This work analyzed DNA methylation and gene expression changes in two Arabidopsis ecotypes and their reciprocal hybrids and found that increased DNA methylation in both hybrid genomes, possibly directed by the RdDM pathway, may play a role in heterosis.
Abstract
Heterosis is a fundamental biological phenomenon characterized by the superior performance of a hybrid over its parents in many traits, but the underlying molecular basis remains elusive. To investigate whether DNA methylation plays a role in heterosis, we compared at single-base-pair resolution the DNA methylomes of Arabidopsis thaliana Landsberg erecta and C24 parental lines and their reciprocal F1 hybrids that exhibited heterosis. Both hybrids displayed increased DNA methylation across their entire genomes, especially in transposable elements. Interestingly, increased methylation of the hybrid genomes predominantly occurred in regions that were differentially methylated in the two parents and covered by small RNAs, implying that the RNA-directed DNA methylation (RdDM) pathway may direct DNA methylation in hybrids. In addition, we found that 77 genes sensitive to methylome remodeling were transcriptionally repressed in both reciprocal hybrids, including genes involved in flavonoid biosynthesis and two circadian oscillator genes CIRCADIAN CLOCK ASSOCIATED1 and LATE ELONGATED HYPOCOTYL. Moreover, growth vigor of F1 hybrids was compromised by treatment with an agent that demethylates DNA and by abolishing production of functional small RNAs due to mutations in Arabidopsis RNA methyltransferase HUA ENHANCER1. Together, our data suggest that genome-wide remodeling of DNA methylation directed by the RdDM pathway may play a role in heterosis.
INTRODUCTION
Heterosis, or hybrid vigor, refers to the phenomenon that progeny of diverse members of a species are superior to their parents in many traits, such as biomass, growth rate, and fertility (Birchler et al., 2003, 2010; Hochholdinger and Hoecker, 2007; Z.J. Chen, 2010). This phenomenon has been extensively exploited to increase agronomic production; however, the underlying biological mechanisms remain largely unknown despite more than a century of study (Birchler et al., 2003; Hochholdinger and Hoecker, 2007; Birchler et al., 2010; Z.J. Chen, 2010). Genetic explanations for heterosis include the classic dominance and overdominance hypotheses, which were proposed over a century ago, and the more recently developed epistasis hypothesis (Crow, 1948; Yu et al., 1997). In brief, the dominance hypothesis postulates that heterosis arises from complementation of defective parental alleles, whereas the overdominance hypothesis postulates that it is due to interactions between parental alleles in hybrids; by contrast, the epistasis hypothesis regards interactions between different parental genes in hybrids as a key component of heterosis (Birchler et al., 2003, 2010; Hochholdinger and Hoecker, 2007; Z.J. Chen, 2010). Although each hypothesis is supported by many lines of evidence, little consensus has yet been reached. Moreover, these hypotheses are largely conceptual and not connected to molecular principles and are therefore far from explaining the molecular basis of heterosis (Birchler et al., 2003; Hochholdinger and Hoecker, 2007; He et al., 2011).
Genetic quantitative trait locus analyses indicate that a large number of genes contribute to heterotic phenotypes (Semel et al., 2006; Frascaroli et al., 2007; Lippman and Zamir, 2007; Radoev et al., 2008; Birchler et al., 2010; Meyer et al., 2010). However, a single heterozygous gene, SINGLE FLOWER TRUSS, was shown to cause hybrid vigor in tomato (Solanum lycopersicum), providing the first molecular example of a single overdominant gene driving heterosis (Krieger et al., 2010). In addition, several studies revealed that epigenetic variation contributed to the molecular mechanisms of complex traits, including hybrid vigor (Cubas et al., 1999; Manning et al., 2006; Shindo et al., 2006; Ni et al., 2009; He et al., 2010). For example, a recent study showed that changes in the expression of a few regulatory genes, such as the circadian oscillators CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), were associated with growth vigor in Arabidopsis thaliana hybrids (Ni et al., 2009). Another report suggested that epigenetic diversity between parental lines, such as differences in levels of 24-nucleotide small interfering RNAs (siRNAs) in two Arabidopsis ecotypes, created allelic variants that influenced the activity of a large number of metabolic and regulatory genes that may contribute to heterotic phenotypes (Groszmann et al., 2011).
Epigenetic regulation of chromatin structure and genomic stability, which is essential for the proper interpretation of genetic information and determination of phenotype, includes DNA methylation, histone modifications, and certain aspects of siRNA pathways (Henderson and Jacobsen, 2007; Zhang, 2008; He et al., 2011). DNA methylation, the addition of a methyl group to a cytosine, primarily serves as an epigenetic silencing mechanism and predominantly occurs in transposons and other repetitive DNA elements in plants (Martienssen and Colot, 2001; Bird, 2002; Chan et al., 2005; Law and Jacobsen, 2010). In contrast with DNA methylation in mammals, which occurs almost exclusively in the symmetric CG context, DNA methylation in plants commonly occurs at cytosine bases in all sequence contexts: the symmetric CG and CHG contexts (where H = A, T, or C) and the asymmetric CHH context (Henderson and Jacobsen, 2007; Law and Jacobsen, 2010). In the Arabidopsis genome, DNA methylation levels of ~24, 6.7, and 1.7% were observed in the CG, CHG, and CHH contexts, respectively (Cokus et al., 2008; Lister et al., 2008). In plants, de novo methylation is catalyzed by DOMAINS REARRANGED METHYLTRANSFERASE2 (DRM2) and maintained by different pathways: CG and CHG methylation is maintained by DNA METHYLTRANSFERASE1 (MET1) and CHROMOMETHYLASE3 (CMT3), respectively, whereas asymmetric CHH methylation is maintained through persistent de novo methylation by DRM2 (Chan et al., 2005; Law and Jacobsen, 2010).
A central question in understanding DNA methylation is how sequences are targeted for silencing. Emerging evidence shows that siRNAs generated by the RNA interference pathway can target homologous genomic DNA sequences for cytosine methylation in all sequence contexts through a process called RNA-directed DNA methylation (RdDM) (Wassenegger et al., 1994; Mathieu and Bender, 2004; Henderson and Jacobsen, 2007; Law and Jacobsen, 2009, 2010). In this process, 24-nucleotide siRNAs guide the de novo methyltransferase DRM2 to homologous loci to establish DNA methylation, which leads to transcriptional silencing (Law and Jacobsen, 2009, 2010). Plants encode multiple homologs of the RNA interference components, some of which function in RdDM, including DICER-LIKE3 (DCL3) and ARGONAUTE4 (AGO4). The endoribonuclease DCL3 generates 24-nucleotide siRNAs that are loaded onto AGO4, and these AGO4-associated siRNAs are proposed to guide the cytosine-methyltransferase activity of DRM2 (Henderson and Jacobsen, 2007; Law and Jacobsen, 2010).
Recently developed microarray and high-throughput sequencing technologies have enabled investigation of the molecular basis of heterosis at the genomic level (Zhang et al., 2008a; Ni et al., 2009; Wei et al., 2009; Lai et al., 2010; Song et al., 2010; Groszmann et al., 2011). Several recent studies using methylation-sensitive amplified polymorphism analysis have shown that cytosine DNA methylation is altered in F1 hybrids (Zhao et al., 2008; Banaei Moghaddam et al., 2010; Nakamura and Hosaka, 2010). However, the overall changes in hybrid methylomes and their possible roles in heterosis remain unclear. In this study, we investigated the DNA methylation landscapes of the entire genomes of two Arabidopsis ecotypes, Landsberg erecta (Ler) and C24, and their reciprocal F1 hybrids at single-base resolution. We show that both hybrids had increased DNA methylation across their entire genomes and that this increase predominantly occurred in regions that were differentially methylated in the parents and covered by small RNAs, implying that RdDM may be responsible for increased DNA methylation in reciprocal hybrids. We further show that growth vigor of F1 hybrids was compromised by treatment with an agent that demethylates DNA and by abolishing production of functional small RNAs due to mutations in an Arabidopsis RNA methyltransferase HUA ENHANCER1 (HEN1). Our data therefore suggest that genome-wide methylome remodeling in hybrids may contribute to their growth vigor.
RESULTS
DNA Methylomes of Ler and C24 Parents and Their Reciprocal Hybrids
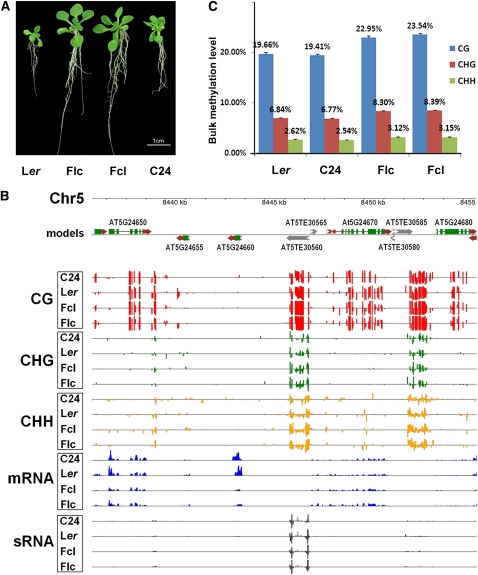
Consistent with a recent report (Groszmann et al., 2011), both reciprocal F1 hybrids of Arabidopsis Ler and C24 ecotypes exhibited significant growth vigor in many characters, including fresh weight, leaf number, root length, and silique number (Figure 1A; see Supplemental Figure 1 and Supplemental Table 1 online). Heterotic phenotypes were evident in the hybrids within 15 d after sowing (Figure 1A). To investigate the potential role of DNA methylation in heterosis, we determined the DNA methylomes of the 15-d-old Ler and C24 parental lines and their reciprocal F1 hybrids by methylC-seq, a next-generation technology enabling direct sequencing of the entire cytosine methylome at single-base resolution (Lister et al., 2008). Single-base resolution maps of the DNA methylomes of Arabidopsis, human, and several other species have recently been reported (Cokus et al., 2008; Lister et al., 2008, 2009, 2011; Hsieh et al., 2009; Lister and Ecker, 2009; Feng et al., 2010; Zemach et al., 2010).
Figure 1.
DNA Methylation Landscapes of the Genomes of Ler and C24 and Their Reciprocal Hybrids.
(A) Heterotic phenotypes of 15-d-old Arabidopsis F1 hybrids relative to their parents. Flc, F1 where maternal line is Ler; Fcl, F1 where maternal line is C24.
(B) An example of DNA methylation profiles at three cytosine contexts and correlated mRNA and sRNA profiles in a representative region of chromosome 5 in Arabidopsis F1 hybrids and their parents. The top panel shows annotated protein-coding genes and TEs in this region based on the TAIR10 release of the Arabidopsis genome. Predicted coding sequences are shown in green, 5′- and 3′-untranslated regions are shown in red, and predicted TEs are shown in black. The directions of both genes and TEs are indicated by arrows. The bottom panel shows the locations of cytosine methylation, mRNAs, and sRNAs detected by high-throughput sequencing. The height of the bar is proportional to the number of reads detected on each strand.
