This work provides evolutionary insight into the acquisition and divergence of leaf-like organs called cladodes in the genus Asparagus. The data suggest that co-option of genes involved in leaf development in axillary shoots led to the acquisition of a flattened, leaf-like form. Alterations in gene expression appear to have led to the evolution of cylindrical cladodes in one clade of Asparagus.
Abstract
The genus Asparagus is unusual in producing axillary, determinate organs called cladodes, which may take on either a flattened or cylindrical form. Here, we investigated the evolution of cladodes to elucidate the mechanisms at play in the diversification of shoot morphology. Our observations of Asparagus asparagoides, which has leaf-like cladodes, showed that its cladodes are anatomically and developmentally similar to leaves but differ in the adaxial/abaxial polarity of the vasculature. In addition to the expression of an ortholog of KNAT1, orthologous genes that are normally expressed in leaves, ASYMMETRIC LEAVES1 and HD-ZIPIII, were found to be expressed in cladode primordia in a leaf-like manner. The cylindrical cladodes of Asparagus officinalis showed largely similar expression patterns but showed evidence of being genetically abaxialized. These results provide evidence that cladodes are modified axillary shoots, suggest that the co-option of preexisting gene networks involved in leaf development transferred the leaf-like form to axillary shoots, and imply that altered expression of leaf polarity genes led to the evolution of cylindrical cladodes in the A. officinalis clade.
INTRODUCTION
Plants display an amazing diversity in shoot morphology and architecture (Tsukaya, 2006; Bell, 2008; Nicotra et al., 2011), and their refined artifice is astonishing. Many studies have addressed the morphological diversification of leaves, which often vary in size, shape, and form (e.g., Blein et al., 2008; Kimura et al., 2008; Berger et al., 2009; Piazza et al., 2010; Yamaguchi et al., 2010). While often more conservative than leaves, extensive morphological diversity is also seen in stems, most dramatically with the production of tendrils, thorns, and flattened, leaf-like stems (Bell, 2008). In contrast with the extensive work on leaf form evolution, relatively few studies have elucidated the genetic mechanisms of shoot morphological diversification.
Generally, plant shoot systems consist of stems bearing leaves, with floral buds and axillary shoots developing only in leaf axils (Figure 1A). In the genus Asparagus, however, foliage leaves are reduced in size to become scale leaves and no longer play a major photosynthetic role. Instead, all species of Asparagus have cladodes, which are leaf-like, photosynthetically active organs that develop in the axils of scale leaves (Figures 1B to 1E; Guo et al., 2002). Cladodes are unusual organs, but not unique, because similar-looking structures have evolved independently in other genera of angiosperms (Ruscus, Danae, and Epiphyllum) and gymnosperms (Phyllocladus) (Cooney-Sovetts and Sattler, 1986; Tomlinson et al., 1987; Bell, 2008).
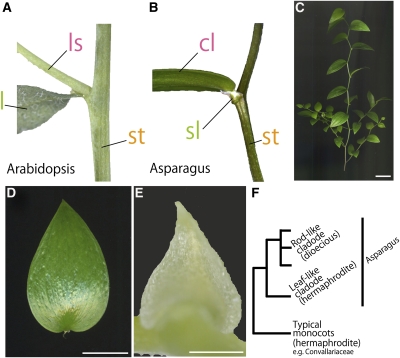
Figure 1.
Gross Morphology of Cladodes.
(A) and (B) Generating position of a lateral shoot in Arabidopsis (A) and a cladode of A. asparagoides (B). Note that the cladode arises at the axil of the scale leaf. cl, cladode; l, leaf; ls, lateral stem; sl, scale leaf; st, stem.
(C) A shoot of A. asparagoides.
(D) A cladode of A. asparagoides.
(E) A scale leaf of A. asparagoides.
(F) Simplified phylogenetic relationship of cladode morphology in the genus Asparagus based on Fukuda et al. (2005).
Bars = 1 cm in (C) and (D) and 1 mm in (E).
Cladodes have so far been considered to be either modified stems, ectopic leaves, or de novo organs (Hirsch, 1977; Cooney-Sovetts and Sattler, 1986; Hirayama et al., 2007). Under any of these interpretations, clarifying the genetic regulation of cladode development could help answer some outstanding questions. If cladodes are modified stems, how did cladodes become flattened and determinate? By contrast, if cladodes are leaves, how did they come to develop in axils where axillary shoots normally emerge? Previous phylogenetic analyses have shown that, within Asparagus, dioecism evolved from hermaphroditism (Fukuda et al., 2005; Kubota et al., 2012). Hence, it has been proposed that cladodes evolved from an ancestral, leaf-like (flattened) form seen only in hermaphrodite species, to a rod-like (cylindrical) form seen among dioecious species of subgenus Asparagus. In particular, Asparagus asparagoides, which is hermaphroditic plant and has leaf-like cladodes, is sister to all other Asparagus in the phylogenetic trees. (Figure 1F; Fukuda et al., 2005). This fact raises the additional question of how the flattened form came to be lost in subgenus Asparagus. In this study, we attempted to answer these questions by studying the expression of genes involved in leaf identity and leaf adaxial/abaxial polarity in Asparagus cladodes.
Recent advances in molecular genetics of various model plant species have expanded our knowledge of the genetic factors controlling the plant body plan (Williams and Fletcher, 2005; Kidner and Timmermans, 2007; Reddy, 2008). The regulatory mechanism of leaf development is one of the most advanced fields (Dengler and Tsukaya, 2001; Eshed et al., 2001; Kessler and Sinha, 2004; Byrne, 2005; Tsukaya, 2006; Braybrook and Kuhlemeier, 2010; Szakonyi et al., 2010). Knowledge of the regulatory mechanism of leaf development provides an essential backdrop for understanding the development of cladodes. Shoot apical meristems (SAMs), on whose flanks leaf primordia develop, require the activity of class I KNOTTED1-LIKE HOMEOBOX (KNOX) genes to establish and maintain undifferentiated cell fate (Long et al., 1996; Vollbrecht et al., 2000; Ha et al., 2010). Localized downregulation of KNOX expression is a landmark of the initiation of leaves or homologous lateral organs, and KNOX repression is maintained throughout leaf development in simple-leaved species (Szakonyi et al., 2010; Uchida et al., 2010). Simultaneously, the ASYMMETRIC LEAVES1 (AS1) gene, encoding a Myb domain protein, is expressed in founder cells of leaves in a pattern complementary to KNOX. Subsequently, AS1 transcripts accumulate around vascular tissues in leaf primordia (Iwakawa et al., 2007; Ikezaki et al., 2010). AS1 genes support leaf differentiation by repressing KNOX genes throughout leaf development by chromatin remodeling (Guo et al., 2008). The mutually exclusive expression patterns of KNOX and AS1 homologs are observed in diverse model species, indicating that the leaf initiation program is conserved among angiosperms (Timmermans et al., 1999; Byrne et al., 2000; Langdale, 2008). Additionally, AS1 in combination with class III homeodomain-leucine zipper (HD-ZIP III) genes, plays an important role in promoting adaxial cell fate, which is need for proper establishment of adaxial/abaxial polarity and leaf laminar outgrowth. In Arabidopsis thaliana, three HD-ZIP III genes, PHABULOSA (PHB), PHAVOLUTA, and REVOLUTA (REV), are expressed in the adaxial domain of leaves and promote adaxial identity in the developing leaf (McConnell et al., 2001; Juarez et al., 2004; Itoh et al., 2008). Moreover, HD-ZIP III genes are strictly regulated by miR166, which is expressed toward the abaxial side of leaf primordia. The miRNA cleaves HD-ZIP III mRNA and limits the expression of HD-ZIP III to the adaxial domain of the leaf primordia. Conversely, AUXIN RESPONSE FACTOR3/ETTIN (Pekker et al., 2005) is expressed abaxially and promotes abaxial identity.
