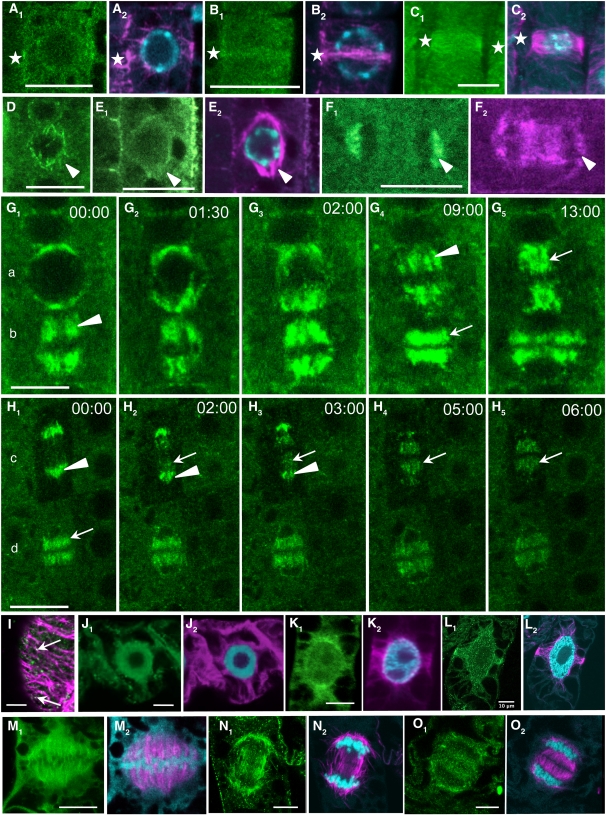
MT immunolabeling (Figures 4A2, 4B2, 4C2, 4E2, and 4F2) and 4′,6-diamidino-2-phenylindole (DAPI) staining (Figures 4A2, 4B2, 4C2, and 4E2) were performed after fixation. A diffuse GFP background could be observed in the cytoplasm of all cells. In such conditions, no significant GFP signal emerged from the fluorescent background in the cortex, which makes these cells inappropriate to the study of cortical MT nucleating sites. However, a significant signal was present within interphase nuclei (n = 150). Twenty percent of cells showed a bright nucleoplasm, 20% had a spotty pattern at the nuclear periphery, and 60% had a nucleoplasmic fluorescence not exceeding that of the cytoplasmic background (see Supplemental Movie 1 online). During mitosis, GIP labeling colocalized with preprophase band (PPB) MTs (Figures 4A1, 4B1, and 4C1, stars), prospindles (Figures 4D and 4E, arrowheads), and mainly kinetochore MTs in anaphase (Figure 4F, arrowheads). When observed in vivo after nuclear envelope breakdown (NEB), GIP2-GFP and GIP1-GFP dynamics could be followed (Figures 4G and 4H, respectively). GIPs localized within spindles during prometaphase to anaphase, and they relocated on interpolar and phragmoplast MTs from mid-anaphase to telophase. During telophase, GIP-GFP also decorated the newly formed NE of daughter cells (see Supplemental Movies 1 and 2 online).
Figure 4.
GIP Localization throughout the Cell Cycle.
(A) to (H) Localization of GIP1- and GIP2-GFP on MT mitotic arrays in dividing meristematic Arabidopsis cells. Root tips were either immunolabeled with anti-α-tubulin antibodies (magenta) and stained with DAPI (cyan) or directly observed in living seedlings ([D], [G], and [H]) using confocal microscopy. Localization of GIP1-GFP ([A1], [B1], [D], [E1], [F1], and [H]) and of GIP2-GFP ([C] and [G]). Images in (G) and (H) correspond to time-lapse observations of mitotic cells from prophase to late anaphase ([G1] to [G5], cell a), prometaphase to mid-telophase ([G1] to [G5], cell b), anaphase to early telophase ([H1] to [H5], cell c), and telophase ([H1] to [H5], cell d). Time lapse in minutes and seconds (see Supplemental Movies 1 and 2 online). Stars indicate the position of PPB in (A) to (C). Arrowheads and arrows in (D) to (H) indicate spindle MTs and interpolar or phragmoplast MTs, respectively.
(I) to (O) Immunolocalization of GIPs in tobacco BY-2 cells. Anti-GIP1 ([I], [J1], to [O1]), anti-α-tubulin antibodies (magenta), and DAPI staining (blue in [J2] to [O2]). Cortex of an interphase cell showing two GIP dots localized on MT arrays (arrows) is shown in (I). Bars = 10 μm.
To study the localization of endogenous GIPs, a polyclonal rabbit antiserum was raised and tested positively against recombinant GIP1 migrating between the 10- and 17-kD positions of protein markers (see Supplemental Figure 2B online), validating the specificity of the anti-GIP antibody. GIP1 was detected when ectopically expressed as a GFP fusion protein (see Supplemental Figure 2B online). Unexpectedly, however, immunoblot analyses using standard conditions could not reveal any endogenous GIPs in protein extracts from either wild-type or P35S:GIP1:Etag:GFP Arabidopsis lines. In addition, no significant signal corresponding to GIP immunolabeling could be observed in wild-type seedling roots. GIPs could only be detected in the analysis of proteins obtained with an improved extraction method (see Supplemental Figure 3C online), which shows that GIPs are present at very low amounts in Arabidopsis tissues. Considering the conservation of GIP in plants, immunomicroscopy experiments were further performed on growing tobacco (Nicotiana tabacum) BY-2 cells (Figures 4I to 4O). In the mitotic cells, GIPs showed the same localization pattern as for GIP-GFP in Arabidopsis. In addition, several cortical dots were observed in interphase cells, some of them coinciding with MT arrays (Figure 4I).
GIP localization studies indicate the presence of GIPs in MT nucleation and organization sites, especially at the onset and during mitosis.
GIP1 Colocalizes with γ-Tubulin GCP3 or GCP4 in the Spindle
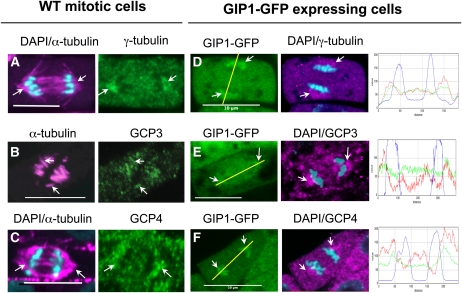
To determine whether GIPs colocalize with γ-tubulin complexes, the distribution of GIPs was compared with that of other GCPs in wild-type plants and plants expressing GIP1-GFP (Figure 5). Anti-GCP3 or anti-γ-tubulin antibodies were used as markers of the plant γ-TuSC. Anti-GCP4 antibodies were used as markers for the γ-TuRC. The labeling was also compared with the microtubular pattern revealed by anti-α-tubulin antibodies. During mitosis, the three GCP proteins were mainly shown to be associated with kinetochore fibers in all dividing cells. This was particularly visible during anaphase with the triangular labeling of polar MT fibers (Figure 5, arrows). When the antibodies were used on GIP-GFP–expressing cells, the GCP labeling coincided with the GIP recombinant protein signal. This was confirmed by superimposed fluorescence intensity profiles (green and red), compared with the chromatid staining (blue). Notably, the GCP3 staining was less visible within the spindle in GIP1-GFP–expressing cells than in wild-type controls, arguing in favor of a steric cluttering limiting the immunoreaction with the presence of GFP. Such a difference was not observed using anti-γ-tubulin or anti-GCP4 antibodies. Altogether, our data show that GIPs colocalize with either GCP3, γ-tubulin, or GCP4 in spindle MTs. These findings are consistent with the association of GIPs with γ-TuSCs and/or γ-TuRCs in Arabidopsis.
Figure 5.
Colocalization of γ-Tubulin, GCP3, or GCP4 with GIP1 in Arabidopsis.
(A) to (C) Wild-type (WT) Arabidopsis anaphase root cells immunolabeled with anti-α-tubulin (magenta) and either anti-γ-tubulin (A), anti-GCP3 (B), or anti-GCP4 antibodies (C). DAPI staining (cyan).