(C) Elevated DNA methylation in F1 hybrids relative to their parents. Columns represent bulk methylation levels at three cytosine contexts in the parental and hybrid genomes as determined by bisulfite sequencing. Data are averages of three independent biological replicates, each consisting of 100 pooled seedlings of the corresponding genotype. Error bars represent sd.
MethylC-seq libraries were constructed from DNAs extracted from the Ler and C24 parents and their reciprocal hybrids, respectively, and subjected to high-throughput Solexa (Illumina) sequencing. Sequencing reads from each library were mapped to the TAIR10 release of the Arabidopsis genome using the MAQ algorithm (Li et al., 2008) to determine the frequency of reads matching each genomic position. To exploit sequencing data for repetitive regions, all reads that could be mapped to multiple genomic locations were assigned to one position at random and retained for further analyses. For both hybrid and parental lines, three independent biological replicates were used for the subsequent genome-wide methylation analyses. When all three sets of data from each genotype were combined, 6.0, 6.4, 6.4, and 8.5 billion base pairs were aligned to the Columbia-0 (Col-0) reference genome for Ler and C24 parents and their reciprocal F1 hybrids Flc (Ler is the maternal line) and Fcl (C24 is the maternal line), respectively, which corresponds to 25- to 36-fold coverage of the Arabidopsis genome (see Supplemental Table 2 online).
We next characterized the DNA methylomes of the parental lines and their reciprocal F1 hybrids at single-base-pair resolution. Figure 1B shows an example of typical DNA methylation profiles at the three cytosine contexts in a region of chromosome 5. Interestingly, we found that some protein-coding gene and transposable element (TE) regions were differently methylated in the parental lines and their reciprocal hybrids (Figure 1B), which will be discussed later. Moreover, we found increased DNA methylation levels in all sequence contexts in both F1 hybrids relative to their parents (Figures 1B and 1C; see Supplemental Figure 2 online). For example, CG methylation levels in Flc and Fcl hybrids were 17.5 and 20.5% higher than the average levels of the parents, respectively (Figure 1C). CHG and CHH methylation levels were also similarly higher in F1 hybrids than in their parents (Figure 1C). However, the cytosine methylation levels of Ler and C24 determined in our study were similar to each other (Figure 1C) and to data previously reported for Col-0 (Cokus et al., 2008; Lister et al., 2008; Hsieh et al., 2009). The DNA methylation density was higher in the pericentromeric regions in all sequence contexts (see Supplemental Figure 3 online), also consistent with several recent reports (Cokus et al., 2008; Lister et al., 2008; Hsieh et al., 2009; Feng et al., 2010). Together, our data indicated that both F1 hybrids had increased cytosine methylation in all three contexts relative to their parents.
Methylation Patterns of Ler and C24 Parents and Their Reciprocal Hybrids
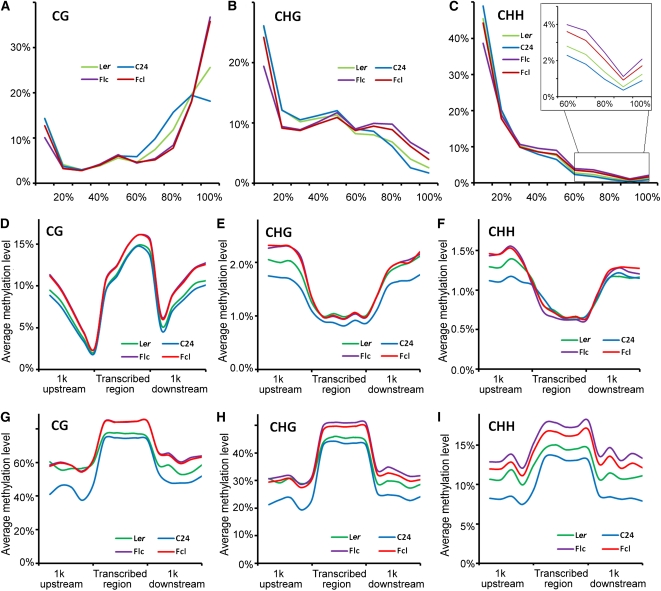
Next, we analyzed the methylation patterns of Ler and C24 parents and their reciprocal hybrids. We first estimated the percentage of methylation of each methyl-cytosine, calculated from the number of sequenced cytosines divided by the total read depth. We found that each cytosine context had distinct methylation profiles. CG sites were predominantly highly methylated, whereas CHG and CHH sites had more unmethylated cytosines (Figures 2A to 2C). Moreover, the proportions of highly methylated cytosines in all three sequence contexts increased in F1 hybrids relative to their parents (Figures 2A to 2C). For example, only 18 and 26% of CGs were methylated in C24 and Ler parents, respectively, while this increased to 36 and 37% in Fcl and Flc hybrids, respectively (Figure 2A). Increased numbers of highly methylated CHG and CHH positions were also observed in the F1 hybrids, although the difference was not as large as the changes in CGs (Figures 2A to 2C).
Figure 2.
Methylation Patterns of Ler and C24 Parents and Their Reciprocal Hybrids.
(A) to (C) Distribution of the percentage methylation at each cytosine context in F1 hybrids and their parents. x axis, percentage methylation in bins of 10%; y axis, percentage of total methylcytosines found in each bin. Data are averages of four independent biological replicates, each consisting of 50 pooled seedlings of the corresponding genotype.
(D) to (F) Methylation distribution in protein-coding genes at each cytosine context. x axis, position relative to the transcribed region; y axis, average methylation level (%). Data are averages of four independent biological replicates, each consisting of 50 pooled seedlings of the corresponding genotype.
(G) to (I) Methylation distribution in TEs at each cytosine context. x axis, position relative to the transcribed region; y axis, average methylation level (%). Data are averages of four independent biological replicates, each consisting of 50 pooled seedlings of the corresponding genotype.
We then characterized the methylation patterns of protein-coding genes and TEs in each cytosine context for parental lines and their reciprocal hybrids. All four genotypes showed a similar pattern of CG, CHG, and CHH methylation in both protein-coding genes and TEs (Figures 2D to 2I). As reported recently (Zhang et al., 2006; Zilberman et al., 2007; Cokus et al., 2008; Feng et al., 2010), CG methylation of protein-coding genes declined starting 1 kb upstream to a minimum at the transcription start site, then increased throughout the transcribed region before abruptly declining at the transcription stop, and again rose downstream of the stop (Figure 2D). By contrast, relatively low levels of CHG and CHH methylation were observed in transcribed regions compared with regions 1 kb upstream or downstream (Figures 2E and 2F). However, all three types of methylation showed a similar pattern along TEs (Figures 2G to 2I). Moreover, F1 hybrids had increased DNA methylation relative to their parents in both protein-coding genes (especially in the regions 1 kb upstream or downstream) and TEs, and the increase in TEs was greater than in protein-coding genes (Figures 2D to 2I). Together, these data confirmed the increased DNA methylation in F1 hybrids and indicated that increased DNA methylation in TEs may predominantly account for the overall increased DNA methylation in F1 hybrids.
sRNAomes of Ler and C24 Parents and Their Reciprocal Hybrids
Previous studies showed that siRNAs can direct DNA methylation at their target loci through the RdDM pathway (Wassenegger et al., 1994; Henderson and Jacobsen, 2007; Law and Jacobsen, 2010). A recent report also showed that changes in 24-nucleotide siRNA levels in Arabidopsis hybrids may contribute to heterosis (Groszmann et al., 2011). To characterize further the potential role of siRNAs in heterosis, we investigated the small RNA (sRNA) expression profiles of Ler and C24 parental lines and their reciprocal hybrids by high-throughput sequencing. Three independent biological replicates of each genotype were used for sRNA analyses. A total of 81.9 million reads (35 bp per read) were obtained from the 12 sequenced libraries, of which 55.5 million reads were mapped to unique locations in the Arabidopsis genome using Model-based Analysis of ChIP-Seq software (Zhang et al., 2008b). After removing reads mapped to rRNAs, tRNAs, and small nuclear and nucleolar RNAs, 30.8 million mapped sRNA reads were finally obtained (see Supplemental Table 3 online). Subsequent analyses showed that the correlation coefficient between each pair of the three biological replicates of each genotype was always >0.99 (see Supplemental Table 4 online). We therefore combined the data from the three replicates for each genotype to improve the sRNA depth and facilitate cluster identification (see below).
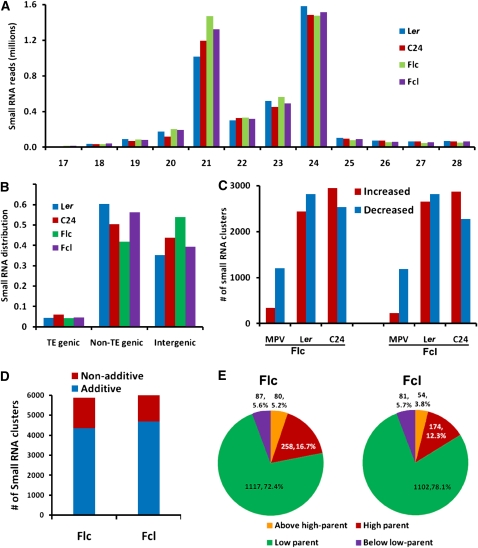
We then investigated the distribution of sRNA classes in the parental lines and their reciprocal hybrids. Our data showed that 21- and 24-nucleotide classes were the most abundant groups of sRNAs (Figure 3A), consistent with two recent reports (Lister et al., 2008; Groszmann et al., 2011). Our data also support the observation by Groszmann et al. that the Ler ecotype had fewer 21-nucleotide but more 24-nucleotide sRNAs than C24 (Figure 3A). However, we observed increased levels of 21-nucleotide sRNAs in both hybrids compared with the parents, but no obvious changes in 24-nucleotide sRNA levels between hybrids and parents (Figure 3A), which is in contrast with the report by Groszmann et al. that both hybrids showed decreased levels of 24-nucleotide sRNAs relative to their parents.
Figure 3.
The Genomic Distribution of sRNAs in Parental Lines and Their Reciprocal Hybrids.
(A) sRNA length distribution in the parental lines and their reciprocal hybrids.
(B) Distribution of sRNAs among TE-genic, non-TE-genic, and intergenic regions in the Arabidopsis genome.
(C) Numbers of sRNA clusters in each hybrid showing differential expression levels compared with the mid-parent value and with each parent. Red bars, increased expression; blue bars, decreased expression.
(D) Numbers of additively and nonadditively expressed sRNA clusters in F1 hybrids.
(E) Expression patterns of nonadditively expressed sRNA clusters in F1 hybrids.
