The cold dependency of pigment formation in blood orange constitutes a major limitation on production worldwide and is due to the cold induction of retrotransposons controlling this gain-of-function trait.
Abstract
Traditionally, Sicilian blood oranges (Citrus sinensis) have been associated with cardiovascular health, and consumption has been shown to prevent obesity in mice fed a high-fat diet. Despite increasing consumer interest in these health-promoting attributes, production of blood oranges remains unreliable due largely to a dependency on cold for full color formation. We show that Sicilian blood orange arose by insertion of a Copia-like retrotransposon adjacent to a gene encoding Ruby, a MYB transcriptional activator of anthocyanin production. The retrotransposon controls Ruby expression, and cold dependency reflects the induction of the retroelement by stress. A blood orange of Chinese origin results from an independent insertion of a similar retrotransposon, and color formation in its fruit is also cold dependent. Our results suggest that transposition and recombination of retroelements are likely important sources of variation in Citrus.
INTRODUCTION
In addition to their striking color, blood oranges are believed to have significant health-promoting properties, combining the high content of vitamin C, carotenoids, and fiber of common blond oranges with the health-promoting properties of anthocyanin pigments (Prior and Wu, 2006; Davies, 2007; de Pascual-Teresa et al., 2010; Paredes-López et al., 2010). The high anthocyanin content of blood oranges underpins their high antioxidant activity (Rapisarda et al., 1999; Jayaprakasha and Patl, 2007; Kelebek et al., 2008; Proteggente et al., 2003). Consumption of blood orange juice has been shown to reduce oxidative stress in diabetic patients (Bonina et al., 2002), protect DNA against oxidative damage (Riso et al., 2005; Guarnieri et al., 2007) and may reduce cardiovascular risk factors more generally, as demonstrated for other high-anthocyanin foods (Toufektsian et al., 2008; de Pascual-Teresa et al., 2010; Paredes-López et al., 2010). Recently, blood orange juice has been shown to limit the development of adipocytes and weight gain in mice and to confer resistance to obesity compared with blond orange juice or water (Titta et al., 2010). In a mouse model of obesity, blood orange juice consumption rescued almost completely the transcriptional reprogramming induced by a high-fat diet.
Despite increasing consumer interest in their high nutritional quality, blood oranges do not have a global market, largely due to a lack of dependability of color development. All blood orange varieties require strong day-night thermal clines for intense color formation in fruit flesh, and varieties such as Moro, with the potential for high pigmentation, are strongly dependent on the prevailing climatic conditions during fruit ripening for full color development. Postharvest storage of fruit in the cold enhances pigmentation, but this is an expensive measure to ensure high levels of pigmentation and can increase postharvest losses (Rapisarda et al., 1999; Latado et al., 2008; Crifò et al., 2011). The dependency of anthocyanin accumulation on environment means that the most reliable blood orange production, on a commercial scale, is limited to Italy, specifically to the Sicilian area around Mount Etna (Zarba and Pulvirenti, 2006) where it remains highly seasonal. Although blood oranges are grown in other countries, in some years entire harvests are lost due to nonoptimal conditions during ripening of fruit, and when they are cultivated in Brazil or Florida (the largest producers of oranges worldwide), coloration is generally weak or absent and unreliable (Hodgson, 1967; Latado et al., 2008).
Anthocyanins are natural pigments found typically in red, purple, and blue fruit as well as flowers (Winkel-Shirley, 2001). Most modern varieties of blood orange have been derived from old Italian varieties, such as Doppio Sanguigno, and include more recently derived varieties, such as Tarocco and Moro, which generally have higher levels of anthocyanin pigmentation of their fruit (Figure 1). The history of these varieties is debated, although authoritative texts suggest three independent derivations: one Italian/Sicilian from Doppio Sanguigno/Maltaise Sanguine, a second in Spain from Doblefina, and a third from Shamouti Orange referred to as Shamouti Blood or Palestinian Blood Jaffa Orange (Hodgson, 1967). In the mid 19th century, it was believed that blood oranges arose by bud mutation in the Mediterranean region following the introduction of sweet orange (Citrus sinensis) in the 16th century (Holmes, 1924). More recently, it was suggested that blood oranges originated much earlier in Asia (Chapot, 1963; Hodgson, 1967). The blood orange was first documented in Italy in ‘Hesperides’ by Ferrari (1646), a Jesuit scholar who wrote of an orange with purple-colored flesh that tasted strangely like a grape (Ferrari, 1646). Ferrari suggested that the blood orange was brought to Sicily by a Genoese missionary after a long journey that started in China. However, claims of Chinese poems referring to scarlet/red oranges, dating from the Tang period to more recent times, are probably based on mistranslation of the term “orange” and likely refer to mandarins, which never accumulate anthocyanins. Some definitively red oranges are portrayed in a picture by Bartolomeo Bimbi, a Florentine artist who painted the Medici Citrus collections early in the 18th century (see Supplemental Figure 1 online).
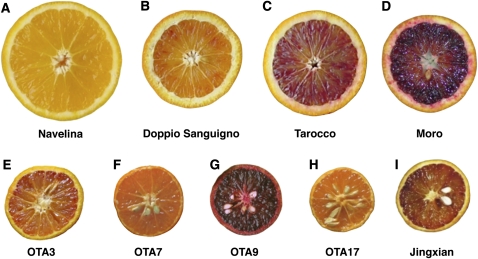
Figure 1.
Phenotypes and Genotypes of Orange Varieties and Hybrids.
Navelina R, r-2 (A); Doppio Sanguigno RD-1, r-2 (B); Tarocco (CRA) RD-1, r-2 (C); Moro (CRA) RD-2, r-2 (D); OTA3 RD-2, r-2 (E); OTA7 r-2, r-2 (F); OTA9 RD-2, r-1 (G); OTA17 r-1, r-2 (H); and Jingxian RD-3, r-2 (I).