(D) to (F) Anaphase cells expressing GIP1-GFP. γ-Tubulin (D), GCP3 (E), and GCP4 (F) colocalize with GIP1-GFP in spindle polar regions (arrows). Fluorescence intensity signals confirming GIP1 (green) and GCPs (red) colocalization. The graphs to the right show the fluorescence intensities along the yellow lines in parts (D) to and (F) for GIP1 (green), GCPs (red), and chromatid staining (blue).
Bars = 10 μm.
Characterization of gip Single Mutants, Sesquimutants, and Double Mutants
To gain insights into the function of plant GIPs, Arabidopsis T-DNA insertion mutants were studied. The mutated gip1 allele carries a T-DNA insertion in the 5′ untranslated region (UTR) intron located upstream of the open reading frame, and the gip2 allele carries a T-DNA insertion in the 5′ UTR, associated with a downstream deletion of 67 bp (see Supplemental Figure 3A online). These positions were both checked using PCR amplification and sequencing of the T-DNA flanking regions. Homozygous gip1 and gip2 lines were crossed to produce the double mutants.
GIP1 and GIP2 mRNA levels were determined in the wild type, [GIP1gip1 gip2gip2] or [gip1gip1 GIP2gip2] sesquimutants, and double mutants. No significant wild-type GIP1 mRNA was detected in gip1 mutants using specific primers located upstream and downstream from the T-DNA insertion. RT PCR using downstream primers indicates that the T-DNA insertion prevents a correct splicing of GIP1 transcripts, suggesting a significant knockdown of GIP1 expression (see Supplemental Figure 3B online). For gip2, due to the T-DNA insertion in the 5′ end of the 5′ UTR, expression levels could only be checked using downstream PCR primers. In gip2 mutants, a chimeric transcript, corresponding to the 3′ end of the T-DNA insertion in fusion with the first exon (5′ UTR) of GIP2, was detected. The accumulation of this aberrant transcript (above the level of wild-type mRNAs) is probably due to the presence of the 35S promoter in the T-DNA. However, the production of functional GIP2 is unlikely according to the classic translation rules of eukaryotic mRNAs. Such a case has already been described for γ-tubulin T-DNA insertion mutants (Pastuglia et al., 2006).
The presence of GIPs in wild-type and mutant lines was checked by immunoblotting of concentrated protein extracts. GIPs were faintly detected in the extracts from wild-type plants (see Supplemental Figure 3C online), and similar low amounts of GIP1 or GIP2 were present in sesquimutants. However, GIPs were undetectable in extracts from double mutants according to our experimental conditions, suggesting that T-DNA insertions impair GIP synthesis from both mutant alleles. The gip double mutants may therefore be considered as strong knockdown mutants.
The gip Double Mutant Shows Defects in Zygote Development and Exhibits an Altered Growth Phenotype
To check whether GIP1 and GIP2 played a role at specific tissue or developmental stages, seedlings, growing and mature plants corresponding to gip1 and gip2 homozygous single mutants, and wild-type lines were compared. Single mutants and sesquimutants had a wild-type-like developmental phenotype and were fertile, suggesting functional redundancy for GIP genes in standard growing conditions.
The combination of gip1 and gip2 homozygous alleles induced severe developmental defects (Figure 6). Eighteen to 23% of seeds were unable to germinate (see Supplemental Table 1 online; n = 446). For comparison, only 2% of ungerminated seeds were observed after control wild-type autofertilization (n = 450). Ten days after germination, a strong phenotype was revealed for 2.5 to 5.8% of seedlings, suggesting that GIP loss induces gametophyte and/or embryo lethality. Indeed, the analysis of the sesquimutant siliques showed either ovules, in which the development of the zygote was blocked at a globular stage, certainly resulting in aborted seeds, or embryos of a much smaller size than that of heterozygotes (Figures 6A to 6F). All the phenotypically affected plantlets exhibited a [gip1gip1 gip2gip2] genotype (Figures 6G to 6K). Their development was strongly delayed and only one-third of the seedlings developed until flowering, indicating that GIPs are required for plant development. Double mutant seedlings had short primary roots that grew like corkscrews (Figures 6G and 6I), very short hypocotyl, and abnormal cotyledons appearing as green callus-like structures (Figures 6G and 6I). Rosette development showed narrow, curved leaves before the formation of short bolts (Figures 6J and 6K). To confirm that the observed phenotypes were linked to gip mutations, DNA sequences corresponding to 2.8-kb GIP1 and 2.4-kb GIP2 genomic fragments were reintroduced into sesquimutants. Both wild-type genomic constructs rescued the developmental phenotype in the double mutant background obtained after sesquimutant autofertilization (Figures 6M and 6N). Moreover, the P35S:GIP1:Etag:GFP construct also restored a wild-type-like phenotype of the gip1 gip2 double mutant (see Supplemental Figure 4 online). A specific GFP signal was observed using confocal microscopy in sporophytic tissues (notably in dividing root cells), in gametophytes, and in dissected developing embryos (see Supplemental Figure 5 online), indicating that in the complemented Arabidopsis mutant lines obtained, GIP1-GFP was expressed throughout the different developmental stages.
Figure 6.
gip Double Mutants Exhibit Developmental Defects.
(A) and (B) Siliques of GIP sesquimutants containing aborted and abnormal seeds (arrows).
(C) to (F) Developing embryos of sesquimutants. Arrows indicate abnormal embryos.
(G) to (I) Ten-day-old seedlings of wild-type (WT) ([I], right), sesquimutants ([I], asterisks, [gip1gip1 GIP2gip2], left; [GIP1gip1 gip2gip2], right), and double mutant ([G], [H], and arrows in [I]) plants.
(J) to (L) Shoots of double mutant ([J] and [K]) and wild-type (l) plants. (J2) is at a higher magnification of (J1).
(M) and (N) The phenotype of the double mutant is rescued by a GIP1 (N) Arabidopsis genomic fragment.
(O) and (P) Flowers of double mutant plants ([O], arrow) have larger sepals and petals compared with flowers of sesquimutant (asterisk) or wild-type plants. Note the absence of ovules and pollen in cleared flowers of double mutants (P). The double mutants are sterile.
Bars = 50 μm in (E) and (F), 500μm in (A) to (D), (G), and (H), 1 cm in (I), (J2), and (K) to (N), and 1 mm in (O) and (P).
[See online article for color version of this figure.]
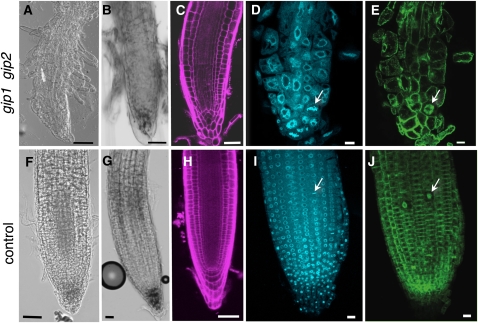
Defects in meristem organization and maintenance were observed in the shoot and the root apical meristems of double mutants. The cellular organization of the root apical meristem was carefully investigated. The pattern of the different cell types was disturbed, and cells were often bulging, irregular, and bigger in size compared with the wild type or sesquimutants (Figures 7A to 7E). Collapsing meristematic, elongating, and differentiating root zones were observed in differential interference contrast (DIC) and after propidium iodide cell wall labeling (Figures 7A and 7C). In double mutants, starch granules stained with Lugol were reduced and laterally mislocated in a few remnant columella cells (Figure 7B). The stem cell niche was disorganized, and it was often difficult to identify the quiescent center. Root tip cells of double mutants showed nuclei that were three times as large as those of sesquimutants (Figures 7D and 7I) in which one wild-type GIP allele was present, suggesting either an increase in DNA content or enhanced chromatin decondensation. These nuclei were distorted in shape, losing their circularity but maintaining a DAPI fluorescence intensity similar to that of control nuclei. Altogether, these data suggest that GIPs may be linked to the regulation of cellular polarity and patterning as well as to the determination of nuclear morphology.
Figure 7.