To explore the evolutionary origin and developmental basis of cladode morphology, we investigated the anatomy and development of the leaf-like (flattened) cladodes of A. asparagoides and examined the expression patterns of genes involved in leaf and shoot development. We showed that the internal anatomy of cladodes differs from that of stems and leaves and that the orthologs involved in leaf development are expressed in cladode primordia, as well as KNOX ortholog expression, which is involved in shoot development. Additionally, we investigated Asparagus officinalis, which has rod-like (cylindrical) cladodes, to reveal the evolutionary processes of morphological diversification in cladodes in the genus (Figure 1F). We demonstrated that gene expression patterns involved in establishing adaxial/abaxial polarity in cladodes of A. officinalis differ from those of A. asparagoides. These results indicate that cladodes in Asparagus could be considered as a modified axillary shoot, that expression of orthologs involved in leaf development confers its leaf-like form, and that the alteration of the expression patterns has led to the cylindrical form of cladodes. Based on these results, we discuss the developmental mechanisms, origin, and evolutionary history of cladodes, a leaf-like organ in the genus Asparagus.
RESULTS
Cladodes in A. asparagoides Have a Typical Adaxial/abaxial Polarity
Analogous to typical axillary shoots that arise in leaf axils (Figure 1A), the cladodes of the Asparagus also develop at leaf axils. In A. asparagoides, the foliage leaves are reduced and scale-like (Figure 1B). Cladodes are flat and resemble foliage leaves (Figures 1C to 1E). To determine the presence or absence of adaxial/abaxial polarity in cladodes, we observed the internal structure of stems, axillary shoots, and cladodes, focusing on the positioning of vascular bundles and mesophyll cell form. The vascular bundles in the stems and axillary shoots of A. asparagoides are aligned in a ring, as is typical in monocot stems (Figures 2A and 2B). By contrast, the vascular bundles in cladodes and scale leaves are arranged in a line, as is typical of bifacial leaves (Figures 2C and 2D, arrowheads). Mesophyll cells in cladodes on the adaxial side are aligned vertically on a central-marginal plane like the palisade tissues of bifacial leaves, whereas cells on the abaxial side are aligned horizontally with intercellular spaces like spongy tissues of leaves (Figure 2E). Distinct differentiation between adaxial and abaxial mesophyll was not observed in scale leaves (Figure 2F), possibly reflecting its immaturity and the early phase of growth arrest. Interestingly the position of the xylem and phloem was inverted in cladodes compared with that of foliage leaves, with phloem being on the adaxial side and xylem on the abaxial side of the cladode (Figure 2G).
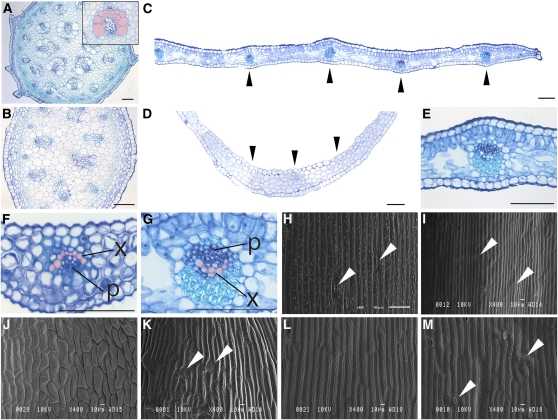
Figure 2.
Comparison of Internal Structures and Epidermal Cell Shapes of the Stem, Axillary Shoot, Cladode, and Scale Leaf in A. asparagoides.
(A) Mature stem. Inset shows close-up view of the vascular bundle; top of the image is the side facing the stem center.
(B) Axillary stem.
(C) and (D) A part of a cladode (C) and a scale leaf (D).
(E) Close-up view of the inner structure of a cladode.
(F) and (G) Close-up views of the vascular bundle of a scale leaf (F) and of a cladode (G).
(H) to (M) Scanning electron micrographs of a stem epidermal cell (H), axillary shoot (I), adaxial side of a scale leaf (J), abaxial side of a scale leaf (K), adaxial side of a cladode (L), and abaxial side of a cladode (M).
(A) to (G) are transverse sections. In (C) to (G), top of the image is the adaxial side of each organ. In (A), (F), and (G), the xylem is colored in pink. Black and white arrowheads indicate vascular bundle and stoma, respectively. p, phloem; x, xylem. Bars = 1 mm in (C) and (D) and 100 μm in (A), (B), and (E) to (G).
Epidermal cell shape and stomatal distribution pattern often show adaxial/abaxial differentiation in leaves. Therefore, we characterized the epidermal cell shape of each A. asparagoides organ using scanning electron microscopy. Stem epidermal cells were rectangular in form and elongated along the apical-basal axis, with interspersed stomata (Figures 2H and 2I). Primary stems also had scattered cell surface protrusions (Figure 2H). The adaxial side of scale leaves was characterized by relatively short, smooth cells and a lack of either stomata or cell-surface protrusions (Figure 2J). By contrast, epidermal cells on the abaxial side of scale leaves were longer with a sculpted cell surface and interspersed with stomata (Figure 2K). Cladode epidermal cells on both surfaces resembled the primary stem, but stomata and cell surface protrusions were restricted to the abaxial side (Figures 2L and 2M).
In summary, the internal anatomy and the distribution of stomata and cell protrusions showed that cladodes have distinct adaxial/adaxial polarity, indicating that cladodes differ from stems. However, the inverted vascular bundle and epidermal cell shape showed a difference from leaves. Therefore, cladodes have unique anatomical features that differ from both stems and leaves.
The Developmental of Cladodes Is Highly Similar to Leaves
Cladodes sometimes appeared on primary shoots, characterized by spiral phyllotaxis (Figure 3A), but most cladodes formed on axillary shoots, which show alternate phyllotaxis. Cladode primordia developed in all axils on the axillary shoot (Figures 3B and 3C). Because a cladode primordium generates within an axil of a scale leaf primordium, we were able to apply a plastochron index to cladodes, as for leaves, Plastochron 1 (P1) representing the youngest primordium and P2 the next youngest, etc. (Itoh et al., 2005). Cladode primordia initiated as a bulge between the SAM and scale leaf soon as the scale leaf developed (Figures 3C and 3D). Initiation of axillary shoots on the primary shoot also occurred just after the development of scale leaves (Figure 3E). These observations indicate that the timing of organ initiation from axils is similar for axillary shoot and cladode primordia. During the development of cladode primordia, differentiation appeared to proceed basipetally (Figures 3F to 3I).
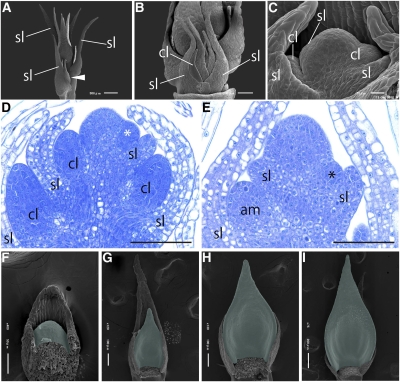
Figure 3.