RESULTS
Identification of a Gene Encoding an R2R3 MYB Transcription Factor Expressed in the Fruit of Blood Orange Varieties
Anthocyanin biosynthesis is regulated mainly at the transcriptional level (Winkel-Shirley, 2001). A regulatory complex, composed of proteins of the Myb, basic helix-loop-helix (bHLH), and WD-repeat families of transcription factors, governs the expression of the structural genes required for anthocyanin biosynthesis, modification, and transport (Ramsay and Glover, 2005; Butelli et al., 2008). In blood orange, several anthocyanin biosynthetic genes show increased expression compared with blond orange (Licciardello et al., 2008; Bernardi et al., 2010; Cultrone et al., 2010). Variation in pigment intensity or tissue specificity in plants is governed largely by the activity of the R2R3 Myb transcription factors in the complex (Kobayashi et al., 2004; Schwinn et al., 2006; Takos et al., 2006; Espley et al., 2007; Geekiyanage et al., 2007; Walker et al., 2007). Therefore, a partial cDNA fragment encoding the conserved Myb DNA binding domain, typical of the R2R3 Myb regulators of anthocyanin biosynthesis, was isolated from RNA from Moro flesh using degenerate PCR. This fragment was extended using 5′ and 3′ rapid amplification of cDNA ends (RACE) PCR on cDNA prepared from the flesh of Moro fruit, to obtain a full-length cDNA and to map the start of transcription. We were unable to amplify an equivalent cDNA from fruit of common blond oranges and found no ESTs for the gene in the database collections. We named this R2R3 Myb gene Ruby.
The Ruby cDNA encodes a 262–amino acid protein containing an R2R3 Myb domain with a signature motif for interaction with bHLH proteins from the clade 3f (DLX2RX3LX6LX3R; Heim et al., 2003; Zimmermann et al., 2004; Lin-Wang et al., 2010; Figure 2A). It also has a conserved sequence motif KPXPR(S/T)F in its C-terminal domain found in other R2R3 Myb regulators of anthocyanin biosynthesis (Stracke et al., 2001; Lin-Wang et al., 2010) Phylogenetic analysis revealed that Ruby clusters with members of R2R3 Myb subgroup 6, known to regulate anthocyanin biosynthesis in dicotyledonous plants (Bailey et al., 2008; Lin-Wang et al., 2010; Figure 2B; see Supplemental Data Set 1 online).
Figure 2.
Features of the Protein Encoded by the Ruby Transcript.
(A) Protein alignment of Ruby with characterized members of the anthocyanin-specific family of Myb factors (http://multalin.toulouse.inra.fr/multalin/). Identical amino acids are shown in pink. The GenBank accession numbers of these proteins are as follows: Ph AN2 (Petunia × hybrida), AAF66727.1; Sl AN1 (tomato [Solanum lycopersicum]), AAQ55181.1; Vv MYBA1 (V. vinifera), ABB87013.1; Ib MYB1 (Ipomoea batatas), BAF45118.1; At PAP1 (Arabidopsis), ABB03877.1; and Am ROSEA1 (snapdragon), ABB83826.1. Arrowheads indicate the region to which degenerate primers were designed. The signature motif for interaction with bHLH proteins within the R3 Myb DNA binding domain and the conserved motif defining members of R2R3 Myb subgroup 6 are boxed in gray.
(B) Phylogenetic analysis showing that the Ruby transcription factor clusters with anthocyanin-specific members of subgroup 6 from the R2R3MYB family. Bootstrap values are shown if significant at or above the 90% confidence limit. Locus identifiers for Arabidopsis proteins are At MYB3 (At1g22640), At MYB4 (At4g38620), At MYB6 (At4g09460), At MYB7 (At2g16720), At MYB8 (At1g35515), At MYB13 (At1g06180), At MYB14 (At2g31180), At MYB15 (At3g23250), At MYB32 (At4g34990), At MYB11 (At3g62610), At MYB12 (At2g47460), At MYB111 (At5g49330), At MYB113 (At1g66370), At MYB114 (At1g66380), At MYB75 (At1g56650), At MYB90 (At1g66390), and At MYB123 (At5g35550). Identifiers for snapdragon proteins are Am MYB308 (P81393), Am MYB330 (P81395), Am ROSEA1 (DQ275529), Am ROSEA2 (DQ275530), and Am VENOSA (DQ275531). Identifiers for maize (Zea mays) proteins are Zm MYB42 (AM156908), Zm MYBC1 (X06333), Zm MYBP (U57002), and Zm MYBPL (L19494). Identifiers for grape proteins are Vv MYBF1 (FJ948477), Vv MYBA1 (AB097923), and Vv MYBA2 (AB097924). Identifiers for tobacco proteins are Nt MYB2 (AB028649) and Nt MYBJS1 (AB236951). Identifiers for tomato proteins are Sl MYB12 (EU419748) and Sl ANT1 (AY348870). Identifiers for other proteins are as follows: Cs Ruby from sweet orange (this study, GenBank/EMBL JN402334); Dc MYB1 from carrot (Daucus carota) (AB218778); Os MYB4 from rice (Oryza sativa) (Y11414); Na MYB8 from Nicotiana attenuata (GU451752); Eg MYB1 from Eucalyptus gunnii (AJ576024); Pt MYB14 from Pinus taeda (DQ399056); Sb MYBY1 from sorghum (Sorghum bicolor) (AY860968); and Ph AN2 from petunia (Petunia hybrida) (AF146702).