Morphology Defects of the Root Apical Meristem in the gip Double Mutant.
(A) to (E) Roots of gip double mutants.
(F) to (H) Wild-type roots.
(I) to (J) Roots of sesquimutant plants.
Roots are observed in phase contrast ([A], [B], [F], and [G]). Confocal optical median sections are shown in (C) to (E) and (H) to (J). Roots were labeled with Lugol ([B] and [G]), propidium iodide ([C] and [H]), DAPI ([D] and [I]), or immunolabeled with anti-α-tubulin antibodies ([E] and [J]). Chromosomes in (D) and (I) and corresponding spindles ([E] and [J]) of metaphase cells are indicated by arrows. Bars = 50 μm in (A) to (C) and (F) to (H) and 20 μm in (D), (E), (I), and (J).
[See online article for color version of this figure.]
Abnormal MT Distribution and Chromosome Instability Are Associated with Cell Division Defects in Double Mutants
To better understand the causes of tissue patterning defects, mitosis was further investigated in double mutants by combining two approaches. First, MTs and chromatin of fixed cells were stained. Then, to correlate these results with GIP functions during cell division, MTs were visualized through the MT binding domain (MBD) of MAP4 fused to GFP, and chromatin was highlighted through the expression of a fluorescent Histone H2A variant (H2AX). Thus, a P35S:MBD:GFP or a P35S:H2AX:RFP (for red fluorescent protein) construct was introgressed in the mutated gip1 gip2 background. This helped to follow MT array dynamics or chromatin segregation in meristematic root cells.
In gip1 gip2 seedlings, the mitotic index was half of that observed in the wild type, indicating that cell division progression was affected in gip double mutants (see Supplemental Figure 6 online). This was in accordance with the simultaneously observed smaller root length.
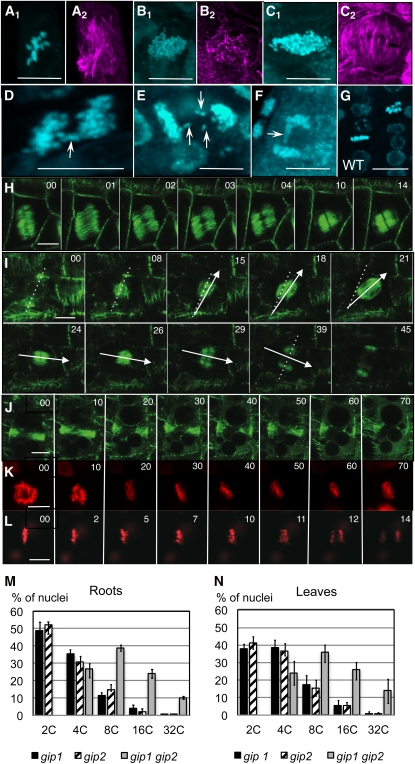
Cytoskeletal and chromosomal abnormalities were found in 86% of the cells analyzed in gip1 gip2 root meristems (Figure 8; n = 50). At the microtubular level, 30% of division figures corresponded to cells in late G2 (attested by the presence of a PPB), 38% were in prometaphase and metaphase, 7.5% in anaphase, and 22% in telophase. In wild-type roots, 28% of cells were in G2, 20% in prometaphase and metaphase, 20% in anaphase, and 26% in telophase (n = 50). These results indicated an anaphase delay during mitosis in gip1 gip2 mutants. At mitotic onset, the PPB formed. However, the positioning of PPBs was altered (n = 15) and either oblique (33%), asymmetric (64%), or double (3%) PPBs were observed. After NEB, defects in spindle polarity (Figure 8A), spindle MT organization (Figure 8B), and/or spindle size (Figures 8C and 8H) occurred. For the 20% of cells that formed a PPB, spindles were either misoriented (Figure 8I; see Supplemental Movie 3 online) or did not form (Figure 8J). Large polygonal cells which were able to form oblique spindles and divide (Figure 8H) could lead to daughter cells losing the straight cellular organization in root files like that shown in Figure 7. Unstable metaphase plates were observed, but as spindle orientation defects could be spontaneously rescued (Figure 8I), the amount of mitotic defects could possibly be underestimated. In case of PPB narrowing and further decondensation without spindle formation (Figure 8J), the cellular DNA content doubled leading to polyploid cells.
Figure 8.
Abnormal MT Array Organization and Higher Ploidy Levels in the gip Double Mutant.
(A) to (G) Anti-α-tubulin and/or DAPI staining in double mutants and wild-type (WT) (G) roots. Abnormal spindles ([A2], [B2], and [C2]) with numerous chromosomes ([A1], [B1], and [C1]). Metaphase plates ([A] and [C1]) and late anaphases ([B1] and [D] to [F]) with lagging chromosomes (arrows). Bars = 10 μm.
(H) to (L) Time-lapse confocal images of dividing cells in young roots of double mutants expressing MBD-GFP ([H] to [J]) or H2AX-RFP ([K] and [L]) showing an oblique spindle (H), spindle orientation rescue ([I]; see Supplemental Movie 3 online), an absence of spindle formation (J), metaphase arrest (K), and anaphase delay ([L]; see Supplemental Movie 4 online). In (I), the dotted line indicates the PPB axis and the arrows underline the spindle rotation. Bars = 10 μm.(M) and (N) Flow cytometry analysis of root (M) or leaf (N) nuclei of [gip1gip1 GIP2gip2] (black) or [GIP1gip1 gip2gip2] sesquimutant (hatched) and double mutant (gray) plantlets. The percentage of 2C, 4C, 8C, 16C, and 32C nuclei are presented, each assayed in biological triplicates.
At the level of chromosomes, metaphase plates and anaphase segregation showed a large increase in the number of chromosomes (Figure 8C) with the presence of lagging chromosomes in anaphase (Figures 8D to 8F) and the appearance of micronuclei in interphase when compared with control wild-type cells (Figure 8G). After NEB, chromosome congression was observed but condensed chromosomes remained blocked in metaphase in 20% of cells (Figure 8K). For the remaining 80% of mitotic cells, anaphase was strongly delayed (Figure 8L; see Supplemental Movie 4 online) and chromatid segregation could lead to aneuploidy linked to chromosome lagging.
All these observations suggested an increase in the ploidy level relative to wild-type cells. To determine ploidy at the DNA level, nuclear DNA contents were compared using flow cytometry. In all tissues analyzed, the nuclei of the wild-type or sesquimutant plants showed a majority of 2C and 4C nuclei corresponding to diploid cells. A complete loss of diploid cells and a shift up to 32C DNA content was detected in double mutants, confirming the presence of polyploid cells (Figures 8M and 8N). This indicates that three replication cycles without division and/or defects in DNA segregation can occur in double mutants.
Altogether, our data suggest a defect in the mechanisms of spindle orientation, which could argue in favor of a role of GIPs in kinetochore fiber formation or stability and in spindle polarity and/or integrity. In addition, anaphase delay and PPB decondensation reveal the activation of the mitotic checkpoint, but lagging chromosomes or misequilibrated anaphases suggest that plant cells can partially overcome chromosome abnormalities. The high number of polyploid and aneuploid cells in the very young root tip also suggests that, early in embryogenesis, the root stem cells were likely impeded in their normal division. During seed germination, a new genetic program may then overcome the higher DNA amount and restart cell division.