Development of Cladodes in A. asparagoides.
(A) to (C) Scanning electron micrographs of the shoot apex.
(A) Axillary shoots arising on the axils of primary shoot scale leaves. White arrowhead indicates the axillary shoot.
(B) Close-up view of an axillary shoot meristem composed of scale leaves and cladode primordia.
(C) Close-up view of cladodes arising at the axil of scale leaves on the axillary shoot meristem.
(D) Longitudinal section of the axillary shoot meristem with emerging cladodes. White asterisk indicates the emerging cladode.
(E) Longitudinal section of the primary shoot with emerging axillary shoot meristem. Black asterisk indicates the emerging axillary shoot meristem.
(F) to (I) Scanning electron micrographs of cladode development in series. Cladodes are colored green.
Unnecessary organs were removed in (B), (C), and (F) to (I). am, axially shoot meristem; cl, cladode; sl, scale leaf. Bars = 100 μm in (D) and (E).
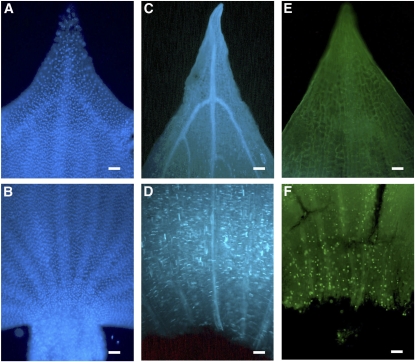
To confirm the growth pattern of cladodes, we observed the cell proliferation in cladodes with 4′,6′-diamino-3-phenylindole (DAPI), aniline blue, and 5′-ethynyl-2′-deoxyuridine (EdU). Nuclear signals stained with DAPI were detected relatively uniformly in distal regions of cladodes (Figure 4A), whereas signals were relatively dense in the basal (proximal) region of the cladode (Figure 4B). This result indicates that the basal regions of cladodes consist of smaller cells. We used aniline blue staining, which marks the septum walls of newly divided cells (Kakimoto and Shibaoka, 1992), to investigate whether the smaller cell size in the basal part is due to proliferation. Positive aniline blue signals were not detected in the distal region of cladodes (Figure 4C), whereas signals were seen in the proximal region (Figure 4D). We obtained a similar result from the EdU detection system, which identifies DNA replication during the S-phase of the cell cycle (Salic and Mitchison, 2008). No signals were observed in the distal region (Figure 4E), but positive signals occurred in the proximal region (Figure 4F). These results indicate that the cell proliferation in cladodes at this stage is concentrated in the proximal region similar to that of leaves (Sylvester et al., 1990).
Figure 4.
Cell Cycle Activity in Cladodes.
Cladodes stained with DAPI ([A] and [B]), aniline blue ([C] and [D]), and the EdU system ([E] and [F]). (A), (C), and (E) are distal regions of a cladode. (B), (D), and (F) are proximal regions of a cladode. Bars = 100 μm.
In summary, these developmental analyses showed that the initiation timing and position of cladodes are similar to those of the axillary shoot but that cell proliferation in cladodes is highly similar to leaves.
Gene Isolation from Asparagus Species
We isolated orthologs involved in leaf and shoot development to clarify the developmental processes that might impact cladode identity. We focused on orthologs of KNAT1, AS1, PHB, and REV. Total RNA was extracted from shoots, including scale leaves and cladodes, and RT-PCR was performed with degenerate primers or target-specific primers. Amplified fragments were cloned and sequenced. BLAST searches revealed that putative amino acid sequences encoded by these fragments were similar to those of each target gene in other species. The 5′ and 3′ regions of the amplified DNA fragments were then isolated by rapid amplification of cDNA ends (RACE). Finally, putative full-length cDNA clones including the complete coding region were obtained by PCR. The fragments isolated from A. asparagoides were named Aas KNAT1, Aas AS1, Aas PHB, and Aas REV; fragments isolated from A. officinalis were named Aof KNAT1, Aof AS1, and Aof PHB. Multiple sequence alignments demonstrated that each putative protein had a characteristic domain(s), which is conserved among these homologs (see Supplemental Figures 1 to 4 online). Phylogenetic analyses also supported that each identified is an ortholog of the targeted genes: Aas KNAT1 and Aof KNAT1 were located in an At (Arabidopsis) KNAT1 clade in class KNOX phylogeny (see Supplemental Figure 5 online), Aas AS1 and Aof AS1 in an At AS1 clade (see Supplemental Figure 6 online), Aas PHB and Aof PHB, and Aas REV in At PHB and REV clades, respectively, in the HD-ZIP III phylogeny (see Supplemental Figure 7 online). Additionally, we confirmed that the target site of miR166 is conserved in the Aas PHB and Aof PHB sequences (see Supplemental Figure 3B online).
AS1, KNOX, HD-ZIPIII, and miR166 Are Expressed in Cladode Primordia in a Leaf-Like Manner
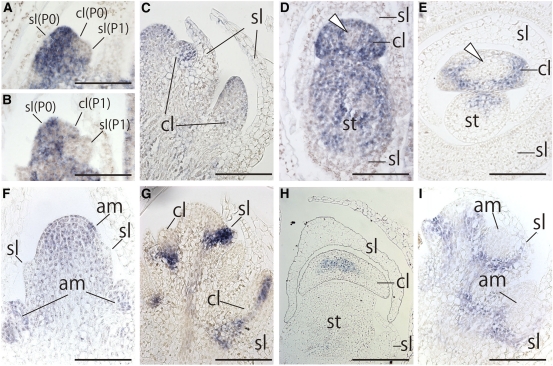
To clarify spatial and temporal expression patterns in the flattened cladodes of A. asparagoides, we performed in situ hybridization with gene-specific probes. In no case was signal detected with sense probe. Aas KNAT1 expression was detected in the SAM, as expected (Figure 5A). By contrast, Aas KNAT1 expression was not observed in P0 or P1 scale-leaf primordia, as known for other species with simple leaves (Figures 5A and 5B). Expression was similarly absent from the P0 or P1 cladode primordia (Figures 5A and 5B). However, Aas KNAT1 was expressed in cladode primordia at later stages (Figures 5C to 5E), especially toward the periphery (Figure 5D and 5E). The overall expression level appeared to become weaker as the cladode developed (Figure 5E). Aas KNAT1 expression was also detected in the axillary shoot meristem (Figure 5F).
Figure 5.
Expression Patterns of Aas KNAT1 and Aas AS1 in A. asparagoides.
(A) to (F) In situ localization of Aas KNAT1 transcripts.
(A) to (C) Longitudinal sections of axillary shoot bearing P0-P1 leaf and P0 cladode primordia (A), P0-P1 leaf primordium and P1 cladode primordia (B), and later stages of leaf and cladode primordium (C).
(D) and (E) Transverse sections of axillary shoot meristem bearing leaf and cladode primordia. Expression patterns of Aas KNAT1 in axillary shoot meristem bearing cladode primordium (D) and in axillary shoot meristem bearing a later stage of a cladode primordium (E). White arrowheads indicate the region where Aas KNAT1 expression was diminished. Aas KNAT1 expression pattern in a longitudinal section of primary stem subtending an axillary shoot (F).
