A large and diverse miRNA superfamily targets mRNAs of disease resistance proteins and initiates phased secondary siRNA that target other mRNAs. Pathogen-encoded suppressor proteins interfere with this process; consequently, infection increases the level of mRNAs for disease resistance and other proteins.
Abstract
Analysis of tomato (Solanum lycopersicum) small RNA data sets revealed the presence of a regulatory cascade affecting disease resistance. The initiators of the cascade are microRNA members of an unusually diverse superfamily in which miR482 and miR2118 are prominent members. Members of this superfamily are variable in sequence and abundance in different species, but all variants target the coding sequence for the P-loop motif in the mRNA sequences for disease resistance proteins with nucleotide binding site (NBS) and leucine-rich repeat (LRR) motifs. We confirm, using transient expression in Nicotiana benthamiana, that miR482 targets mRNAs for NBS-LRR disease resistance proteins with coiled-coil domains at their N terminus. The targeting causes mRNA decay and production of secondary siRNAs in a manner that depends on RNA-dependent RNA polymerase 6. At least one of these secondary siRNAs targets other mRNAs of a defense-related protein. The miR482-mediated silencing cascade is suppressed in plants infected with viruses or bacteria so that expression of mRNAs with miR482 or secondary siRNA target sequences is increased. We propose that this process allows pathogen-inducible expression of NBS-LRR proteins and that it contributes to a novel layer of defense against pathogen attack.
INTRODUCTION
MicroRNAs (miRNAs) are versatile regulators of gene expression in plants and animals. They are 21 to 24 nucleotides long and processed by a Dicer nuclease from long RNA precursors with base paired foldback structures (Baulcombe, 2004). The single-stranded form of the miRNA forms a ribonucleoprotein complex with Argonaute (AGO) that can bind by base pairing to a target RNA (Bartel, 2009; Voinnet, 2009). In plants, the successful targeting reaction requires complementarity of the miRNA at most of the residues (Mallory and Bouché, 2008).
The consequence of the targeting reaction depends on the nature of the targeted RNA and the extent of complementarity with the miRNA. The target RNA is cleaved and the level of the protein product is reduced if there is near complete complementarity, including positions 9 and 10 of the miRNA. Translational suppression without turnover of the target RNA is mediated by miRNAs with incomplete complementarity to their target (Brodersen et al., 2008; Lanet et al., 2009). In addition, there may be miRNA-mediated targeting of chromatin-associated RNAs that leads directly or indirectly to targeted epigenetic modification (Khraiwesh et al., 2010; Wu et al., 2010).
In some instances, miRNA-mediated gene silencing is a simple negative switch: whenever the miRNA gene is active the target mRNA is silent. However, these versatile RNA regulators may also participate in feedback loops and carry out more subtle roles in genetic regulation. They might dampen fluctuations in target gene expression, for example, or influence temporal changes (Voinnet, 2009). In some instances, the miRNAs or their precursors may move through plasmodesmata and different stages in the feedback system occur in adjacent cells or in separate roots and shoots (Bari et al., 2006; Pant et al., 2008).
miRNAs may also initiate regulatory cascades with multiple mRNA targets (MacLean et al., 2010). These cascades involve secondary small interfering RNAs (siRNAs) that associate with AGO proteins, similarly to miRNAs. The first step in these cascades requires an RNA-dependent RNA polymerase (RDR; RDR6 in Arabidopsis thaliana) and it takes place when the initiator miRNA duplex structure is asymmetrical (Manavella et al., 2012), if the initiator miRNA is 22 nucleotides rather than 21 nucleotides long (Chen et al., 2010; Cuperus et al., 2010), or if there are two target sites for 21-nucleotide RNAs (Axtell et al., 2006). The initiator miRNA stimulates the RDR to convert the targeted RNA into long, double-stranded RNA that is then processed by Dicer into secondary siRNAs. A high proportion of the secondary siRNAs are in a 21-nucleotide phased register in which the first position is the cleavage target of the initiator miRNA (Chen et al., 2007).
The best characterized secondary siRNAs are known as trans-acting siRNAs, and their targets include a pentatricopeptide (PPR) and auxin response factor (ARF) gene mRNAs (Axtell et al., 2006). These secondary siRNAs may move between cells and establish developmental gradients (Chitwood et al., 2009). Both PPR and ARF mRNAs are targeted separately by secondary siRNAs and by miR161 (Rhoades et al., 2002; Allen et al., 2005; Yoshikawa et al., 2005), miR160, or miR167 (Rhoades et al., 2002; Allen et al., 2005). Therefore, it is likely that the secondary siRNAs play a role in either reinforcing or coordinating the action of primary sRNAs or miRNAs.
The secondary siRNA loci, like miRNA genes, have evolved independently at different times (Cuperus et al., 2011). The TAS3-derived secondary siRNAs targeting ARF mRNAs, for example, are conserved in distantly related plants (Axtell and Bartel, 2005), and it is likely that the TAS3 loci evolved in a common ancestor of seed plants. Other secondary siRNA loci in rice (Oryza sativa) and Brachypodium are species specific and are likely to have arisen more recently (Johnson et al., 2009; Vogel et al., 2010). Similarly, the TAS1 secondary siRNAs that target PPR mRNAs are specific to Arabidopsis and they are also likely to be relatively modern.
Both ancient and modern secondary siRNA loci are targeted by miR2118. In Medicago (Zhai et al., 2011), the miR2118 family sequences initiate secondary siRNAs on the mRNAs for nucleotide binding site–Leu-rich repeat (NBS-LRR) proteins in a process that is likely to have originated in a common ancestor of monocots and dicots. The miR2118 family also initiates secondary siRNAs on uncharacterized mRNAs expressed in rice flowers (Song et al., 2012). However, these alternative secondary siRNAs are not found in other species and so their origin is likely to be in the rice evolutionary lineage and more recent than a monocot/dicot common ancestor.
Here, we characterize secondary siRNA loci and the initiator miRNAs in Solanum species. The loci correspond to the genes for NBS-LRR disease resistance proteins, and we demonstrate that at least one of the secondary siRNAs targets the mRNA that, in a related form, has been implicated in basal immunity in Arabidopsis. The initiators of secondary siRNAs on NBS-LRR mRNAs comprise an unusually diverse superfamily comprising miR482 and miR2118 in other species. The sequence of this miRNA superfamily is variable and a high proportion of the NBS-LRR mRNAs are targeted. The miR482-mediated silencing of NBS-LRR mRNAs is lost in virus- and bacteria-infected tissues most likely due to the action of pathogen-encoded proteins. Based on these data, we propose a model in which the plant exploits pathogen-derived suppressors of RNA silencing to achieve inducible expression of defense-related genes.
RESULTS
An Unusually Diverse miRNA Superfamily
The miR482 family is unusual in that the members are 22 nucleotides rather than 21 nucleotides long and they have more variable sequences than other miRNA families. In different plant species, the miR482 sequences vary at nine positions (Figures 1A to 1C) and there are at least 31 isoforms. Other miRNAs, even when they exist as families, have fewer differences and generally fewer isoforms (Felippes et al., 2008; Meyers et al., 2008; Cuperus et al., 2011).
Figure 1.
Sequence Diversity of miR482.
(A) Multiple alignment of diverse miR482 family members. Alignment of six unique miR482 sequences identified from tomato small RNA data sets.
(B) Alignment of a tomato miR482 isoform with atmiR472.
(C) Alignment of all known miR482 isoforms.
Alignments were made with ClustalX2, and miR482 sequences were taken from miRBase (http://www.mirbase.org/). Sequences shown in red are not conserved among miRNAs. sl, S. lycopersicum; ath, Arabidopsis; gh, G. hirsutum; vv, V. vinifera; pab, Picea abies; ag, Aquilegia caerulea; cs, Citrus sinensis; gma, G. max; gsi, G. soja; pt, Populus trichocarpa; pta, Pinus taeda; stu, S. tuberosum; pv, Phaseolus vulgaris; mdm, Malus domestica; gr, G. raimondii.
The miR482 family could be extended to include miR472 in Arabidopsis by allowing an additional variation at position 2 (Figure 1B). In addition, if 22-nucleotide miRNAs with a first position that aligns with position 3 of miR482 are also allowed, a superfamily would form that includes the miR2118 subgroup (Zhai et al., 2011). The nomenclature of these miRNAs in the online miRBase (Griffiths-Jones et al., 2008) is inconsistent because there are miR2118-like sequences named as miR482, miR5300, or miR2809. In this article we refer to the 22-nucleotide miRNAs that align with position 1 of miR482 as “miR482 type” and those that align with position 3 or 4 of miR482 as “miR2118 type” members of the miR482/2118 superfamily (see Supplemental Figure 1 online).
Our sRNA data sets from tomato seedlings (Solanum lycopersicum cv M82) include six isoforms of mature miR482 with variation at seven of the nine variable sites (Figure 1A). Two other tomato sequences are of the miR2118 type (Mohorianu et al., 2011). One of these is similar to Medicago miR2118a and it is named as slmiR482 in miRBase. The second is named as miR5300. The miR2118 family in rice is even more diverse. There are 18 pre-miRNA variants corresponding to 16 different mature miRNAs (Johnson et al., 2009; Vogel et al., 2010).