As expected, sRNAs were transcribed from a wide range of locations on all five chromosomes in the parental lines and their reciprocal hybrids (see Supplemental Figure 4 online). In both parents and hybrids, non-TE genes showed more enrichment of sRNAs than did TE-related genes, and a large number of sRNA reads mapped to intergenic regions (Figure 3B). In addition, sRNAs were differentially distributed in the Ler and C24 parental lines: Ler had more sRNAs in non-TE regions, whereas C24 had more sRNAs in intergenic regions (Figure 3B).
A siRNA cluster was defined as a genomic region matched by at least three sRNA reads. If one cluster resided within 200 nucleotides of another, they were merged and regarded as a single cluster. Based on these definitions, the sRNA reads (mostly siRNA) from all four genotypes were pooled and used to quantitate siRNA abundance. In total, 80,553 siRNA clusters were identified in all four genotypes, among which 64,550, 66,294, 69,354, and 69,440 clusters were found in Ler, C24, Flc, and Fcl, respectively. In most cases, clusters that were found in only one parent were found in both hybrids, which is why there were more clusters in the hybrids than in the parents.
The expression level of a siRNA cluster was estimated by summing the number of sRNA reads mapped to it. Differences in expression of siRNA clusters were determined by DEGseq (Wang et al., 2010) with a P value cutoff of 0.001. Our results revealed that most (>93%) siRNA clusters were not differentially expressed between parents and hybrids. However, >5000 siRNA clusters were identified that did show differential expression in pairwise comparisons of parents and hybrids (Figure 3C). The expression of these clusters in the hybrids was often close to the mid-parent value (MPV; average of the two parents), so few clusters showed increased expression when compared with the MPV (Figure 3C). Nearly equal numbers of clusters with increased and decreased expression were found in pairwise comparisons of parents and hybrids, and more than 3 times as many clusters were differentially expressed when hybrids were compared with the parents individually than when the hybrids were compared with the MPV (Figure 3C).
The majority of the differentially expressed clusters (73.8% in Flc and 76.8% in Fcl) showed additive expression (i.e., their levels were equal to the MPV) (Figure 3D). For the remaining nonadditively expressed clusters, four expression patterns were identified based on previous studies (Birchler et al., 2003; Springer and Stupar, 2007; He et al., 2010). High parent or low parent patterns were inferred when the expression level in hybrids was similar to the higher or lower parent, respectively. Above high parent or below low parent patterns were inferred when the expression level in hybrids was above that of the higher parent or below that of the lower parent, respectively. Based on these modes, we characterized the expression patterns of nonadditively expressed sRNA clusters in both F1 hybrids. Of the 1542 and 1411 nonadditively expressed sRNA clusters in Flc and Fcl, respectively, the majority (72.4% in Flc and 78.1% in Fcl) exhibited a low parent pattern in reciprocal hybrids (Figure 3E), a phenomenon also observed in rice (Oryza sativa) hybrids (He et al., 2010).
sRNAs Were Associated with Increased DNA Methylation in F1 Hybrids
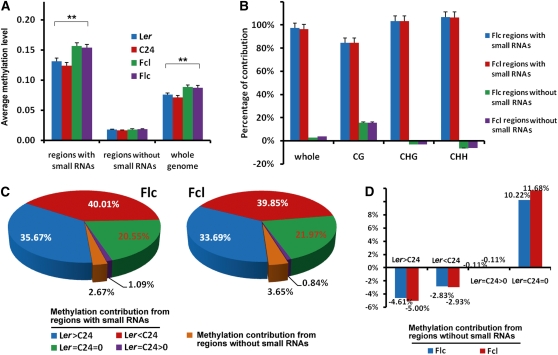
To investigate the relationship between sRNAs and DNA methylation, we divided the genome into regions with and without sRNAs and calculated the DNA methylation levels in each type of region. Interestingly, we found that DNA methylation levels were significantly higher in regions with sRNAs than those without in both the parental lines and their reciprocal hybrids (Figure 4A). Moreover, in regions with sRNAs, DNA methylation levels in all three cytosine contexts were significantly higher in hybrids than in parents (Figure 4A; see Supplemental Figure 5 online). Further analyses showed that regions covered by sRNAs were the main contributors to increased DNA methylation in hybrids both in overall methylation and individual cytosine contexts (Figure 4B). Interestingly, for CHG and CHH contexts, methylation in regions without sRNAs was always lower in both hybrids (shown as negative contribution; Figure 4B). Taken together, these data indicate that increased DNA methylation in the reciprocal hybrids predominantly occurred in regions covered by sRNAs.
Figure 4.
Contributions to Increased DNA Methylation in F1 Hybrids by the Regions with and without sRNAs.
(A) Average DNA methylation levels in regions with and without sRNAs in parents and hybrids. Error bars represent sd. **P < 0.01 (Student’s t test) for comparisons between parents and hybrids.
(B) The regions covered by sRNAs account for most of the methylation increase in all three cytosine contexts in F1 hybrids. Error bars represent sd.
(C) Contribution to increased DNA methylation in F1 hybrids by the regions with or without sRNAs. The cytosine positions of the genome were divided into four categories based on the methylation levels of the two parents: Ler > C24, positions with higher methylation levels in Ler than in C24; Ler < C24, positions with higher methylation levels in C24 than in Ler; Ler = C24 > 0; positions where methylation was detected but levels were equal in Ler and C24; Ler = C24 = 0, positions without detectable methylation in both Ler and C24.
(D) Contribution to increased DNA methylation in F1 hybrids by the regions without sRNAs.
To characterize how differences in DNA methylation in the parental lines affected DNA methylation in the reciprocal hybrids, we grouped the cytosines into four categories based on the methylation levels of the parental lines: positions more highly methylated in Ler than in C24 (Ler > C24), positions more highly methylated in C24 than in Ler (Ler < C24), positions where methylation was detected but levels were equal in Ler and C24 (Ler = C24 > 0), and positions lacking detectable methylation in both Ler and C24 (Ler = C24 = 0). Our results showed that regions covered by sRNAs that were differentially methylated in the parents (either Ler > C24 or Ler < C24) contributed >70% of the increased methylation in the reciprocal hybrids (Figure 4C). By contrast, the regions that were not covered by sRNAs only contributed ~3% of the increased methylation in the hybrids (Figures 4B and 4C). The contribution from this part largely came from regions where there was no detectable methylation in either parent (Figure 4D). If methylation was detected in either parent in regions that were not covered by sRNAs, methylation was always lower in the hybrids (shown as negative contribution) regardless of the difference between the parents (Figure 4D). Together, these data indicate that the regions that were differentially methylated between the parents and covered by sRNAs were primarily responsible for the increased methylation in the reciprocal hybrids.
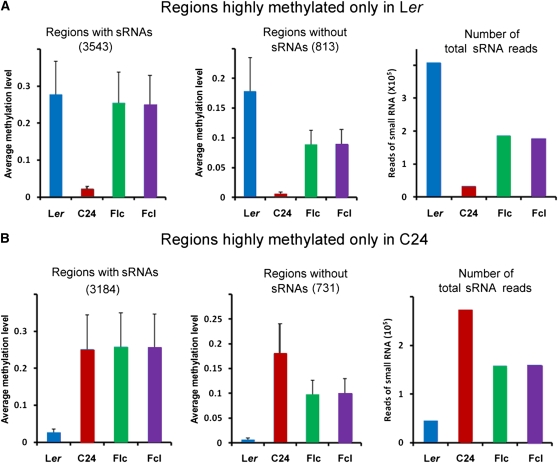
To gain more insights into the relationship between DNA methylation and sRNA expression, we next analyzed the regions where one parent was highly methylated while the other was not (for examples, see Supplemental Figure 6 online). Approximately 1.04 and 0.92% of cytosines were highly methylated only in Ler or C24, respectively. These cytosines were therefore subjected to further sRNA analyses. Our data showed that in the highly methylated regions only found in Ler, DNA methylation levels in both reciprocal hybrids were close to that of Ler but not C24 in regions covered by sRNAs (Figure 5A, left panel). However, in regions not covered by sRNAs, DNA methylation levels in both hybrids were close to the MPV (Figure 5A, middle panel). Moreover, in the regions that were differentially methylated in Ler and C24, the numbers of sRNA reads in both hybrids were close to the MPV (Figure 5A, right panel). Similar results were also obtained for the highly methylated regions only found in C24 (Figure 5B) and from a similar analysis that compared all the regions showing differential methylation between the two parents (see Supplemental Figure 7 online). Thus, our data suggest that sRNAs in the regions that were differentially methylated in the parents may direct DNA methylation of homologous sequences in hybrids through the RdDM pathway, which may account for the increased DNA methylation in reciprocal hybrids (see Supplemental Figure 8 online).
Figure 5.
sRNA Analyses of Regions That Are Differentially Methylated in the Parental Lines.
(A) sRNA analyses of regions highly methylated only in Ler. Left, average DNA methylation levels of 3543 regions covered by sRNAs (1.11 × 106 bp) that were highly methylated only in Ler. Middle, average DNA methylation levels of 813 regions not covered by sRNAs (8.45 × 104 bp) that were highly methylated only in Ler. Right, numbers of total sRNA reads in regions that were highly methylated only in Ler. Error bars represent sd.
(B) sRNA analyses of regions highly methylated only in C24. Left, average DNA methylation levels of 3184 regions covered by sRNAs (0.98 × 106 bp) that were highly methylated only in C24. Middle, average DNA methylation levels of 731 regions not covered by sRNAs (7.64 × 104 bp) that were highly methylated only in C24. Right, numbers of total sRNA reads in regions that were highly methylated only in C24. Error bars represent sd.