(G) to (I) In situ localization of Aas AS1 transcripts. Longitudinal sections of an axillary shoot bearing leaf and cladode primordia (G) and transverse section of an axillary shoot bearing leaf and cladode primordia (H). Aas AS1 expression pattern in a longitudinal section of primary stem subtending the axillary shoot (I).
All panels are the same magnification. am, axially meristem; cl, cladode; sl, scale leaf; st, stem. Bars = 100 μm.
Next, we examined expression pattern of Aas AS1 to determine whether genes involved in leaf morphogenesis are expressed in cladode primordia. Aas AS1 expression was detected in young leaf primordia as expected (Figure 5G), but disappeared during later stages of development (Figure 5H). Aas AS1 was expressed at the base of cladode primordia (Figure 5G), but subsequent expression was restricted to the central region of the cladodes, including the presumptive region of vascular bundle development (Figure 5H). Aas AS1 was not detected in the axillary shoot meristem (Figure 5I). These results indicate that developmental processes of cladodes involve genes that are typically involved in leaf development.
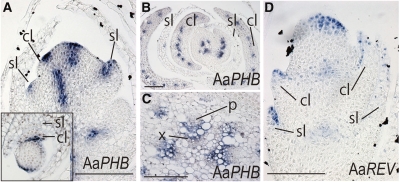
In addition to the observation that cladodes show morphological abaxial/adaxial polarity, we detected Aas PHB expression in the adaxial regions of leaf primordia and cladodes (Figure 6A). During later stages of cladode development, expression was restricted to the presumptive vascular bundles (Figure 6B). In stems, Aas PHB was only detected in xylem tissue (Figure 6C). Aas REV expression was also observed in the adaxial region of leaf and cladode primordia, similar to Aas PHB (Figure 6D). We detected miR166 in the abaxial region of leaves and cladode primordia, especially toward the base, as expected from the HD-ZIP III expression pattern in these primordia (see Supplemental Figure 8 online).
Figure 6.
Expression Patterns of Aas PHB and Aas REV in A. asparagoides.
(A) to (C) In situ localization of Aas PHB transcripts. Longitudinal section of axillary shoot bearing leaf and cladode primordia. Inset: transverse section (A). Transverse section of axillary shoot bearing later stages of leaf and cladode primordia (B) and vascular bundle of a primary shoot (C).
(D) In situ localization of Aas REV in a longitudinal section of an axillary shoot bearing leaf and cladode primordia.
cl, cladode; p, phloem; sl, scale leaf; x, xylem. Bars = 100 μm.
In summary, in addition to Aas KNAT1 expression, orthologous genes involved in leaf development, particularly those involved in abaxial/adaxial polarity, such as AS1, HD-ZIP III, and miR166, were expressed in A. asparagoides cladodes in a leaf-like manner.
A. officinalis Shows Altered Expression Patterns of Genes Involved in Establishing Adaxial/Abaxial Polarity
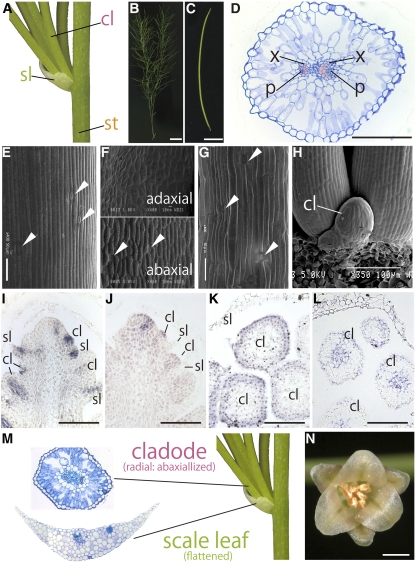
Cladode morphology shows remarkable diversity in the genus Asparagus. To reveal the process of diversification, we performed an anatomical analysis of cylindrical cladodes in A. officinalis (Figures 7A to 7C). These cladodes had parenchymatous cells arranged radially with many spaces, as seen on the abaxial side of bifacial leaves. Vascular bundles were arranged in a ring with the xylem of each bundle directed toward the center of the cladode (Figure 7D), suggesting a lack of abaxial/adaxial polarity. The shapes of epidermal cells and the distribution of stomata in stems and leaves were almost identical to A. asparagoides (Figures 7E and 7F). The cladode epidermis was similar to that of stems, and stomata were observed all around the cladode. This suggests that the cylindrical cladodes of A. officinalis have an abaxialized identity (Figure 7G). Cladode primordia were cylindrical from just after emergence (Figure 7H).
Figure 7.
Morphology, Anatomy, and Ortholog Expression Patterns Associated with Shoot and/or Leaf Morphogenesis Genes in A. officinalis.
Generating position of cladodes (A), gross morphology of a shoot (B), and cylindrical cladode of A. officinalis (C).
(D) Internal structure of the cylindrical cladode. Top of the image is the adaxial side of the cladode.
(E) to (G) Scanning electron micrographs of epidermal cells from the stem (E), scale leaf (F), and cladode (G).
(H) Close-up view of arising cladodes.
(I) to (L) In situ localization of Aof AS1 and Aof KNAT1 transcripts in an axillary shoot bearing leaf and cladode primordia and of miR166 and Aof PHB transcripts in cladode primordia. Aof AS1 transcripts (I) and Aof KNAT1 transcripts (J) in a longitudinal section of an axillary stem subtending leaf and cladode primordia. In situ localization of miR166 (K) and Aof PHB transcripts (L) in a transverse section of cladode primordia.
(M) Comparison of the internal structure of an A. officinalis cladode and scale leaf.
(N) Gross morphology of an A. officinalis male flower.
In (D), the xylem is colored pink. cl, cladode; p, phloem; sl, scale leaf; st, stem; x, xylem. White arrowhead indicates stoma. Bars = 1 cm in (B), 0.5 cm in (C), 100 μm in (D) and (I) to (L), and 1 mm in (N).
Then we performed in situ hybridization analyses using A. officinalis to understand how the difference between flattened and cylindrical cladodes could arise. Identically to A. asparagoides, Aof AS1 was detected in leaf and cladode primordia at the early stage (Figure 7I) and Aof KNAT1 was expressed in the SAM and cladode primordia (Figure 7J). However, miR166 was seen throughout the outermost region of cladode primordia (Figure 7K), whereas Aof PHB was not detected in cladode primordia except in the central region, including the presumptive region of the vascular bundle (Figure 7L).
In summary, the tested orthologs involved in shoot and leaf development were also expressed in A. officinalis cladode primordia, as observed in A. asparagoides. However, we observed differences in the expression patterns for microRNA and genes involved in establishing leaf adaxial/abaxial polarity between A. asparagoides and A. officinalis.
DISCUSSION
We investigated the basic molecular mechanism underlying cladodes development in the genus Asparagus. We also analyzed the mechanisms responsible for the difference between flattened and cylindrical cladodes by comparing A. asparagoides and A. officinalis. We showed that cladodes shared characteristics of both leaves and stems in terms of anatomical features, developmental patterns, and the expression patterns of developmental genes. These results indicate that ectopic expression of genes involved in leaf morphogenesis in axillary shoots may have led to the acquisition of leaf-like features. Moreover, we showed that alteration of the expression pattern correlates with the cylindrical form of cladodes seen A. officinalis. Our results suggest that co-opting preexisting gene networks and subsequent modifications can explain the acquisition and diversification of these unusual leaf-like organs through plant evolution, as discussed more fully below.
