This article shows that the small signaling molecule CLE8 plays an important role in regulating cell division patterns in the Arabidopsis thaliana embryo, as well as the proliferation of the surrounding nutrient-rich endosperm. CLE8 promotes seed size and induces expression of the WOX8 transcription factor gene, acting in a novel CLE-WOX regulatory cassette that coordinates seed formation.
Abstract
The plant seed is a major nutritional source for humans as well as an essential embryo development and dispersal unit. To ensure proper seed formation, fine spatial and temporal coordination between the embryo, endosperm, and maternal seed components must be achieved. However, the intercellular signaling pathways that direct the synchronous development of these tissues are poorly understood. Here we show that the Arabidopsis thaliana peptide ligand CLAVATA3/EMBRYO SURROUNDING REGION-RELATED8 (CLE8) is exclusively expressed in young embryos and endosperm, and that it acts cell and noncell autonomously to regulate basal embryo cell division patterns, endosperm proliferation, and the timing of endosperm differentiation. CLE8 positively regulates expression of the transcription factor gene WUSCHEL-LIKE HOMEOBOX8 (WOX8), and together CLE8 and WOX8 form a signaling module that promotes seed growth and overall seed size. These results demonstrate that seed development is coordinated by a secreted peptide ligand that plays a key early role in orchestrating cell patterning and proliferation in the embryo and endosperm.
INTRODUCTION
Angiosperm seed formation is characterized by the process of double fertilization, during which the female gametophyte receives a pollen tube that delivers two sperm cells to the ovule. One sperm cell fertilizes the egg cell to produce a zygote, which develops into an embryo, whereas the second sperm cell targets the central cell polar nuclei, leading to endosperm formation. The endosperm surrounds and nourishes the embryo as it develops (Dumas and Rogowsky, 2008), and although both tissues seem to develop autonomously during early seed development, the endosperm plays an important role in regulating embryo growth at later stages (Weijers et al., 2003; Kondou et al., 2008). Both the diploid embryo and the triploid endosperm are enclosed by the maternally derived integument layers that form the seed coat, and successful seed formation requires the simultaneous development of all three of these genetically distinct tissues (Garcia et al., 2005; Berger et al., 2006; Ingouff et al., 2006).
Although little is known about the molecular mechanisms that coordinate the genetic programs of the three seed components, evidence indicates that intercellular signal transduction pathways play major roles (Saulsberry et al., 2002; Nodine et al., 2011). In Arabidopsis thaliana, nearly 400 kinases are expressed in embryos (Nodine et al., 2011), and among these, several receptor-like kinases (RLKs) have been shown to be required for proper endosperm and/or embryo development. HAIKU2 (IKU2) encodes a Leu-rich repeat RLK that is expressed in the endosperm of developing seeds (Luo et al., 2005). IKU mutants display precocious endosperm cellularization as well as decreased endosperm size, which ultimately results in a smaller embryo and a decrease in seed size. Several other RLKs, including RPK1/TOAD2 and ACR4/ALE2, are required during embryogenesis for pattern formation and cell fate maintenance (Nodine et al., 2011). In addition, two cytoplasmic kinases regulate early embryonic patterning (Lukowitz et al., 2004; Bayer et al., 2009). An important unresolved question is the identity of the extracellular signals that trigger these kinase pathways, because despite the presence of >1000 putative peptide ligand genes in the Arabidopsis genome (Lease and Walker, 2006), no small signaling molecule has yet been shown to influence seed development.
Candidates for polypeptide signaling molecules that could regulate seed formation include members of the CLAVATA3/EMBRYO SURROUNDING REGION-RELATED (CLE) family. The CLE genes encode small proteins that consist of a signal peptide or membrane anchor sequence, a 40- to 90-amino-acid variable domain, and a highly conserved 14-amino-acid motif near their carboxyl termini, called the CLE domain (Cock and McCormick, 2001). Full-length Arabidopsis CLV3 is proteolytically processed (Ni and Clark, 2006) to a mature 12- or 13-amino-acid arabinosylated glycopeptide consisting of the CLE domain, which has full biological function (Kondo et al., 2006; Ohyama et al., 2009). Synthetic peptides corresponding to the CLE domains of other CLE proteins also exhibit activity in various bioassays (Fiers et al., 2005; Ito et al., 2006), suggesting that this region represents the bioactive form of the protein.
Thirty-two members of the CLE gene family have been identified in Arabidopsis, yet the biological functions of only three are known. CLV3 is specifically expressed in the stem cell reservoirs of shoot and floral meristems (Fletcher et al., 1999) but has a noncell autonomous effect on the interior meristem cells, where the CLV3 mature polypeptide is perceived by three transmembrane receptor complexes (Ogawa et al., 2008; Guo et al., 2010; Kinoshita et al., 2010). Intercellular signaling through the CLV3 pathway limits to the organizing center the expression domain of the WOX family transcription factor gene WUSCHEL (Laux et al., 1996), which in turn promotes stem cell activity (Schoof et al., 2000) and directly activates CLV3 transcription (Yadav et al., 2011). Thus, this signal transduction pathway functions as part of a negative feedback loop that modulates stem cell accumulation during plant development (Brand et al., 2000). A comparable signaling module involving CLE40, the receptor kinase ACR4, and the WUS-related protein WOX5 regulates stem cell homeostasis in the root apical meristem (Stahl et al., 2009), whereas a module consisting of CLE41, PXY/TDR, and WOX4 regulates vascular meristem activity (Hirakawa et al., 2008; Etchells and Turner, 2010; Hirakawa et al., 2010).
Although the activities of CLV3, CLE40, and CLE41 are restricted to meristems, the expression patterns of the other CLE family members suggest that the polypeptides may play diverse signaling roles during the plant life cycle. At least one CLE gene is expressed in each Arabidopsis tissue and at each stage of development (Jun et al., 2010), including more than six during embryogenesis (Fiers et al., 2004; Fiume et al., 2011). Most tissues and cell types express multiple CLE genes (Sharma et al., 2003; Jun et al., 2010; Fiume et al., 2011). These data, along with the observation of CLE overexpression phenotypes ranging from dwarfism to anthocyanin overproduction (Ito et al., 2006; Ni and Clark, 2006; Strabala et al., 2006; Whitford et al., 2008; Jun et al., 2010), indicate that CLE-mediated signaling pathways are likely to function in many biological processes. Here we show that the CLE8 gene, which is expressed in early embryos and in endosperm, regulates embryo and suspensor proliferation. It also promotes seed size by regulating both endosperm proliferation and the timing of endosperm differentiation. We additionally demonstrate that CLE8 positively regulates expression of the WUS-related gene WOX8, uncovering a novel CLE-WOX signaling module that directs Arabidopsis suspensor and endosperm growth.