Ruby Is a Regulator of Anthocyanin Biosynthesis
The ability of Ruby to activate anthocyanin biosynthesis was verified by its ectopic expression under the control of the constitutive cauliflower mosaic virus (CaMV) 35S promoter in tobacco (Nicotiana tabacum), where it resulted in visible purple-red pigmentation in undifferentiated callus and in developed tissues of regenerated plants (Figures 3A and 3B). Pigmentation in leaves of adult plants was particularly strong in older, senescent leaves (see Supplemental Figure 2 online). Coexpression of Ruby with the Delila and Mutabilis genes encoding bHLH transcription factors of clade 3f from snapdragon (Antirrhinum majus) (Schwinn et al., 2006), known to interact with anthocyanin-specific Myb factors, enhanced pigmentation in tobacco (Figure 3C). Ruby promoted stronger pigmentation in combination with Mutabilis than with Delila, suggesting some selectivity in the ability of the Ruby MYB protein to interact with different bHLH partners (Figure 3D). These bioassays confirmed the functionality of Ruby as a regulator of anthocyanin biosynthesis. A cDNA encoding a clade 3f bHLH protein (Cs MYC2) has been identified in orange (Cultrone et al., 2010). We showed, using transfection assays, that this bHLH protein will induce expression from the promoters of anthocyanin biosynthetic genes in combination with the Rosea1 R2R3 Myb regulator of anthocyanin biosynthesis from snapdragon (see Supplemental Figure 3 online). These data support the idea that it is the expression of Ruby that limits anthocyanin biosynthesis in blood oranges because the Cs MYC2 gene (encoding the bHLH partner in the MBW complex) is expressed at detectable levels in both blond and blood oranges (Cultrone et al., 2010).
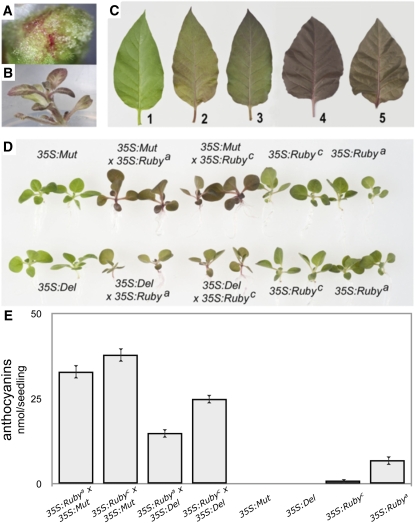
Figure 3.
Ruby Is a Regulator of Anthocyanin Biosynthesis.
(A) and (B) Constitutive expression of Ruby cDNA under the control of the 35S promoter in tobacco results in strong anthocyanin-based pigmentation in undifferentiated callus (A) and regenerating shoots (B).
(C) Coexpression of the bHLH regulatory genes Delila and Mutabilis from snapdragon increases further anthocyanin accumulation in young leaves: (1) 35S:Ruby, (2) 35S:Ruby × 35S:Delila, (3) 35S:Ruby × 35S:Mutabilis, (4) 35S:Rosea1 × 35S:Delila, and (5) 35S:Rosea1 × 35S:Mutabilis.
(D) Ruby partners more successfully with the bHLH protein Mutabilis than with the bHLH protein Delila to activate anthocyanin biosynthesis in tobacco. Anthocyanin accumulation in two seedlings of each genotype is shown. 35S:Rubya and 35S:Rubyc represent two independent transformants.
(E) Anthocyanin content in tobacco seedlings transformed with different regulatory genes (n = 9). Error bars show se of the mean.
Expression of Ruby in Blood and Blond Orange Accessions
In blood oranges, Ruby expression was limited to the fruit (Figure 4A). High levels of Ruby expression were detected in flesh and rind, including the white, spongy inner lining (albedo). Expression of Ruby was not detected in blond oranges (Figure 4A).
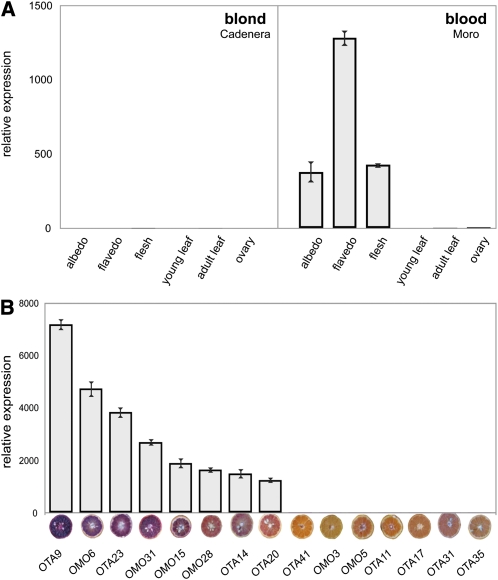
Figure 4.
Expression Analysis of Ruby.
(A) Expression of Ruby in different tissues of blond Cadenera and blood Moro oranges. Albedo is the spongy white layer of the orange peel. The flavedo is the peripheral surface of the pericarp.
(B) Expression of Ruby in diploid hybrids obtained crossing clementine (C. clementina cv Oroval) with C. sinensis cv Moro (OMO series) or C. sinensis cv Tarocco (OTA series). Hybrids lacking anthocyanins (OMO41, 3, and 5; OTA11, 17, 31, and 35) appear slightly different colors due to varying carotenoid levels in the fruit flesh. Error bars show se of the mean.
We tested the levels of Ruby expression in different accessions of blood orange and in hybrids between clementine and Moro (OMO) and clementine and Tarocco (OTA), produced in the experimental orchard of the Centro di Ricerca per l'Agrumicoltura e le Colture Mediterranee (CRA; Rapisarda et al., 2009). The different orange accessions and the hybrids displayed a range of pigmentation levels when grown under equivalent conditions, from very high pigmentation (OTA9), medium pigmentation (OMO6, OTA23, OMO31, and OMO15), and low pigmentation (OMO28, OTA14, and OTA20) through to no pigmentation (OTA41, OMO3, OMO5, OTA11, OTA17, OTA31, and OTA35). The levels of Ruby transcript in the flesh of fruit grown under the same conditions at the field station were determined by quantitative RT-PCR (qRT-PCR). There was a clear correlation between the levels of Ruby transcript and the levels of anthocyanin in fruit flesh (Figure 4B). Expression of Ruby was also detected in ancient varieties belonging to the Sanguigno/Sanguinello group, generally characterized by light pigmentation of their flesh (see Supplemental Figure 4 online).