Analysis of Differentially Methylated Regions and sRNAs in Hybrids and Parental Lines
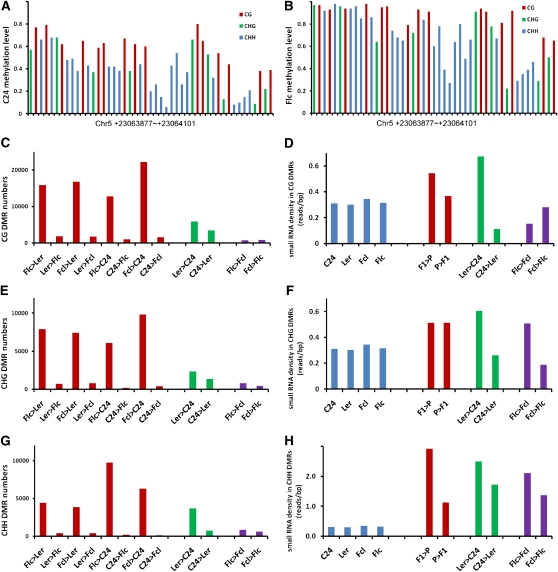
We next sought to identify sequences that were differentially methylated in the four genotypes. We calculated fractional methylation in each cytosine context by measuring the percentage methylation within 50-bp windows and defined differentially methylated regions (DMRs) as regions where the difference in DNA methylation in a particular context was at least 10% (P < 0.05) in pairwise comparisons among the four genotypes. A typical DMR between the C24 parent and the Flc hybrid is shown in Figures 6A and 6B, in which methylation levels in all three cytosine contexts were obviously higher in the Flc hybrid than in the C24 parent. More DMRs were identified between hybrids and parents than between the parents or between the hybrids in all cytosine contexts (Figures 6C, 6E, and 6G). At least 90% of DMRs were more highly methylated in hybrids than in either parent, consistent with the overall higher methylation levels in hybrids. Notably, very few DMRs were detected between the hybrids, whereas more DMRs were observed between the parents (Figures 6C, 6E, and 6G; see Supplemental Table 5 online). By mapping the DMRs to protein-coding genes and TEs, we estimated that 44% of the CG sites that were more highly methylated in hybrids were located in genic regions. By contrast, only 8 and 11% of CHG and CHH sites, respectively, that were more highly methylated in hybrids were located in genic regions (see Supplemental Figure 9 online), consistent with the low methylation pattern of non-CG sites in these regions (Figures 2E and 2F). More than 84% of the regions that showed differential CG methylation between the parents also showed differential CG methylation between parents and hybrids. Similarly, at least 72 and 52% of the regions that showed differential CHG and CHH methylation, respectively, between the parents were also differentially methylated between parents and hybrids (see Supplemental Figure 10 online). These results indicate that differences in methylation between the parents may be a major cause of the differences in methylation between parents and hybrids.
Figure 6.
Analysis of DMRs and sRNAs in F1 Hybrids and Their Parents.
(A) and (B) A typical DMR between C24 parent (A) and Flc hybrid (B). Each column represents the methylation level of the corresponding cytosine residue.
(C) Numbers of CG DMRs in pairwise comparisons of parents and hybrids.
(D) sRNA densities in CG DMRs of parental and hybrid genomes (blue bars) and in pairwise comparisons of parents and hybrids.
(E) Numbers of CHG DMRs in pairwise comparisons of parents and hybrids.
(F) sRNA densities in CHG DMRs of parental and hybrid genomes (blue bars) and in pairwise comparisons of parents and hybrids.
(G) Numbers of CHH DMRs in pairwise comparisons of parents and hybrids.
(H) sRNA densities in CHH DMRs of parental and hybrid genomes (blue bars) and in pairwise comparisons of parents and hybrids.
We also compared our DMR data with sRNA profiles. In pairwise comparisons of parental and hybrid CG and CHG DMRs, we observed only mild, if any, differences in sRNA density (Figures 6D and 6F). By contrast, significantly higher sRNA densities were always observed for CHH DMRs (Figure 6H). Moreover, in pairwise comparisons among parents and hybrids, we found that more sRNAs were often generated from the more methylated CG, CHG, and CHH DMRs of one genotype compared with the less methylated DMRs of the other (see Supplemental Table 6 online). Together, these results further support that sRNAs likely play a role in altering DNA methylation and may be responsible for the increased DNA methylation in reciprocal hybrids.
Transcriptomes of Hybrids and Parental Lines
Previous reports showed that DNA methylation affects the downregulation and silencing of transposons and some genes (Lippman et al., 2004; Zhang et al., 2006; Lister et al., 2008). To investigate whether the observed changes in DNA methylation lead to altered gene expression in hybrids, we measured the transcriptomes of hybrids and their parents by mRNA sequencing (mRNA-seq). Approximately 25 to 31 million mRNA-seq reads were obtained from the libraries of each genotype, which were mapped to over 80% of all predicted protein-coding genes (see Supplemental Figure 11 and Supplemental Table 7 online). Most reads (>90%) were mapped to predicted exons (including some mapped to 5′- or 3′-untranslated regions), and very few reads were mapped to intergenic regions or predicted introns (see Supplemental Figure 11C online). We validated our mRNA-seq data by real-time quantitative RT-PCR (qRT-PCR) analysis of 78 genes and obtained a correlation coefficient of 0.93 (P < 10−23), indicating a strong linear relationship between our qRT-PCR and mRNA-seq results (see Supplemental Figure 12 online).
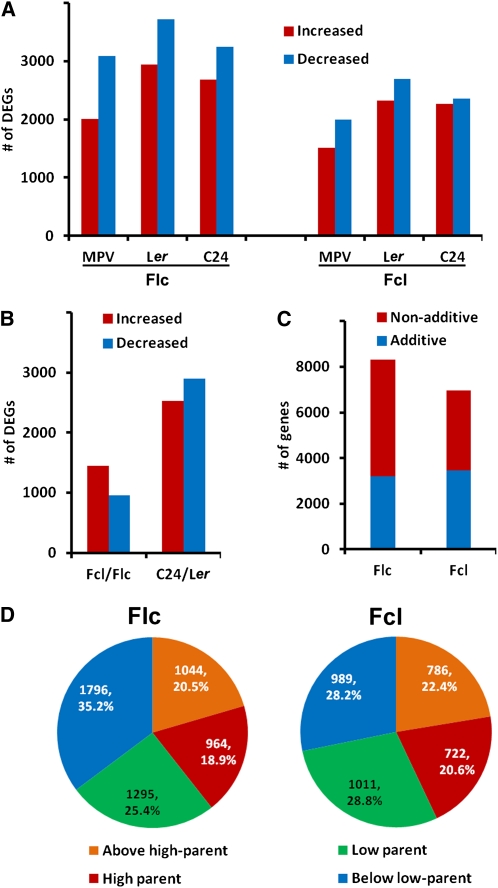
Next, we analyzed differentially expressed genes (DEGs) among parents and hybrids. We found that more genes were down- rather than upregulated in hybrids compared with either parent or the MPV (Figure 7A). Further comparisons found many more DEGs between the parents than between the hybrids (Figure 7B). Most genes whose expression levels were not equal to the MPV (i.e., nonadditively expressed genes) were expressed at low parent or below low parent levels (Figures 7C and 7D), consistent with more genes being down- than upregulated in both hybrids.
Figure 7.
Transcriptomes of Hybrid and Parental Lines.
(A) Numbers of DEGs in each hybrid compared with either parent or with the MPV.
(B) Numbers of DEGs whose expression was increased or decreased in Fcl compared with that in Flc and in C24 compared with that in Ler.
(C) Numbers of additively and nonadditively expressed genes in each F1 hybrid.
(D) Expression patterns of nonadditively expressed genes in each hybrid.
Finally, we generated an integrated map of DNA methylomes, sRNAomes, and transcriptomes of the Ler and C24 parental lines and their reciprocal hybrids. A representative example of a region on chromosome 5 showing predicted gene models with DNA methylation (in all three cytosine contexts), mRNA, and sRNA profiles in all four genotypes is shown in Figure 1B. The entire data set has been uploaded to the National Institutes of Health Gene Expression Omnibus database (see accession numbers below).
DNA Methylation Changes in Hybrids Correlate with Altered Gene Expression
Alteration of the DNA methylome may significantly affect plant development, since several mutants deficient in DNA methylation exhibited retarded growth (Finnegan et al., 1996; Ronemus et al., 1996; Cao and Jacobsen, 2002). Recent whole-genome tiling array studies have identified several hundreds of genes with altered expression levels in DNA methyltransferase null mutants, such as drm1 drm2 cmt3 (ddc) and met1 mutants (Zhang et al., 2006; Kurihara et al., 2008). However, although 215 and 226 genes were discovered to be upregulated in ddc mutants by Zhang et al. and Kurihara et al., respectively, only 21 genes overlapped in these two studies (Kurihara et al., 2008). A major factor responsible for the variation in these data sets may be different growth stages, as Zhang et al. used 5-week-old plants, whereas Kurihara et al. studied 15-d-old plants. Since we used 15-d-old plants in our study (see Methods), we selected the genes identified by Kurihara et al. for further analyses.
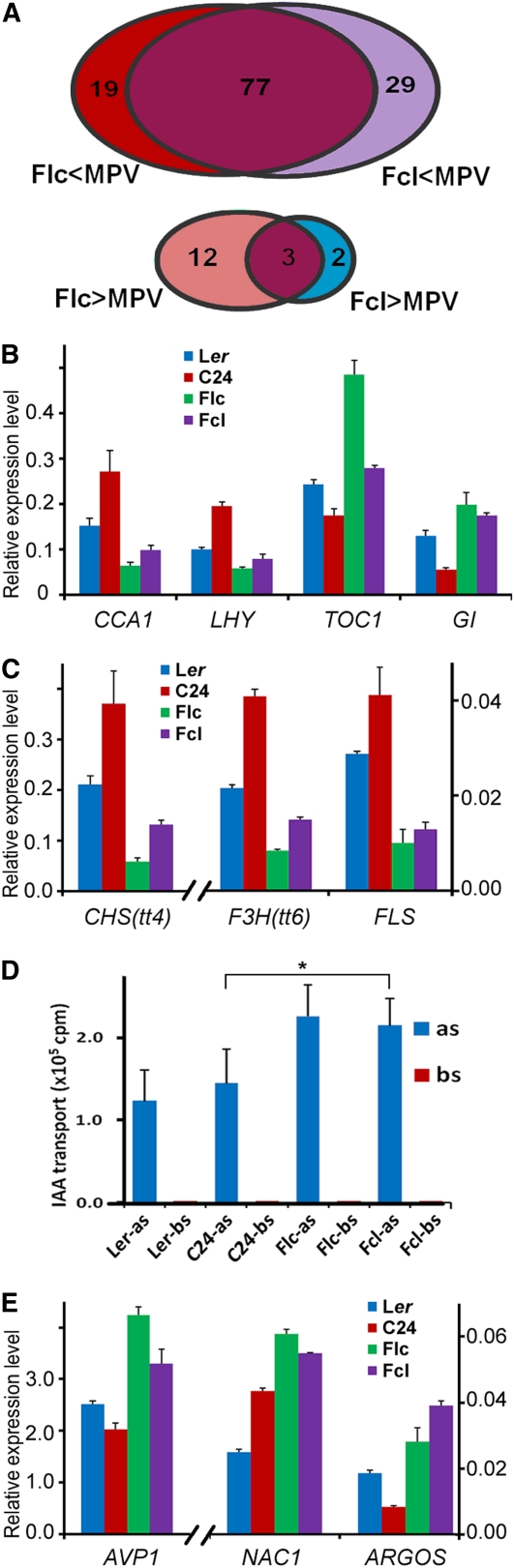
We first examined whether the expression of the 226 genes upregulated in ddc mutants was altered in reciprocal F1 hybrids. Our analysis revealed that 77 of these genes were downregulated in both Flc and Fcl hybrids; by contrast, only three genes were upregulated in both F1 hybrids (Figure 8A). Therefore, our data indicate that upregulation of DNA methylation in both F1 hybrids had the opposite effect on gene expression as the ddc mutations. It has been previously shown that the vast majority of non-CG methylation, but not CG methylation, was eliminated in ddc mutants (Cao and Jacobsen, 2002; Tariq et al., 2003). Thus, downregulation of these 77 genes in both F1 hybrids was possibly due to elevated non-CG methylation.