If variation in the miR482/2118 superfamily had been generated in an ancient ancestor, we would expect different plants to share similar isoforms of miR482. However, most of the 31 isoforms of miR482 (Figure 1A) are species specific and a more likely scenario is that most of the variation was generated since the divergence of even closely related species. There are only a few instances of identical sequence variants in more than one plant; thus, stumiR482 (potato [Solanum tuberosum]) is identical to slmiR482b (tomato), and gmmiR482a (soybean [Glycine max]) is the same as pvmiR482 (Phaseolus) (Xie et al., 2011; Zhai et al., 2011). In these examples, the common form may have evolved before the divergence of the two species but not in an ancient ancestor.
There are only four examples of distantly related plant species with an identical isoform of miR482. These are tomato with slmiR482f and cotton (Gossypium hirsutum) with ghmiR482a; grape (Vitis vinifera) with vvmiR482 and Picea with pabmiR482c; and poplar (Populus spp) with miR482c and Picea with pabmiR482b. In addition, there is one variant that is shared by three distantly related species in the Pinaceae (Pinus-ptamiR482a/b and Picea-pabmiR482a) and the Salicaceae (Populus) (ptmiR482a/b).These exceptions to the species-specific pattern are probably due to independent evolution of identical isoforms in different plant lineages.
Diversification of the miR2118 subfamily is similarly species specific. The pre-miR2118 sequences from rice, for example, are all more similar to each other than they are to the corresponding sequences from other species.
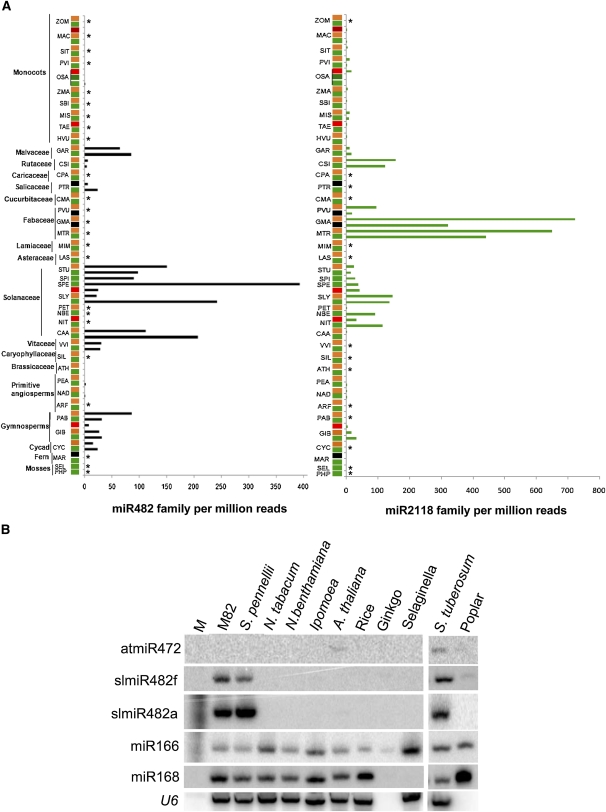
The abundance of miR482/2118 superfamily RNAs varies greatly between species and families of plants. These RNAs are rare in monocots and infrequent in Caricaceae, Asteraceae, Cucurbitaceae, Lamiaceae, Brassicaceae, Caryophyllaceae, basal angiosperms, ferns, and mosses (Figure 2A). The miR482 members are abundant in cycads, gymnosperms, members of the Malvaceae and Vitaceae, and the Solanum and Capsicum genera in the Solanaceae. In seedling data sets of tomato and in its wild relative Solanum pennellii, the miR482 type are predominant with miR482f represented 100-fold more frequently than miR482a (see Supplemental Figure 2 online). The miR2118 family members, by contrast, are most abundant in the Rutaceae, Solanaceae, and, particularly, in the Fabaceae. The miR5300 that is assigned as miR2118 type is most abundant in the Solanaceae.
Figure 2.
Abundance of miR2118/482 Superfamily Members in Different Plants.
(A) Small RNA data sets were accessed through GEO, and miRNA abundance was analyzed through miRProf (Moxon et al., 2008) and expressed as counts per million reads. miR482 and 2118 sequences are defined in the text. Samples were from leaf/shoot tissue (green), floral tissue (orange), panicle/tassel (red), and other (black). Asterisk indicates those plant species where miRNA family members were not cloned. PHP, Physcomitrella patens; SEL, Selaginella; MAR, Marsilea; CYC, Cycas; GIB, Gingko biloba; PAB, Picea abies; ARF, Aristolochia fimbriata; NAD, Nuphar advena; PEA, Persea americana; ATH, Arabidopsis; SIL, Silene latifolia; VVI, V. vinifera; CAA, Capsicum annum; NIT, Nicotiana tabacum; NBE, N. benthamiana; PET, Petunia hybrida; SLY, S. lycopersicum; SPE, S. pennellii; SPI, S. pimpinellifolium; STU, S. tuberosum; LAS, Lactuca sativus; MIM, Mimulus; MTR, Medicago truncatula; GMA, G. max; PVU, Phaseolus vulgaris; CMA , Cucurbita maxima; PTR, Populus trichocarpa; CPA, Carica papaya; CSI, Citrus sinensis; GAR, Gossypium arboreum; HVU, Hordeum vulgare; TAE, Triticum aestivum; MIS, Miscanthes; SBI, Sorghum bicolor; ZMA, Zea mays; OSA, O. sativa; PVI, Panicum virgatum; SIT, Setaria italica; MAC, Musa acuminata; ZOM, Zostera marina.
(B) RNA gel blot analysis of tomato miR482 isoforms and cross-hybridizing homologs in different species. Fifteen micrograms of total RNA from young seedlings was used for analysis of each sample. Total RNA was electrophoresed in a 15% polyacrylamide gel, transferred to membrane, and probed with corresponding DNA oligonucleotides (see Supplemental Table 1 online) labeled with [γ-32P]ATP. U6 serves as loading control. M, decade (Ambion) size marker.
RNA gel blot analysis was consistent with Solanum species having unusually high levels of miR482: probes for isoforms miR482a and f detected abundant RNAs in S. lycopersicum (tomato), S. pennellii, and S. tuberosum but not in Nicotiana or non-Solanaceous species (Figure 2B; see Supplemental Table 1 online). A probe for Arabidopsis miR472 hybridized with an RNA in potato but only weakly with tomato miRNAs.
Together, these various analyses indicate that the miR482/2118 superfamily is unusually variable both in sequence and in expression level. The pattern of variability indicates that this superfamily is in constant evolutionary flux with different species and genera having specific isoforms or combinations of isoforms.
miR482/2118 Superfamily Members as Potential Regulators of NBS-LRR Disease Resistance Genes
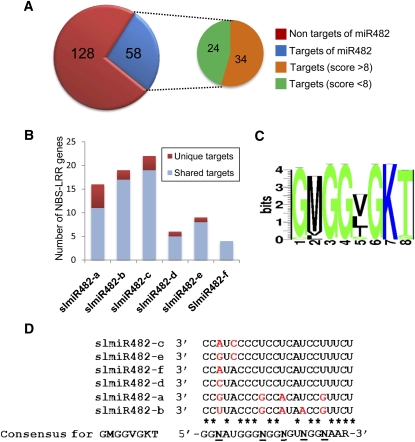
The psRNATarget algorithm that predicts targets of plant miRNAs (Dai and Zhao, 2011) (see Supplemental Tables 2 and 3 online) identified the mRNAs of disease resistance proteins as having binding sites for miR482 species. These proteins have NBS and LRR domains (Meyers et al., 2003), and, of the 186 predicted NBS-LRR proteins encoded in the tomato genome, there were 58 with miR482 target motifs as assessed by the TAPIR protocol (Bonnet et al., 2010) (Figure 3A). It is highly likely that these are bona fide targets because they have a high degree of complementarity to the miR482 sequences with more than half having a TAPIR algorithm score of >8. The mRNAs of NBS-LRR proteins have also been identified recently as targets of the miR2118 family in Medicago truncatula (Zhai et al., 2011).
Figure 3.
Predicted Targets of miR482 Isoforms in Tomato.
(A) Predicted miR482 targets among the 186 NBS-LRR sequences in tomato.
(B) Representation of miR482 targets in the tomato genome as either unique or repeated sequence motifs.
(C) Coding sequence of the miR482 targets in the mRNAs of NBS-LRR mRNAs.
(D) Variable residues in the miR482 family correspond to wobble or variable sequences in NBS-LRR mRNAs.
Target sequences were predicted using the TAPIR algorithm.
A minority of the miR482 isoforms had unique NBS-LRR targets in tomato (Figure 3B). However, most miR482 family members had multiple NBS-LRR targets (see Supplemental Table 4 online). This pattern arises because the target sequence encodes a variant of the P-loop or Walker A motif that is specific to R proteins (Figure 3C), and, of the seven variable positions in the tomato miR482 family (Figure 1A), there are six that are complementary to the wobble position of conserved amino acids in the NBS domain of R proteins or to amino acid position 5 in this motif (Meyers et al., 2003) (Figures 3C and 3D) that is not conserved. The miR2118 family shows a similar pattern of variation, and we conclude that the miR482/2118 superfamily has the potential to control expression of many NBS-LRR genes and to be a master regulator of disease resistance in tomato.