RESULTS
CLE8 Is Expressed in Embryos and Endosperm
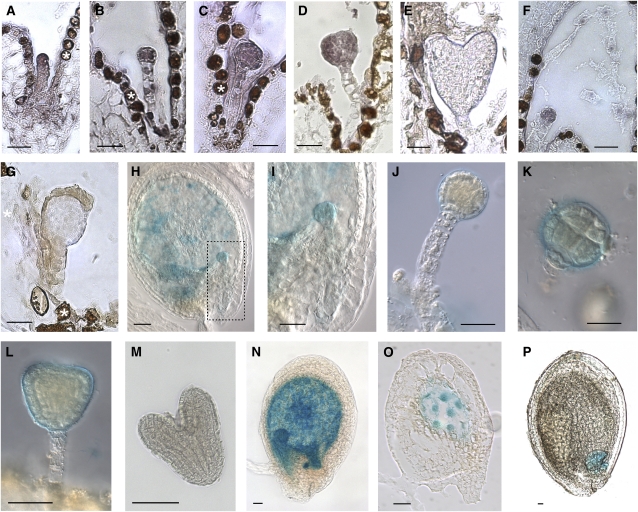
CLE8 is the only Arabidopsis CLE gene that is expressed exclusively in seed-bearing siliques (Sharma et al., 2003), and thus we hypothesized that CLE8 might play a specific role in seed development. RNA in situ hybridization experiments detected CLE8 mRNA expression in young embryos (Figures 1A to 1D) and endosperm (Figure 1F). CLE8 mRNA was expressed throughout the embryo proper, beginning at the one-cell stage (Figure 1A) and continuing through the globular and the triangular stages (Figures 1B to 1D). By the heart stage, CLE8 expression was no longer detectable (Figure 1E). In contrast with the strong signal in the embryo proper, the weak staining observed in developing suspensor cells was indistinguishable from the surrounding background signal, indicating that CLE8 is not highly transcribed in the suspensor.
Figure 1.
CLE8 Expression Analysis in Developing Seed Tissues.
(A) to (F) CLE8 mRNA expression pattern in a one-cell stage embryo (A), a four-cell stage embryo (B), a globular stage embryo (C), a triangular stage embryo (D), a heart stage embryo (E), and in the endosperm nuclei (F).
(G) Control hybridization with a CLE8 sense probe.
(H) to (P) CLE8 promoter-driven GUS activity in embryos ([H] to [M]) and endosperm ([N] to [P]). (I) shows a higher magnification image of the embryo boxed with the dotted lines in (H).
White stars indicate background browning of the seed coat. Bars = 20 μm.
We verified the CLE8 expression pattern by generating transgenic lines carrying the CLE8 2.9-kb putative promoter region fused to the uid-A reporter gene. This ProCLE8:GUS construct drove expression solely in seeds of stably transformed Arabidopsis plants. We detected β-glucuronidase (GUS) activity in developing embryos (Figures 1H to 1L) prior to the heart stage (Figure 1M). To investigate whether ProCLE8:GUS activity was present in the suspensor cells that connect the embryo to the maternal seed coat, we modified our staining procedure to prevent the solution from completely penetrating the tissues (see Methods). This experiment resulted in preferential GUS staining in the outer layers of the isolated embryos and confirmed the absence of ProCLE8:GUS activity in the suspensor (Figures 1I, 1J, and 1L). We also detected ProCLE8:GUS activity in all endosperm compartments at early stages of seed development (Figures 1H, 1N, and 1O), whereas from the torpedo stage onward, it became restricted to the chalazal endosperm in the nucellar region of the seed (Figure 1P). This expression analysis suggested a role for CLE8 in the initial stages of seed formation, potentially acting as a signal coordinating the early development of the various seed tissues.
CLE8 Acts Noncell Autonomously to Regulate Early Embryo Development
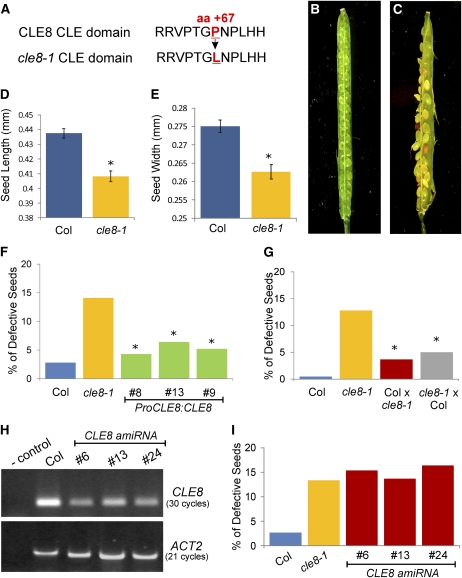
To functionally characterize CLE8, we obtained an ethyl methanesulfonate–induced allele via TILLING (McCallum et al., 2000). The cle8-1 mutation is a C to T nucleotide substitution at position 200 in the CLE8 coding region, which results in a nonconservative amino acid substitution of the second Pro residue of the CLE domain with a Leu residue (Figure 2A). This Pro is believed to be important for CLE peptide function, because it undergoes posttranslational modifications in CLV3 (Kondo et al., 2006; Ohyama et al., 2009) and is conserved in all Arabidopsis A-type CLE genes except CLE40 (Jun et al., 2008). This Pro is adjacent to another functionally crucial amino acid, a highly conserved Gly that, when mutated in two CLV3 alleles, causes a moderate phenotype (Fletcher et al., 1999).
Figure 2.
cle8-1 and CLE8 amiRNA Seed Phenotypes.
(A) Wild-type CLE8 CLE domain sequence (Top) and cle8-1 CLE domain sequence (Bottom). The mutated amino acid and its position in the protein sequence are indicated in red.
(B) and (C) A wild-type (B) and a cle8-1 (C) open silique containing mature seeds. This cle8-1 silique contains more than the average number of abnormal seeds but illustrates the range of seed defects observed.
(D) and (E) Length (D) and width (E) of wild-type Col and cle8-1 seeds (n = 100).
(F) cle8-1 complementation experiment (n = 118 to 186). All transgenic lines are in the cle8-1 homozygous background. Graph values are means ± se, * indicates the difference is statistically significant (Z test, P < 0.05) when compared with the cle8-1 value but not when compared with the wild-type value.
(G) Reciprocal crosses between wild-type Col and cle8-1 plants (n = 179 to 226). * indicates the values are statistically different from wild-type and cle8-1 values (Z test, P < 0.05). The difference in the percentage of defective seeds between the two reciprocal crosses is not significant.
(H) RT-PCR analysis performed on RNA extracts from siliques of Col and three independent transgenic CLE8 amiRNA lines, using two technical replicates.
(I) Quantification of defective seeds in Col, cle8-1, and the CLE8 amiRNA transgenic lines (n = 122 to 275). The difference between wild-type Col and the other genotypes is statistically significant (Z test, P < 0.01), whereas the CLE8 amiRNA transgenic line values are not significantly different from the cle8-1 values.