Figure 8.
Increased DNA Methylation in F1 Hybrids Altered the Expression of DNA Methylation-Sensitive Genes.
(A) Number of genes that were shown to be upregulated in ddc mutants (Kurihara et al., 2008) but were downregulated (top panel) or upregulated (bottom panel) in 15-d-old F1 hybrids of Ler and C24.
(B) Real-time qRT-PCR analysis showing downregulation of CCA1 and LHY and upregulation of TOC1 and GI expression in F1 hybrids of Ler and C24. Seedlings grown 15 d under white light conditions (16 h light/8 h dark) were collected and subjected to real-time qRT-PCR analysis. Error bars represent sd of triplicate experiments.
(C) Real-time qRT-PCR analysis showing downregulation of three genes (CHS, F3H, and FLS) involved in flavonoid biosynthesis in 15-d-old F1 hybrids of Ler and C24. Error bars represent sd of triplicate experiments.
(D) Auxin transport in Ler and C24 parental lines and their reciprocal hybrids tested by 3H-IAA assays. Error bars represent sd (n = 4). *P < 0.05 (Student’s t test) for the indicated pair of samples. as, apical side; bs, basal side.
(E) Real-time qRT-PCR analysis showing upregulation of three auxin-related genes (AVP1, NAC1, and ARGOS) in 15-d-old F1 hybrids of Ler and C24. Error bars represent sd of triplicate experiments.
A recent study reported that altered expression of circadian oscillator genes, CCA1 and LHY, is associated with growth vigor in Arabidopsis hybrids (Ni et al., 2009). Interestingly, we found that CCA1 and LHY were upregulated in ddc mutants (see Supplemental Table 3 in Kurihara et al., 2008) and downregulated in both F1 hybrids (Figure 8B). Previous studies discovered that the expression of TIMING OF CAB EXPRESSION1 (TOC1) and GIGANTEA (GI) was negatively regulated by CCA1 and LHY (Park et al., 1999; Strayer et al., 2000; Alabadí et al., 2001). Consistent with this, our data showed that TOC1 and GI were upregulated in both F1 hybrids (Figure 8B). Thus, these data imply that increased DNA methylation in F1 hybrids may cause the repression of CCA1 and LHY, which ultimately contributes to hybrid vigor through circadian-mediated physiological and metabolic pathways, as suggested by Ni et al. (2009).
In addition, three genes involved in the flavonoid synthetic pathway, CHALCONE SYNTHASE (CHS), FLAVANONE 3-HYDROXYLASE (F3H) and FLAVONOL SYNTHASE (FLS) (see Supplemental Figure 13 online), were also upregulated in ddc mutants (see Supplemental Table 3 in Kurihara et al., 2008) and downregulated in both F1 hybrids (Figure 8C). Flavonoids are a class of plant secondary metabolites and perform numerous physiological functions (Koes et al., 1994; Shirley, 1996; Lepiniec et al., 2006). Interestingly, the flavonoid pathway has been implicated in heterosis for cold stress (Korn et al., 2008). One function of flavonoids is to act as negative regulators of auxin transport in vivo (Brown et al., 2001; Peer et al., 2004); therefore, we tested whether auxin transport increased in the F1 hybrids. As shown in Figure 8D, indole-3-acetic acid (IAA) was transported considerably faster in both F1 hybrids than in either parent, indicating that auxin transport was indeed upregulated in both hybrids. Arabidopsis vacuolar H+-pyrophosphatase (AVP1) was shown to regulate auxin transport and consequently auxin-dependent development (Li et al., 2005). Plants overexpressing AVP1 showed increased auxin transport and grew larger than the wild type (Gaxiola et al., 2001; Li et al., 2005; Pasapula et al., 2011). Interestingly, we observed that both Flc and Fcl hybrids expressed AVP1 at higher levels than their parents (Figure 8E). In addition, NAC1 and ARGOS are two important downstream genes in auxin signaling, and plants overexpressing NAC1 or ARGOS also grew larger than the wild type (Xie et al., 2000; Hu et al., 2003). Our real-time qRT-PCR data also showed that NAC1 and ARGOS were both upregulated in Flc and Fcl hybrids (Figure 8E). Taken together, these results suggest that the growth vigor of F1 hybrids was at least partially due to increased auxin signaling.
Reduction of DNA Methylation in Hybrids Compromised Growth Vigor
To obtain more evidence supporting our hypothesis that changes in DNA methylation can affect hybrid vigor, we treated both reciprocal hybrids and their parents with 5-aza-deoxycytidine (5′-Aza-dC), a DNA methylation inhibitor (Bäurle et al., 2007). We first confirmed that treatment with 5′-Aza-dC indeed caused demethylation of genomic DNA in Arabidopsis seedlings (see Supplemental Figure 14 online). Moreover, 5′-Aza-dC treatment also inhibited the growth of both parents and hybrids, and the hybrids were more sensitive to 5′-Aza-dC than their parents (see Supplemental Figure 15 online). We further tested the expression profiles of circadian oscillator genes and flavonoid biosynthetic genes in both F1 seedlings treated with 5′-Aza-dC. Our results showed that the expression of these genes was upregulated in treated seedlings compared with untreated seedlings (see Supplemental Figures 16A to 16E online). Moreover, the expression of auxin-related genes was downregulated in 5′-Aza-dC–treated hybrid seedlings (see Supplemental Figures 16F and 16G online), consistent with the opposite changes in expression of flavonoid synthetic and auxin-related genes mentioned above. Taken together, these results support that DNA methylation plays a role in hybrid vigor.
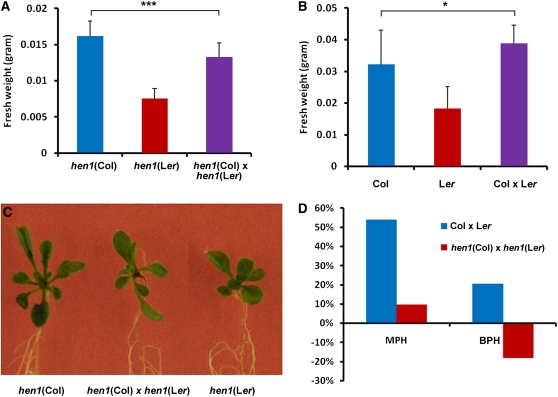
HEN1, an Arabidopsis RNA methyltransferase, is a crucial factor in the biogenesis of plant sRNAs, including miRNAs and siRNAs, and therefore plays an important role in the RdDM pathway (X. Chen, 2009, 2010; Vilkaitis et al., 2010). Arabidopsis hen1 mutants exhibited pleiotropic phenotypes, such as reduced size of aerial organs and reduced male and female fertility (Chen et al., 2002). To test the role of sRNAs in heterosis, we generated hen1 hybrids by crossing an ethyl methanesulfonate mutant line in the Ler background with a T-DNA insertion line in the Col background. The DNA methylation levels of both hen1 parental lines and their hybrids were lower than the wild type (see Supplemental Figure 17 online). Moreover, growth vigor was compromised in F1 hybrids of hen1 (Col) and hen1 (Ler) (Figures 9A to 9C). Mid-parent heterosis of hen1 hybrids was only about one-fifth that of the wild-type hybrids; moreover, best-parent heterosis of hen1 hybrids showed an opposite trend to that of the wild-type hybrids (Figure 9D).
Figure 9.
Heterosis Was Compromised in F1 Hybrids of hen1 (Col) and hen1 (Ler) Mutants.
(A) Fresh weight of 15-d-old hen1 (Col) and hen1 (Ler) mutants and their F1 hybrids. Error bars represent sd (n = 30). ***P < 0.001 (Student’s t test) for the indicated pair of samples.
(B) Fresh weight of 15-d-old wild-type Col and Ler plants and their F1 hybrids. Error bars represent sd (n = 30). *P < 0.05 (Student’s t test) for the indicated pair of samples.
(C) Phenotypes of 15-d-old hen1 (Col) and hen1 (Ler) mutants and their F1 hybrids.
(D) Mid-parent heterosis (MPH) and best-parent heterosis (BPH) of F1 hybrids of hen1 (Col) and hen1 (Ler) mutants and of wild-type Col and Ler plants.
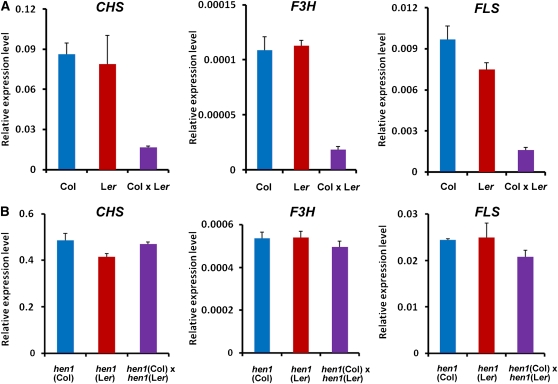
We further tested the expression profiles of circadian oscillator genes (CCA1 and LHY) and flavonoid synthetic genes (CHS, F3H, and FLS) in F1 hybrids of the wild type and of the hen1 mutants. As mentioned above, all these genes were upregulated in ddc mutants but downregulated in both Flc and Fcl hybrids (Figure 8). Downregulation of these genes was also confirmed in the hybrids of Col and Ler ecotypes (Figure 10A; see Supplemental Figure 18A online). However, these genes were not downregulated in the hybrids of hen1 mutants (Figure 10B; see Supplemental Figure 18B online). Together, these data further support that sRNAs play an important role in heterosis, possibly by directing DNA methylation through the RdDM pathway.
Figure 10.
Expression of CHS, F3H, and FLS Genes Was Downregulated in F1 Hybrids of Wild-Type Col and Ler Plants but Not in Those of hen1 (Col) and hen1 (Ler) Mutants.
(A) Real-time qRT-PCR analysis showing the relative expression levels of CHS, F3H, and FLS in 15-d-old wild-type Col and Ler plants and their F1 hybrids. Error bars represent sd of triplicate experiments.
(B) Real-time qRT-PCR analysis showing the relative expression levels of CHS, F3H, and FLS in 15-d-old hen1 (Col) and hen1 (Ler) mutants and their F1 hybrids. Error bars represent sd of triplicate experiments.