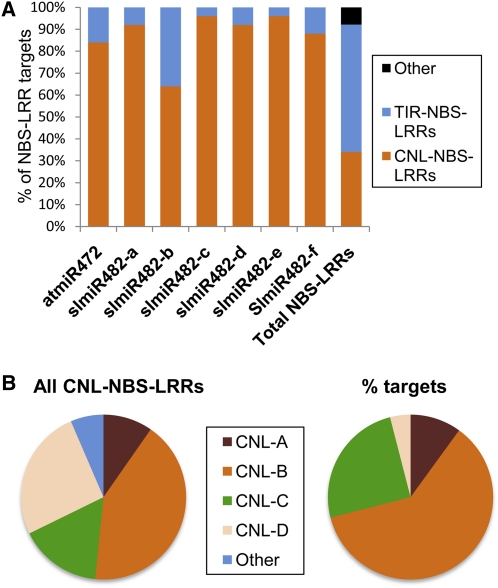
To find out whether the miR482 family targets a particular type of NBS-LRR protein, we aligned its sequences against the well annotated resistance (R) gene sequences in the Arabidopsis genome. Using the TAPIR rules for miRNA targeting, we established that mRNAs of coiled-coil NBS-LRR (CNL)-type proteins (Meyers et al., 2003), with a coiled-coiled domain at the N terminus rather than a Toll/Interleukin Repeat (TIR) motif, were the preferred targets of miR482 (Figure 4A). There was further differentiation because the CNL–D-type mRNAs are less represented in the targets than in the total CNL-type NBS-LRRs (Figure 4B).
Figure 4.
CNL-Type NBS-LRR mRNAs Are Preferred Targets of miR482 Family Members.
(A) The targets for tomato miR482 were identified among the Arabidopsis NBS-LRR mRNA sequences and classified as either CNL or TIR type.
(B) The CNL-type NBS-LRRs were classified as CNL types A to D according to Meyers et al. (2003). Each type is represented similarly in the total and miR482 targets all except that the CNL-D types are underrepresented in the targets (left panel).
The miR2118 family from M. truncatula also targets the P-loop of NBS-LRR proteins as revealed by alignment with sequences of M. truncatula (Zhai et al., 2011). The miR2118b variant, like miR482, targets CNL-NBS-LRR mRNAs (see Supplemental Figure 3 online). However, targets of Medicago miR2118a and c as well as tomato miR482 (a miR2118 type sequence) include both TIR- and CNL-NBS-LRR proteins. This targeting prediction is consistent with the high sequence homology of slmiR482 (slmiR2118) with Medicago miR2118a (see Supplemental Figure 1 online).
Therefore, this computational analysis indicates that the miR482 and miR2118 families constitute an unusually diverse miRNA superfamily with sequence variation that corresponds to the mRNAs of NBS-LRR proteins. The presence of the miR482/2118 superfamily would have the potential to suppress expression of NBS-LRR proteins to an extent that varies between plants. Those with abundant and diverse miR482/2118 would silence these disease resistance proteins more extensively than plants like Arabidopsis in which this superfamily is represented by a single nonabundant miRNA (miR472).
Secondary siRNAs Initiated by miR482 Family Members
A property of 22-nucleotide miRNAs is the ability to initiate the synthesis of secondary siRNAs from the 3′ side of the target RNA sequence (Chen et al., 2010; Cuperus et al., 2010). These secondary siRNAs align in a phased register with respect to the target site of the 22-nucleotide miRNA and, in some instances, they also have mRNA targets. In our tomato sRNA data sets, we identified 15 genomic RNA loci with phased siRNAs (Chen et al., 2007, 2010). Of these, three corresponded to the conserved TAS3 RNA that has been well characterized in Arabidopsis, one matched AGO1 mRNA that is also known in Arabidopsis to have secondary siRNAs initiated by miR168, five had unidentified miRNA initiators, and the remaining six were predicted targets of miR482 and encoded NBS-LRR proteins (see Supplemental Tables 5 and 6 online).
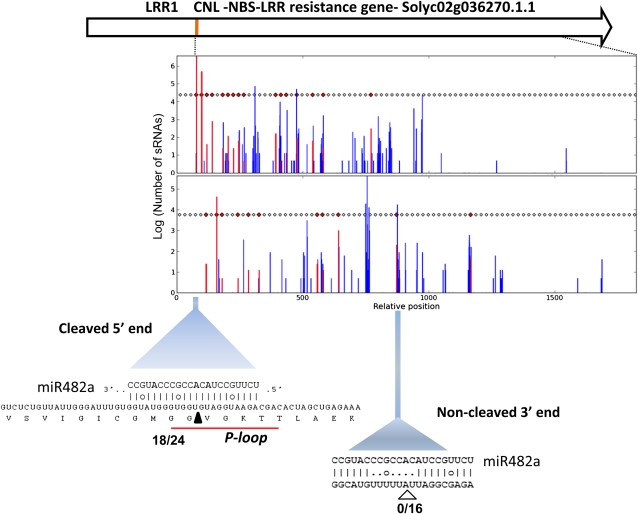
Figure 5 illustrates the distribution of phased siRNAs that target an NBS-LRR mRNA named LRR1 (Solyco02g036270.1.1). Most of the abundant siRNAs are in the phased register, and they align to a region close to the 3′ side of the miR482a target site. It is likely that the targeted mRNA would be cleaved by an AGO/miR482 nucleoprotein because there is complete miRNA/mRNA complementarity at positions 9 and 10 of the target site (Figure 5). This cleavage site defines the first position in the phased registers. Also consistent with this proposed cleavage site, we used 5′ rapid amplification of cDNA ends (RACE) analysis to detect cleavage of LRR1 mRNA (18/24 clones) between positions 10 and 11 in the predicted miR482a target site (Figure 5). This cleavage site matches well with a previous analysis of the tomato degradome (13 out of 22 clones; Li et al., 2012a). The degradome corresponds to mRNA fragments resulting from miRNA-directed cleavage.
Figure 5.
Phased Secondary siRNAs Initiated by miR482 Target Sites in an NBS-LRR mRNA.
Genome view of a CNL-type NBS-LRR locus (LRR1) that aligns with phased secondary siRNAs. Most of the secondary siRNAs align between miR482 target sites. Red bars indicate the start position of sRNAs in phase; blue bars indicate the start position for those out of phase. Rectangles indicate expected phased positions and those in red indicate phased sequences. Orange box indicates position of P-loop. Numbers next to the miR482 target site indicate frequency of 5′ RACE clones matching this prediction out of total clones used for analysis. Arrow color code: red, 21 nucleotides; blue, 24 nucleotides; green, 22 nucleotides. The black triangle indicates cleaved site, and the white triangle indicates proposed cleavage site.
In at least two of the NBS-LRR protein mRNAs (Figure 5; see Supplemental Figure 4 online), there is a second miR482 target site at the 3′ end of the region with phased siRNAs. This second target site has less complementary to the mRNA, and, as expected, 5′ RACE analysis did not identify cleaved products matching this region (Figure 5). However, the final position in the register aligns opposite to position 10 of miR482 and so the target site is in phase with the secondary siRNAs. These secondary siRNAs along with the accumulation of cleaved RNAs provide direct evidence that NBS-LRR protein mRNAs are targeted by miR482.
miR482 Silences NBS-LRR Protein mRNAs
The sequence of miR482 family members and the miR482-initiated secondary siRNAs aligned to NBS-LRR protein mRNAs suggest that miR482 is a suppressor of R protein biosynthesis. In line with this possibility, tomato CNL-NBS-LRRs (LRR1, Solyco02g036270.1.1; LRR2, Solyc04g005540.1.1) that are good predicted targets of miR482 members are less abundant than the Hero and Mi NBS-LRR mRNAs that are not miR482 targets (see Supplemental Figure 5 online). It is likely that this difference is due to miR482 expression because the wild tomato relative S. pennellii, in which miR482a is more abundant than M82, has lower expression of LRR1 and LRR2 than M82 but similar levels of Hero and Mi mRNAs. Similarly in F1 and F2 lines from an M82 × S. pennellii cross, the LRR1 and LRR2 mRNAs are less abundant than in M82 but expression in an introgression line (IL8-3; Eshed and Zamir, 1995) resembles that of M82 tomato.
We next used a transient assay system in Nicotiana benthamiana to test the contribution of miR482 and secondary siRNAs as mediators of NBS-LRR mRNA silencing. This species has only low levels of miR482 (Figures 2A and 2B), and we used plants that were either wild type or with reduced expression of RDR6 (RDR6i; Schwach et al., 2005). RDR6 is an essential factor in secondary siRNA production (Yoshikawa et al., 2005).