Molecular Constitution of Ruby in Sweet Oranges
Blood orange is a derivative of sweet orange, which is believed to be an interspecific hybrid between pummelo (Citrus maxima) and mandarin (Scora, 1975; Barrett and Rhodes, 1976; Mabberley, 1997; Nicolosi et al., 2000; Moore, 2001; Li et al., 2010). To establish the genotypic constitution of different orange varieties at the Ruby locus, genomic DNA was extracted from leaves of two different accessions of pummelo (Chandler and Reinking), two accessions of mandarin (Citrus reticulata Blanco cv Ponkan and Willowleaf mandarin Citrus deliciosa cv Avana), clementine (Citrus clementina cv Oroval), and several sweet orange accessions. The genomic DNA from the Citrus accessions was mapped using a range of restriction enzymes (Figure 5), amplified by PCR and inverse PCR, and characterized by sequencing. Both pummelo accessions contain two similar potentially functional alleles of Ruby, one of which showed complete sequence identity over 1.7 kb with the R allele identified in blond orange varieties. Both mandarin accessions were heterozygous at the Ruby locus. One allele contains a stop codon in the third exon of the gene, which is predicted to result in an inactive Ruby protein. We termed this allele r-1. To characterize the second Ruby allele, we used inverse PCR to identify a 2006-bp deletion, which encompasses the first two exons of the Ruby gene and 1.4 kb of the region upstream (Figure 6), creating a nonfunctional allele, which we named r-2. This second Ruby allele was present in mandarin and all orange varieties (Figure 6). Our data from the Ruby locus are consistent with the finding that sweet orange is a hybrid between pummelo and mandarin and has inherited the R allele from pummelo and the r-2 allele from mandarin. Diploid mandarin carries two nonfunctional alleles of Ruby (r-1 and r-2), implying that the generation of a blood mandarin would be highly improbable. The haploid sequenced genome of clementine (http://phytozome.net) carries the r-1 allele. Oroval clementine carries the same alleles of Ruby as the two mandarin accessions. Consequently, it was impossible to confirm or refute the suggestion that clementine is the result of a chance hybridization between Willowleaf mandarin and sweet orange from the sequence information available for the Ruby locus (Nicolosi et al., 2000).
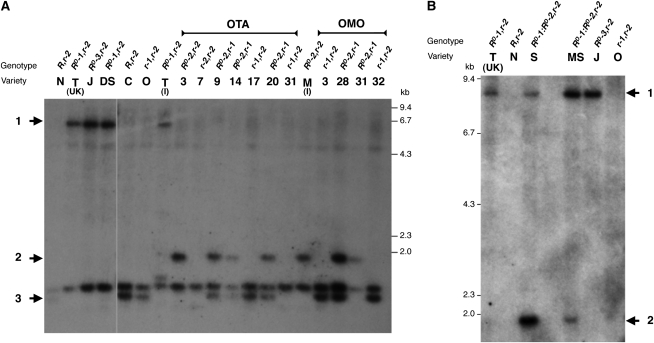
Figure 5.
Genomic Characterization of the Ruby Locus.
(A) DNA gel blot of genomic DNA from different blood and blond orange varieties and from hybrids between Tarocco and mandarin (OTA) and Moro and mandarin (OMO) digested with AseI and probed with a 32P-labeled probe of the Ruby gene from 465 bp upstream of the ATG to the stop codon of the gene. Varieties: C, Cadenera; DS, Doppio Sanguigno; J, Jingxian; M (I), Moro from CRA; N, Navelina; O, Oroval Clementine; T (UK), Tarocco from Reeds (UK); T (I), Tarocco from CRA, Italy. OTA hybrids 3, 9, 14, and 20 are pigmented with anthocyanin; OMO hybrids 28 and 31 are pigmented with anthocyanin. Band 1 is the AseI fragment at the 5′ end of the Ruby gene with the full retroelement insertion (Tcs1 for Tarocco and Doppio Sanguigno and Tcs2 for Jingxian). Band 2 is the AseI fragment at the 5′ end of the Ruby gene containing the solo LTR insertion. Band 3 is the AseI fragment at the 5′ end of the Ruby gene without any insertion (i.e., the wild-type R allele) present in blond oranges.
(B) DNA gel blot of genomic DNA from different blood and blond orange varieties. Genomic DNA was digested with AseI and probed with a 32P-labeled probe of the Ruby gene from 465 bp upstream of the ATG to the stop codon of the gene. The gel was run for longer than that shown in Figure 2A so that only the 5′ fragments of the Ruby gene with insertions remained on the gel. Varieties: J, Jingxian; MS, Maltaise Sanguine; N, Navelina; O, Oroval; S, Sanguinelli; T, Tarocco (Reeds, UK). The presence of DNA containing only the solo LTR (band 2) as well as DNA containing the full Tcs1 retroelement (band 1) is apparent in Sanguinelli and Maltaise Sanguine accessions.
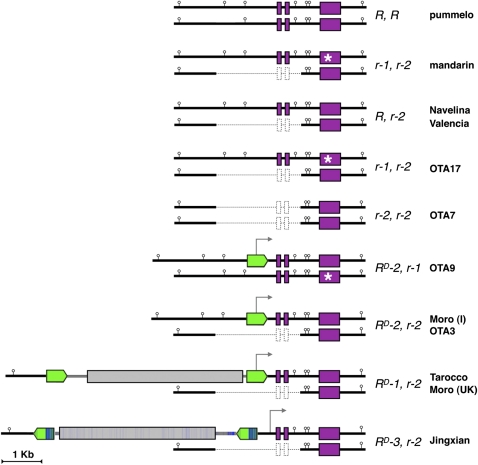
Figure 6.