DISCUSSION
The two leading hypotheses for heterosis, dominance and overdominance, are both based on differences between the parental genomes (Birchler et al., 2003, 2010). However, mechanistic connections between differences in parental genomes and growth vigor in F1 hybrids are still poorly understood. It was reported that the promoter regions of genes that are differentially expressed in two heterotic rice crosses were enriched in insertion/deletion polymorphisms (Zhang et al., 2008a). Another recent report suggested that epigenetic diversity between parental lines, such as differences in 24-nucleotide siRNA levels between two Arabidopsis ecotypes, may contribute to hybrid vigor (Groszmann et al., 2011). In our study, we investigated DNA methylomes, sRNA transcriptomes, and mRNA transcriptomes of reciprocal F1 hybrids and their parents by high-throughput sequencing and found distinct differences between the hybrids and their parents in all three aspects. Though it is very complicated to link these differences between hybrids and parents with specific heterotic traits, some interesting alterations in reciprocal hybrids relative to their parents were consistent with the observed heterosis. We observed increased DNA methylation in the reciprocal hybrids relative to their parents, and about three-quarters of this increased methylation in both hybrids originated from regions that were differentially methylated in their parents. We provide further evidence supporting that the increased DNA methylation in F1 hybrids contributed to growth vigor. Therefore, our data suggest that DNA methylation changes in F1 hybrids may play a role in heterosis.
The fact that 96 to 97% of the increased methylation in reciprocal hybrids was on cytosines covered by sRNAs (Figure 4B) suggests that this likely originated from the trans-acting effects of sRNAs, possibly through the RdDM pathway. Up to three-quarters of the increased methylation resided in regions that were differentially methylated in the parents (Figure 4C), suggesting that sRNAs generated from these regions may act in trans to increase cytosine methylation to the higher parental level (see Supplemental Figures 6 and 8 online). It is interesting to note that ~20% of the increased methylation came from regions where no methylation was detected in either parent but sRNAs were present (Figure 4C), indicating that in those regions sRNAs directed de novo DNA methylation in hybrids. By contrast, very little contribution (only 2.67% in Flc and 3.65% in Fcl) was made to increased methylation in reciprocal hybrids by the regions without sRNAs (Figure 4C). In addition, in regions without sRNAs, methylation changes in hybrids depended on the cytosine context. Methylation levels in the CG context increased slightly in hybrids, whereas methylation levels in the CHG and CHH contexts decreased slightly (Figure 4B). It is possible that in CHG and CHH contexts, the lack of sRNAs may be a reason for the loss of methylation. Together, all our data indicate that sRNAs are critical for the increased methylation in hybrid genomes.
The genome-wide increase in DNA methylation in F1 hybrids was most dramatic in TEs that also generate abundant sRNAs. Because TEs cause genomic instability, they need to be highly methylated to preserve genomic integrity (Teixeira and Colot, 2010). On the other hand, we showed that many genes sensitive to methylome remodeling, including genes involved in flavonoid biosynthesis and two circadian oscillator genes CCA1 and LHY, were transcriptionally down- or upregulated in both reciprocal hybrids. Thus, our data, together with the existing knowledge, may lead to an unintended scenario: a mechanism for preserving genomic integrity results in hybrid vigor by altering the expression of genes that normally limit growth.
Our observation that distinct epigenetic modifications between two parents can give rise to increased DNA methylation and heterosis in their reciprocal hybrids suggests that increased DNA methylation in F1 hybrids resulting from differences in parental epigenomes may be a common mechanism in other organisms. In fact, data in several published reports support this assumption. For example, in a cross between two rice subspecies, Nipponbare (O. sativa ssp japonica) and 93-11 (O. sativa ssp indica), 82.1 and 70.8% of DMRs in the hybrid genomes exhibited high parent or above high parent levels of methylation, clearly an overall increase in DNA methylation in both hybrids (see Figure 4D in He et al., 2010). Another report showed that in two Arabidopsis allotetraploid lines, Allo733 and Allo738, 40 and 56 genes upregulated in ddc mutants were differentially expressed, respectively, compared with their parents, among which, 38 genes in Allo733 and 52 genes in Allo738 (>90% in both lines) were downregulated (Wang et al., 2006). These results are similar to what was observed in our reciprocal F1 hybrids (Figure 8A), suggesting that similar alteration of DNA methylation may have happened in the genomes of Arabidopsis allotetraploids. In addition, in F1 hybrids of Col-0 and C24, genome-wide methylation-sensitive amplified polymorphism analysis indicated that 3% of MspI/HpaII recognition sites showed signs of increased methylation when compared with the parental lines, suggesting a slight increase of DNA methylation in F1 hybrids (Banaei Moghaddam et al., 2010).
It was reported that levels of sRNAs were decreased in F1 hybrids of rice (He et al., 2010). However, a careful reexamination of our data and hybrid rice data did not support this conclusion. First, nearly all (99.2%) sRNA clusters in hybrid rice were not differentially expressed between parents and hybrids. Of the 235,257 sRNA clusters identified in rice by He et al. (2010), only 1892 sRNA clusters were differentially expressed in F1 hybrids compared with the MPV, similar to the data in this study (Figure 3C). Second, in this study, more sRNA clusters were found in F1 hybrids than in either parent, due to the fact that sRNA clusters found in only one parent are always present in both F1 hybrids (see Results). In fact, nearly equal numbers of these sRNA clusters exhibited significant down- or upregulation in F1 hybrids compared with either parent (Figure 3C). However, as the expression of these sRNA clusters in F1 hybrids was often close to the MPV, much fewer sRNA clusters showing differential expression were ultimately identified, among which, more downregulated sRNA clusters were observed (Figure 3C). A similar phenomenon was also observed in hybrid rice: Of the 1892 differentially expressed sRNA clusters mentioned above, 1269 and 623 clusters were down- and upregulated, respectively (He et al., 2010). We therefore argue that the changes in this small fraction of sRNA clusters (<0.8%) may not represent the overall alteration of the sRNA clusters in the whole genome, let alone the fact that actually no obvious changes were observed when this small fraction of sRNA clusters was compared with either parent.
A recent study by Groszmann et al. (2011) reported that sRNA levels (primarily 24-nucleotide siRNAs) were decreased in Arabidopsis hybrids, suggesting that these changes may contribute to hybrid vigor. In that study, both reciprocal hybrids showed an average decrease in 24-nucleotide siRNA levels in the range of 17% (23/24 nucleotides) to 27% (24 nucleotides) from the MPV (Groszmann et al., 2011). However, the total sRNA clusters obtained in that study only covered 1.60 to 2.08% of the genome, far below our total sRNA clusters, which covered 47.27% of the genome. It was therefore doubtful whether conclusions from such a small sample can truly reflect the changes in the whole genome. Moreover, if decreased sRNA levels in F1 hybrids indeed contribute to heterosis, mutations in hen1 would confer positive effects on growth vigor of F1 hybrids. However, our results indicated that this is not the case (Figure 9).
In this study, we show that 77 genes sensitive to methylome remodeling were transcriptionally repressed in both F1 hybrids (Figure 8A). One would assume that downregulation of these genes should be directly associated with increase of their DNA methylation levels. However, our results did not support this assumption. We failed to detect any significant correlation between differential methylation and gene expression in these 77 genes. In fact, the relationship between DNA methylation and gene expression is complicated in Arabidopsis, and the expression of a gene is usually not directly correlated with its methylation level. From our sequencing data, two-thirds of Arabidopsis genes (>20,000 genes) were methylated within the immediate context, including 1 kb upstream and downstream of the annotated gene model. However, in met1 and ddc mutants, although nearly all CG and non-CG methylation was eliminated, respectively (Cao and Jacobsen, 2002; Tariq et al., 2003; Zhang et al., 2006; Lister et al., 2008), only 319 and 215 genes were overexpressed, respectively, in these two mutants (Zhang et al., 2006). Therefore, in most cases we cannot deduce a specific gene’s expression from the DNA methylation state of its context. Instead, it is more likely that a small but significant group of genes are affected by the DNA methylation state of the entire genome.
Several reports have shown that altering the expression of certain specific genes, such as circadian oscillator genes, is associated with heterosis (Zhang et al., 2008a; Ni et al., 2009; Krieger et al., 2010). Our data showed that the expression of CCA1 and LHY was indeed downregulated in both F1 hybrids (Figure 8B). In addition, genes involved in flavonoid biosynthesis are among the key genes that are downregulated in reciprocal hybrids. The flavonoid pathway has been suggested to play a role in heterosis for cold stress (Korn et al., 2008). Flavonoids can also repress auxin-promoted growth by inhibiting auxin transport (Brown et al., 2001; Peer et al., 2004), and our expression analysis showed that several genes involved in flavonoid biosynthesis and auxin transport (and signaling) were correspondingly changed in both hybrids (Figures 8C and 8E). Interestingly, flavonoid biosynthesis and auxin signaling are all regulated by the circadian clock (Yanovsky and Kay, 2001; Yakir et al., 2007). Therefore, altered circadian rhythms may be a fundamental molecular mechanism for hybrid vigor (Ni et al., 2009; Z.J. Chen, 2010). Together, our results suggest that increased genome-wide DNA methylation in F1 hybrids, possibly due to RdDM, may cause altered circadian rhythms, which change gene expression in F1 hybrids and ultimately leads to hybrid vigor.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana Ler and C24 ecotypes and their reciprocal F1 hybrids Flc (Ler is the maternal line) and Fcl (C24 is the maternal line) were used in this study. Plants were grown on Murashige and Skoog plates containing 1% Suc at 22°C under white light (100 μmol m−2 s−1) conditions (16 h light/8 h dark). The 15-d-old seedlings were used for various assays, unless otherwise indicated. Around 50 to 100 seedlings were pooled in each sample for genomic DNA and total RNA extractions.
Generation of F1 Hybrid Lines
The respective F1 hybrid lines were generated by crossing the indicated parental lines. The F1 hybrids of hen1 mutants were generated by crossing hen1 in Col (SALK_049197) with hen1 in Ler background (CS6583).
Library Generation for MethylC-seq
Libraries for high-throughput bisulfite sequencing were generated as described previously (Lister et al., 2008). Briefly, 5 μg of purified genomic DNAs were first isolated, sonicated, and ligated to Illumina methylated DNA adaptors, and then adaptor-ligated molecules of 50 to 500 bp were isolated by agarose gel electrophoresis and subjected to two successive treatments of sodium bisulfite conversion to obtain a high conversion rate based on a recent report (Lister et al., 2008). Five nanograms of bisulfite-converted, adapter-ligated DNA molecules were enriched by 15 cycles of PCR. Finally, the enriched libraries were purified with the PCR purification kit (Qiagen) and subjected to high-throughput Illumina sequencing.