A. asparagoides Cladodes Are Modified Axillary Shoots
Morphological and anatomical studies showed the presence of abaxial/adaxial polarity in A. asparagoides cladodes. Additionally, unlike primary and axillary shoots, which show apical growth, mitotic cells were distributed basipetally in developing cladodes, as is typical of leaves. These observations indicate anatomical and developmental similarities between cladodes and leaves. Consistent with this pattern, Aas AS1 was expressed in both leaves and in cladode primordia, and orthologs of HD-ZIP III genes (Aas PHB and Aas REV) and miR166 were expressed in the adaxial and abaxial side of cladode primordia, respectively. These expression patterns suggest that the planar form and the adaxial/abaxial polarity of A. asparagoides cladodes is regulated in much the same way as in leaves.
One obvious difference between cladodes and leaves is the inversion of vascular bundles. The inversion seems to occur around the base of cladodes (see Supplemental Figure 9 online) because inversion of vascular bundles was not observed in the stem. Inverted vascular bundles were observed also in the leaves of Cladium, which is a member of Poales (Fisher, 1971), suggesting that, at least in monocots, the regulation of abaxial/adaxial polarity in vasculature is not tightly linked to that of the organ as a whole.
Class I KNOX genes play a crucial role in the indeterminate fate of SAMs by promoting cell division (Scofield and Murray, 2006; Veit, 2009). Aas KNAT1 was expressed in cladode primordia from the early to middle stages in A. asparagoides, but expression diminished at later stages. Cladodes are determinate organ, and it seems likely that diminished expression of Aas KNAT1 is responsible for cladode determinacy. However, while Aas KNAT1 was expressed in cladodes, there was still a noticeable downregulation of class I KNOX genes, indicating that the mechanisms for initiating cladodes may be essentially identical to that of leaves. Aas KNAT1 was also expressed in SAM and axillary shoot meristems but was not expressed in leaf primordia. Hence, Aas KNAT1 expression in cladode primordia could be considered a vestige of the axillary shoot (Figure 8).
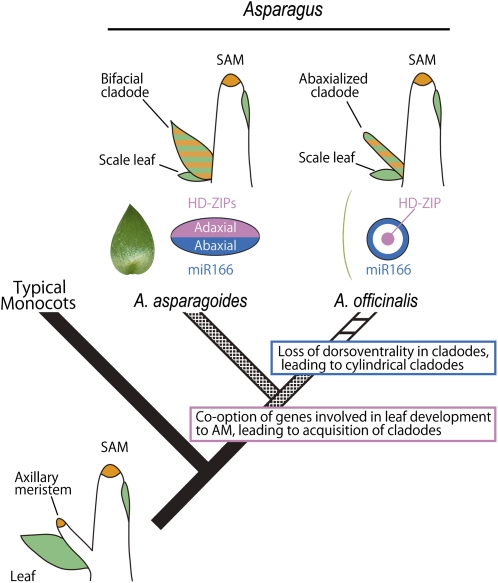
Figure 8.
Schematic Diagram of the Evolutionary Path of Cladodes in the Genus Asparagus.
Simplified phylogenetic tree (Fukuda et al., 2005) with illustrations of shoots and expression patterns in transverse section of cladodes indicate evolutionary processes of acquisition and diversification of cladodes, respectively. The expression patterns of orthologs of HD-ZIPIII (pink) and miR166 (blue) in a transverse section of cladodes and expression patterns of class I KNOX (orange) and AS1 (green) in shoots. Boxes indicate presumed key events during evolution. See text for details.
Acquisition of Leaf-Like Organs in Asparagaceae and the Diversification of Cladode Morphology in Asparagus Might Be Caused by Altered Leaf Polarity Gene Expression Patterns
Besides Asparagus, another genus of Asparagaceae, Ruscus, also develops a leaf-like organs, termed a phylloclades, in the axils of scale leaves (Cooney-Sovetts and Sattler, 1986). Phylloclades differ from cladodes in consisting of a stem with multiple internodes (Bell, 2008). Expression analysis by PCR showed that the STM and YABBY orthologs are expressed in phylloclades (Hirayama et al., 2007). However, both anatomical (Conran and Tamura, 1998; Rudall et al., 1998) and molecular phylogenetic analyses (Kim et al., 2010) suggest that the evolution of leaf-like axillary shoots occurred independently in Asparagus and Ruscus. The fact that acquisition of leaf-like organs has occurred at least twice in the evolution of Asparagaceae suggests that members of this family have a genetic background that allows easy modification of axillary shoots into leaf-like organs.
AS1 is critical for the repression of class I KNOX genes in developing leaf primordia (Szakonyi et al., 2010) and development of a symmetrical leaf lamina (Iwakawa et al., 2007). However, ectopic expression of AS1 is not sufficient to convert axillary shoots into leaf-like structures in Arabidopsis (Xu et al., 2003). Thus, in addition to KNOX repression and symmetrical lamina formation of AS1, other genes were almost certainly involved in the conversion of axillary shoots into leaf-like organs. Nevertheless, an investigation into the causes of expression of AS1 ortholog in axillary shoots in Asparagus could greatly improve our understanding of the origin of cladodes.
The cylindrical form of cladodes seen in A. officinalis represents a synapomorphy of a major subclade of Asparagus (Figure 1F; Fukuda et al., 2005). The evolutionary transition from leaf-like to cylindrical cladodes is thought to be an adaptation to dry conditions (Fukuda et al., 2005), as the surface area should enhance resistance to drought stress (Robinchaux and Canfield, 1985; Robinchaux et al., 1990; Nicotra et al., 2011). Internal structures and the distribution of stomata indicate that A. officinalis cladodes are anatomically abaxialized. Because Aof KNAT1 and Aof AS1 are similarly expressed in cylindrical and laminar cladodes, the basic genetic network seems to be conserved between A. asparagoides and A. officinalis. However, there were differences in the gene expression patterns of genes involved in establishing adaxial/abaxial polarity: miR166 was expressed all around the cladode primordia in A. officinalis but just on the abaxial face in A. asparagoides; Aof PHB was expressed only in the central region, including the presumptive xylem region of cladode primordia in A. officinalis, but just on the adaxial face in A. asparagoides. These results confirm that the cladode in A. officinalis is an abaxialized derivative of the flattened cladodes such as those found in A. asparagoides (Figure 8). However, this is unlikely to be due to a simple loss-of-function mutation in the coding region because no plausible mutations were observed in the isolated genes, and leaves, petals, and other floral organs of A. officinalis seem to develop with adaxial/abaxial polarity (Figures 7M and 7N). It seems most likely that abaxialization of cladodes in A. officinalis occurred via cis-regulatory changes, since these have been implicated in explaining morphological differences between closely related taxa of several organisms (McGregor et al., 2007; Chan et al., 2010; Piazza et al., 2010; Yamaguchi et al., 2010).
Our study implies that the co-option and modification of a deeply conserved genetic module involved in leaf development may be responsible for the acquisition and subsequent evolution of a leaf-like novel organ. However, it seems that only part of the leaf developmental program has been co-opted into cladode development because Aa DL, which is expressed in the phloem of leaves, is not expressed in the phloem of cladodes throughout development (Nakayama et al., 2010). In addition to understanding such genetic modularity, further elucidation of a key genetic alteration(s) that gives rise to the evolution of cladodes could provide new insights into the genetic changes underlying the evolutionary origin of novel and unique organs.