Maps of Structures of the Ruby Locus in the Different Citrus Species, Orange Accessions, and Hybrids.
Thick green arrows, retrotransposon LTRs; thick gray bars, open reading frame encoding functional Gag-Pol polyprotein; thick purple bars, open reading frame encoding the Ruby protein; thin bars, noncoding regions. The asterisk indicates a stop codon. The differences in sequence between Tcs1 and Tcs2 are indicated by blue lines on the map of Tcs2. The start of transcription of the Ruby mRNA is shown by a gray arrow for each accession. The AseI sites (AseI does not cut in either Tcs1 or Tcs2) in the Ruby locus are also shown. Moro (I) indicates the accession of Moro from CRA, Sicily, and Moro (UK) indicates the accession of Moro from Reeds, UK.
Molecular Differences at the Ruby Locus between Blood and Blond Oranges
The Ruby gene was cloned from three blood (Sanguinelli, Maltaise Sanguine, and Moro) and three blond (Navelina, Salustiana, and Cadenera) varieties by PCR of genomic DNA. Sequence analysis revealed 100% nucleotide identity in the three exons and two introns that constitute the gene among the six varieties. The dramatic differences in expression of Ruby between blood and blond oranges suggested that they resulted from differences in the regulation of Ruby transcription. The upstream regulatory regions of Ruby from Moro and Cadenera were isolated by chromosome walking and sequencing revealed an insertion of 501 nucleotides in Moro, 254 bp upstream of the initiating ATG in Ruby, compared with the otherwise identical Ruby promoter from the blond Cadenera orange (Figures 5 and 6). The insertion showed sequence similarity to the long terminal repeats (LTRs) of the Copia family of retrotransposons and included a 5-bp direct repeat, typical of LTR retroelement insertion sites (Kim et al., 1998).
DNA from the different blood orange accessions and from hybrids derived from the crosses between clementine and Moro (OMO hybrids) or Tarocco (OTA hybrids) (Rapisarda et al., 2009) was then mapped, amplified, and sequenced (Figures 5A and 6). An accession of Moro from the UK National Citrus collection (Reeds Nursery Loddon, UK) had a larger insertion in the region upstream of the Ruby gene than the Moro accession from CRA, Acireale, Sicily. The same, larger insertion (termed RD-1) was present in accessions of Tarocco from both CRA and the Reeds collection, in Maltaise Sanguine, and Doppio Sanguigno, representing older varieties of Italian blood oranges (Chapot, 1963; Hodgson, 1967) as well as in the Spanish Sanguinelli derived from Doblefina (Figures 5A and 5B). The large insertions in Moro (Reeds), Tarocco, Maltaise Sanguine, Doppio Sanguigno, and Sanguinelli were identical and corresponded to a complete retrotransposon, which we named Tcs1.
Tcs1 is 5413 nucleotides in length and shows all the features of a typical Copia-like LTR retrotransposon. It contains an open reading frame, which encodes the proteins (Gag and Pol) required for the reverse transcription of the element and integration into the host genome, flanked by two identical LTRs of 496 nucleotides, identical to the solo LTR insertion in the Sicilian Moro (CRA). Because Tcs1 encodes complete Gag and Pol proteins, it is likely an active retrotransposon, and because the LTRs of Tcs1 at the Ruby locus are identical, it is likely that this is a very recent insertion.
A precise copy of Tcs1 is not present in the recently released sequence of the diploid sweet orange genome (http://phytozome.net). Two closely related elements are present in the more accurately sequenced genome of a haploid C. reticulata (clementine). They have identical LTRs to Tcs1 but differ in the noncoding region downstream of the 5′ LTR. Several hundred Tcs1-like sequences can be identified in both species, but the vast majority of these elements are predicted to be inactive. In sweet orange, LTR retrotransposons have been estimated to constitute ~23% of the genome (Rico-Cabanas and Martínez-Izquierdo, 2007), but we calculate full-length Tcs1-like copies, which have two LTRs available for recombination, constitute only ~0.23% of the genome, and we could find no active Tcs1-like copies with complete open reading frames in the available sequence.
Recombination between the LTRs of Tcs1 Gives Plants Chimeric at the Ruby Locus or Progeny with Just the LTR Insertion at the Ruby Locus
In addition to the insertion of the full Tcs1 element at the Ruby locus (which generated the dominant RD-1 allele), the DNA from the leaves of Maltaise Sanguine and Sanguinelli accessions also contained versions of the Ruby locus with just the LTR inserted upstream, as shown by DNA gel blots (Figure 5B) and PCR. We deduced that these accessions are chimeric for insertions of the full element and the solo LTR. The intensity of the signal from the fragment containing the solo LTR from both Maltaise Sanguine and Sanguinelli suggests that these accessions are periclinal chimeras for the Ruby locus; a recombination could have occurred in the L1 or L2/L3 layers and then have been maintained over long periods of time because these varieties are propagated largely by grafting, as has been reported to occur commonly in other clonally propagated crops like grape (Vitis vinifera) (Pelsy, 2010).
Among the hybrids between clementine and Tarocco (OTA hybrids), all those with pigmented fruit flesh had an insertion of the solo LTR at the Ruby locus, although a vegetative clone of the Tarocco parent plant used for the crosses had the full Tcs1 insertion (Figure 5A). This showed that recombination can occur in the germline and suggested that it might be induced by meiosis. We also identified two accessions of Moro, one (Reeds, UK) with the full Tcs1 retroelement insertion and the other (CRA) with just the solo LTR at Ruby, indicating that unequal crossing over can also occur somatically (because blood orange is propagated by cuttings) and give rise to nonchimeric progeny.