Careful morphological analysis of cle8-1 homozygous plants throughout vegetative and reproductive growth did not reveal any aberrant phenotype. However, analysis of open mature cle8-1 siliques revealed the presence of a variety of defective seeds, ranging from wrinkled or misshapen seeds to discolored seeds to seeds aborted at early stages of development (Figures 2B and 2C). Although cle8-1 seeds that aborted early were generally not recovered during seed harvesting, we also observed a significant germination defect (see Supplemental Table 1 online), indicating that morphologically normal-looking seeds were among those that failed to germinate.
Further examination of wild-type and mutant seeds revealed a low frequency of defective wild-type accession Columbia (Col) seeds (1.21%), whereas ~15% of cle8-1 seeds displayed morphological defects by the early globular to triangular stage (see Supplemental Table 2 online). In addition, we determined using quantitative analysis that mature cle8-1 seeds with a morphologically normal appearance were significantly smaller than wild-type seeds (Figures 2D and 2E). CLE8 full-length genomic sequence rescued the cle8-1 phenotype (Figure 2F), confirming that the observed aberrant seeds arose solely because of the mutation in CLE8. Furthermore, reciprocal crosses between cle8-1 and wild-type plants indicated that the cle8-1 mutation did not produce a maternal sporophytic effect (Figure 2G).
We tested whether cle8-1 is a hypomorphic allele by generating a constitutively expressed artificial microRNA (amiRNA) that specifically targeted the CLE8 sequence (Ossowski et al., 2008). The phenotypes of three independent CLE8 amiRNA transgenic lines showing substantially reduced CLE8 transcript levels (Figure 2H) were analyzed. The frequency of aberrant seeds in CLE8 amiRNA plants was significantly higher than that observed in wild-type plants but was the same as in cle8-1 plants (Figure 2I), consistent with cle8-1 acting as a hypomorphic allele. However, reciprocal crosses using cle8-1 resulted in a significant number of defective F1 seeds (Figure 2G). These data suggest that although the seed phenotype results from reduced CLE8 expression levels, the cle8-1 allele causes haploinsufficiency; thus, plants are sensitive to the dosage of CLE8.
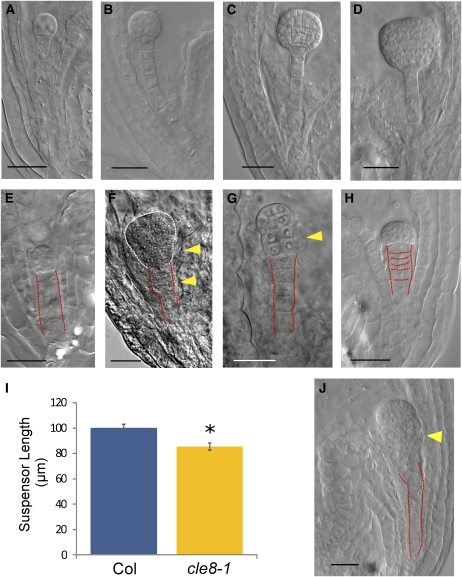
We isolated cle8-1 seeds containing developing embryos from the two-cell to the early globular stage and analyzed their morphology using Nomarski microscopy (Figures 3A to 3H; see Supplemental Table 2 online). Among the aberrant seeds, 33.9% showed embryo defects, 38.8% showed suspensor defects, and 21.8% showed endosperm defects. Approximately 5.5% of defective seeds showed more than one defect. All wild-type embryos progressed through early development with normal cell division patterns, and their boundary with the suspensor was always clearly recognizable (Figures 3A to 3D). By contrast, cle8-1 embryos prematurely ceased developing (Figure 3E) or underwent uncontrolled cell divisions in their basal portion (Figures 3F and 3G). We did not observe any aberrant embryo morphology at later stages of development, suggesting that these early defects lead to seed abortion. Wild-type embryos were subtended by suspensors consisting of a single file of elongated cells (Figures 3A to 3D), whereas in cle8-1 embryos, the uppermost suspensor cell often acquired the wrong plane of cell division. This produced suspensors with their uppermost portion consisting of more than one tier of cells (Figure 3F) and occasional embryos with an indistinct boundary between the embryo proper and the suspensor (Figure 3G). On average, wild-type and cle8-1 embryos at the dermatogen stage had the same number of suspensor cells (Col = 6.37 and cle8-1 = 6.33; n = 20). However, in most of the cle8-1 defective suspensors, the cells failed to elongate properly (Figure 3H), and suspensor length was significantly shorter in cle8-1 than in wild-type embryos (Figure 3I). We likewise observed uncontrolled division of the uppermost suspensor cells and the basal cells of the embryo proper in CLE8 amiRNA transgenic seeds (Figure 3J). We conclude that CLE8 plays a key role in embryo development by regulating cell proliferation in the basal domain of the embryo proper, as well as organizing suspensor cell division planes and regulating suspensor cell proliferation and elongation. Because CLE8 is not transcribed in suspensor cells, CLE8 plays a noncell autonomous role in this tissue.
Figure 3.
cle8-1 and CLE8 amiRNA Embryo Phenotypes.
(A) to (D) Wild-type embryos at the two-cell (A), 16-cell (B), globular (C), and triangular (D) stage.
(E) to (H) cle8-1 aberrant embryos in the same developmental window.
(I) Wild-type and cle8-1 suspensor length measured at the dermatogen embryo stage (n = 30). Graph values are means ± se, * indicates statistically significant values (Student’s t test, P < 0.05).
(J) CLE8 amiRNA aberrant embryo.
Red dashed lines outline the suspensors; white dashed lines outline the embryo proper; yellow arrowheads indicate unusual planes of cell division. Bars = 20 μm.
CLE8 Regulates Endosperm Proliferation and Differentiation
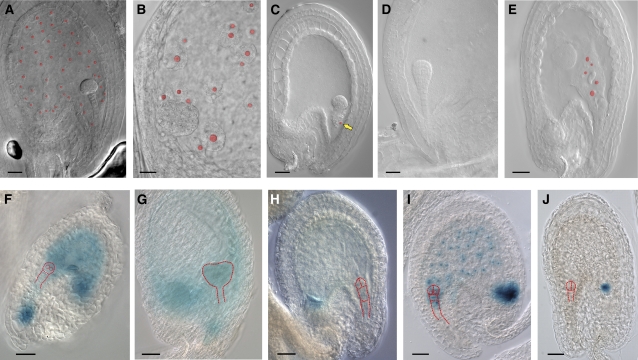
More than 24% of defective cle8-1 seeds showed aberrations in the developing endosperm. In wild-type seeds containing globular stage embryos, the syncytial cytoplasm of the embryo-surrounding region (ESR) surrounded the developing embryo, and the multinucleate peripheral endosperm syncytium was a thin layer with evenly spaced nuclei (Figure 4A). In corresponding cle8-1 seeds, we observed endosperm tissue that lacked this organization and showed uneven distribution of the peripheral endosperm nuclei (Figure 4B) or seeds that altogether lacked the peripheral endosperm nuclei and contained only a few endosperm nuclei at the micropylar and chalazal pole of the embryo sac (Figure 4C). We determined that wild-type seeds with embryos at either the one-cell or two-cell stage contained an average of 22.3 endosperm nuclei, whereas cle8-1 seeds that had fewer endosperm nuclei contained an average of 7.4. Seeds from transgenic CLE8 amiRNA plants likewise either lacked peripheral endosperm (Figure 4D) or showed aberrant distribution of the endosperm nuclei (Figure 4E).