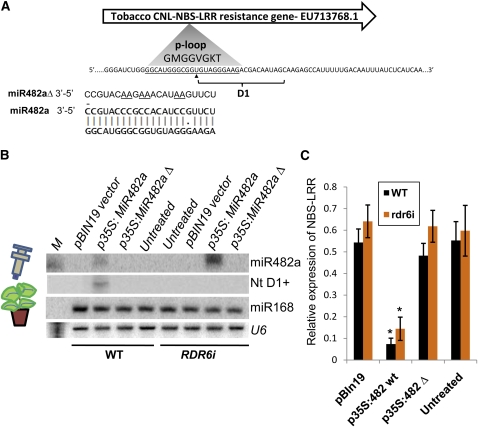
Tomato miR482a was expressed transiently using Agrobacterium infiltration, and we assayed the levels of a tobacco NBS-LRR mRNA-EU713768.1 (Figure 6A) that is conserved in N. benthamiana and that has a good target site for miR482a. The expression construct had the miR482a precursor sequence coupled to the cauliflower mosaic virus 35S promoter. We also expressed miR482aΔ that contains alterations to the mature miR482 sequence so that the complementarity with the NBS-LRR mRNA is lost.
Figure 6.
Transient Expression of Tomato miR482a Targets an NBS-LRR mRNA in N. benthamiana Resulting in RDR6-Dependant Phased siRNA Biogenesis.
(A) Targeting of Nicotiana EU713768.1 NBS-LRR by miR482a. The alignment shows the target site of miR482a in a tobacco NBS-LRR mRNA, and the predicted sequences of phased secondary siRNAs are shown at an adjacent site D1. Δ indicates modified sequences underlined in the miR482aΔ construct.
(B) RNA gel blot analysis of miR482a and D1 siRNA in Agrobacterium tumefaciens infiltrated zones of N. benthamiana wild-type (WT) and RDR6i lines. Analyses of miR168 and U6 RNAs are included as controls. M, decade marker indicating a 20-nucleotide RNA.
(C) U713768.1 mRNA accumulation 3 d after inoculation by quantitative PCR. See Methods for construct design. Error bars indicate sd (n = 3), and asterisks indicate a significant difference from corresponding control samples (t test, P value < 0.05).
Expression of the native miR482a but not miR482aΔ in this assay initiated the expression of secondary siRNA D1 (Figure 6B) and silencing of EU713768.1 mRNA by 10-fold or greater (Figure 6C). In N. benthamiana wild-type and RDR6i lines, transient expression of miR482a also targeted mRNA from an EU713768.1 mRNA (Figure 6C), but there was no secondary small RNA production at the D1 register in RDR6i lines (Figure 6B). Therefore, in this system, the secondary siRNAs were not required for suppression of the NBS-LRR mRNA; miR482 primary targeting was sufficient.
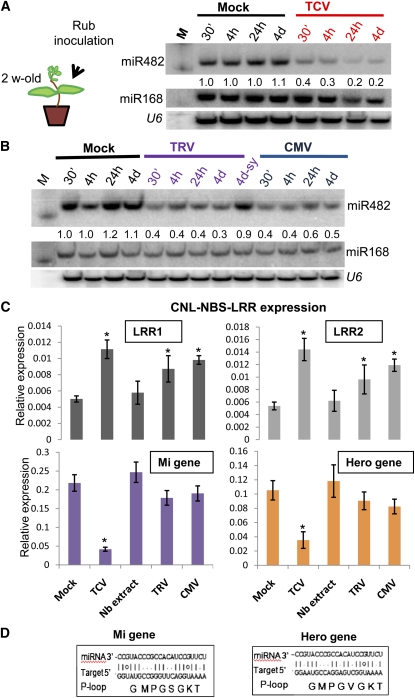
A second assay exploited the potential of plant viruses to produce suppressors of RNA silencing. We predicted that these suppressor proteins in virus-infected plants would interfere with either the production or targeting of miR482 so effectively that NBS-LRR mRNAs with target motifs would increase in abundance in virus-infected tissue. NBS-LRR proteins without the target motifs should not be affected in the same way.
The level of miR482 was reduced slightly in the inoculated leaves of tomato plants infected with Turnip crinkle virus (TCV), Cucumber mosaic virus (CMV), and Tobacco rattle virus (TRV), whereas miR168 was unaffected (Figures 7A and 7B). The LRR1 and LRR2 mRNAs with miR482 targets were both more abundant in plants infected with these three viruses, with the increase in both instances being most pronounced with TCV (Figure 7C). Neither of two NBS-LRR mRNAs (Mi and Hero) without miR482 targets was more abundant after virus infection (Figures 7C and 7D). TRV and CMV had no significant effect on Mi and Hero, whereas TCV led to a decrease in the level of these mRNAs.
Figure 7.
miR482 Silencing of NBS-LRR mRNAs Is Suppressed in Virus-Infected Plants.
(A) RNA gel blot analysis of miR482 in TCV-infected tomato leaves. Total RNA from TCV infected and noninfected leaves of N. benthamiana was inoculated to M82 tomato (TCV and Mock). RNAs isolated at the indicated times after inoculation were separated on a 15% polyacrylamide gel. The RNA was transferred to a membrane and probed with radiolabeled DNA oligonucleotides for miR482 with miR168 and U6 probes as loading controls. M, decade marker indicating a 20-nucleotide RNA.
(B) RNA gel blot analysis of miR482 in TRV- and CMV-infected tomato leaves. Total sap from TRV infected, CMV infected, and noninfected leaves of N. benthamiana were inoculated to M82 tomato (TRV, CMV, and Mock, respectively). RNAs isolated at the indicated times after inoculation were separated on a 15% polyacrylamide gel. The RNA was transferred to a membrane and probed with radiolabeled DNA oligonucleotides for miR482 with miR168 and U6 probes as loading controls. 4d sy, tissue from systemic uninoculated leaf; M, decade marker (Ambion) indicating a 20-nucleotide RNA.
(C) Quantitative PCR analysis for the abundance of two miR482 target mRNAs (LRR1 and LRR2) (Top) and Mi and Hero NBS-LRRs that are not targets of miR482 (Bottom; see [D]). RNA was extracted 4 h after inoculation. Error bars indicate sd (n = 3), and asterisks indicate a significant difference from corresponding control samples (t test, P value < 0.05).
(D) The best target sequences of miR482 with Mi and Hero mRNAs.
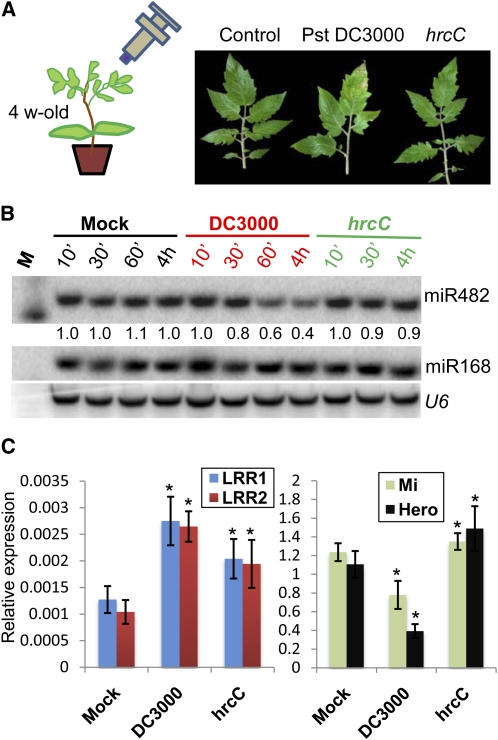
In a similar experiment, we exploited the finding that the bacterial pathogen Pseudomonas syringae DC3000 encodes suppressors of RNA silencing (Navarro et al., 2008). These suppressors are transported into infected plant cells via a type III secretion system. We predicted that, as in virus-infected plants, the DC3000 infection of tomato would induce LRR1 and LRR2 mRNAs but not Mi and Hero mRNAs. We further predicted that any increase in LRR1 and LRR2 mRNAs would be reduced in a hrcC mutant of P. syringae DC3000 in which the type III secretion system would be less active. The results were fully consistent with these predictions (Figures 8A to 8C). There was a slight induction of LRR1, LRR2, Mi, and Hero mRNAs in hrcC mutants independent of miR482 levels, indicating that the observed effects are partially independent of the delivery of effectors.
Figure 8.
miR482 Silencing of NBS-LRR mRNAs Is Suppressed in Plants Infected with P. syringae DC3000.
(A) Symptoms produced by the wild type or hrcC mutant P. syringae DC3000 in 4-week-old leaves of tomato at 2 weeks after inoculation.
(B) RNA gel blot analysis of miR482 accumulation in mock-, DC3000-, and hrcC-inoculated leaves at various time points. M, decade size marker.
(C) Quantitative PCR analysis for the abundance of two miR482 target mRNAs (LRR1 and LRR2) (top panel) and Mi and Hero NBS-LRRs that are not targets of miR482 (bottom panel). RNA was extracted 4 h after inoculation. Error bars indicate sd (n = 4), and asterisks indicate a significant difference from corresponding control samples (t test, P value < 0.05).
From these data with viruses and bacteria, we conclude that miR482-mediated silencing of NBS-LRR mRNAs is relieved in pathogen-infected tomato plants. In principle, there could be a similar loss of miR2118 type silencing in other plants following infection with bacteria and viruses resulting in increased abundance of resistance proteins. However, the expression of miR2118 is specific to panicles in monocots (Song et al., 2012), whereas tomato miR482 is expressed at similar levels in every tissue tested (see Supplemental Figure 6 online). Hence, the induction of NBS-LRR mRNAs in infected plants may be specific to Solanum species and members of the Fabaceae (Figure 2A) in which the 482/2118 superfamily miRNAs are abundant.