Library Generation for mRNA and sRNA Sequencing
Total RNAs were isolated from 15-d-old seedlings using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions and were treated with RNase-free DNase I (Promega) to remove contaminating genomic DNA. mRNAs were extracted from the total RNAs using Dynabeads oligo(dT) (Invitrogen Dynal) following the manufacturer’s instructions. First- and second-strand cDNAs were generated using SuperScript II reverse transcriptase (Invitrogen) and random hexamer primers. Double-stranded cDNAs were fragmented by nebulization and used for mRNA library construction following the standard Illumina protocol. sRNAs separated from total RNAs by polyacrylamide gel electrophoresis were extracted and used to create libraries for Illumina sequencing as described previously (Mi et al., 2008; Wang et al., 2009).
Auxin Transport Assay
IAA transport activity was assayed as described previously (Okada et al., 1991; Lewis and Muday, 2009). Briefly, 2 μL of 3H-IAA at 50 Ci mmol−1 was added to 1 mL of 0.05% MES, pH 5.5 to 5.7, to yield a 100 nM 3H-IAA solution. The inflorescence axes of parental and hybrid plants were cut into 2.5-cm-long segments and put into 1.5-mL Eppendorf tubes in normal or inverted orientation. Thirty microliters of 100 nM 3H-IAA solution was supplied at the bottom of each tube. After incubation at room temperature for 9 h, small slices of ~5 mm thickness were excised at the nonsubmerged end of the segments. Radioactivity of the small slices was counted by a liquid scintillation counter.
Real-Time qRT-PCR Assay
Total RNAs were extracted as described above. After DNase I (Promega) digestion, cDNAs were synthesized from 2 μg total RNAs using the SuperScript II first-strand cDNA synthesis system (Invitrogen) according to the manufacturer’s instructions. The primers used for real-time qRT-PCR are listed in Supplemental Table 8 online. Real-time PCR was performed using the respective pair of primers and Power SYBR Green PCR Master Mix (Applied Biosystems) with a Bio-Rad CFX96 real-time PCR detection system as described previously (Li et al., 2010). PCR reactions were performed in triplicate for each sample, and the expression levels were normalized to that of an Actin gene (At5g09810).
5′-Aza-dC Treatment Assay
Seeds were germinated on Murashige and Skoog plates containing varying concentrations of 5′-Aza-dC (Sigma-Aldrich) and grown for 15 d before measurement. Three independent batches with more than 30 plants per batch were assayed.
Bioinformatics Analyses
Please see Supplemental Table 9 and Supplemental Methods 1 online for details about bioinformatics analyses.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: CCA1 (At2g46830), LHY (At1g01060), TOC1 (At5g61380), GI (At1g22770), CHS (At5g13930), F3H (At3g51240), FLS (At5g08640), AVP1 (At1g15690), NAC1 (At1g56010), ARGOS (At3g59900), and HEN1 (At4g20910). All original data sets have been deposited in the Gene Expression Omnibus database under the accession number GSE34658.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Heterotic Phenotypes of 35-d-Old Arabidopsis F1 Hybrids (Flc and Fcl) Relative to Their Parents (Ler and C24).
Supplemental Figure 2. Bulk Methylation Levels of Ler and C24 Parental Lines and Their Reciprocal Hybrids When 0 Mismatch Mapping Was Allowed.
Supplemental Figure 3. Chromosomal Distribution of DNA Methylation at Three Cytosine Contexts in the C24 Parent.
Supplemental Figure 4. Chromosomal Distribution of Small RNAs in the C24 Parent.
Supplemental Figure 5. Differential DNA Methylation at Three Cytosine Contexts in Regions with and without Small RNAs.
Supplemental Figure 6. Examples of Differentially Methylated Regions between Two Parents.
Supplemental Figure 7. Differential DNA Methylation in F1 Hybrids Is Related to the Methylation Patterns in Their Parents and the Prevalence of Small RNAs.
Supplemental Figure 8. A Model Showing How Small RNAs Contribute to Upregulation of DNA Methylation in F1 Hybrids.
Supplemental Figure 9. Distribution of DMRs in Genic, TE-Related, and Other Regions.
Supplemental Figure 10. Percentage of DMRs between Two Parents That Are Also Differentially Methylated between Parents and Hybrids.
Supplemental Figure 11. Analysis of Mapped mRNA-seq Reads in Parents and Hybrids.
Supplemental Figure 12. Correlation Analysis of the Expression of 78 Genes Assayed by Real-Time qRT-PCR and mRNA-seq.
Supplemental Figure 13. Flavonoid Biosynthesis Pathway.
Supplemental Figure 14. PCR Assay of the Effect of 5′-Aza-dC on DNA Methylation of At SN1.
Supplemental Figure 15. Treatment with 5′-Aza-dC Inhibited the Growth of Both Parental and Hybrid Seedlings.
Supplemental Figure 16. Relative Expression Levels of Genes in Flc and Fcl Seedlings after Demethylation Treatment with 5′-Aza-dC.
Supplemental Figure 17. PCR Assay of the Effect of hen1 Mutation on DNA Methylation of SN1.
Supplemental Figure 18. Relative Expression Levels of CCA1 and LHY in the Wild-Type (Col and Ler) and hen1 (Col and Ler) Parental Lines and Their F1 Hybrids.
Supplemental Table 1. Phenotypes Measured for Parental Lines and Their Reciprocal Hybrids.
Supplemental Table 2. Total Reads Obtained by MethylC-seq.
Supplemental Table 3. Total Small RNA Reads Obtained by Small RNA Sequencing.
Supplemental Table 4. Correlation Analyses of Different Replicates in MethylC-seq, sRNA-seq, and mRNA-seq.
Supplemental Table 5. Pairwise Comparisons of DMRs among Parents and Hybrids.
Supplemental Table 6. Total Small RNA Reads in DMRs among Parents and Hybrids.
Supplemental Table 7. Total mRNA-seq Reads in Parental Lines and Their Reciprocal Hybrids.
Supplemental Table 8. Summary of Primers Used in Real-Time qRT-PCR Assays.
Supplemental Table 9. Summary of MethylC-seq Reads in Three Biological Replicates.
Supplemental Methods 1. Bioinformatics Analysis Methods.
Acknowledgments
We thank Mingqiu Dai, Gang Li, Xinhao Ouyang, and Bosheng Li for their help and suggestions on the project. This work was supported by grants from the National Basic Research Program of China (973 Program; 2012CB910900), the U.S. National Science Foundation Plant Genome Program (DBI0922604), the Ministry of Agriculture of China (2010ZX08010-003), and the Peking University 985 Program.
AUTHOR CONTRIBUTIONS
X.W.D. conceived the project. X.W.D. and H.S. designed the experiments. H.S., L.G., G.H., S.Z., and Y.Q. performed the experiments. H.H., W.C., X.W., and Z.P. conducted bioinformatics analyses. J.L., H.S., W.T., and X.W.D. prepared the article.
References
- Alabadí D., Oyama T., Yanovsky M.J., Harmon F.G., Más P., Kay S.A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 [DOI] [PubMed] [Google Scholar]
- Banaei Moghaddam A.M., Fuchs J., Czauderna T., Houben A., Mette M.F. (2010). Intraspecific hybrids of Arabidopsis thaliana revealed no gross alterations in endopolyploidy, DNA methylation, histone modifications and transcript levels. Theor. Appl. Genet. 120: 215–226 [DOI] [PubMed] [Google Scholar]
- Bäurle I., Smith L., Baulcombe D.C., Dean C. (2007). Widespread role for the flowering-time regulators FCA and FPA in RNA-mediated chromatin silencing. Science 318: 109–112 [DOI] [PubMed] [Google Scholar]
- Birchler J.A., Auger D.L., Riddle N.C. (2003). In search of the molecular basis of heterosis. Plant Cell 15: 2236–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J.A., Yao H., Chudalayandi S., Vaiman D., Veitia R.A. (2010). Heterosis. Plant Cell 22: 2105–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. (2002). DNA methylation patterns and epigenetic memory. Genes Dev. 16: 6–21 [DOI] [PubMed] [Google Scholar]
- Brown D.E., Rashotte A.M., Murphy A.S., Normanly J., Tague B.W., Peer W.A., Taiz L., Muday G.K. (2001). Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiol. 126: 524–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Jacobsen S.E. (2002). Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc. Natl. Acad. Sci. USA 99(suppl. 4): 16491–16498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S.W., Henderson I.R., Jacobsen S.E. (2005). Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat. Rev. Genet. 6: 351–360 [DOI] [PubMed] [Google Scholar]
- Chen X. (2009). Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. Biol. 25: 21–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. (2010). Small RNAs - Secrets and surprises of the genome. Plant J. 61: 941–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Liu J., Cheng Y., Jia D. (2002). HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development 129: 1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.J. (2010). Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant Sci. 15: 57–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus S.J., Feng S., Zhang X., Chen Z., Merriman B., Haudenschild C.D., Pradhan S., Nelson S.F., Pellegrini M., Jacobsen S.E. (2008). Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452: 215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow J.F. (1948). Alternative hypotheses of hybrid vigor. Genetics 33: 477–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas P., Vincent C., Coen E. (1999). An epigenetic mutation responsible for natural variation in floral symmetry. Nature 401: 157–161 [DOI] [PubMed] [Google Scholar]
- Feng S., et al. (2010). Conservation and divergence of methylation patterning in plants and animals. Proc. Natl. Acad. Sci. USA 107: 8689–8694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan E.J., Peacock W.J., Dennis E.S. (1996). Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc. Natl. Acad. Sci. USA 93: 8449–8454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frascaroli E., Canè M.A., Landi P., Pea G., Gianfranceschi L., Villa M., Morgante M., Pè M.E. (2007). Classical genetic and quantitative trait loci analyses of heterosis in a maize hybrid between two elite inbred lines. Genetics 176: 625–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaxiola R.A., Li J., Undurraga S., Dang L.M., Allen G.J., Alper S.L., Fink G.R. (2001). Drought- and salt-tolerant plants result from overexpression of the AVP1 H+-pump. Proc. Natl. Acad. Sci. USA 98: 11444–11449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszmann M., Greaves I.K., Albertyn Z.I., Scofield G.N., Peacock W.J., Dennis E.S. (2011). Changes in 24-nt siRNA levels in Arabidopsis hybrids suggest an epigenetic contribution to hybrid vigor. Proc. Natl. Acad. Sci. USA 108: 2617–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G., Elling A.A., Deng X.W. (2011). The epigenome and plant development. Annu. Rev. Plant Biol. 62: 411–435 [DOI] [PubMed] [Google Scholar]
- He G., et al. (2010). Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell 22: 17–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson I.R., Jacobsen S.E. (2007). Epigenetic inheritance in plants. Nature 447: 418–424 [DOI] [PubMed] [Google Scholar]
- Hochholdinger F., Hoecker N. (2007). Towards the molecular basis of heterosis. Trends Plant Sci. 12: 427–432 [DOI] [PubMed] [Google Scholar]