Figure 4.
cle8-1 and CLE8 amiRNA Endosperm Phenotypes.
(A) to (C) Wild-type seed (A) and cle8-1 seeds ([B] and [C]). Free endosperm nuclei are highlighted in red, and the yellow arrow indicates a solitary endosperm nucleus near the suspensor.
(D) and (E) CLE8 amiRNA seeds.
(F) to (H) Wild-type (F) and (G) and cle8-1 (H) seeds expressing the ProMEA:GUS marker.
(I) and (J) Wild-type (I) and cle8-1 (J) seeds expressing the ProFIS2:GUS marker. (F) to (J) Red dashed lines outline the embryos.
Bars = 20 μm.
To further characterize these defects, we used ProMEA:GUS (Luo et al., 2000) as a marker for early endosperm development. In wild-type seeds, this marker is expressed throughout the developing endosperm (Figure 4F) until 2 d after fertilization, when its expression fades in the peripheral endosperm but persists at the posterior pole (Figure 4G). At early stages of development, even cle8-1 seeds that did not exhibit morphological endosperm defects displayed weak ProMEA:GUS activity throughout the peripheral endosperm, whereas the chalazal endosperm exhibited stronger marker activity characteristic of endosperm at later stages of development (Figure 4H). This result suggests that cle8-1 endosperm undergoes an accelerated acquisition of characteristics of more mature endosperm. We monitored endosperm proliferation using the ProFIS2:GUS marker, which is specifically expressed in the central cell and endosperm upon fertilization (Luo et al., 2000). In wild-type seeds containing two-cell stage embryos, the endosperm had undergone many rounds of nuclear division and completely filled the seed cavity (Figure 4I), whereas comparable cle8-1 seeds with normal or abnormal morphology had fewer endosperm nuclei, indicative of a delay in endosperm proliferation (Figure 4J). Thus, CLE8 promotes the proliferation and organized distribution of the free endosperm nuclei and prevents premature endosperm differentiation.
CLE8 Is Sufficient to Regulate Arabidopsis Seed Size
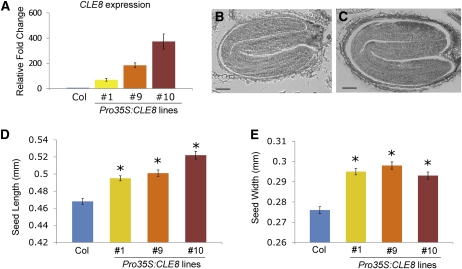
Our results demonstrate that CLE8 function is necessary to produce normal-sized seeds. To determine whether increasing CLE8 activity is sufficient to enhance seed size, we generated CLE8-overexpressing plants under the control of a double cauliflower mosaic virus 35S promoter. We obtained multiple independent lines that displayed significantly increased levels of CLE8 mRNA expression (Figure 5A). We examined the endosperm of Pro35S:CLE8 seeds at early stages of development and detected no difference from wild-type endosperm in terms of proliferation or differentiation. Because of technical limitations, quantifying endosperm nuclei at later stages was not feasible, so it remains an open question whether CLE8 overexpression can induce excess endosperm proliferation. Nonetheless, we observed that Pro35S:CLE8 plants contained 6 to 11.5% longer mature embryos than wild-type plants (Figures 5B and 5C) and produced 6 to 7% larger seeds (Figures 5D and 5E). These data indicate that Arabidopsis seed size is positively influenced by the level of the CLE8 signal.
Figure 5.
Pro35S:CLE8 Seed Phenotypes.
(A) CLE8 mRNA transcription levels in wild-type and three independent Pro35S:CLE8 transgenic lines determined by real-time quantitative RT-PCR. Graph values are means of three biological replicates ± se. All experimental values are statistically significant (Student’s t test, P < 0.05).
(B) and (C) Mature wild-type (B) and Pro35S:CLE8 transgenic (C) seeds.
(D) and (E) Length (D) and width (E) of wild-type and Pro35S:CLE8 seeds from three independent transgenic lines. Graph values are means ± se, * indicates statistically significant values (n = 100, Student’s t test, P < 0.01).
Bars = 50 μm.
[See online article for color version of this figure.]
A CLE8-WOX8 Signaling Pathway Functions in Suspensor and Endosperm Development
Although little is known about the molecular targets of CLE signal transduction pathways, several Arabidopsis CLE peptides act upstream of WOX family transcription factor genes (Brand et al., 2000; Stahl et al., 2009; Hirakawa et al., 2010). WOX8 and WOX9 function redundantly during seed development to regulate patterning and cell proliferation in the embryo. wox8 wox9 embryos form irregular cell division planes or enlarged, misshapen cells in the suspensor (Wu et al., 2007; Breuninger et al., 2008), phenotypes strikingly similar to those of cle8-1 embryos. We focused on WOX8 as a likely CLE8 target gene candidate, because it is the only WOX family member expressed in both the endosperm and the embryo basal lineage after the zygotic division (Haecker et al., 2004; Breuninger et al., 2008).
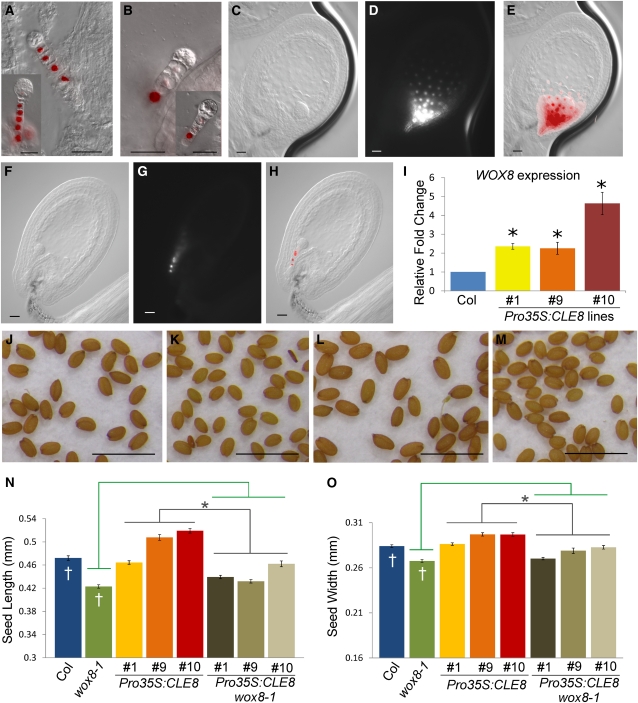
We analyzed WOX8 expression in cle8-1 seeds using a WOX8gΔ:NLS-venusYFP3 marker line that recapitulates the endogenous WOX8 expression pattern (Breuninger et al., 2008). In early-stage wild-type embryos, WOX8 is expressed in all suspensor cells except the hypophysis (Figure 6A) and, at later stages, is also detected in the hypophysis (Figure 6A, inset). In cle8-1 seeds, we observed strong WOX8 expression only in the most basal suspensor cell, whereas its expression in the other suspensor cells was dramatically reduced or absent (Figure 6B). To exclude the possibility that reduced CLE8 function generally perturbs gene expression levels in suspensor cells, we analyzed ProWOX2:NLS-venusYFP3 expression and observed no significant difference between wild-type and cle8-1 seeds (see Supplemental Figure 1 online). Therefore, CLE8 acts upstream of WOX8 and is required for high levels of WOX8 transcription in the upper suspensor cells.