METHODS
Plant Materials
Asparagus asparagoides and Asparagus officinalis were grown in a greenhouse at the Hongo Campus, University of Tokyo, Japan. Seedlings of both species were planted in pots containing a soil-peat-sand mixture with a bimonthly application of plant fertilizer. Watering was performed every 2 d. The shoots, including scale leaves and cladodes, were frozen in liquid nitrogen just after sampling and then stored at –80°C until use for DNA and RNA extraction.
Morphological Observations
Shoots, lateral branches, scale leaves, and cladodes were dissected and fixed overnight in 45% ethanol, 2.5% acetic acid, and 2.5% formaldehyde at 4°C. The fixed samples were dehydrated in an ethanol series (50, 60, 70, 80, 90, 95, 99.5, and 100% [v/v]; 30 min per step) and stored overnight in 100% (v/v) ethanol at 4°C. The ethanol was replaced with isopentyl acetate, and the samples were dried in a critical-point dryer (JCPD-5; JEOL), sputter-coated with carbon using an anion sputter (JFC-1300; JEOL), and examined under a JSM-6510LV scanning electron microscope (JEOL). Fixed specimens embedded in Technovit resin (Kulzer and Co.) were sectioned as described previously (Tsukaya et al., 1995, 2002; Tsuge et al., 1996). Samples were photographed as described previously (Tsukaya and Uchimiya, 1997).
Detecting the Cell Proliferation Zone in Cladodes with DAPI, Aniline Blue, and EdU
To identify the cell nuclei and dividing cells of cladodes, we stained with DAPI and aniline blue or an EdU incorporation assay. For DAPI staining, shoot apices fixed in 2.5% formalin in 20 mM Tris-HCl were stained with 1 μg/mL DAPI after washing with 50 mM sodium phosphate buffer, pH 7.0. Aniline blue staining enabled us to visualize the septum walls in newly divided cells (Kakimoto and Shibaoka, 1992). Shoot apices were fixed in ethanol and acetic acid (4:1 [v/v]) and then rinsed in ethanol (95%) and in a mixture of ethanol and 100 mM phosphate buffer (pH 9.0; 1:1; [v/v]) for 30 min. The samples were then immersed in 100 mM phosphate buffer, pH 9.0. The samples were immersed in a 0.02% (w/v) aniline blue solution in 100 mM phosphate buffer, pH 9.0, for 7 d at 4°C. EdU is a terminal alkyne-containing nucleotide analog of thymidine and is incorporated into DNA during active DNA synthesis (S-phase) (Kotogány et al., 2010). For EdU incorporation assays, dissected shoot apices were incubated in water containing 10 μM EdU (Click-iT EdU Alexa Fluor 488 imaging kit; Invitrogen). After 3 h, the samples were fixed in 3.7% formaldehyde in PBS, pH 7.4, for 30 min. Samples were then washed in PBS three times with shaking. Coupling of EdU to the Alexa Fluor substrate was performed in the dark in the Click-iT reaction mixture, according to the manufacturer’s instructions. Photographs were taken using a Leica DM-4500 (Leica).
RNA Extraction and Degenerate PCR
Total RNA was extracted from shoots including scale leaves and cladodes using an RNeasy plant mini kit (Qiagen) with DNase I treatment. The cDNA was synthesized from 5 μg total RNA using SuperScript III reverse transcriptase (Invitrogen). A fraction of the reverse-transcribed cDNA (0.5 μL) was subjected to 40 cycles of PCR amplification (94°C for 30 s, 55°C for 30 s, and 72°C for 30 s) using degenerate primers, which were designed using the CODEHOP program (Rose et al., 2003) to amplify each conserved sequence. The PCR products were cloned into the pGEM-T Easy vector (Promega) and sequenced using an automated sequencer (model 3100; Applied Biosystems) for at least eight clones per fragment, according to the manufacturer’s protocol. Primer sequences are listed in Supplemental Table 1 online.
5′ and 3′ RACE
To determine 5′ and 3′ sequences of homologous genes, both 5′ and 3′ RACE were performed using the GeneRacer kit (Invitrogen) according to manufacturer’s instructions. The primers were designed to amplify the 5′ and 3′ regions of the isolated fragment (see Supplemental Table 1 online), and PCR was performed as follows: 94°C for 2 min; five cycles at 94°C for 30 s and 72°C for 2 min; five cycles at 94°C for 30 s and 70°C for 2 min; 20 cycles at 94°C for 30 s, 65°C for 30 s, and 68°C for 2 min; and a final extension at 68°C for 10 min. Nested PCR using the specific primers was then performed as follows: 94°C for 2 min, followed by 25 cycles at 94°C for 30 s, 65°C for 30 s, and 68°C for 2 min, with a final extension at 68°C for 10 min. The PCR products were cloned into a pGEM-T Easy vector (Promega) and sequenced.
Phylogenetic Analyses
The amino acid sequences of isolated genes were aligned using ClustalW and readjusted manually. Phylogenetic trees were reconstructed using MEGA5 (Tamura et al., 2011) using the neighbor-joining method (Perrière and Gouy, 1996). Bootstrap values were derived from 1000 replicate runs.
In Situ Hybridization
The partial fragments of genes isolated in the pGEM-T Easy vector were amplified by PCR using universal primers, M13-Fw (5′-CGCCAGGGTTTTCCCAGTCACGAC-3′) and M13-Rv (5′-TCACACAGGAAACAGCTATGAC-3′). The amplified fragment was then used to produce digoxigenin (DIG)-labeled sense and antisense RNA probes using a DIG RNA labeling kit (Roche). Shoots were fixed in 100 mM sodium phosphate, pH 7.5, containing 3% (w/v) paraformaldehyde, 0.25% (w/v) glutaraldehyde, and 1 M NaOH. Fixed samples were dehydrated in an ethanol series and embedded in Paraplast Plus (Sigma-Aldrich). The embedded samples were sectioned to a thickness of 8 μm, and the sections were affixed to glass slides by an overnight incubation at 45°C. DIG-labeled sense and antisense RNA probes were synthesized with SP6 RNA polymerase or T7 RNA polymerase (Roche). Hybridizations were performed overnight in a moist chamber at 50°C, followed by washing twice in 0.5× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) at 50°C for 20 min. The slides were blocked with 0.5% blocking reagent for 90 min, and anti-DIG antibody in 0.1% BSA was added to detect the DIG-labeled RNA probes. Slides were washed three times in 0.1 M Tris containing 150 mM NaCl, pH 7.5, for 10 min and then transferred into 0.1 M Tris, 0.1 M NaCl, pH 9.5, and 50 mM MgCl2 for 5 min. The slides were incubated in detection buffer containing nitro-blue tetrazolium chloride and 5-bromo-4-chloro-3-indolyl phosphate, toluidine salt (Roche) at room temperature overnight. Photographs were taken using a Leica DM-4500. The experiments were performed more than twice.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: AB673047 (Aas KNAT1), AB673048 (Aof KNAT1), AB673049 (Aas AS1), AB673050 (Aof AS1), AB673051 (Aas REV), AB673052 (Aas PHB), and AB673053 (Aof PHB).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alignments of KNAT1-Like Proteins.
Supplemental Figure 2. Alignment of AS1-Like Proteins.
Supplemental Figure 3. Alignment of PHB-Like Proteins and mR166 with Putative Target Sites.
Supplemental Figure 4. Alignment of REV-Like Proteins.
Supplemental Figure 5. Phylogenetic Tree of Class I KNOX Genes.