Natural Variation in Secondary Silencing
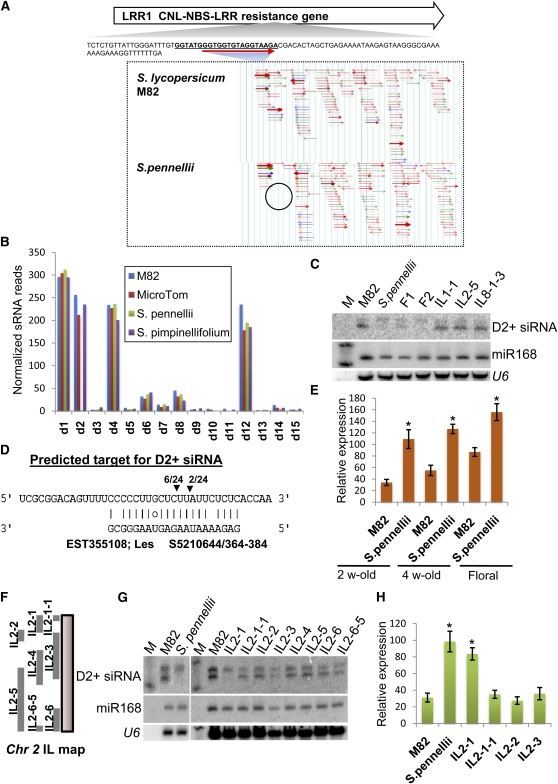
The miR173 and miR390 initiate secondary siRNAs in Arabidopsis (Allen et al., 2005; Chen et al., 2007) that target mRNA for ARF or PPR proteins. To find out whether miR482 has siRNA-mediated secondary mRNA targets, we exploited a difference between tomato and S. pennellii in the profile of phased secondary siRNAs on the LRR1 mRNA. Tomato has abundant LRR1 secondary siRNAs in the D2 register (Figure 9A), whereas, in S. pennellii, this secondary siRNA is absent (Figures 9B and 9C). The introgression line IL2-1, in which the LRR1 locus is derived from S. pennellii, also lacks the 21-nucleotide siRNA from D2 (Figures 9F and 9G). The predicted target of the D2 siRNA is the mRNA for an unknown protein (Figure 9D). Consistent with this prediction, we detected mRNA cleavage products with a 5′ end that corresponds to position 10 of the D2 siRNA in extracts of tomato but not S. pennellii and that this mRNA is more abundant in S. pennellii and IL2-1 than in M82 (Figures 9E and 9H).
Figure 9.
Natural Variation in Secondary siRNAs Derived from LRR1.
(A) Genome browser view of secondary siRNAs aligned to LRR1 mRNA. The miR482 target site at the 5′ end of the gene is indicated. The circled region shows the absence of D2+siRNA in S. pennellii.
(B) Bar chart showing accumulation of phased secondary siRNAs in two cultivars of tomato (M82 and MicroTom) and two wild species (S. pennellii and S. pimpinellifolium). D1 to D15 indicate the different positions in the phased register that is established by the miR482 targeting event.
(C) RNA gel blot analysis of D2+ siRNA in tomato, S. pennellii, their F1 and F2 hybrids, and in introgression lines (ILs). The D2+ probe detected a 24- and 21-nucleotide species (top and bottom signal) of which only the 21-nucleotide species was absent in S. pennellii and the F1 and F2 samples. M, decade size marker.
(D) Predicted target for the D2+ siRNA derived from LRR1.
(E) Quantitative PCR analysis of abundance of D2+ target gene between M82 and S. pennellii. Error bars indicate sd (n = 6), and asterisks indicate a significant difference from corresponding control samples in S. pennellii (t test, P value < 0.05).
(F) Genetic map of IL2 series introgression lines (Eshed and Zamir, 1995). The S. pennellii genomic regions in each IL are marked.
(G) RNA gel blot analysis of D2+ siRNA in tomato, S. pennellii, and the IL2 series introgression lines.
(H) Accumulation of the D2+ target mRNA in tomato, S. pennellii, and the IL2 series introgression lines. Error bars indicate sd (n = 3), and asterisks indicate a significant difference from corresponding control samples in S. pennellii and IL2-1 (t test, P value < 0.05).
DISCUSSION
Variation in the miR482/2118 Superfamily
Defense against pathogens is one of the most potent drivers of evolutionary change in host organisms (Nilsson et al., 1997). It accounts for a high degree of variation both within and between species in host defense molecules and for diversifying selection in the corresponding gene sequences. In this analysis of miR482 in tomato and S. pennellii, we provide evidence for a different type of variation that would be associated with the pathogen-inducible expression of NBS-LRR mRNAs and possibly other mRNAs associated with defense.
The NBS-LRR proteins are normally associated with effector-triggered immunity against pathogens. From these and other data (Figures 5 to 8; Zhai et al., 2011), it is evident that their mRNAs are targeted by members of the miR482/2118 superfamily. Other mRNAs may also be linked to miR482 if they are secondary siRNA targets (Figure 9). In this study, for example, we identify the mRNA for an unidentified protein as a secondary target of miR482. This unidentified protein has similarity to PEN3 that is involved for basal immunity in Arabidopsis (Stein et al., 2006). In addition, a predicted target of the highly abundant D4-siRNA (normalized reads of ~200 in our data sets; Figure 9B) derived from LRR1 is a proteosome subunit (Les#S38989204/290-3111; TAPIR targeting score 4) that could also be involved in resistance gene function and disease resistance (Beers et al., 2000; Peart et al., 2002; Tör et al., 2003). We envision that miR482 regulates defense mechanisms in tomato via effects on NBS-LRR and other defense proteins.
A Role for NBS-LRR Proteins in Non-Race-Specific Immunity
The NBS-LRR proteins are normally associated with effector-triggered immunity in which there is a gene-for-gene relationship between the host and the pathogen (Jones and Dangl, 2006). The host gene encodes an NBS-LRR protein that mediates recognition, either direct or indirect, of a pathogenesis effector that is encoded by the pathogen. This effector-triggered immunity is normally specific for some but not all races or strains of a pathogen. However, if the NBS-LRR proteins are overexpressed (Bendahmane et al., 1999), defense can also be induced independently of protein-based recognition mechanisms. Based on the effect of NBS-LRR protein overexpression and on the analysis presented here, we propose a role of miRNA regulated NBS-LRR proteins in non-race-specific resistance against viral and bacterial pathogens that is linked to miRNAs and secondary siRNAs.
Our proposal involves an NBS-LRR induction mechanism that is mediated by suppressors of RNA silencing encoded by plant pathogenic viruses and bacteria (Brigneti et al., 1998; Voinnet et al., 1999; Navarro et al., 2008). In some instances, these proteins block the biogenesis of siRNA or miRNA, and in others, they target the RNA silencing effector complexes (Voinnet, 2005). Once the pathogen has become established, the suppressor would be produced in or transported into the infected plant cell and, as observed (Figures 7 and 8), the miRNA-mediated silencing of NBS-LRR proteins would be relieved. Presumably there would be the same release of secondary siRNA silencing resulting in increased levels of targets of NBS-LRR–derived siRNAs (Figure 9).
The release of silencing described here results in a two- to threefold increase of individual NBS-LRR mRNAs in infected plants (Figures 7 and 8). However, as the induction may involve many tens of mRNAs with miR482 targets (Figures 3 and 4), the net effect on the cell would be equivalent to induced expression of any one NBS-LRR mRNA by 100-fold or more. Thus, the release of miR482/2118 silencing in infected cells would have the same effect as overexpressing an individual NBS-LRR protein and the level of immunity in the plant would be enhanced. An effect on defense would be reinforced through loss of secondary siRNAs affecting other proteins with roles in defense (Figure 9).
The increased NBS-LRR proteins in infected cells would also potentiate effector-mediated immunity due to secondary infection. With an elevated level of NBS-LRR proteins, there would be accelerated activation of the defense pathways against secondary pathogens and the plants would be protected against double infection.
This proposal that expression of defense genes is coupled to the RNA silencing system adds another layer to the defense and counterdefense interactions of pathogens and their plant hosts. In the context of viruses, the RNA silencing system is a first layer of defense. The viral suppressors are a counterdefense system (Voinnet, 2005), and miR482 is then a counter-counterdefense system that is dependent on the counterdefense system.
There is also an involvement of miRNA-mediated silencing in the first layer of defense against bacterial pathogens of plants. Correspondingly, several of the bacterial effectors of disease are suppressor of RNA silencing (Navarro et al., 2008). This finding explains how there could also be induction of miR482 targets in plants infected with bacteria (Figure 8) and, possibly, with other pathogens if they encode RNA silencing suppressors.