Figure 6.
WOX8 Activity in cle8-1 and Pro35S:CLE8 Plants.
(A) to (H) Merged Nomarski and fluorescence images of wild-type ([A] and [C] to [E]) and cle8-1 ([B] and [F] to [H]) seeds expressing WOX8gΔ:NLS-venusYFP3. Insets show slightly older embryos displaying the same expression patterns.
(I) WOX8 mRNA transcription levels in three independent Pro35S:CLE8 transgenic lines. Graph values are means of three biological replicates ± se, * indicates statistically significant values (Student’s t test, P < 0.05).
(J) to (M) Wild-type (J), wox8-1 (K), Pro35S:CLE8 (L), and Pro35S:CLE8 wox8-1 (M) seeds.
(N) and (O) Length (N) and width (O) of wild-type, wox8-1, Pro35S:CLE8, and Pro35S:CLE8 wox8-1 seeds from three independent transgenic lines. * indicates that the values of the Pro35S:CLE8 wox8-1 seeds are significantly different (n = 100, Student’s t test, P < 0.05) from those of the Pro35S:CLE8 seeds (gray bars) but not from those of the wox8-1 seeds (green bars). In this generation, Pro35S:CLE8 line 1 transgenic plants were silenced (n = 100).
Bars in (A) to (H) = 20 μm; bars in (J) to (M) = 1 mm.
In early-stage wild-type seeds, WOX8 was also strongly expressed in the ESR endosperm cells (Figures 6C to 6E). However, in cle8-1 seeds with wild-type endosperm morphology, WOX8 expression was restricted to a few cells of the micropylar endosperm (Figures 6F to 6H). This reveals that CLE8 establishes the proper gene expression programs in both the embryo basal lineage and the ESR endosperm. Conversely, CLE8-overexpressing lines showed a significant elevation of WOX8 mRNA levels (Figure 6I), demonstrating that CLE8 is sufficient to induce WOX8 expression. The biological relevance of WOX8 regulation by CLE8 was demonstrated by the observations that wox8-1 seeds were smaller than wild-type seeds (Figures 6J, 6K, 6N, and 6O) and that the wox8-1 mutation suppressed the enlarged seed phenotype of CLE8-overexpressing lines (Figures 6L to 6O). Together, these data indicate that CLE8 and WOX8 form a signaling module that regulates suspensor and endosperm development, with CLE8 acting upstream in the pathway to positively regulate WOX8 and promote seed growth.
DISCUSSION
Angiosperm seed development is a complex process that requires the coordinated growth and development of the embryo, endosperm, and seed coat tissues. Here we have shown that in wild-type Arabidopsis plants, CLE8 peptide is produced by the embryo proper and the endosperm, and that it is involved in essential signaling–mediated regulation of transcriptional programs that regulate fundamental cell proliferation and cell division processes. CLE8 is the only Arabidopsis CLE gene whose expression is restricted to the developing seed. We detect CLE8 mRNA throughout the embryo proper from the one-cell stage to the early heart stage, making it, to our knowledge, the earliest expressed polypeptide signaling molecule identified to date (De Smet et al., 2010; Katsir et al., 2011). Previous work indicated that CLE genes are generally not expressed at high levels (Sharma et al., 2003), and we likewise observed low GUS activity in ProCLE8:GUS transgenic seeds. Nonetheless, the pattern of GUS activity driven by CLE8 promoter sequences that complement the cle8-1 phenotype confirms the in situ analysis and demonstrates that CLE8 is expressed in both young embryos and in the endosperm. By contrast, CLE8 expression is low in or absent from the suspensor cells.
Most Arabidopsis CLE genes lack introns and consist of small open reading frames of 240 to 360 nucleotides (Cock and McCormick, 2001; Jun et al., 2008), making them small targets for mutagenesis. To date, no CLE8 insertion mutants have been identified. We were able to obtain an ethyl methanesulfonate–induced cle8-1 allele via TILLING that altered a conserved amino acid in the CLE domain. The seed phenotype caused by the cle8-1 missense mutation shows incomplete penetrance and is indistinguishable from that conditioned by our CLE8 amiRNA construct, which substantially reduces but does not completely abolish CLE8 expression. The most parsimonious interpretation of these results is that cle8-1 represents a hypomorphic, partial loss-of-function allele. Residual activity of cle8-1 mutant peptide may allow most cle8-1 seeds to develop normally, explaining why only 15% display aberrant phenotypes. However, we cannot rule out that cle8-1 is a null allele that produces a nonfunctional protein, in which case other embryo-expressed CLE genes (Fiers et al., 2004; Fiume et al., 2011) may act partially redundantly with CLE8.
Although the CLE proteins constitute the largest known family of plant-specific signaling peptides, only a few members have been assigned a clear function. Our study reveals a seed-specific role for CLE8 in regulating the development of the embryo proper, the suspensor, and the endosperm. We observe that nearly one-quarter of cle8 aberrant seeds contain defective endosperm. In most of these seeds, endosperm development initiates but fails to proliferate beyond the globular stage of embryogenesis. In other instances, the distribution of the free endosperm nuclei in the embryo sac lacks organization, and the nuclei are enlarged. Analysis of the early endosperm-specific marker ProMEA:GUS indicates that early-stage cle8 seeds displayed a GUS staining pattern similar to that of later-stage wild-type seeds (Luo et al., 2000), suggesting that cle8 endosperm undergoes a faster rate of maturation. This premature endosperm differentiation may limit nutrient availability to the developing embryo.
We additionally find that CLE8 regulates cell division and proliferation in the embryo proper as well as cell elongation and patterning in the suspensor. Most cle8 aberrant seeds show defects only in basal embryo cells, although CLE8 transcripts accumulate throughout the embryo proper, from the one-cell stage until the heart stage. This suggests either that completely eliminating CLE8 function is required to affect the apical embryo domain or that other CLE genes act redundantly with CLE8 in apical embryo cells. The identification of null alleles in CLE8 and other embryo-expressed CLE genes will be necessary to resolve this issue. It has been observed that some mutations that cause embryo defects also lead to abnormalities in the suspensor (Hamann et al., 1999; Lukowitz et al., 2004; Bureau et al., 2010). It is therefore possible that the cle8 suspensor phenotype may be a secondary effect of a primary embryo defect, although both are a consequence of reduced CLE8 function.