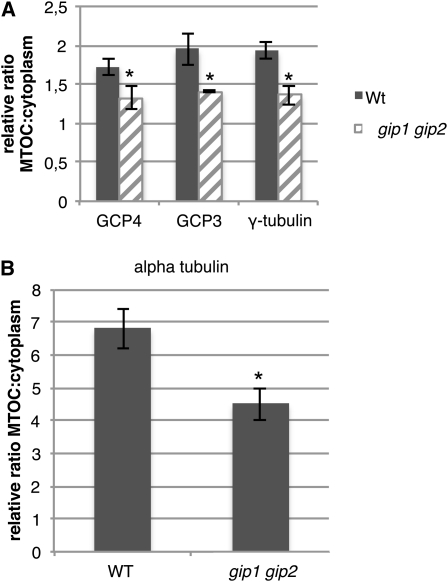
GIP Depletion Affects γ-Tubulin, GCP3, and GCP4 Localization at MT Nucleation Sites
GIP localization was highly similar to either γ-tubulin, GCP3, or GCP4 distribution, suggesting a molecular interaction among GIPs and γ-TuSCs/γ-TuRCs. To test this hypothesis, the amount of γ-tubulin, GCP3, and GCP4 present at NE, spindles, and phragmoplasts was compared between wild-type and gip double mutant cells. A significant decrease in γ-tubulin, GCP3, or GCP4 labeling was observed in mutant cells (Figure 9A; P < 0.05, n = 40; see Supplemental Figure 7 online). A simultaneous reduction in the α-tubulin fluorescence intensity at the nuclear periphery and within spindles and phragmoplasts was observed in mutants compared with wild-type cells (Figure 9B; n = 120), indicating that GIP depletion affects the density of perinuclear and mitotic MT arrays. These results show that GIPs play a role in the proper localization of γ-TuSC or γ-TuRC components and in MT array organization.
Figure 9.
The Decrease in GCP Fluorescence Intensity at MT Nucleation Sites in gip Double Mutants Is Associated with a Reduction of the α-Tubulin Signal.
(A) Relative ratio of fluorescence between MT nucleation sites (NE, spindle, and phragmoplast) and the cytoplasmic background after GCP4, GCP3, and γ-tubulin immunolabeling in wild-type cells (Wt) (gray) and gip mutants (hatched).
(B) α-Tubulin intensity variation in corresponding cells. Asterisk indicates a statistically significant difference from the wild type (WT) using Student’s t tests; P < 0.05.
The data represent the average of three independent biological replicates. Error bars represent sd.
DISCUSSION
GIP Homologs Are Present in Most Eukaryotes
First identified in Arabidopsis (Janski et al., 2008), GIP-related sequences were found in a large variety of eukaryotes from algae to humans and are ubiquitously expressed in plants. They are significantly smaller than the GCPs characterized so far. They do not share sequence similarities with them either.
Tandem affinity purification experiments and mass spectrometry analyses of proteins isolated from HeLa cells arrested in mitosis revealed that Homo sapiens GIP/MZT1 copurified with either Hs γ-tubulin, Hs GCP3, or Hs GCP6 (Hutchins et al., 2010). Hs GIP/MZT1 was also associated with purified γ-TuRCs (Teixidó-Travesa et al., 2010). These data, in addition to our results showing that Arabidopsis GIPs associate with γ-tubulin complexes, suggest that eukaryotic GIP homologs may be key factors for the basic mechanisms involving MT-dependent functions.
GIP Subcellular Localization in Higher Plant Cells
The dynamic distribution of GIP-GFP (Figures 10A and 10B) revealed the nuclear surface that was shown to act as an MTOC in plant cells (Stoppin et al., 1994; Seltzer et al., 2007), especially in late G2 (Erhardt et al., 2002). As GCP3 is located at the NE, the perinuclear GIP pattern may correspond to the direct interaction of GIP and GCP3 in vivo.
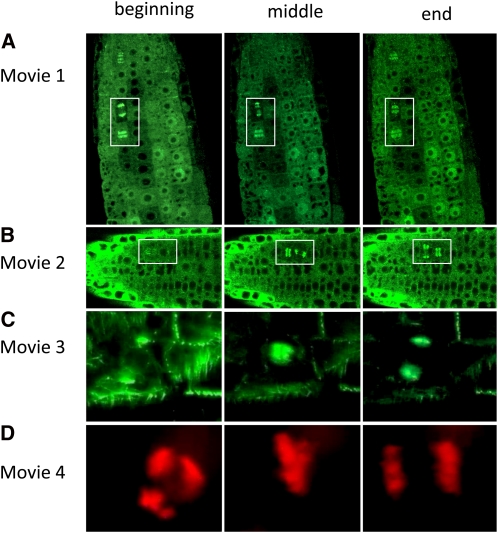
Figure 10.
Still Images from the Beginning, Middle, and End of Supplemental Movies 1 to 4 Online.
See Figures 4 and 8 for additional still images. Boxes in (A) and (B) are details shown in Figures 4H and 4G, respectively.
[See online article for color version of this figure.]
The presence of GIPs is required for the specific localization of GCP3 and other GCPs at the NE. However, as the analysis of GIP sequences does not highlight any transmembrane domain, it is likely that other partners are involved in the anchorage of γ-TuRCs at the NE. Its small size may argue in favor of a regulatory role of GIP that could modulate γ-tubulin complex anchorage.
At preprophase, GIP is observed in the PPB and as polar caps around the nucleus. The latter localization overlaps that of γ-tubulin, NEDD1, and AUG3 (Liu et al., 1993; Zeng et al., 2009; Ho et al., 2011). During mitosis, Hs GIP/MZT1 was detected at centrosomes and on the mitotic spindle (Hutchins et al., 2010). In plants such as Arabidopsis and tobacco, GIP localized along kinetochore MTs—it was similar to that described for γ-tubulin (Liu et al., 1993) and for GFP-tagged GCP2 and GCP3 (Nakamura et al., 2010)—and along phragmoplast MTs as shown for GCP2, 3, and 4 (Kong et al., 2010; Nakamura et al., 2010). The subcellular distribution of plant and human GIPs is therefore compatible with a role of these proteins in MT assembly and organization, especially at the entry and during progression of mitosis.
GIP Depletion Induces Major Cell Morphogenesis and Developmental Defects Associated with Misorganization of MT Arrays and Chromosome Instability
The absence of obvious growth phenotypes in gip1 or gip2 single mutants indicates that GIP1 and GIP2 have some overlapping functions in standard growing conditions. GIP depletion in gip1 gip2 double mutant lines was linked to MT array disorganization and chromosome mis-segregation (Figures 10C and 10D). The regular parallel cell files generated from the quiescent center in wild-type roots (De Tullio et al., 2010) is altered in gip double mutants with MTs disorganization similar to the alterations described for BY-2 lines expressing TUA-GFP (Yoneda et al., 2005). Defects in spindle positioning and chromosome segregation, associated with abnormally oriented cell plates, were also reported after MT End Binding (EB1c) protein depletion (Komaki et al., 2010). Alterations of cell divisions and polarity may therefore affect cell fate and patterning in gip mutants. It is well established that the PPB is an early mark of division polarity (Lloyd and Chan, 2006). The formation of double PPBs was linked to high perturbations of MT arrays and further spindle defects (Yoneda et al., 2005). As GIP-depleted cells showed an increased number of mispositioned PPBs, GIPs may be involved in the stabilization of these MT arrays. It would be interesting to determine whether GIPs participate in the mechanisms that regulate the establishment of the division plane in addition to TANGLED (Rasmussen et al., 2011) and to RanGAP (Xu et al., 2008). The spindle formation and position are also linked to the Ran pathway, as many spindle assembly factors, such as Targeting Protein for Xklp2 (Vos et al., 2008) and Ribonucleic acid export 1 (Lee et al., 2009), are regulated by RanGTP (Kalab and Heald, 2008).
The conservation of GCPs and GIPs among eukaryotes argues in favor of functional similarities in the molecular mechanisms of spindle assembly in plant and animal cells. In Drosophila melanogaster S2 cells, >200 genes contribute to spindle assembly (Goshima et al., 2007). GCP2-RNA interference (RNAi) delays prometaphase and induces polyploidy and cells escape prematurely from the mitotic checkpoint due to the defect in γ-TuSC integrity (Colombié et al., 2006). In human cells, GIP/MZT1 RNAi leads to strong mitotic defects, such as monopolar spindles (Hutchins et al., 2010). In fungi, γ-tubulin complexes are involved in the spindle assembly checkpoint (Vardy and Toda, 2000), establishment of spindle polarity (Prigozhina et al., 2004), chromosome segregation, and cytokinesis (Hendrickson et al., 2001). Defects in mitosis progression are observed in the Arabidopsis gip double mutants, suggesting that, in addition to their role in MT nucleation, the GCPs and their associated proteins carry out other functions essential for mitosis, MT organization and dynamics, and the control of the mitotic checkpoint (Jung et al., 2001; Colombié et al., 2006; Bouissou et al., 2009).