An additional potential layer of regulation in this system could involve numerous truncated genes for NBS-LRR proteins in plant genomes (Meyers et al., 2003). Their transcripts could sequester miR482/2118 and thereby fine-tune the suppression of NBS-LRR mRNAs. There are precedents for regulation of miRNAs by target mimics in both plant (Franco-Zorrilla et al., 2007) and animal systems (Cesana et al., 2011; Karreth et al., 2011; Sumazin et al., 2011; Tay et al., 2011).
The Costs and Benefits of the miR482/2118 Superfamily
Disease resistance proteins may have a cost to the plant (Tian et al., 2003). The miR482/2118 system described here would minimize this cost because the NBS-LRR would be at a low level in the absence of a pathogen. In this connection, it is striking that NBS-LRR genes are unusually numerous (Pan et al., 2000; Xu et al., 2011; Zhai et al., 2011) in the members of Solanaceae and Fabaceae, in which the NBS-LRR mRNAs are regulated by miR482/2118 (Zhai et al., 2011). Perhaps the low-level expression of NBS-LRR protein mRNAs in these plants reduces any cost associated with individual gene family members and creates a background in which there can be diversification of the NBS-LRR gene family without there being a penalty for the plant. We envision that plants would benefit from low levels of NBS-LRR proteins due to miR482/2118 if infection pressure is low or if they have other layers of defense against pathogens.
However, for plants under high infection pressure or without alternatives to the NBS-LRR defense system, the benefit of low expression of NBS-LRR proteins could be offset by the other costs. For example, it could be that low level of NBS-LRR proteins results in a delay to effector-triggered immunity that would be damaging for plants that are frequently attacked by pathogens. We speculate that the variation in the sequences and expression level of the miR482/2118 superfamily (Figures 1 and 2) is likely to reflect the shift in the balance of the costs and benefits in different plants depending on the environment and other defense systems.
Postscript
In this report, we focus on the link of the miR482/2118 superfamily and disease resistance. Further testing of the hypotheses and speculation based on our data will require Medicago and tomato lines in which RDR6 and other RNA silencing proteins are knocked down or knocked out. Unfortunately, such lines are not yet available. There is also an outstanding question about the possible alternative roles of the miR482/2118 superfamily. In rice, for example, TIR-NBS-LRR proteins are absent (Zhou et al., 2004), but the miR2118 subfamily that targets their mRNAs is particularly diverse. We would therefore expect that there are other, as yet unidentified, targets of these miRNAs. While this article was under revision, 2118 family miRNAs targeting the mRNA for a tobacco TIR-NBS-LRR (N) were described (Li et al., 2012b). When these miRNAs were transiently expressed in N. benthamiana, they interfered with N-mediated resistance to Tobacco mosaic virus. Li et al. proposed that shutdown of miRNAs and siRNAs upon pathogen infection could enhance R gene activity.
METHODS
Plant Growth
Tomato (Solanum lycopersicum) plants were raised from seed in compost (Levington M3) and maintained in a growth room at 23°C with 16-h-light and 8-h-dark periods with 60% relative humidity and at light intensity of 300 μmol/m2/s. Two-week-old seedlings taken for RNA analysis generally had two simple leaves and one compound leaf and a leaf bud.
RNA Analysis and Cloning
RNA gel blot analysis was performed as described previously (Shivaprasad et al., 2012). Briefly, total RNA was extracted from 100 mg plant tissue (2-week-old seedlings) and ground into fine powder in liquid nitrogen using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. About 12 or 15 μg total RNA was resuspended in 10 μL loading buffer (0.10% bromophenol blue and 0.10% xylene cyanol in 100% de-ionized formamide), heated at 95°C for 1 min, and loaded on 15% polyacrylamide denaturing gel (a 19:1 ratio of acrylamide to bisacrylamide, 8 M urea). The gel was run using electrophoresis apparatus (Bio-Rad) at 110 V for 2 h, and then RNAs were transferred to a Hybond N+ membrane by electroblotting in 1× tris-boric acid EDTA buffer at 10 V overnight. The hybridization was performed at 35°C for 18 to 24 h in UltraHyb-oligo buffer (Ambion) using short DNA oligos as a probe (see Supplemental Table 1 online) end-labeled with 32P by polynucleotide kinase (New England Biolabs) and purified through MicroSpin G-25 columns (GE Healthcare) according to the manufacturers’ recommendations. The blot was washed twice with 2× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) and 0.5% SDS for 30 min at 35°C. The signal was detected after exposure to a phosphor imager screen using a molecular imager (Bio-Rad).
Small RNA cloning was performed using an Illumina sRNA cloning kit. About 20 μg of total RNA was separated on a 15% 1× tris-boric acid EDTA polyacrylamide gel, and a small RNA region between 15 and 40 nucleotides was excised and purified. This small RNA fraction was used for ligation of adapters and amplification of fragments. Sequencing was performed at Cancer Research UK (Cambridge, UK).
Bioinformatics
Reference DNA Sequence
The small RNA libraries were aligned to the tomato genome sequence (Mueller et al., 2005, 2009) downloaded from The International Tomato Genome Sequencing Consortium (http://solgenomics.net/), release bacs v340, as well as the EST collection from solgenomics.net website. For identifying tomato NBS-LRRs, the latest version (SL2.40) of the genome as well as entries in the National Center for Biotechnology Information were used to collect sequences.
Alignment
The small RNA high-throughput sequencing libraries were aligned to the reference sequence using the PatMaN (Prüfer et al., 2008) alignment program. Only reads with 100% match to the genome were used in further analysis.
Identifying miR482 Diversity, Sequence Alignments, and Its Targets
The UEA small RNA analysis toolkit (Moxon et al., 2008) was used to identify members of a given miRNA family (miRProf and miRCat). Sequences of miR482 members were obtained from miRBase release 18 (Kozomara and Griffiths-Jones, 2011) and aligned using ClustalW and ClustalX2 (Larkin et al., 2007). Protein sequence logos were generated using seqLogos (http://imed.med.ucm.es/Tools/seqlogo.html). Targets of miRNA were identified using two different algorithms, namely, psRNATarget algorithm (Dai and Zhao, 2011) and TAPIR algorithm (Bonnet et al., 2010). To find targets of miR482 family in the Arabidopsis thaliana genome, target NBS-LRR sequences were taken from the NIBLRRS Project website (http://niblrrs.ucdavis.edu/index.php) that was supported by the National Science Foundation Plant Genome Program Award 9975971. Images of small RNAs mapped to NBS-LRRs were generated using the UEA small RNA toolkit function SiLoMa (Moxon et al., 2008).
Analysis of abundance of miR482/2118 superfamily was performed through miRProf analysis of published large-scale data sets derived from various plant species available through the Gene Expression Omnibus (GEO) platform (Edgar et al., 2002). These libraries have been described previously (Pilcher et al., 2007; Havecker et al., 2010; Ma et al., 2010; Molnar et al., 2010; Wang et al., 2011; Zhai et al., 2011; Shivaprasad et al., 2012; Song et al., 2012). miRNA sequences were checked to compensate for the misannotation of miR482 type and miR2118-type sequences in miRBase.
Phasing Analysis of Small RNAs
The identification of phased segments in the genomes was performed by calculating the log P value from the hypergeometric distribution adapting the method used previously (Chen et al., 2007, 2010).
Quantitative PCR
cDNA synthesis was performed according to the manufacturer’s protocol using random hexamers or oligo(dT) (Superscript II or III; Invitrogen). Quantitative PCR was performed using the CFX96 real-time system (Bio-Rad) using SYBR Green JumpStartTaq ReadyMix (Sigma-Aldrich). Data were analyzed as described (Schmittgen and Livak, 2008). Primers used for amplification are as follows: LRR1 (forward 5′-TGATGTGGTCCGAGACGTGGCT-3′ and reverse 5′-TCCCAGCAACTCGGTCAGCAC-3′: 814 bp), LRR2 (forward 5′-TGGTAGGCACAGAGAAACAGAGGT-3′ and reverse 5′-TCCACACATCATCCAGGGCAACA-3′; 298 bp), Mi (forward 5′-GCTGGAGTCATTGCTGGGAGGG-3′ and reverse 5′-GCACCCACGGACAGCACTCG-3′; 857 bp), Hero (forward 5′-TCCAAGGACGCATGGTAGCCGA-3′ and reverse 5′-AGCCGGTGACTTGTGCCACG-3′; 633 bp), Target of D2+ mRNA (forward 5′-TGCAGATGTGGACTTGGAGCAATCG-3′ and reverse 5′-GCCGTTGCGGAAAATGCCCC-3′; 325 bp); and Nicotiana tabacum NBS-LRR (forward 5′-GGGCATGGGCGGTGTAGGGA-3′ and reverse 5′-ACGCAGCAAAGAACCCCACACTT-3′; 526 bp). GAPDH (forward 5′-GGAGGAGGGAACAACAAGAGG-3′ and reverse 5′-AGATGCCGTCAGTGCCGA-3′; 238 bp) was used as internal control.
Validation of miRNA Cleavage
miRNA target validation was performed using the GeneRacer kit (Invitrogen) as per the manufacturer’s instructions. Briefly, total RNA derived from 2-week-old seedlings was used for RNA extraction using Trizol (Invitrogen). RNA was ligated with an adapter and reverse transcribed using gene-specific primers that annealed 150 to 300 nucleotides downstream of the predicted cleavage site within the corresponding target mRNAs. The 5′ RACE amplification products were gel purified (Qiagen), cloned into TOPO TA cloning vector (Invitrogen), and sequenced.