Our results from loss-of-function and gain-of-function analyses demonstrate that CLE8 has a qualitative, positive effect on seed size. cle8 seeds are smaller on average than wild-type seeds, even those in which the embryos inside are morphologically normal. We observed that in cle8 seeds, the endosperm fails to sufficiently proliferate and seems to undergo accelerated maturation. Endosperm proliferation and growth are key determinants of embryo size and ultimately seed size (Garcia et al., 2005; Kang et al., 2008), and therefore reduced proliferation and premature differentiation of the endosperm is likely to be one cause of the cle8 small seed phenotype. We note that CLE8 expression during later stages of wild-type seed development is restricted to the chalazal endosperm, the compartment responsible for nutrient uptake from the maternal tissue (Costa et al., 2004). CLE8 activity in this compartment may be important to positively regulate nutrient uptake at the maternal–filial interface and may indirectly promote embryo growth. Conversely, transgenic Pro35S:CLE8 seeds are significantly larger than wild-type seeds. In this case, the embryos within them are clearly larger than wild-type embryos, whereas early endosperm development seems to proceed normally. Given that CLE8 acts to promote proliferation and prevent premature differentiation of the endosperm, we predict that broader and more prolonged CLE8 expression may delay endosperm maturation and cause it to overproliferate at later developmental stages, thereby supporting excess embryo growth. These specific effects on endosperm and embryo development make CLE8 an important determinant of seed size.
Many studies have shown that ectopic overexpression of almost every CLE gene results in the termination of the shoot and/or root meristems or other pleiotropic phenotypes (reviewed in Jun et al., 2008, 2010). These data imply that multiple CLE peptides have the capacity to activate the same receptors, and thus that the tissue distribution of CLE gene expression may be an important factor in determining the specificity of CLE function. Interestingly, CLE8 behaves differently than other CLE genes in this respect. Ectopic CLE8 expression does not generate any aberrant phenotypes in tissues other than seeds, where it is normally transcribed. Because CLE8 is the only member of the family whose expression is restricted to the seed (Sharma et al., 2003; Jun et al., 2010), it can be speculated that CLE8 evolved to specifically fulfill a role in seed development and that it diverged from other CLE proteins to exclusively interact with its receptor(s). Therefore, top candidates for a CLE8 cognate receptor(s) would be those that are specifically and uniquely expressed in the developing seed.
Finally, it has been well established that signaling pathways comprising CLE polypeptides and WOX transcription factors regulate aspects of shoot, root, and vascular meristem function. Our findings that CLE8 signaling positively regulates WOX8 expression in endosperm and suspensor cells and that WOX8 is required to condition the CLE8 overexpression phenotype indicates that CLE-WOX regulatory module activity extends beyond the meristems and likely has been repeatedly recruited during evolution to regulate a variety of developmental processes. It is important to emphasize that downregulation of the WOX8gΔ:NLS-venusYFP3 marker in cle8 seeds was also observed in morphologically wild-type–looking suspensors, indicating that it is not merely a secondary consequence of a physical defect, but rather an informative molecular characteristic of cle8 seeds. Interestingly, WOX8 is a target of a cell-autonomous CLE8 function in the endosperm as well as potentially also a noncell autonomous function in the suspensor. A role for WOX8 in endosperm formation has not been defined, but because expression and functional studies suggest that WOX genes are generally involved in promoting cell division and/or preventing differentiation (van der Graaff et al., 2009), it is reasonable to speculate that WOX8 might play a role in promoting endosperm proliferation. WOX8 regulates embryo and suspensor development by activating WOX2, which in turn induces expression of the auxin transport gene PINFORMED1 (PIN1) and formation of auxin gradients during early embryogenesis (Breuninger et al., 2008). Thus, in embryos, CLE8 functions as a key upstream component of a regulatory cascade that drives hormone-mediated developmental patterning.
METHODS
Plant Material
All Arabidopsis thaliana genotypes were in the Col accession. Plants were grown either on Murashige and Skoog medium or on soil (1:1:1 mixture of perlite:vermiculite:topsoil), under continuous light (120 μmol·m−2·s−1) at 22°C. Seeds were stratified at 4°C for 4 d prior to light exposure. Prior to in vitro culture, seeds were surface-sterilized for 40 min with chlorine gas. Control seeds were collected and grown side by side with experimental seeds for each experiment.
Plant Transformation
Agrobacterium tumefaciens strain GV3101 was used to stably transform Arabidopsis plants by the floral dip method (Clough and Bent, 1998). Transient transformation was performed by particle bombardment of onion epidermis cells using a Biolistic PDS-1000/He unit (Bio-Rad), as previously described (Sanford et al., 1993).
Cloning
PCR-amplified products were cloned into the pENTR/D-TOPO vector (Invitrogen) and then moved into the appropriate destination vector from the pMDC series (Curtis and Grossniklaus, 2003) or pBGWFS7 (Karimi et al., 2002) through an LR reaction (Invitrogen), according to the manufacturer’s instructions. The CLE8 promoter sequence, consisting of 2959 bp from the stop codon of the nearest upstream gene to the CLE8 start codon, was PCR-amplified and cloned into the binary vector pBGWFS7 for expression analysis. The CLE8 coding sequence, which does not contain introns, was PCR-amplified and cloned into the binary vector pMDC32 under the control of a double cauliflower mosaic virus 35S promoter for overexpression analysis. An Arabidopsis genomic sequence consisting of the CLE8 promoter described above and the CLE8 coding region was PCR-amplified and cloned into the binary vector pMDC107 for cle8-1 complementation analysis. The CLE8 amiRNA was designed using the publicly available online tool WMD3 (http://wmd3.weigelworld.org) and cloned into the binary vector pMDC32. CLE8 mRNA transcripts were PCR-amplified and cloned into the pGEM-T Easy vector (Promega) for in vitro transcription of an in situ hybridization probe. Primer sequences are listed in Supplemental Table 3 online.
Plant DNA and RNA Extraction
Preparation of genomic DNA for PCR analysis was conducted as previously described (Edwards et al., 1991). Genomic DNA was extracted for cloning using the DNeasy Plant Kit (Qiagen). Total RNA isolated using the RNeasy Plant Kit (Qiagen) was treated with RNase-free DNase (Roche) for 30 min at 37°C and then purified with phenol/chloroform.