Retroelement Expression Controls the Expression of Ruby in Blood Orange
Retrotransposons can insert within or near transcriptionally active regions and can cause mutations by disrupting genes, altering their expression or driving genomic rearrangements (McClintock, 1984; Feschotte et al., 2002; Shapiro, 2005). We mapped the start of Ruby transcription by 5′ RACE PCR on cDNA prepared from RNA from Moro fruit flesh. The start of transcription mapped to an A, 551 nucleotides upstream of the initiating ATG of the Ruby gene. A TATA box was identified 32 bp upstream of the start of transcription within the LTR, whereas it was not possible to identify a TATA box in the sequence upstream of the Ruby gene in the R allele from blond oranges. The LTR also provided a 5′ donor splice site for the first intron in the Ruby transcript, the 3′ acceptor site being located within the sequences upstream of the Ruby open reading frame. A second intron detected in the Ruby transcript from Moro blood orange was defined by donor and acceptor sites within the 5′ untranslated region of the Ruby sequence (Figure 7). We concluded that when Tcs1 is inserted at the Ruby locus, it must provide the regulatory sequences for initiating expression of the Ruby gene, since the transcription start site maps to the LTR.
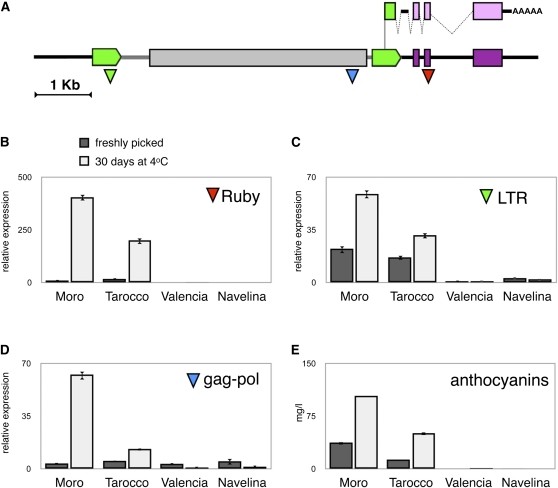
Figure 7.
Expression of Tcs1, Ruby, and Anthocyanin Production in Blood and Blond Oranges in Response to Cold Storage.
(A) Detailed map of the Ruby locus with the full Tcs1 retroelement insertion (the RD-1 allele), showing the start of transcription of the Ruby gene. The map of the Ruby transcript is shown above; the thick green bars indicating sequence from the LTR in the 5′ untranslated region, the thick purple bars indicating the open reading frame encoding the Ruby protein, also indicated by thick pink bars in the transcript; the splice sites for the transcript are shown. The positions of the oligonucleotides used for measuring transcript levels of Ruby and Tcs1 are also indicated by color-coded inverted triangles.
(B) Levels of Ruby transcripts in Moro and Tarocco blood orange accessions and in Valencia and Navelina blond orange accessions from CRA in fruit picked fresh and following storage for 30 d at 4°C, determined by qRT-PCR. Error bars show se of the mean.
(C) Levels of Tcs1 transcripts from the LTR in Moro and Tarocco blood orange accessions and in Valencia and Navelina blond orange accessions from CRA in fruit picked fresh and following storage for 30 d at 4°C, determined by qRT-PCR.
(D) Levels of Tcs1 and Tcs2 transcripts from the Gag-Pol region of the element in Moro and Tarocco blood orange accessions and in Valencia and Navelina blond orange accessions from CRA in fruit picked fresh and following storage for 30 d at 4°C, determined by qRT-PCR.
(E) Levels of anthocyanins in Moro and Tarocco blood orange accessions and in Valencia and Navelina blond orange accessions from CRA in fruit picked fresh and following storage for 30 d at 4°C.
Transcription of active retroelements, a prerequisite for transposition, is usually repressed by the host. However, activation in response to a variety of biotic and abiotic stresses is a common feature of most retrotransposons (Wessler, 1996). McClintock’s theory of genome shock suggested that enhanced transposition under stress might represent an evolutionary strategy to increase the chances of survival under unfavorable conditions (McClintock, 1984). We therefore investigated whether the expression pattern of Ruby in blood oranges was a function of the expression of Tcs1 particularly in response to low temperature.
To assess the activity of Tcs1 and Tcs1-like elements, two sets of primers corresponding to the transcribed portion of the LTR or internal to the Gag-Pol region of the element (Figure 7) were used for qRT-PCR. The downstream primer for the transcript of the LTR corresponded to a sequence that is spliced from the Ruby transcript in the first intron in blood oranges (Figure 7A). Consequently, the Tcs1 LTR transcript levels we measured did not include those from the Ruby locus.
Tcs1 transcripts could be detected in fruit, but not in leaves, of blood oranges. Elevated levels of Tcs1 transcripts were observed in Tarocco and Moro blood varieties following storage of fruit in the cold, indicating that Tcs1 and Tcs1-like elements are activated by cold and ultimately are responsible for temperature-dependent anthocyanin accumulation in blood oranges (Figure 7). The expression of Tcs1 matched well the change in transcript levels of Ruby in blood orange varieties in response to cold storage (Figure 7). Since in Moro (CRA) the solo LTR controls expression of Ruby, cold-dependent transcriptional activation appears to be an intrinsic feature of Tcs1-like elements, irrespective of their association with the Ruby locus. Consequently, in the Sicilian blood orange varieties, the fruit-specific, cold-inducible production of anthocyanin (Figure 7) is a feature of the regulation of the Tcs1 element, which controls expression of Ruby as a result of the positioning of the strong promoter provided by the 3′ LTR adjacent to the Ruby gene. Moro produces higher levels of anthocyanins than Tarocco. The higher anthocyanin production in Moro fruit is accompanied by higher expression of Tcs1. Consequently, the selection of sports producing more deeply pigmented fruit flesh likely reflects selection for higher levels of Tcs1 transcription during the derivation of improved blood orange cultivars.