Construct Design and Mobilization to Agrobacterium tumefaciens
miR482 was PCR amplified from tomato genomic DNA. The following primer combinations were used to amplify 35Spro:miR482: 482_35spro_BamHI_For (5′-CCGGggatccTCTACACTTTTCCACCATATCC-3′) and 482_wtpro/35spro_BamHI_Rev (5′-GGCCggatccTCGTGTCATGCAATTTTAAATTCTTGCC-3′). To introduce mismatches into the miR482 sequence (miR482Δ), a PCR ligation method (Trinks et al., 2005) was used. For 35Spro:miR482Δ, the following primer combinations were used for PCR #1: 482_35spro_BamHI_For and 482_Δ_Rev (5′-GCATTTATGGCATGTTCTTTGTATTCAAGACCACTCTTGCG-3′); PCR#2: 482_Δ_For (5′-AGTGGTCTTGAATACAAAGAACATGCCATAAATGCAGAGG-3′) and 482_wtpro/35spro_BamHI_Rev. PCR ligation for 35Spro:miR482Δ was performed with 482_35spro_BamHI_For and 482_wtpro/35spro_BamHI_Rev. pBIN61 (Bendahmane et al., 2002) was used as binary vector. For 35Spro:miR482 and 35Spro:miR482Δ, pBIN61 was linearized with BamHI and miR482pro:miR482 and miR482pro:miR482Δ were cloned downstream of the 35S promoter. Constructs were confirmed with standard sequencing, and correct constructs were transformed into Agrobacterium strain C58C1 (Sciaky et al., 1978) by electroporation using the Gene Pulser Xcell Electroporation System (Bio-Rad).
Bacterial and Viral Inoculation
Pseudomonas syringae DC3000 and hrcC was inoculated into 4-week-old tomato leaves as described (Sohn et al., 2007). TCV and CMV infections were performed as described (Harvey et al., 2011). TRV was a gift from S.P. Dinesh-Kumar (Liu et al., 2002). Tomato plants were infected with sap from Nicotiana benthamiana infected with TRV and CMV, while sap from uninfected N. benthamiana was used as control.
Accession Numbers
RNA sequencing data are available from Gene Expression Omnibus GSE23562.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Sequence Alignment of the miR482/2118 Superfamily.
Supplemental Figure 2. Abundance of miR482 Family Members in Total Extracts of Tomato (M82) and S. pennellii.
Supplemental Figure 3. NBS-LRR Targets of Medicago miR2118 Family Members and slmiR482 in Arabidopsis.
Supplemental Figure 4. Secondary siRNAs Derived from the LRR2 NBS-LRR mRNA.
Supplemental Figure 5. Abundance of miR482a and Two Target mRNAs in Tomato (M82) and S. pennellii.
Supplemental Figure 6. Accumulation of miR482 in Different Tissues of M82 and S. pennellii.
Supplemental Table 1. Oligonucleotides Used for RNA Gel Blot Analysis.
Supplemental Table 2. Targets of miR482 among Tomato Unigenes.
Supplemental Table 3. Targets of miR482 among Tomato Genomic Sequences.
Supplemental Table 4. Targeting Abilities for 33 Tomato NBS-LRRs by miR482 Isoforms.
Supplemental Table 5. Phased sRNA Loci in Tomato Matching Genomic Sequences.
Supplemental Table 6. Phased sRNA Loci in Tomato Matching Unigenes.
Acknowledgments
We thank Dani Zamir and the Tomato Genetics Resources Center at the University of California, Davis, for providing various tomato lines. We thank the Genomics and Bioinformatics Core Facilities (Cancer Research UK Cambridge Research Institute) for Illumina sequencing and Jonathan Jones for Pst DC3000 and hrcC strains. We thank Sol Genomics Network and the International Tomato Genome Sequencing Consortium for sequences. We also thank Krystyna Kelly and Risha Narayan for helping with accessing GEO data sets. We thank Gail Preston, Natasha Elina, and D.C.B. laboratory members for fruitful discussions. We thank Shuoya Tang for plant care and James Barlow for media preparations. This work was supported by the Biotechnology and Biological Science Research Council (Grant BB/E006981/2), the Gatsby Charitable Foundation, and European Research Council Advanced Investigator Grant 233325 REVOLUTION. H.-M.C.’s visit to the Baulcombe lab was supported by NSC 97-2311-B-001-003-MY3. D.C.B. is a Royal Society Research Professor.
AUTHOR CONTRIBUTIONS
P.V.S. and D.C.B. designed the study and drafted the article. P.V.S. carried out the bioinformatic, RNA, and phenotypic analyses. K.P. and D.M.B. made constructs. H.-M.C. and B.A.C.M.S. carried out phasing analysis.
References
- Allen E., Xie Z., Gustafson A.M., Carrington J.C. (2005). MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 [DOI] [PubMed] [Google Scholar]
- Axtell M.J., Bartel D.P. (2005). Antiquity of microRNAs and their targets in land plants. Plant Cell 17: 1658–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell M.J., Jan C., Rajagopalan R., Bartel D.P. (2006). A two-hit trigger for siRNA biogenesis in plants. Cell 127: 565–577 [DOI] [PubMed] [Google Scholar]
- Bari R., Datt Pant B., Stitt M., Scheible W.-R. (2006). PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 141: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. (2009). MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe D. (2004). RNA silencing in plants. Nature 431: 356–363 [DOI] [PubMed] [Google Scholar]
- Beers E.P., Woffenden B.J., Zhao C.S. (2000). Plant proteolytic enzymes: Possible roles during programmed cell death. Plant Mol. Biol. 44: 399–415 [DOI] [PubMed] [Google Scholar]
- Bendahmane A., Farnham G., Moffett P., Baulcombe D.C. (2002). Constitutive gain-of-function mutants in a nucleotide binding site-leucine rich repeat protein encoded at the Rx locus of potato. Plant J. 32: 195–204 [DOI] [PubMed] [Google Scholar]
- Bendahmane A., Kanyuka K., Baulcombe D.C. (1999). The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 11: 781–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet E., He Y., Billiau K., Van de Peer Y. (2010). TAPIR, a web server for the prediction of plant microRNA targets, including target mimics. Bioinformatics 26: 1566–1568 [DOI] [PubMed] [Google Scholar]
- Brigneti G., Voinnet O., Li W.X., Ji L.H., Ding S.W., Baulcombe D.C. (1998). Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17: 6739–6746 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Brodersen P., Sakvarelidze-Achard L., Bruun-Rasmussen M., Dunoyer P., Yamamoto Y.Y., Sieburth L., Voinnet O. (2008). Widespread translational inhibition by plant miRNAs and siRNAs. Science 320: 1185–1190 [DOI] [PubMed] [Google Scholar]
- Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., Chinappi M., Tramontano A., Bozzoni I. (2011). A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147: 358–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.-M., Chen L.-T., Patel K., Li Y.-H., Baulcombe D.C., Wu S.-H. (2010). 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc. Natl. Acad. Sci. USA 107: 15269–15274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.M., Li Y.H., Wu S.H. (2007). Bioinformatic prediction and experimental validation of a microRNA-directed tandem trans-acting siRNA cascade in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 3318–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood D.H., Nogueira F.T.S., Howell M.D., Montgomery T.A., Carrington J.C., Timmermans M.C.P. (2009). Pattern formation via small RNA mobility. Genes Dev. 23: 549–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus J.T., Carbonell A., Fahlgren N., Garcia-Ruiz H., Burke R.T., Takeda A., Sullivan C.M., Gilbert S.D., Montgomery T.A., Carrington J.C. (2010). Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat. Struct. Mol. Biol. 17: 997–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus J.T., Fahlgren N., Carrington J.C. (2011). Evolution and functional diversification of MIRNA genes. Plant Cell 23: 431–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X.B., Zhao P.X. (2011). psRNATarget: A plant small RNA target analysis server. Nucleic Acids Res. 39(Web Server issue): W155–W159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A.E. (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed Y., Zamir D. (1995). An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141: 1147–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felippes F.F., Schneeberger K., Dezulian T., Huson D.H., Weigel D. (2008). Evolution of Arabidopsis thaliana microRNAs from random sequences. RNA 14: 2455–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla J.M., Valli A., Todesco M., Mateos I., Puga M.I., Rubio-Somoza I., Leyva A., Weigel D., García J.A., Paz-Ares J. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39: 1033–1037 [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S., Saini H.K., van Dongen S., Enright A.J. (2008). miRBase: Tools for microRNA genomics. Nucleic Acids Res. 36(Database issue): D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey J.J.W., Lewsey M.G., Patel K., Westwood J., Heimstädt S., Carr J.P., Baulcombe D.C. (2011). An antiviral defense role of AGO2 in plants. PLoS ONE 6: e14639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havecker E.R., Wallbridge L.M., Hardcastle T.J., Bush M.S., Kelly K.A., Dunn R.M., Schwach F., Doonan J.H., Baulcombe D.C. (2010). The Arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell 22: 321–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C., Kasprzewska A., Tennessen K., Fernandes J., Nan G.L., Walbot V., Sundaresan V., Vance V., Bowman L.H. (2009). Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Res. 19: 1429–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Karreth F.A., et al. (2011). In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell 147: 382–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khraiwesh B., Arif M.A., Seumel G.I., Ossowski S., Weigel D., Reski R., Frank W. (2010). Transcriptional control of gene expression by microRNAs. Cell 140: 111–122 [DOI] [PubMed] [Google Scholar]