Robust spindle assembly is dependent on the establishment of kinetochore fibers and de novo chromosome-based MT assembly (Akiyoshi et al., 2010). The spindle disorganization and the presence of lagging chromosomes in double mutants suggest that GIPs are key components favoring spindle fiber stability and the establishment of proper chromosome/MT connections leading to efficient chromatid segregation. During telophase, the formation of a dense phragmoplast is rapid, and these MTs also certainly need to become stably oriented to favor Golgi vesicle migration. Such self-organization of MT arrays (Zhang and Dawe, 2011) may be conditioned by the activity of GIPs.
GIPs May Modulate γ-Tubulin Nucleating Complex Localization and Activity
Considering their small size, compared with structural GCPs (GCP1 to GCP6), GIP proteins could play a specific role in γ-tubulin complex formation, recruitment, and/or cellular distribution.
In the gip double mutants, a significant decrease in γ-tubulin complex recruitment at MT nucleation sites was observed. A similar loss of γ-tubulin at centrosomes was observed after MZT1 depletion in human cells (Hutchins et al., 2010), suggesting that GIPs and MZT1 share conserved functions. T-DNA insertions in the Arabidopsis γ-tubulin genes or direct γ-tubulin depletion by RNAi led to cell file perturbations and defects in the formation and organization of MT arrays (Binarová et al., 2006; Pastuglia et al., 2006). These phenotypes are very similar to those observed for gip mutants, indicating a functional link between GIPs and γ-tubulin. The absence of GIPs may lead to a defect in the localization of γ-tubulin complexes. Consequently, fewer MTs may be nucleated or stabilized, resulting in MT array mispolarity and instability. Ploidy defects may then become a consequence of the abortion of functional spindles.
In Arabidopsis, GIPs may therefore participate in the recruitment of γ-TuSC or γ-TuRC complexes to the NE before mitotic entry and may maintain the proper localization of MT nucleation complexes throughout mitosis. Such a hypothesis could be reinforced by determining the relative stoichiometry of GIPs in plant γ-tubulin complexes and by establishing whether GIPs are new core components or more transient interactors.
METHODS
Plant Materials, Transformation, and Growth Conditions
The Arabidopsis thaliana gip1 T-DNA insertion line (GABI_213D01) was obtained from the Gabi-Kat collection (Rosso et al., 2003) via the Nottingham Arabidopsis Stock Centre. The gip2 line (FLAG_36406) was obtained from FLAGdb/FST at the Institut de la Recherche Agronomique (Versailles, France; Samson et al., 2002). Homozygous insertion lines identified through PCR genotyping (see Supplemental Table 2 online) were crossed to produce double mutant lines. The GFP-MBD marker (Camilleri et al., 2002) or the H2AX-RFP marker in Arabidopsis was introduced into both GIP sesquimutant backgrounds through crossing.
Arabidopsis transformation was performed using the floral dipping method (Clough and Bent, 1998) and the Agrobacterium tumefaciens strain GV3101. Wild-type and transgenic Arabidopsis lines were grown in vitro on Murashige and Skoog medium (Serva) at 20°C in 12 h per day of 70 μmol m−2 s−1 fluorescent lighting, subcultured on soil, and transferred to a growth chamber with light/dark cycles of 16 h/8 h for 2 weeks and then to the greenhouse. Tobacco BY-2 cells (Nicotiana tabacum cv Bright Yellow 2) were grown according to Nagata et al. (1992).
Construction of Recombinant Plasmids
Arabidopsis GIP1, GIP2, GCP1, GCP2, and GCP3 cDNA fragments were generated by PCR using primers derived from the gene sequences (see Supplemental Table 2 online). To express GST fusion proteins in Escherichia coli, the full coding sequences of GIP1, GIP2, GCP1, GCP2, and GCP3 were cloned in pGEX-2TK (GE Healthcare).
To generate antibodies, the Arabidopsis GIP1 coding sequence was introduced into pQE60 (Qiagen). For in vitro–coupled transcription/translation and yeast two-hybrid assays, GIP1 and GIP2 coding sequences were cloned in pGBKT7 (Clontech). The GIP2 and GCP3 coding sequences were inserted into pAS2ΔΔ and pACTIIst (Institut Pasteur, Centre National de la Recherche Scientifique). The DNA fragments corresponding to GCP3 regions 1 to 5 (Seltzer et al., 2007) were subcloned in pGBKT7. pGAD10-GIP1 (Janski et al., 2008) was also used.
GIP1 and GIP2 coding sequences were cloned under the control of the cauliflower mosaic virus 35S promoter into the NcoI site of the plant expression vector pNEG-X1 (Vos et al., 2008) to produce C-terminal E-tag peptide (GE Healthcare) and GFP fusion proteins. For stable transformation of Arabidopsis, the plant expression cassette was excised from recombinant pNEG-X1 vectors with HindIII and EcoRI and subcloned into the binary vector pGreenII 0029 or pGreenII 0179 (Hellens et al., 2000). The genomic fragments used for complementation experiments were based on the annotated At4g09550 (GIP1) and the At1g73790 (GIP2) loci. The GIP1 2.8-kb or GIP2 2.4-kb DNA fragments encompassed the entire coding sequences, a 2- or 1.8-kb promoter sequence upstream of the start codon, and a 0.6- or 0.4-kb sequence downstream of the stop codon, respectively. GIP1 or GIP2 PCR products were cloned into the EcoRI site of pGreenII 0179.
In Vitro Translation and Pull-Down Experiments
GST pull-down assays were performed as described (Janski et al., 2008). [35S]Met-labeled GIP1 or GIP2 was produced by incubating pGBKT7-GIP1 or pGBKT7-GIP2 in the TNT T7 Coupled Reticulocyte Lysate System (Promega). GST and GST fusions proteins were expressed in E. coli BL21 Rosetta-pRARE (Novagen).
Yeast Two-Hybrid Assays
The two-hybrid analysis of interacting proteins was performed using yeast strain Y187 and a cotransformation procedure according to the Clontech yeast protocols handbook PT-3024-1. Positive protein interactions were detected in a β-galactosidase colony-lift filter assay.
Coimmunoprecipitation
Fifteen-day-old transgenic Arabidopsis seedlings (500 mg fresh weight) expressing GFP (a gift from C. Himber, Institut de Biologie Moléculaire des Plantes) or GIP1-GFP were frozen in liquid nitrogen and ground to powder. The extraction buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Nonidet P-40, and 5% glycerol), supplemented with Complete Protease Inhibitor cocktail (Roche Diagnostics), was added to the powder. The supernatant was collected after centrifugation at 15,000g. Protein complexes, containing GCP3 and/or γ-tubulin associated with GIP1-GFP, were enriched with polyclonal anti-GFP antibodies (Invitrogen) bound to the Dynabeads Protein A (Invitrogen) or with the μMACS GFP isolation kit (Miltenyi Biotec), respectively, according to the manufacturer’s instructions. Protein fractions were separated with SDS-PAGE and transferred to Immobilon membranes (Millipore) for immunoblotting. GFP and GIP1-GFP recombinant proteins were detected using the polyclonal anti-GFP antibody (Invitrogen; 1:5000 dilution). Native proteins were detected by the monoclonal anti-γ-tubulin (Exbio Praha, Academy of Sciences) and the polyclonal anti-GCP3 (Seltzer et al., 2007) antibodies (1:5000).
Antibodies and Immunolocalization
To generate polyclonal antibodies directed against Arabidopsis GIP1, six-His-tagged GIP1 was produced in E. coli BL21 (DE3) cells transformed with pQE60-GIP1, purified on a nickel-nitrilotriacetic acid agarose column according to the manufacturer’s protocol (Qiagen), and injected into rabbits at the Polyclonal Antibodies Service (Institut de Génétique et Biologie Moléculaire et Cellulaire, Illkirch, France).