Supplemental Figure 6. Phylogenetic Tree of AS1 Genes.
Supplemental Figure 7. Phylogenetic Tree of HD-ZIP III Genes.
Supplemental Figure 8. Expression Patterns of miR166 in Asparagus asparagoides.
Supplemental Figure 9. Inner structure between Proximal Region and Base of a Cladode.
Supplemental Table 1. List of Oligonucleotide Primers Used in This Study.
Acknowledgments
We thank Atsushi Komamine, Ryoko Imaichi, Jun Yokoyama, Tatsuya Fukuda, and Takuro Ito for their support during the course of this study. This work was partially supported by the following sources: Grants-in-Aid for Scientific Research (A) to H.T. and for Creative Scientific Research to H.T. (18GS0313) from the Japan Society for the Promotion of Science and for Scientific Research on Priority Areas to H.T. (19060002); a grant for Young Scientists (B) to T.Y. (19770041) from the Ministry of Education, Culture, Sports, Science and Technology, Japan; a grant from the Sumitomo Foundation to T.Y.; and a Research Fellowship from the Japan Society for the Promotion of Science to H.N.
AUTHOR CONTRIBUTIONS
H.N. conceived and designed the project, performed the experiments, and wrote the article. T.Y. and H.T. designed the project and wrote the article.
References
- Bell A.D. (2008). Plant Form: An Illustrated Guide to Flowering Plant Morphology. (Portland, OR: Timber Press; ). [Google Scholar]
- Berger Y., Harpaz-Saad S., Brand A., Melnik H., Sirding N., Alvarez J.P., Zinder M., Samach A., Eshed Y., Ori N. (2009). The NAC-domain transcription factor GOBLET specifies leaflet boundaries in compound tomato leaves. Development 136: 823–832 [DOI] [PubMed] [Google Scholar]
- Blein T., Pulido A., Vialette-Guiraud A., Nikovics K., Morin H., Hay A., Johansen I.E., Tsiantis M., Laufs P. (2008). A conserved molecular framework for compound leaf development. Science 322: 1835–1839 [DOI] [PubMed] [Google Scholar]
- Braybrook S.A., Kuhlemeier C. (2010). How a plant builds leaves. Plant Cell 22: 1006–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne M.E. (2005). Networks in leaf development. Curr. Opin. Plant Biol. 8: 59–66 [DOI] [PubMed] [Google Scholar]
- Byrne M.E., Barley R., Curtis M., Arroyo J.M., Dunham M., Hudson A., Martienssen R.A. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408: 967–971 [DOI] [PubMed] [Google Scholar]
- Chan Y.F., et al. (2010). Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science 327: 302–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conran J.G., Tamura M.N. (1998). Convallariaceae. In The Families and Genera of Vascular Plants, Vol. 3, K. Kubitzki, ed (Berlin: Springer-Verlag), pp. 125–128
- Cooney-Sovetts C., Sattler R. (1986). Cladode development in the Asparagaceae: An example of homeostasis. Bot. J. Linn. Soc. 94: 327–371 [Google Scholar]
- Dengler N.G., Tsukaya H. (2001). Leaf morphogenesis in dicotyledons: Current issues. Int. J. Plant Sci. 162: 459–464 [Google Scholar]
- Eshed Y., Baum S.F., Perea J.V., Bowman J.L. (2001). Establishment of polarity in lateral organs of plants. Curr. Biol. 11: 1251–1260 [DOI] [PubMed] [Google Scholar]
- Fisher J. (1971). Inverted vascular bundles in the leaf of Cladium (Cyperaceae). Bot. J. Linn. Soc. 64: 277–293 [Google Scholar]
- Fukuda T., Ashizawa H., Suzuki R., Ochiai T., Nakamura T., Kanno A., Kameya T., Yokoyama J. (2005). Molecular phylogeny of the genus Asparagus (Asparagaceae) inferred from plastid petB intron and petD-rpoA intergenic spacer sequences. Plant Species Biol. 20: 121–132 [Google Scholar]
- Guo J., Jermyn W.A., Turnbull M.H. (2002). Carbon assimilation, partitioning and export in mature cladophylls of two asparagus (Asparagus officinalis) cultivars with contrasting yield. Physiol. Plant. 115: 362–369 [DOI] [PubMed] [Google Scholar]
- Guo M., Thomas J., Collins G., Timmermans M.C. (2008). Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell 20: 48–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha C.M., Jun J.H., Fletcher J.C. (2010). Shoot apical meristem form and function. Curr. Top. Dev. Biol. 91: 103–140 [DOI] [PubMed] [Google Scholar]
- Hirayama Y., Yamada T., Oya Y., Ito M., Kato M., Imaichi R. (2007). Expression patterns of class I KNOX and YABBY genes in Ruscus aculeatus (Asparagaceae) with implications for phylloclade homology. Dev. Genes Evol. 217: 363–372 [DOI] [PubMed] [Google Scholar]
- Hirsch A.M. (1977). A developmental study of the cladodes of Ruscus aculeatus L. Bot. J. Linn. Soc. 74: 355–365 [Google Scholar]
- Ikezaki M., Kojima M., Sakakibara H., Kojima S., Ueno Y., Machida C., Machida Y. (2010). Genetic networks regulated by ASYMMETRIC LEAVES1 (AS1) and AS2 in leaf development in Arabidopsis thaliana: KNOX genes control five morphological events. Plant J. 61: 70–82 [DOI] [PubMed] [Google Scholar]
- Itoh J., Nonomura K., Ikeda K., Yamaki S., Inukai Y., Yamagishi H., Kitano H., Nagato Y. (2005). Rice plant development: From zygote to spikelet. Plant Cell Physiol. 46: 23–47 [DOI] [PubMed] [Google Scholar]
- Itoh J.I., Sato Y., Nagato Y. (2008). The SHOOT ORGANIZATION2 gene coordinates leaf domain development along the central-marginal axis in rice. Plant Cell Physiol. 49: 1226–1236 [DOI] [PubMed] [Google Scholar]
- Iwakawa H., Iwasaki M., Kojima S., Ueno Y., Soma T., Tanaka H., Semiarti E., Machida Y., Machida C. (2007). Expression of the ASYMMETRIC LEAVES2 gene in the adaxial domain of Arabidopsis leaves represses cell proliferation in this domain and is critical for the development of properly expanded leaves. Plant J. 51: 173–184 [DOI] [PubMed] [Google Scholar]
- Juarez M.T., Kui J.S., Thomas J., Heller B.A., Timmermans M.C. (2004). microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428: 84–88 [DOI] [PubMed] [Google Scholar]
- Kakimoto T., Shibaoka H. (1992). Synthesis of polysaccharides in phragmoplasts isolated from tobacco BY-2 cells. Plant Cell Physiol. 33: 353–361 [Google Scholar]
- Kessler S., Sinha N. (2004). Shaping up: The genetic control of leaf shape. Curr. Opin. Plant Biol. 7: 65–72 [DOI] [PubMed] [Google Scholar]
- Kidner C.A., Timmermans M.C.P. (2007). Mixing and matching pathways in leaf polarity. Curr. Opin. Plant Biol. 10: 13–20 [DOI] [PubMed] [Google Scholar]
- Kim J.-H., Kim D.-K., Forest F., Fay M.F., Chase M.W. (2010). Molecular phylogenetics of Ruscaceae sensu lato and related families (Asparagales) based on plastid and nuclear DNA sequences. Ann. Bot. (Lond.) 106: 775–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S., Koenig D., Kang J., Yoong F.Y., Sinha N. (2008). Natural variation in leaf morphology results from mutation of a novel KNOX gene. Curr. Biol. 18: 672–677 [DOI] [PubMed] [Google Scholar]