An Independent Blood Orange Accession from Jingxian, China
Today, blood oranges are grown in places as far apart as Japan, Australia, South Africa, Pakistan, California, China, and Iran. However, the unreliable production associated with existing commercial blood orange varieties due to their cold dependency means that the availability and consumption of blood orange on a global scale has declined in recent years (Zarba and Pulvirenti, 2006). There is considerable interest in identifying independent blood orange types that might be free from these production problems. We therefore looked among existing blood orange accessions for any independent events. Because of their long juvenile phase, oranges, like most Citrus species, are almost exclusively propagated by grafting on selected rootstocks. Striking phenotypic diversity mainly is due to the selection of superior or unusual branches derived from bud mutations. Our molecular analyses showed that Sanguinelli, which is a derivative of the Spanish Doblefina, shared a common origin with Sicilian blood oranges. Shamouti Blood Orange has been reported to be a chimera (Spiegel-Roy, 1979), possibly derived from grafting with Maltaise Sanguine (Hodgson, 1967), and is therefore also unlikely to represent an independent event.
Most of the blood orange cultivars grown in China are of direct or indirect Sicilian origin. However, one old variety, Jingxian, has been retained and is believed to be the only blood orange of Chinese origin (Yuan et al., 2008). Jingxian blood orange was first recorded in the Official Record for Huaihua Region in 1996. In 1965, when people began to select out this variety, the oldest tree they found in the countryside was ~70 years old. Consequently, the Jingxian blood orange has existed for at least 110 years.
DNA gel blot analysis indicated that Jingxian contains a DNA insertion in the Ruby locus of similar size to Tcs1 (Figure 5A). However, inverse PCR showed this to be a different element, which we named Tcs2 (Figure 6). Tcs2 is 5454 bp long, generates a 5-bp direct duplication upon insertion, and is similar in sequence to Tcs1, except for the first half of the two LTRs, which differ considerably. Like Tcs1, Tcs2 appears to be an active retroelement. In Jingxian blood orange, Tcs2 is inserted in the Ruby locus just 196 nucleotides upstream of the Tcs1 insertion site and 450 bp upstream of the Ruby ATG, but it is inserted in the opposite orientation to Tcs1 relative to the Ruby gene (Figure 6).
The blood phenotypes in both Jingxian and the Sicilian group of blood oranges are therefore of independent origin, showing that different members of the same family of retrotransposons may alter the expression of nearby genes through parallel but distinct mechanisms.
Despite smaller fruit size and higher seed content, confirming that Jingxian is not a derivative of Sicilian blood oranges currently cultivated in China, Jingxian fruit display the same pattern of cold-induced, fruit-specific anthocyanin accumulation as Sicilian and Spanish blood oranges, suggesting Tcs2 to be cold inducible as well. For the LTR of Tcs2 to drive expression of Ruby, it must provide a bidirectional activator sequence. We mapped the start of transcription of Ruby in Jingxian juice to a position 321 nucleotides downstream of the Tcs2 insertion (Figure 6). No TATA box was evident in the sequence ~30 bp upstream of this transcriptional start site. Our data suggest that Tcs2 provides an upstream activating sequence that controls the expression of Ruby and the production of anthocyanin concordant with its own expression.
Activity of Copia-Like Retroelements in Citrus
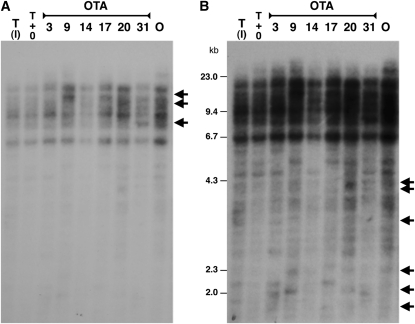
A priori, the probability of independent gain-of-function mutations involving the same family of retroelements is low, especially given that Citrus varieties are almost exclusively vegetatively propagated. However, genome shock of the type caused by interspecific hybridization does induce retroelement expression and transposition. It may be that the relatively recent origin (in terms of meiotic cycles) of sweet orange through interspecific hybridization between pummelo and mandarin (Scora, 1975; Barrett and Rhodes, 1976; Mabberley, 1997; Nicolosi et al., 2000; Moore, 2001; Li et al., 2010) induced accompanying high levels of retroelement activity that have been further selected during breeding of blood orange. Indeed, active retroelements may represent an important source of variation available to breeders of Citrus who depend on mutation-based differences (presumably often gain-of-function mutations) that arise in buds (Tao et al., 2005; De Felice et al., 2008). Comparison of the DNA of the OTA hybrids and their parental lines showed new insertions of Tcs1-like elements following this interspecific cross (Figure 8A). However, more striking was the high frequency of unequal crossing over between the LTRs of the full element to leave solo LTR insertions (Figure 8B). This has also occurred at the Ruby locus in our accessions of Sanguinelli, Maltaise Sanguine, Moro, and Tarocco. Sweet orange is a relatively recent derivative from an interspecific hybridization, and the genome shock resulting from such hybridization may have stimulated higher levels of unequal crossing over. Interestingly, Parisod et al. (2004) highlighted significant structural changes occurring as a result of recombination involving retroelements, rather than transposition, during interspecific hybridization and allopolyploidization in plants.
Figure 8.
DNA Gel Blots of Genomic DNA from Oroval (Mandarin) and Tarocco Parents and the OTA Hybrids.
Genomic DNA was digested with AseI and probed with a 32P-labeled probe of the Tcs1 LTR. AseI does not cut within Tcs1; therefore, fragments larger than 5.4 kb likely represent full-length retroelement insertions. O, Oroval; T (I), Tarocco (CRA); T+O, equimolar mixture of Tarocco and Oroval DNA.
(A) Short exposure shows large, strongly hybridizing bands (two copies of the LTR). New bands present in the hybrids but not in the parents are arrowed and likely represent new transpositions.