Arabidopsis seedlings and BY-2 cells were fixed for 40 min in 1.5% paraformaldehyde and 0.5% glutaraldehyde in PEMT buffer (50 mM PIPES, 2 mM EGTA, 2 mM MgSO4, and 0.05% Triton X-100, pH 7.2) and then treated as described by Erhardt et al. (2002). Nonspecific binding was blocked by incubation in PBS (3.2 mM Na2HPO4, 0.5 mM KH2PO4, 1.3 mM KCl, and 135 mM NaCl, pH 7.4) containing 2% BSA and 0.1% Triton X-100 (IF buffer) for 10 min. Primary and secondary antibodies were diluted in IF buffer and incubated for 1 h at room temperature. The primary antibodies used in this study were the rabbit polyclonal anti-GIP1 (1/1000), monoclonal anti-α-tubulin (clone DM1A; Sigma-Aldrich) (1/6000), polyclonal anti-γ-tubulin (1/4000) (Seltzer et al., 2007), anti-GCP3 (1/1000) (Erhardt et al., 2002), and anti-GCP4 (1/300) generously given by B. Liu (University of California, Davis, CA). Alexa 488– and Alexa 568–conjugated goat anti-rabbit IgG and goat anti-mouse IgG secondary antibodies (1:300) were from Molecular Probes. DNA was stained using 0.1 μg/mL DAPI.
Fluorescence Microscopy
The MAP4 MBD fused to GFP (GFP-MBD) was used as reporter protein to visualize dynamic changes in the organization of the MT cytoskeleton in living seedlings mounted in water. Seedlings and immunostained cells were observed with a Zeiss LSM 700 confocal microscope in multitracking mode using 405-, 488-, or 555-nm laser excitation.
Anatomical Analysis of Arabidopsis Seedlings and Gametophytes
To observe ovules and embryos, Arabidopsis flowers and developing seeds were cleared in 8/2/1 (w/v/v) chloral hydrate/water/glycerol. Bright-field, phase contrast, or DIC images were acquired using a Zeiss AxioImager Z1 microscope and an Axiocam MRm digital camera. For the analysis of root apical meristem morphology, Arabidopsis seedlings were treated with 10 μg/mL propidium iodide and observed using confocal microscopy and DIC.
Sequence Alignments and Databases
Available GenBank EST libraries were screened with Arabidopsis and human GIPs using the TBLASTN algorithm. Putative positive hits were processed in MacVector to extract full-coding frames and translation products. New identified GIP homolog sequences were then reused for iterative searches as long as new hits were obtained. Final alignments were performed using ClustalW in MacVector software.
Flow Cytometry
The nuclear DNA content was estimated through flow cytometry as previously described (Marie and Brown, 1993). In brief, various Arabidopsis tissues were chopped with a razor blade in Galbraith buffer (45 mM MgCl2, 30 mM sodium citrate, 20 mM MOPS, and 1% Triton X-100), filtered through a 48-μm mesh, and stained using propidium iodide (50 μg/mL). Ploidy levels of 5000 to 10,000 stained nuclei were determined using a CyFlow SL cytometer (Partec SARL) with a 532-nm solid-state laser (100 mW) excitation filter. The emission was collected through a 990-nm long-pass filter.
RT-PCR
Total RNA was extracted from plant organs with the Nucleospin RNA plant kit (Macherey-Nagel) according to the manufacturer’s instructions after grinding with glass beads (1.7/2 mm) in a Precellys 24 grinder (Bertin Technologies) at 5500 rpm, 2 × 30 s. After treatment with DNase I (Fermentas), RNAs were stored at −80°C. One microgram of total RNA was then reverse transcribed with the Superscibed III reverse transcriptase (Invitrogen) using oligo(dT) as primers.
PCR amplification was performed with 1 μL of cDNA in a final volume of 25 μL with the qPCR MasterMIX Plus for SYBER Green I with fluorescein (Eurogentec France) and gene-specific primers (see Supplemental Table 2 online). As a reference for PCR quantification, the actin gene was amplified with specific primers. Three quantifications were performed for each sample as described previously (Roa et al., 2009).
Protein Extractions and Immunoblotting
Total protein extracts were prepared from Arabidopsis leaf disks, ground in SDS-PAGE loading buffer (Evrard et al., 2002), and separated using SDS-PAGE. Concentrated protein extracts from 3-week-old Arabidopsis seedlings were prepared according to the protocol described by Wang et al. (2006). The proteins were finally dissolved in SDS-PAGE loading buffer and separated with Tricine-SDS-PAGE (Schägger, 2006). The protein concentration of the extracts was determined on Coomassie Brilliant Blue–stained SDS-PAGE gels and the Bradford protein assay (Bio-Rad) following the manufacturer’s protocol. After SDS-PAGE, the proteins were transferred to Immobilon-PSQ membranes (Millipore) and processed for immunoblotting analysis as described by Evrard et al. (2002). The membranes were treated with the anti-GIP1 rabbit polyclonal antibody (1:5000 dilution) or the anti E-tag peptide (GE Healthcare) monoclonal antibody (1:5000) overnight at 4°C and horseradish peroxidase–conjugated IgG (Molecular Probes; 1:5000) 1 h at room temperature. Signals were revealed using Immobilon Western detection reagents (Millipore).
Accession Numbers
Sequence data from this study can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: Arabidopsis (At) GIP1, At4g09550; At GIP2, At1g73790; At GCP1, At3g61650; At GCP2, At5g17410; At GCP3, At5g06680; Oryza sativa (Os) GIP1, CF989340; Os GIP2, NP_001050438; Physcomitrella patens (Pp) GIP1, DC931782; Pp GIP2, FC379761; Schizosaccharomyces pombe (Sp) GIP, P0CF96; Caenorhabditis elegans (Ce) GIP, NP_001021663; Drosophila melanogaster (Dm) GIP, HDC08084; Xenopus laevis (Xl) GIP, BG162997; Homo sapiens (Hs) GIP, NP_001065243; gip1 T-DNA insertion line, GABI_213D01; and gip2 T-DNA insertion line, FLAG_36406.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Analysis of GIP1 and GIP2 Expression with Quantitative RT-PCR.
Supplemental Figure 2. Immunoblot Analysis of Recombinant GIP Proteins.
Supplemental Figure 3. Molecular Characterization of GIP1 and GIP2 T-DNA Insertion Alleles.
Supplemental Figure 4. Immunolabeling of γ-Tubulin and GCP3 in Wild-Type and GIP Double Mutant Meristematic Cells.
Supplemental Figure 5. Expression of P35S:GIP1:GFP in an Arabidopsis Mutated [gip1gip1 GIP2gip2] Background.
Supplemental Figure 6. Mitotic Index in the Root Apical Meristems of the GIP Double Mutant.
Supplemental Figure 7. Immunolocalization of GCPs in gip Mutants.
Supplemental Table 1. Segregation of GIP Mutants after Sesquimutant Self-Pollination and Growth under Nonselective Medium.
Supplemental Table 2. Primers Used for Genotyping and RT-PCR Experiments.
Supplemental Movie 1. Arabidopsis Root Expressing GIP1-GFP.
Supplemental Movie 2. Mitotic Arabidopsis Root Cells Expressing GIP2-GFP.
Supplemental Movie 3. Expression of MBD-GFP in a gip1 gip2 Genetic Background Showing Spindle Instability.
Supplemental Movie 4. Expression of H2AX-GFP in a gip1 gip2 Genetic Background Showing Anaphase Delay.
Acknowledgments
This work was supported by the Centre National de la Recherche Scientifique. Microscopy experiments were carried out at the Strasbourg-Esplanade cellular imaging facilities sponsored by the Centre National de la Recherche Scientifique, the Université de Strasbourg, and the Région Alsace, the Association de la Recherche sur le Cancer, and the Ligue Nationale contre le Cancer. We thank Bo Liu for the gift of anti-GCP4 antibodies and Christophe Himber for providing the GFP-expressing Arabidopsis line. We also thank Jérôme Mutterer and Malek Alouia for their help. We thank Spencer C. Brown for his help and constructive discussion and Lucien Blech for his valuable advice. Part of this work used the facilities of the cell biology unit of the Imagif platform at the Centre de Recherche de Gif-sur-Yvette, supported by the Conseil Général de l’Essonne.