RT-PCR and Quantitative PCR
The first-strand cDNA synthesis was performed on 1 μg of total RNA using Superscript III RNase H- reverse transcriptase (Invitrogen), according to the manufacturer's instructions. The annealing temperature was 58 to 60°C for all primer pairs. For the RT-PCR amplification from the amiRNA lines, 30 cycles of PCR were used to amplify CLE8 from all samples, and 21 cycles of PCR were used to amplify ACT2 from all samples. Quantification of CLE8 and WOX8 cDNAs by real-time PCR was performed using the SYBR Green PCR master mix (Applied Biosystems) and a MyIQ Thermal Cycler (Bio-Rad). Three technical replicates were run for each sample. The specificity of the amplification was determined by performing a dissociation curve analysis. Relative quantification values were calculated using the 2−ΔCt method (Livak and Schmittgen, 2001). The Ct was calculated using TUB4 as the endogenous control. Three biological replicates were performed for each genotype. Primer sequences are listed in Supplemental Table 3 online.
RNA in Situ Hybridization
RNA in situ hybridization analyses were conducted as described in Carles et al. (2010), using the CLE8 full-length coding sequence as the antisense probe.
GUS Histochemical Assays
GUS activity was assayed as described in Jefferson (1989), with the modification that 1 mM potassium ferrocyanide and 1 mM potassium ferricyanide were used. Whole mount incubation times ranged from 8 to 12 h after vacuum infiltration. Subsequent tissue embedding and sectioning were performed as previously described (Sieburth and Meyerowitz, 1997). To stain isolated young embryos, developing seeds were placed in 100 μL of staining solution on a microscope slide. A cover slip was put over the slide, and pressure was applied to squeeze the embryos out of the seed coats. The slides were incubated overnight at 37°C in a tight container to prevent drying.
Microscopy
Nomarski microscopy analysis was performed by placing seeds in a chloral hydrate solution (containing 1:8:3 glycerol:chloral hydrate:water). After a 16-h incubation at room temperature, the seeds were placed onto slides and visualized on a Zeiss Imager A1 microscope equipped with differential interference contrast optics. For fluorescent analysis, ovules were placed on slides containing 10% glycerin and analyzed using the Zeiss filter set 46HE for yellow fluorescent protein. Mature dry seeds were digitally photographed using a Canon D-40 camera attached to a Zeiss Stemi SV11 dissecting microscope. Seed length and width were measured using National Center for Biotechnology Information Image J software for image processing and analysis (http://rsbweb.nih.gov/ij/).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: CLE8, At1g67775; WOX2, At5g59340; WOX8, At5g45980.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. WOX2 Expression Analysis.
Supplemental Table 1. Seed Germination Defect.
Supplemental Table 2. Percentage of Seeds Showing Morphological Defects.
Supplemental Table 3. Primers Used in the Experimental Procedures.
Acknowledgments
We thank Robert Fischer, Frederic Berger, Frans Tax, Adriana Racolta, Enrico Magnani, Mona Monfared, Robert Blanvillain, and Cristel Carles for helpful comments and discussions, Thomas Laux and Frederic Berger for marker lines, and Minna Mahonen for assistance with genotyping. This article was funded by grants to J.C.F. from the National Science Foundation (Molecular and Cellular Biosciences 0313546) and the U.S. Department of Agriculture–Agricultural Research Service (Current Research Information System 5335-21000-029-00D).
AUTHOR CONTRIBUTIONS
E.F. and J.C.F. designed the research; E.F. performed the research; E.F. and J.C.F. analyzed the data and wrote the article.
References
- Bayer M., Nawy T., Giglione C., Galli M., Meinnel T., Lukowitz W. (2009). Paternal control of embryonic patterning in Arabidopsis thaliana. Science 323: 1485–1488 [DOI] [PubMed] [Google Scholar]
- Berger F., Grini P.E., Schnittger A. (2006). Endosperm: An integrator of seed growth and development. Curr. Opin. Plant Biol. 9: 664–670 [DOI] [PubMed] [Google Scholar]
- Brand U., Fletcher J.C., Hobe M., Meyerowitz E.M., Simon R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617–619 [DOI] [PubMed] [Google Scholar]
- Breuninger H., Rikirsch E., Hermann M., Ueda M., Laux T. (2008). Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Dev. Cell 14: 867–876 [DOI] [PubMed] [Google Scholar]
- Bureau M., Rast M.I., Illmer J., Simon R. (2010). JAGGED LATERAL ORGAN (JLO) controls auxin dependent patterning during development of the Arabidopsis embryo and root. Plant Mol. Biol. 74: 479–491 [DOI] [PubMed] [Google Scholar]
- Carles C.C., Ha C.M., Jun J.H., Fiume E., Fletcher J.C. (2010). Analyzing shoot apical meristem development. In Methods in Molecular Biology, Hennig L., Kohler C., (Totowa, NJ: Humana Press; ), pp. 105–129 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cock J.M., McCormick S. (2001). A large family of genes that share homology with CLAVATA3. Plant Physiol. 126: 939–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa L.M., Gutièrrez-Marcos J.F., Dickinson H.G. (2004). More than a yolk: The short life and complex times of the plant endosperm. Trends Plant Sci. 9: 507–514 [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I., Lau S., Mayer U., Jürgens G. (2010). Embryogenesis - the humble beginnings of plant life. Plant J. 61: 959–970 [DOI] [PubMed] [Google Scholar]
- Dumas C., Rogowsky P.M. (2008). Fertilization and early seed formation. C. R. Biol. 331: 715–725 [DOI] [PubMed] [Google Scholar]
- Edwards K., Johnstone C., Thompson C. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19: 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchells J.P., Turner S.R. (2010). The PXY-CLE41 receptor ligand pair defines a multifunctional pathway that controls the rate and orientation of vascular cell division. Development 137: 767–774 [DOI] [PubMed] [Google Scholar]
- Fiers M., Golemiec E., Xu J., van der Geest L., Heidstra R., Stiekema W., Liu C.-M. (2005). The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. Plant Cell 17: 2542–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers M., Hause G., Boutilier K., Casamitjana-Martinez E., Weijers D., Offringa R., van der Geest L., van Lookeren Campagne M., Liu C.-M. (2004). Mis-expression of the CLV3/ESR-like gene CLE19 in Arabidopsis leads to a consumption of root meristem. Gene 327: 37–49 [DOI] [PubMed] [Google Scholar]
- Fiume E., Monfared M., Jun J.H., Fletcher J.C. (2011). CLE polypeptide signaling gene expression in Arabidopsis embryos. Plant Signal. Behav. 6: 443–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher J.C., Brand U., Running M.P., Simon R., Meyerowitz E.M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283: 1911–1914 [DOI] [PubMed] [Google Scholar]
- Garcia D., Fitz Gerald J.N., Berger F. (2005). Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell 17: 52–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Han L., Hymes M., Denver R., Clark S.E. (2010). CLAVATA2 forms a distinct CLE-binding receptor complex regulating Arabidopsis stem cell specification. Plant J. 63: 889–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haecker A., Gross-Hardt R., Geiges B., Sarkar A., Breuninger H., Herrmann M., Laux T. (2004). Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 131: 657–668 [DOI] [PubMed] [Google Scholar]