- Kotogány E., Dudits D., Horváth G.V., Ayaydin F. (2010). A rapid and robust assay for detection of S-phase cell cycle progression in plant cells and tissues by using ethynyl deoxyuridine. Plant Methods 6: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota S., Konno I., Kanno A. (2012). Molecular phylogeny of the genus Asparagus (Asparagaceae) explains interspecific crossability between the garden asparagus (A. officinalis) and other Asparagus species. Theor. Appl. Genet. 124: 345–354 [DOI] [PubMed] [Google Scholar]
- Langdale J.A. (2008). Evolution of developmental mechanisms in plants. Curr. Opin. Genet. Dev. 18: 368–373 [DOI] [PubMed] [Google Scholar]
- Long J.A., Moan E.I., Medford J.I., Barton M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69 [DOI] [PubMed] [Google Scholar]
- McConnell J.R., Emery J., Eshed Y., Bao N., Bowman J., Barton M.K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411: 709–713 [DOI] [PubMed] [Google Scholar]
- McGregor A.P., Orgogozo V., Delon I., Zanet J., Srinivasan D.G., Payre F., Stern D.L. (2007). Morphological evolution through multiple cis-regulatory mutations at a single gene. Nature 448: 587–590 [DOI] [PubMed] [Google Scholar]
- Nakayama H., Yamaguchi T., Tsukaya H. (2010). Expression patterns of AaDL, a CRABS CLAW ortholog in Asparagus asparagoides (Asparagaceae), demonstrate a stepwise evolution of CRC/DL subfamily of YABBY genes. Am. J. Bot. 97: 591–600 [DOI] [PubMed] [Google Scholar]
- Nicotra A.B., Leigh A., Boyce C.K., Jones C.S., Niklas K.J., Royer D.L., Tsukaya H. (2011). The evolution and functional significance of leaf shape in the angiosperms. Funct. Plant Biol. 38: 535–552 [DOI] [PubMed] [Google Scholar]
- Pekker I., Alvarez J.P., Eshed Y. (2005). Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 17: 2899–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrière G., Gouy M. (1996). WWW-query: An on-line retrieval system for biological sequence banks. Biochimie 78: 364–369 [DOI] [PubMed] [Google Scholar]
- Piazza P., et al. (2010). Arabidopsis thaliana leaf form evolved via loss of KNOX expression in leaves in association with a selective sweep. Curr. Biol. 20: 2223–2228 [DOI] [PubMed] [Google Scholar]
- Reddy G.V. (2008). Live-imaging stem-cell homeostasis in the Arabidopsis shoot apex. Curr. Opin. Plant Biol. 11: 88–93 [DOI] [PubMed] [Google Scholar]
- Robinchaux R.H., Canfield J.E. (1985). Tissue elastic properties of mesic forest Hawaiian Dubautia species that differ in habitat and diploid chromosome number. Oecologia 66: 77–80 [DOI] [PubMed] [Google Scholar]
- Robinchaux R.H., Carr G.D., Liebman M., Pearcy R.W. (1990). Adaptive radiation of the Hawaiian silversword allianve (Compositae-Maddiinae): Ecological, morphological, and physiological diversity. Ann. Mo. Bot. Gard. 77: 64–72 [Google Scholar]
- Rose T.M., Henikoff J.G., Henikoff S. (2003). CODEHOP (COnsensus-DEgenerate Hybrid Oligonucleotide Primer) PCR primer design. Nucleic Acids Res. 31: 3763–3766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudall P.J., Engelman E.M., Hanson L., Chase M.W. (1998). Systematics of Hemiphylacus, Anemarrhena, and Asparagus. Plant Syst. Evol. 211: 181–199 [Google Scholar]
- Salic A., Mitchison T.J. (2008). A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl. Acad. Sci. USA 105: 2415–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield S., Murray J.A. (2006). KNOX gene function in plant stem cell niches. Plant Mol. Biol. 60: 929–946 [DOI] [PubMed] [Google Scholar]
- Sylvester A.W., Cande W.Z., Freeling M. (1990). Division and differentiation during normal and liguleless-1 maize leaf development. Development 110: 985–1000 [DOI] [PubMed] [Google Scholar]
- Szakonyi D., Moschopoulos A., Byrne M.E. (2010). Perspectives on leaf dorsoventral polarity. J. Plant Res. 123: 281–290 [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans M.C.P., Hudson A., Becraft P.W., Nelson T. (1999). ROUGH SHEATH2: A Myb protein that represses knox homeobox genes in maize lateral organ primordia. Science 284: 151–153 [DOI] [PubMed] [Google Scholar]
- Tomlinson P.B., Takaso T., Rattenbury J.A. (1987). Developmental shoot morphology in Phyllocladus (Podocarpaceae). Bot. J. Linn. Soc. 99: 223–248 [Google Scholar]
- Tsuge T., Tsukaya H., Uchimiya H. (1996). Two independent and polarized processes of cell elongation regulate leaf blade expansion in Arabidopsis thaliana (L.) Heynh. Development 122: 1589–1600 [DOI] [PubMed] [Google Scholar]
- Tsukaya H. (2006). Mechanism of leaf-shape determination. Annu. Rev. Plant Biol. 57: 477–496 [DOI] [PubMed] [Google Scholar]
- Tsukaya H., Naito S., Rèdai G.P., Komeda Y. (1995). A new class of mutants in Arabidopsis thaliana, acaulis1, affecting the development of both inflorescences and leaves. Development 118: 751–764 [Google Scholar]
- Tsukaya H., Kozuka T., Kim G.T. (2002). Genetic control of petiole length in Arabidopsis thaliana. Plant Cell Physiol. 43: 1221–1228 [DOI] [PubMed] [Google Scholar]
- Tsukaya H., Uchimiya H. (1997). Genetic analyses of developmental control of serrated margin of leaf morphogenesis with characterization of a specific marker gene, which expresses in hydathodes and stipules in Arabidopsis. Mol. Genet. Genomics 256: 231–238 [DOI] [PubMed] [Google Scholar]
- Uchida N., Kimura S., Koenig D., Sinha N. (2010). Coordination of leaf development via regulation of KNOX1 genes. J. Plant Res. 123: 7–14 [DOI] [PubMed] [Google Scholar]
- Veit B. (2009). Hormone mediated regulation of the shoot apical meristem. Plant Mol. Biol. 69: 397–408 [DOI] [PubMed] [Google Scholar]
- Vollbrecht E., Reiser L., Hake S. (2000). Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene, knotted1. Development 127: 3161–3172 [DOI] [PubMed] [Google Scholar]
- Williams L., Fletcher J.C. (2005). Stem cell regulation in the Arabidopsis shoot apical meristem. Curr. Opin. Plant Biol. 8: 582–586 [DOI] [PubMed] [Google Scholar]
- Xu L., Xu Y., Dong A., Sun Y., Pi L., Xu Y., Huang H. (2003). Novel as1 and as2 defects in leaf adaxial-abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development 130: 4097–4107 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T., Yano S., Tsukaya H. (2010). Genetic framework for flattened leaf blade formation in unifacial leaves of Juncus prismatocarpus. Plant Cell 22: 2141–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]