AUTHOR CONTRIBUTIONS
N.J. and K.M. performed the main research, obtained the tools, and analyzed the data. E.H., M.-E.C., and J.-L.E. designed the research part concerning the molecular approaches, supervised the obtaining of data, and analyzed the results with the PhD students. J.-L.E. performed the sequence comparisons. G.H. contributed to the part concerning the expression levels of all the compared genes. M. Batzenschlager contributed to coimmunoprecipitation and cloning experiments. J.-L.E. designed and performed the biochemical approaches. A.-C.S. performed and supervised the microscopy approaches. M. Bourge performed the flow cytometry experiments. E.H. and A.-C.S. wrote the article.
References
- Akiyoshi B., Sarangapani K.K., Powers A.F., Nelson C.R., Reichow S.L., Arellano-Santoyo H., Gonen T., Ranish J.A., Asbury C.L., Biggins S. (2010). Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature 468: 576–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binarová P., Cenklová V., Procházková J., Doskocilová A., Volc J., Vrlík M., Bögre L. (2006). Gamma-tubulin is essential for acentrosomal microtubule nucleation and coordination of late mitotic events in Arabidopsis. Plant Cell 18: 1199–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouissou A., Vérollet C., Sousa A., Sampaio P., Wright M., Sunkel C.E., Merdes A., Raynaud-Messina B. (2009). Gamma-tubulin ring complexes regulate microtubule plus end dynamics. J. Cell Biol. 187: 327–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilleri C., Azimzadeh J., Pastuglia M., Bellini C., Grandjean O., Bouchez D. (2002). The Arabidopsis TONNEAU2 gene encodes a putative novel protein phosphatase 2A regulatory subunit essential for the control of the cortical cytoskeleton. Plant Cell 14: 833–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.K., Liu P., Sze S.K., Dai C., Qi R.Z. (2010). CDK5RAP2 stimulates microtubule nucleation by the gamma-tubulin ring complex. J. Cell Biol. 191: 1089–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colombié N., Vérollet C., Sampaio P., Moisand A., Sunkel C., Bourbon H.M., Wright M., Raynaud-Messina B. (2006). The Drosophila gamma-tubulin small complex subunit Dgrip84 is required for structural and functional integrity of the spindle apparatus. Mol. Biol. Cell 17: 272–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Tullio M.C., Jiang K., Feldman L.J. (2010). Redox regulation of root apical meristem organization: Connecting root development to its environment. Plant Physiol. Biochem. 48: 328–336 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P., Vischer N., Gadella T.W.J., Jr (2006). Contribution of microtubule growth polarity and flux to spindle assembly and functioning in plant cells. J. Cell Sci. 119: 3193–3205 [DOI] [PubMed] [Google Scholar]
- Erhardt M., Stoppin-Mellet V., Campagne S., Canaday J., Mutterer J., Fabian T., Sauter M., Muller T., Peter C., Lambert A.M., Schmit A.C. (2002). The plant Spc98p homologue colocalizes with gamma-tubulin at microtubule nucleation sites and is required for microtubule nucleation. J. Cell Sci. 115: 2423–2431 [DOI] [PubMed] [Google Scholar]
- Evrard J.L., Nguyen I., Bergdoll M., Mutterer J., Steinmetz A., Lambert A.M. (2002). A novel pollen-specific alpha-tubulin in sunflower: Structure and characterization. Plant Mol. Biol. 49: 611–620 [DOI] [PubMed] [Google Scholar]
- Goshima G., Wollman R., Goodwin S.S., Zhang N., Scholey J.M., Vale R.D., Stuurman N. (2007). Genes required for mitotic spindle assembly in Drosophila S2 cells. Science 316: 417–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichard P., Chrétien D., Marco S., Tassin A.M. (2010). Procentriole assembly revealed by cryo-electron tomography. EMBO J. 29: 1565–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillet V., Knibiehler M., Gregory-Pauron L., Remy M.H., Chemin C., Raynaud-Messina B., Bon C., Kollman J.M., Agard D.A., Merdes A., Mourey L. (2011). Crystal structure of γ-tubulin complex protein GCP4 provides insight into microtubule nucleation. Nat. Struct. Mol. Biol. 18: 915–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens R.P., Edwards E.A., Leyland N.R., Bean S., Mullineaux P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Hendrickson T.W., Yao J., Bhadury S., Corbett A.H., Joshi H.C. (2001). Conditional mutations in gamma-tubulin reveal its involvement in chromosome segregation and cytokinesis. Mol. Biol. Cell 12: 2469–2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C.M., Hotta T., Kong Z., Zeng C.J., Sun J., Lee Y.R., Liu B. (2011). Augmin plays a critical role in organizing the spindle and phragmoplast microtubule arrays in Arabidopsis. Plant Cell 23: 2606–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins J.R., et al. (2010). Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science 328: 593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janski N., Herzog E., Schmit A.C. (2008). Identification of a novel small Arabidopsis protein interacting with gamma-tubulin complex protein 3. Cell Biol. Int. 32: 546–548 [DOI] [PubMed] [Google Scholar]
- Johmura Y., Soung N.K., Park J.E., Yu L.R., Zhou M., Bang J.K., Kim B.Y., Veenstra T.D., Erikson R.L., Lee K.S. (2011). Regulation of microtubule-based microtubule nucleation by mammalian polo-like kinase 1. Proc. Natl. Acad. Sci. USA 108: 11446–11451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung M.K., Prigozhina N., Oakley C.E., Nogales E., Oakley B.R. (2001). Alanine-scanning mutagenesis of Aspergillus gamma-tubulin yields diverse and novel phenotypes. Mol. Biol. Cell 12: 2119–2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P., Heald R. (2008). The RanGTP gradient - A GPS for the mitotic spindle. J. Cell Sci. 121: 1577–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe A., Matsunaga S., Nakagawa K., Kurihara D., Yoneda A., Hasezawa S., Uchiyama S., Fukui K. (2005). Characterization of plant Aurora kinases during mitosis. Plant Mol. Biol. 58: 1–13 [DOI] [PubMed] [Google Scholar]
- Kollman J.M., Polka J.K., Zelter A., Davis T.N., Agard D.A. (2010). Microtubule nucleating gamma-TuSC assembles structures with 13-fold microtubule-like symmetry. Nature 466: 879–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komaki S., Abe T., Coutuer S., Inzé D., Russinova E., Hashimoto T. (2010). Nuclear-localized subtype of end-binding 1 protein regulates spindle organization in Arabidopsis. J. Cell Sci. 123: 451–459 [DOI] [PubMed] [Google Scholar]
- Kong Z., Hotta T., Lee Y.R., Horio T., Liu B. (2010). The gamma-tubulin complex protein GCP4 is required for organizing functional microtubule arrays in Arabidopsis thaliana. Plant Cell 22: 191–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawo S., et al. (2009). HAUS, the 8-subunit human Augmin complex, regulates centrosome and spindle integrity. Curr. Biol. 19: 816–826 [DOI] [PubMed] [Google Scholar]
- Lee J.Y., Lee H.S., Wi S.J., Park K.Y., Schmit A.C., Pai H.S. (2009). Dual functions of Nicotiana benthamiana Rae1 in interphase and mitosis. Plant J. 59: 278–291 [DOI] [PubMed] [Google Scholar]
- Liu B., Joshi H.C., Wilson T.J., Silflow C.D., Palevitz B.A., Snustad D.P. (1994). Gamma-tubulin in Arabidopsis: Gene sequence, immunoblot, and immunofluorescence studies. Plant Cell 6: 303–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Marc J., Joshi H.C., Palevitz B.A. (1993). A gamma-tubulin-related protein associated with the microtubule arrays of higher plants in a cell cycle-dependent manner. J. Cell Sci. 104: 1217–1228 [DOI] [PubMed] [Google Scholar]