- Kozomara A., Griffiths-Jones S. (2011). miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39(Database issue): D152–D157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanet E., Delannoy E., Sormani R., Floris M., Brodersen P., Crété P., Voinnet O., Robaglia C. (2009). Biochemical evidence for translational repression by Arabidopsis microRNAs. Plant Cell 21: 1762–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Li F., Orban R., Baker B. (January 24, 2012a). SoMART, a web server for plant miRNA, tasiRNA and target gene analysis. Plant J. http://dx.doi.org/10.1111/j.1365-313X.2012.04922.x. [DOI] [PubMed]
- Li F., Pignatta D., Bendix C., Brunkard J.O., Cohn M.M., Tung J., Sun H., Kumar P., Baker B. (2012b). MicroRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. USA 109: 1790–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.L., Schiff M., Dinesh-Kumar S.P. (2002). Virus-induced gene silencing in tomato. Plant J. 31: 777–786 [DOI] [PubMed] [Google Scholar]
- Ma Z.R., Coruh C., Axtell M.J. (2010). Arabidopsis lyrata small RNAs: Transient MIRNA and small interfering RNA loci within the Arabidopsis genus. Plant Cell 22: 1090–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean D., Elina N., Havecker E.R., Heimstaedt S.B., Studholme D.J., Baulcombe D.C. (2010). Evidence for large complex networks of plant short silencing RNAs. PLoS ONE 5: e9901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory A.C., Bouché N. (2008). MicroRNA-directed regulation: To cleave or not to cleave. Trends Plant Sci. 13: 359–367 [DOI] [PubMed] [Google Scholar]
- Manavella P.A., Koenig D., Weigel D. (2012). Plant secondary siRNA production determined by microRNA-duplex structure. Proc. Natl. Acad. Sci. USA 109: 2461–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers B.C., et al. (2008). Criteria for annotation of plant microRNAs. Plant Cell 20: 3186–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers B.C., Kozik A., Griego A., Kuang H., Michelmore R.W. (2003). Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15: 809–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohorianu I., Schwach F., Jing R.C., Lopez-Gomollon S., Moxon S., Szittya G., Sorefan K., Moulton V., Dalmay T. (2011). Profiling of short RNAs during fleshy fruit development reveals stage-specific sRNAome expression patterns. Plant J. 67: 232–246 [DOI] [PubMed] [Google Scholar]
- Molnar A., Melnyk C.W., Bassett A., Hardcastle T.J., Dunn R., Baulcombe D.C. (2010). Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science 328: 872–875 [DOI] [PubMed] [Google Scholar]
- Moxon S., Schwach F., Dalmay T., Maclean D., Studholme D.J., Moulton V. (2008). A toolkit for analysing large-scale plant small RNA datasets. Bioinformatics 24: 2252–2253 [DOI] [PubMed] [Google Scholar]
- Mueller L.A., et al. (2009). A anapshot of the emerging tomato genome sequence. Plant Genome 2: 78–92 [Google Scholar]
- Mueller L.A., et al. (2005). The Tomato Sequencing Project, the first cornerstone of the International Solanaceae Project (SOL). Comp. Funct. Genomics 6: 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L., Jay F., Nomura K., He S.Y., Voinnet O. (2008). Suppression of the microRNA pathway by bacterial effector proteins. Science 321: 964–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson J., Ståhl S., Lundeberg J., Uhlén M., Nygren P.A. (1997). Affinity fusion strategies for detection, purification, and immobilization of recombinant proteins. Protein Expr. Purif. 11: 1–16 [DOI] [PubMed] [Google Scholar]
- Pan Q.L., Wendel J., Fluhr R. (2000). Divergent evolution of plant NBS-LRR resistance gene homologues in dicot and cereal genomes. J. Mol. Evol. 50: 203–213 [DOI] [PubMed] [Google Scholar]
- Pant B.D., Buhtz A., Kehr J., Scheible W.R. (2008). MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 53: 731–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart J.R., et al. (2002). Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc. Natl. Acad. Sci. USA 99: 10865–10869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilcher R.L.R., Moxon S., Pakseresht N., Moulton V., Manning K., Seymour G., Dalmay T. (2007). Identification of novel small RNAs in tomato (Solanum lycopersicum). Planta 226: 709–717 [DOI] [PubMed] [Google Scholar]
- Prüfer K., Stenzel U., Dannemann M., Green R.E., Lachmann M., Kelso J. (2008). PatMaN: Rapid alignment of short sequences to large databases. Bioinformatics 24: 1530–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades M.W., Reinhart B.J., Lim L.P., Burge C.B., Bartel B., Bartel D.P. (2002). Prediction of plant microRNA targets. Cell 110: 513–520 [DOI] [PubMed] [Google Scholar]
- Schmittgen T.D., Livak K.J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Schwach F., Vaistij F.E., Jones L., Baulcombe D.C. (2005). An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 138: 1842–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciaky D., Montoya A.L., Chilton M.D. (1978). Fingerprints of Agrobacterium Ti plasmids. Plasmid 1: 238–253 [DOI] [PubMed] [Google Scholar]
- Shivaprasad P.V., Dunn R.M., Santos B.A., Bassett A., Baulcombe D.C. (2012). Extraordinary transgressive phenotypes of hybrid tomato are influenced by epigenetics and small silencing RNAs. EMBO J. 31: 257–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn K.H., Lei R., Nemri A., Jones J.D.G. (2007). The downy mildew effector proteins ATR1 and ATR13 promote disease susceptibility in Arabidopsis thaliana. Plant Cell 19: 4077–4090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., et al. (2012). Roles of DCL4 and DCL3b in rice phased small RNA biogenesis. Plant J. 69: 462–474 [DOI] [PubMed] [Google Scholar]
- Stein M., Dittgen J., Sánchez-Rodríguez C., Hou B.H., Molina A., Schulze-Lefert P., Lipka V., Somerville S. (2006). Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell 18: 731–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumazin P., Yang X., Chiu H.-S., Chung W.-J., Iyer A., Llobet-Navas D., Rajbhandari P., Bansal M., Guarnieri P., Silva J., Califano A. (2011). An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell 147: 370–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y., et al. (2011). Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 147: 344–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D., Traw M.B., Chen J.Q., Kreitman M., Bergelson J. (2003). Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 423: 74–77 [DOI] [PubMed] [Google Scholar]
- Tör M., Yemm A., Holub E. (2003). The role of proteolysis in R gene mediated defence in plants. Mol. Plant Pathol. 4: 287–296 [DOI] [PubMed] [Google Scholar]
- Trinks D., Rajeswaran R., Shivaprasad P.V., Akbergenov R., Oakeley E.J., Veluthambi K., Hohn T., Pooggin M.M. (2005). Suppression of RNA silencing by a geminivirus nuclear protein, AC2, correlates with transactivation of host genes. J. Virol. 79: 2517–2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J.P., et al. ; International Brachypodium Initiative (2010). Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763–768 [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2005). Induction and suppression of RNA silencing: Insights from viral infections. Nat. Rev. Genet. 6: 206–220 [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Voinnet O., Pinto Y.M., Baulcombe D.C. (1999). Suppression of gene silencing: A general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. USA 96: 14147–14152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Wang X.C., Kibet N.K., Song C.N., Zhang C.Q., Li X.Y., Han J., Fang J.G. (2011). Deep sequencing of grapevine flower and berry short RNA library for discovery of novel microRNAs and validation of precise sequences of grapevine microRNAs deposited in miRBase. Physiol. Plant. 143: 64–81 [DOI] [PubMed] [Google Scholar]
- Wu L., Zhou H., Zhang Q., Zhang J., Ni F., Liu C., Qi Y. (2010). DNA methylation mediated by a microRNA pathway. Mol. Cell 38: 465–475 [DOI] [PubMed] [Google Scholar]
- Xie F.L., Frazier T.P., Zhang B.H. (2011). Identification, characterization and expression analysis of MicroRNAs and their targets in the potato (Solanum tuberosum). Gene 473: 8–22 [DOI] [PubMed] [Google Scholar]
- Xu X., et al. ; Potato Genome Sequencing Consortium (2011). Genome sequence and analysis of the tuber crop potato. Nature 475: 189–195 [DOI] [PubMed] [Google Scholar]
- Yoshikawa M., Peragine A., Park M.Y., Poethig R.S. (2005). A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19: 2164–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J., et al. (2011). MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 25: 2540–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Wang Y., Chen J.Q., Araki H., Jing Z., Jiang K., Shen J., Tian D. (2004). Genome-wide identification of NBS genes in japonica rice reveals significant expansion of divergent non-TIR NBS-LRR genes. Mol. Genet. Genomics 271: 402–415 [DOI] [PubMed] [Google Scholar]