- Hsieh T.F., Ibarra C.A., Silva P., Zemach A., Eshed-Williams L., Fischer R.L., Zilberman D. (2009). Genome-wide demethylation of Arabidopsis endosperm. Science 324: 1451–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Xie Q., Chua N.H. (2003). The Arabidopsis auxin-inducible gene ARGOS controls lateral organ size. Plant Cell 15: 1951–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koes R.E., Quattrocchio R., Mol J.N.M. (1994). The flavonoid biosynthetic pathway in plants: Function and evolution. Bioessays 16: 123–132 [Google Scholar]
- Korn M., Peterek S., Mock H.P., Heyer A.G., Hincha D.K. (2008). Heterosis in the freezing tolerance, and sugar and flavonoid contents of crosses between Arabidopsis thaliana accessions of widely varying freezing tolerance. Plant Cell Environ. 31: 813–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger U., Lippman Z.B., Zamir D. (2010). The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat. Genet. 42: 459–463 [DOI] [PubMed] [Google Scholar]
- Kurihara Y., Matsui A., Kawashima M., Kaminuma E., Ishida J., Morosawa T., Mochizuki Y., Kobayashi N., Toyoda T., Shinozaki K., Seki M. (2008). Identification of the candidate genes regulated by RNA-directed DNA methylation in Arabidopsis. Biochem. Biophys. Res. Commun. 376: 553–557 [DOI] [PubMed] [Google Scholar]
- Lai J., et al. (2010). Genome-wide patterns of genetic variation among elite maize inbred lines. Nat. Genet. 42: 1027–1030 [DOI] [PubMed] [Google Scholar]
- Law J.A., Jacobsen S.E. (2009). Molecular biology. Dynamic DNA methylation. Science 323: 1568–1569 [DOI] [PubMed] [Google Scholar]
- Law J.A., Jacobsen S.E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11: 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepiniec L., Debeaujon I., Routaboul J.M., Baudry A., Pourcel L., Nesi N., Caboche M. (2006). Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 57: 405–430 [DOI] [PubMed] [Google Scholar]
- Lewis D.R., Muday G.K. (2009). Measurement of auxin transport in Arabidopsis thaliana. Nat. Protoc. 4: 437–451 [DOI] [PubMed] [Google Scholar]
- Li H., Ruan J., Durbin R. (2008). Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18: 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Li G., Gao S., Martinez C., He G., Zhou Z., Huang X., Lee J.H., Zhang H., Shen Y., Wang H., Deng X.W. (2010). Arabidopsis transcription factor ELONGATED HYPOCOTYL5 plays a role in the feedback regulation of phytochrome A signaling. Plant Cell 22: 3634–3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., et al. (2005). Arabidopsis H+-PPase AVP1 regulates auxin-mediated organ development. Science 310: 121–125 [DOI] [PubMed] [Google Scholar]
- Lippman Z., et al. (2004). Role of transposable elements in heterochromatin and epigenetic control. Nature 430: 471–476 [DOI] [PubMed] [Google Scholar]
- Lippman Z.B., Zamir D. (2007). Heterosis: Revisiting the magic. Trends Genet. 23: 60–66 [DOI] [PubMed] [Google Scholar]
- Lister R., Ecker J.R. (2009). Finding the fifth base: Genome-wide sequencing of cytosine methylation. Genome Res. 19: 959–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R., O’Malley R.C., Tonti-Filippini J., Gregory B.D., Berry C.C., Millar A.H., Ecker J.R. (2008). Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133: 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R., et al. (2011). Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 471: 68–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R., et al. (2009). Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning K., Tör M., Poole M., Hong Y., Thompson A.J., King G.J., Giovannoni J.J., Seymour G.B. (2006). A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 38: 948–952 [DOI] [PubMed] [Google Scholar]
- Martienssen R.A., Colot V. (2001). DNA methylation and epigenetic inheritance in plants and filamentous fungi. Science 293: 1070–1074 [DOI] [PubMed] [Google Scholar]
- Mathieu O., Bender J. (2004). RNA-directed DNA methylation. J. Cell Sci. 117: 4881–4888 [DOI] [PubMed] [Google Scholar]
- Meyer R.C., Kusterer B., Lisec J., Steinfath M., Becher M., Scharr H., Melchinger A.E., Selbig J., Schurr U., Willmitzer L., Altmann T. (2010). QTL analysis of early stage heterosis for biomass in Arabidopsis. Theor. Appl. Genet. 120: 227–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S., et al. (2008). Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133: 116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S., Hosaka K. (2010). DNA methylation in diploid inbred lines of potatoes and its possible role in the regulation of heterosis. Theor. Appl. Genet. 120: 205–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z., Kim E.D., Ha M., Lackey E., Liu J., Zhang Y., Sun Q., Chen Z.J. (2009). Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 457: 327–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K., Ueda J., Komaki M.K., Bell C.J., Shimura Y. (1991). Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 3: 677–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D.H., Somers D.E., Kim Y.S., Choy Y.H., Lim H.K., Soh M.S., Kim H.J., Kay S.A., Nam H.G. (1999). Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285: 1579–1582 [DOI] [PubMed] [Google Scholar]
- Pasapula V., et al. (2011). Expression of an Arabidopsis vacuolar H+-pyrophosphatase gene (AVP1) in cotton improves drought- and salt tolerance and increases fibre yield in the field conditions. Plant Biotechnol. J. 9: 88–99 [DOI] [PubMed] [Google Scholar]
- Peer W.A., Bandyopadhyay A., Blakeslee J.J., Makam S.N., Chen R.J., Masson P.H., Murphy A.S. (2004). Variation in expression and protein localization of the PIN family of auxin efflux facilitator proteins in flavonoid mutants with altered auxin transport in Arabidopsis thaliana. Plant Cell 16: 1898–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radoev M., Becker H.C., Ecke W. (2008). Genetic analysis of heterosis for yield and yield components in rapeseed (Brassica napus L.) by quantitative trait locus mapping. Genetics 179: 1547–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronemus M.J., Galbiati M., Ticknor C., Chen J., Dellaporta S.L. (1996). Demethylation-induced developmental pleiotropy in Arabidopsis. Science 273: 654–657 [DOI] [PubMed] [Google Scholar]
- Semel Y., Nissenbaum J., Menda N., Zinder M., Krieger U., Issman N., Pleban T., Lippman Z., Gur A., Zamir D. (2006). Overdominant quantitative trait loci for yield and fitness in tomato. Proc. Natl. Acad. Sci. USA 103: 12981–12986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo C., Lister C., Crevillen P., Nordborg M., Dean C. (2006). Variation in the epigenetic silencing of FLC contributes to natural variation in Arabidopsis vernalization response. Genes Dev. 20: 3079–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley B.W. (1996). Flavonoid biosynthesis: “New” functions for an “old” pathway. Trends Plant Sci. 1: 377–382 [Google Scholar]
- Song G.S., et al. (2010). Comparative transcriptional profiling and preliminary study on heterosis mechanism of super-hybrid rice. Mol. Plant 3: 1012–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer N.M., Stupar R.M. (2007). Allelic variation and heterosis in maize: How do two halves make more than a whole? Genome Res. 17: 264–275 [DOI] [PubMed] [Google Scholar]
- Strayer C., Oyama T., Schultz T.F., Raman R., Somers D.E., Más P., Panda S., Kreps J.A., Kay S.A. (2000). Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289: 768–771 [DOI] [PubMed] [Google Scholar]
- Tariq M., Saze H., Probst A.V., Lichota J., Habu Y., Paszkowski J. (2003). Erasure of CpG methylation in Arabidopsis alters patterns of histone H3 methylation in heterochromatin. Proc. Natl. Acad. Sci. USA 100: 8823–8827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira F.K., Colot V. (2010). Repeat elements and the Arabidopsis DNA methylation landscape. Heredity (Edinb.) 105: 14–23 [DOI] [PubMed] [Google Scholar]
- Vilkaitis G., Plotnikova A., Klimasauskas S. (2010). Kinetic and functional analysis of the small RNA methyltransferase HEN1: The catalytic domain is essential for preferential modification of duplex RNA. RNA 16: 1935–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Tian L., Lee H.S., Wei N.E., Jiang H., Watson B., Madlung A., Osborn T.C., Doerge R.W., Comai L., Chen Z.J. (2006). Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics 172: 507–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Feng Z., Wang X., Wang X., Zhang X. (2010). DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26: 136–138 [DOI] [PubMed] [Google Scholar]
- Wang X., Elling A.A., Li X., Li N., Peng Z., He G., Sun H., Qi Y., Liu X.S., Deng X.W. (2009). Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell 21: 1053–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenegger M., Heimes S., Riedel L., Sänger H.L. (1994). RNA-directed de novo methylation of genomic sequences in plants. Cell 76: 567–576 [DOI] [PubMed] [Google Scholar]
- Wei G., et al. (2009). A transcriptomic analysis of superhybrid rice LYP9 and its parents. Proc. Natl. Acad. Sci. USA 106: 7695–7701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q., Frugis G., Colgan D., Chua N.H. (2000). Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 14: 3024–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakir E., Hilman D., Harir Y., Green R.M. (2007). Regulation of output from the plant circadian clock. FEBS J. 274: 335–345 [DOI] [PubMed] [Google Scholar]
- Yanovsky M.J., Kay S.A. (2001). Signaling networks in the plant circadian system. Curr. Opin. Plant Biol. 4: 429–435 [DOI] [PubMed] [Google Scholar]
- Yu S.B., Li J.X., Xu C.G., Tan Y.F., Gao Y.J., Li X.H., Zhang Q., Saghai Maroof M.A. (1997). Importance of epistasis as the genetic basis of heterosis in an elite rice hybrid. Proc. Natl. Acad. Sci. USA 94: 9226–9231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A., McDaniel I.E., Silva P., Zilberman D. (2010). Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328: 916–919 [DOI] [PubMed] [Google Scholar]
- Zhang H.Y., He H., Chen L.B., Li L., Liang M.Z., Wang X.F., Liu X.G., He G.M., Chen R.S., Ma L.G., Deng X.W. (2008a). A genome-wide transcription analysis reveals a close correlation of promoter INDEL polymorphism and heterotic gene expression in rice hybrids. Mol. Plant 1: 720–731 [DOI] [PubMed] [Google Scholar]
- Zhang X. (2008). The epigenetic landscape of plants. Science 320: 489–492 [DOI] [PubMed] [Google Scholar]
- Zhang X., Yazaki J., Sundaresan A., Cokus S., Chan S.W., Chen H., Henderson I.R., Shinn P., Pellegrini M., Jacobsen S.E., Ecker J.R. (2006). Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126: 1189–1201 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. (2008b). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Yu S., Xing C., Fan S., Song M. (2008). [DNA methylation in cotton hybrids and their parents]. Mol. Biol. (Mosk.) 42: 195–205 [PubMed] [Google Scholar]
- Zilberman D., Gehring M., Tran R.K., Ballinger T., Henikoff S. (2007). Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 39: 61–69 [DOI] [PubMed] [Google Scholar]