- Lloyd C., Chan J. (2006). Not so divided: The common basis of plant and animal cell division. Nat. Rev. Mol. Cell Biol. 7: 147–152 [DOI] [PubMed] [Google Scholar]
- Manning A.L., Ganem N.J., Bakhoum S.F., Wagenbach M., Wordeman L., Compton D.A. (2007). The kinesin-13 proteins Kif2a, Kif2b, and Kif2c/MCAK have distinct roles during mitosis in human cells. Mol. Biol. Cell 18: 2970–2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie D., Brown S.C. (1993). A cytometric exercise in plant DNA histograms, with 2C values for 70 species. Biol. Cell 78: 41–51 [DOI] [PubMed] [Google Scholar]
- Mishra R.K., Chakraborty P., Arnaoutov A., Fontoura B.M., Dasso M. (2010). The Nup107-160 complex and gamma-TuRC regulate microtubule polymerization at kinetochores. Nat. Cell Biol. 12: 164–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz M., Braunfeld M.B., Guénebaut V., Heuser J., Agard D.A. (2000). Structure of the gamma-tubulin ring complex: A template for microtubule nucleation. Nat. Cell Biol. 2: 365–370 [DOI] [PubMed] [Google Scholar]
- Murata T., Hasebe M. (2007). Microtubule-dependent microtubule nucleation in plant cells. J. Plant Res. 120: 73–78 [DOI] [PubMed] [Google Scholar]
- Murphy S.M., Preble A.M., Patel U.K., O’Connell K.L., Dias D.P., Moritz M., Agard D., Stults J.T., Stearns T. (2001). GCP5 and GCP6: Two new members of the human gamma-tubulin complex. Mol. Biol. Cell 12: 3340–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata T., Nemoto Y., Hasezawa S. (1992). Tobacco BY-2 cell-line as the HeLa-cell in the cell biology of higher-plants. Int. Rev. Cytol. 132: 1–30 [Google Scholar]
- Nakamura M., Ehrhardt D.W., Hashimoto T. (2010). Microtubule and katanin-dependent dynamics of microtubule nucleation complexes in the acentrosomal Arabidopsis cortical array. Nat. Cell Biol. 12: 1064–1070 [DOI] [PubMed] [Google Scholar]
- Nakamura M., Hashimoto T. (2009). A mutation in the Arabidopsis gamma-tubulin-containing complex causes helical growth and abnormal microtubule branching. J. Cell Sci. 122: 2208–2217 [DOI] [PubMed] [Google Scholar]
- Pastuglia M., Azimzadeh J., Goussot M., Camilleri C., Belcram K., Evrard J.L., Schmit A.C., Guerche P., Bouchez D. (2006). Gamma-tubulin is essential for microtubule organization and development in Arabidopsis. Plant Cell 18: 1412–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigozhina N.L., Oakley C.E., Lewis A.M., Nayak T., Osmani S.A., Oakley B.R. (2004). Gamma-tubulin plays an essential role in the coordination of mitotic events. Mol. Biol. Cell 15: 1374–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen C.G., Sun B., Smith L.G. (2011). Tangled localization at the cortical division site of plant cells occurs by several mechanisms. J. Cell Sci. 124: 270–279 [DOI] [PubMed] [Google Scholar]
- Raynaud-Messina B., Merdes A. (2007). Gamma-tubulin complexes and microtubule organization. Curr. Opin. Cell Biol. 19: 24–30 [DOI] [PubMed] [Google Scholar]
- Roa H., Lang J., Culligan K.M., Keller M., Holec S., Cognat V., Montané M.H., Houlné G., Chabouté M.E. (2009). Ribonucleotide reductase regulation in response to genotoxic stress in Arabidopsis. Plant Physiol. 151: 461–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso M.G., Li Y., Strizhov N., Reiss B., Dekker K., Weisshaar B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Samson F., Brunaud V., Balzergue S., Dubreucq B., Lepiniec L., Pelletier G., Caboche M., Lecharny A. (2002). FLAGdb/FST: A database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res. 30: 94–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H. (2006). Tricine-SDS-PAGE. Nat. Protoc. 1: 16–22 [DOI] [PubMed] [Google Scholar]
- Seltzer V., Janski N., Canaday J., Herzog E., Erhardt M., Evrard J.L., Schmit A.C. (2007). Arabidopsis GCP2 and GCP3 are part of a soluble gamma-tubulin complex and have nuclear envelope targeting domains. Plant J. 52: 322–331 [DOI] [PubMed] [Google Scholar]
- Smertenko A.P., Chang H.Y., Sonobe S., Fenyk S.I., Weingartner M., Bögre L., Hussey P.J. (2006). Control of the AtMAP65-1 interaction with microtubules through the cell cycle. J. Cell Sci. 119: 3227–3237 [DOI] [PubMed] [Google Scholar]
- Smertenko A.P., Piette B., Hussey P.J. (2011). The origin of phragmoplast asymmetry. Curr. Biol. 21: 1924–1930 [DOI] [PubMed] [Google Scholar]
- Stoppin V., Vantard M., Schmit A.C., Lambert A.M. (1994). Isolated plant nuclei nucleate microtubule assembly: The nuclear surface in higher plants has centrosome-like activity. Plant Cell 6: 1099–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixidó-Travesa N., Villén J., Lacasa C., Bertran M.T., Archinti M., Gygi S.P., Caelles C., Roig J., Lüders J. (2010). The gammaTuRC revisited: A comparative analysis of interphase and mitotic human gammaTuRC redefines the set of core components and identifies the novel subunit GCP8. Mol. Biol. Cell 21: 3963–3972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara R., Nozawa R.S., Tomioka A., Petry S., Vale R.D., Obuse C., Goshima G. (2009). The augmin complex plays a critical role in spindle microtubule generation for mitotic progression and cytokinesis in human cells. Proc. Natl. Acad. Sci. USA 106: 6998–7003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardy L., Toda T. (2000). The fission yeast gamma-tubulin complex is required in G(1) phase and is a component of the spindle assembly checkpoint. EMBO J. 19: 6098–6111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos J.W., Pieuchot L., Evrard J.L., Janski N., Bergdoll M., de Ronde D., Perez L.H., Sardon T., Vernos I., Schmit A.C. (2008). The plant TPX2 protein regulates prospindle assembly before nuclear envelope breakdown. Plant Cell 20: 2783–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Vignani R., Scali M., Cresti M. (2006). A universal and rapid protocol for protein extraction from recalcitrant plant tissues for proteomic analysis. Electrophoresis 27: 2782–2786 [DOI] [PubMed] [Google Scholar]
- Xie Q., Sanz-Burgos A.P., Hannon G.J., Gutiérrez C. (1996). Plant cells contain a novel member of the retinoblastoma family of growth regulatory proteins. EMBO J. 15: 4900–4908 [PMC free article] [PubMed] [Google Scholar]
- Xu X.M., Zhao Q., Rodrigo-Peiris T., Brkljacic J., He C.S., Müller S., Meier I. (2008). RanGAP1 is a continuous marker of the Arabidopsis cell division plane. Proc. Natl. Acad. Sci. USA 105: 18637–18642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda A., Akatsuka M., Hoshino H., Kumagai F., Hasezawa S. (2005). Decision of spindle poles and division plane by double preprophase bands in a BY-2 cell line expressing GFP-tubulin. Plant Cell Physiol. 46: 531–538 [DOI] [PubMed] [Google Scholar]
- Zeng C.J., Lee Y.R., Liu B. (2009). The WD40 repeat protein NEDD1 functions in microtubule organization during cell division in Arabidopsis thaliana. Plant Cell 21: 1129–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Dawe R.K. (2011). Mechanisms of plant spindle formation. Chromosome Res. 19: 335–344 [DOI] [PubMed] [Google Scholar]
- Zheng Y., Wong M.L., Alberts B., Mitchison T. (1995). Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature 378: 578–583 [DOI] [PubMed] [Google Scholar]
- Zhu H., Fang K., Fang G. (2009). FAM29A, a target of Plk1 regulation, controls the partitioning of NEDD1 between the mitotic spindle and the centrosomes. J. Cell Sci. 122: 2750–2759 [DOI] [PMC free article] [PubMed] [Google Scholar]