- Hamann T., Mayer U., Jürgens G. (1999). The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development 126: 1387–1395 [DOI] [PubMed] [Google Scholar]
- Hirakawa Y., Kondo Y., Fukuda H. (2010). TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. Plant Cell 22: 2618–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa Y., Shinohara H., Kondo Y., Inoue A., Nakanomyo I., Ogawa M., Sawa S., Ohashi-Ito K., Matsubayashi Y., Fukuda H. (2008). Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc. Natl. Acad. Sci. USA 105: 15208–15213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingouff M., Jullien P.E., Berger F. (2006). The female gametophyte and the endosperm control cell proliferation and differentiation of the seed coat in Arabidopsis. Plant Cell 18: 3491–3501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y., Nakanomyo I., Motose H., Iwamoto K., Sawa S., Dohmae N., Fukuda H. (2006). Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 313: 842–845 [DOI] [PubMed] [Google Scholar]
- Jefferson R.A. (1989). The GUS reporter gene system. Nature 342: 837–838 [DOI] [PubMed] [Google Scholar]
- Jun J.H., Fiume E., Fletcher J.C. (2008). The CLE family of plant polypeptide signaling molecules. Cell. Mol. Life Sci. 65: 743–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun J.H., Fiume E., Roeder A.H.K., Meng L., Sharma V.K., Osmont K.S., Baker C., Ha C.M., Meyerowitz E.M., Feldman L.J., Fletcher J.C. (2010). Comprehensive analysis of CLE polypeptide signaling gene expression and overexpression activity in Arabidopsis. Plant Physiol. 154: 1721–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang I.H., Steffen J.G., Portereiko M.F., Lloyd A., Drews G.N. (2008). The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell 20: 635–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Katsir L., Davies K.A., Bergmann D.C., Laux T. (2011). Peptide signaling in plant development. Curr. Biol. 21: R356–R364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita A., Betsuyaku S., Osakabe Y., Mizuno S., Nagawa S., Stahl Y., Simon R., Yamaguchi-Shinozaki K., Fukuda H., Sawa S. (2010). RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development 137: 3911–3920 [DOI] [PubMed] [Google Scholar]
- Kondo T., Sawa S., Kinoshita A., Mizuno S., Kakimoto T., Fukuda H., Sakagami Y. (2006). A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313: 845–848 [DOI] [PubMed] [Google Scholar]
- Kondou Y., Nakazawa M., Kawashima M., Ichikawa T., Yoshizumi T., Suzuki K., Ishikawa A., Koshi T., Matsui R., Muto S., Matsui M. (2008). RETARDED GROWTH OF EMBRYO1, a new basic helix-loop-helix protein, expresses in endosperm to control embryo growth. Plant Physiol. 147: 1924–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laux T., Mayer K.F.X., Berger J., Jürgens G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122: 87–96 [DOI] [PubMed] [Google Scholar]
- Lease K.A., Walker J.C. (2006). The Arabidopsis unannotated secreted peptide database, a resource for plant peptidomics. Plant Physiol. 142: 831–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lukowitz W., Roeder A.H.K., Parmenter D., Somerville C. (2004). A MAPKK kinase gene regulates extra-embryonic cell fate in Arabidopsis. Cell 116: 109–119 [DOI] [PubMed] [Google Scholar]
- Luo M., Bilodeau P., Dennis E.S., Peacock W.J., Chaudhury A. (2000). Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc. Natl. Acad. Sci. USA 97: 10637–10642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Dennis E.S., Berger F., Peacock W.J., Chaudhury A. (2005). MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc. Natl. Acad. Sci. USA 102: 17531–17536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum C.M., Comai L., Greene E.A., Henikoff S. (2000). Targeting induced local lesions IN genomes (TILLING) for plant functional genomics. Plant Physiol. 123: 439–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J., Clark S.E. (2006). Evidence for functional conservation, sufficiency, and proteolytic processing of the CLAVATA3 CLE domain. Plant Physiol. 140: 726–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodine M.D., Bryan A.C., Racolta A., Jerosky K.V., Tax F.E. (2011). A few standing for many: Embryo receptor-like kinases. Trends Plant Sci. 16: 211–217 [DOI] [PubMed] [Google Scholar]
- Ogawa M., Shinohara H., Sakagami Y., Matsubayashi Y. (2008). Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319: 294. [DOI] [PubMed] [Google Scholar]
- Ohyama K., Shinohara H., Ogawa-Ohnishi M., Matsubayashi Y. (2009). A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat. Chem. Biol. 5: 578–580 [DOI] [PubMed] [Google Scholar]
- Ossowski S., Schwab R., Weigel D. (2008). Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J. 53: 674–690 [DOI] [PubMed] [Google Scholar]
- Sanford J.C., Smith F.D., Russell J.A. (1993). Optimizing the biolistic process for different biological applications. Methods Enzymol. 217: 483–509 [DOI] [PubMed] [Google Scholar]
- Saulsberry A., Martin P.R., O’Brien T., Sieburth L.E., Pickett F.B. (2002). The induced sector Arabidopsis apical embryonic fate map. Development 129: 3403–3410 [DOI] [PubMed] [Google Scholar]
- Schoof H., Lenhard M., Haecker A., Mayer K.F.X., Jürgens G., Laux T. (2000). The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644 [DOI] [PubMed] [Google Scholar]
- Sharma V.K., Ramirez J., Fletcher J.C. (2003). The Arabidopsis CLV3-like (CLE) genes are expressed in diverse tissues and encode secreted proteins. Plant Mol. Biol. 51: 415–425 [DOI] [PubMed] [Google Scholar]
- Sieburth L.E., Meyerowitz E.M. (1997). Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell 9: 355–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl Y., Wink R.H., Ingram G.C., Simon R. (2009). A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr. Biol. 19: 909–914 [DOI] [PubMed] [Google Scholar]
- Strabala T.J., O’Donnell P.J., Smit A.-M., Ampomah-Dwamena C., Martin E.J., Netzler N., Nieuwenhuizen N.J., Quinn B.D., Foote H.C.C., Hudson K.R. (2006). Gain-of-function phenotypes of many CLAVATA3/ESR genes, including four new family members, correlate with tandem variations in the conserved CLAVATA3/ESR domain. Plant Physiol. 140: 1331–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Graaff E., Laux T., Rensing S.A. (2009). The WUS homeobox-containing (WOX) protein family. Genome Biol. 10: 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D., Van Hamburg J.-P., Van Rijn E., Hooykaas P.J., Offringa R. (2003). Diphtheria toxin-mediated cell ablation reveals interregional communication during Arabidopsis seed development. Plant Physiol. 133: 1882–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford R., Fernandez A., De Groodt R., Ortega E., Hilson P. (2008). Plant CLE peptides from two distinct functional classes synergistically induce division of vascular cells. Proc. Natl. Acad. Sci. USA 105: 18625–18630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Chory J., Weigel D. (2007). Combinations of WOX activities regulate tissue proliferation during Arabidopsis embryonic development. Dev. Biol. 309: 306–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav R.K., Perales M., Gruel J., Girke T., Jönsson H., Reddy G.V. (2011). WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev. 25: 2025–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]