(B) Long exposure shows smaller, more weakly hybridizing bands. New bands present in the hybrids but not in the parents are arrowed and likely represent unequal crossing over events between the LTRs to leave solo LTR insertions.
DISCUSSION
The molecular analysis of the Ruby locus indicates that the genomes of pummelo and mandarin combined to generate the genome of sweet orange, confirming its reported hybrid origin (Scora, 1975; Barrett and Rhodes, 1976; Mabberley, 1997; Nicolosi et al., 2000; Moore, 2001; Li et al., 2010). At the Ruby locus the genetic contributions of the two parental species were equal, and the functional allele of Ruby was provided by the pummelo parent. Although encoding a functional protein, this allele appears to be inactive in common blond orange because we were unable to amplify any transcripts from this locus from any tissues of blond orange plants. Confirming its apparent lack of expression, no ESTs are available in the Citrus EST databases. Lack of Ruby expression may explain why anthocyanins, common pigments in most plant species, are rare in sweet orange and why mandarins, which carry only nonfunctional alleles of Ruby, never produce anthocyanins.
Our results indicate that all commercial blood orange varieties have a common origin. Anthocyanin pigmentation of fruit must have originated once either in a Mediterranean sweet orange or in a Chinese sweet orange, which has since been lost. Citrus breeders have derived all the diversity in modern blood orange varieties from this original event. The molecular basis of the blood orange trait is retrotransposon-mediated transcriptional activation of the Ruby Myb gene. This is particularly clear in Sicilian blood oranges where the start of transcription of Ruby lies within the 3′ LTR of Tcs1. This provides a striking example of the role of transposable elements as controlling elements in the regulation of gene expression and genome evolution. Our discovery of a second independent insertion of a retroelement in the Jingxian blood orange giving the same gain-of-function phenotype as in Sicilian blood oranges illustrates the strength of the LTR as a promoter and also as an upstream activating sequence. Both Tcs1 and Tcs2 insertions in Ruby give rise to induction of anthocyanin biosynthesis specifically in fruit, which is heavily influenced by environment. The cold dependency of anthocyanin production in blood orange results from the cold induction of retroelement transcription. The expression of Copia-like retrotransposons is determined by sequences within the LTRs that, in blood oranges, provide either a surrogate promoter with a TATA box and a transcriptional start as seen in commercial blood oranges or an upstream activator sequence as seen in Jingxian orange. Consequently, Ruby expression mirrors retroelement expression and is fruit specific and cold inducible.
Different accessions of blood orange demonstrate the high levels of recombination and transposition associated with the retroelements and suggest that they may be responsible for generating much of the diversity available to Citrus breeders (Tao et al., 2005; Rico-Cabanas, and Martínez-Izquierdo, 2007; De Felice et al., 2009). However, recombination between Tcs1 LTRs at the Ruby locus does not result in phenotypic changes in the levels of anthocyanins produced, confirming that the solo LTR carries all the information for the control of Ruby expression in Sicilian blood oranges. This is unlike the situation in grape. In grape, insertion of the Gypsy-like retrotransposon, Gret1, suppresses expression of a Myb gene (MYBA1), and it is believed that this insertion underpinned the development of white-skinned berries (Kobayashi et al., 2004; Fournier-Level et al., 2010). Recombination between Gret1 LTRs results in some restoration of Myb gene function and blush-skinned sports, such as Chardonnay Rose and Flame Muscat (Pelsy, 2010).
The two independent blood orange derivatives, Jingxian and Sicilian blood oranges, represent parallel gains of function; therefore, our results offer little hope of generating or identifying new varieties of blood orange that are free from the major limitation of cold dependency by conventional Citrus breeding methods. However, our improved understanding of the genetic and molecular basis of the blood orange trait could offer relatively straightforward solutions to the requirement for blood orange varieties with dependable production in warmer climates through genetic engineering. Such strategies could provide new blood orange varieties suitable for the major areas of Citrus cultivation and could contribute significantly to increasing production of health-promoting blood oranges.
METHODS
Plant Material
Sweet orange (Citrus sinensis cv Moro, Tarocco, Doppio Sanguigno, Cadenera, Navelina, and Valencia) was grown at the CRA experimental farm (Palazzelli, Sicily, Italy). Different accessions of Moro, Tarocco, and Navelina and the varieties Maltaise Sanguine and Sanguinelli (Spanish) were obtained from the UK National Citrus collection (Reeds Nursery). Fruit and leaves of C. sinensis cv Jingxian were obtained from Jingzhou, Huannan Province, China. OTA and OMO hybrids grown in Palazzelli were obtained by conventional Citrus breeding methods using controlled pollinations between Oroval mandarin (Citrus clementina), used as the female parent, and Tarocco 57-1E-1 or Moro NL 58-8D-1 (C. sinensis), used as the male parent.
Isolation of Ruby cDNA
Total RNA was extracted from Moro (CRA, Sicily) fruit flesh and reverse transcribed using a T7-oligo5(dT) primer and SuperScript III RT (Invitrogen). First-strand cDNA was amplified by PCR using degenerate primers BUT-F3 and BUT-R3. The full-length cDNA was isolated using 5′ and 3′ SMART RACE amplification kit (Clontech) and gene-specific primers PMC-F1 and PMC-R1. For Jingxian, total RNA was extracted from juice and reverse transcribed using the gene-specific primer PMC-Z and SuperScript III RT. Ruby cDNA was obtained using a 5′ RACE kit (Invitrogen) and gene-specific primers PMC-R1 and LeLe-R1. Primers sequences are listed in Supplemental Table 1 online.
Phylogenetic Analysis
Protein sequences from Arabidopsis thaliana and selected Myb proteins from other species belonging to subgroups 2, 4, 5, 6, and 7 were aligned using PRANK (Löytynoja and Goldman, 2008). The alignment of the DNA binding domain only was used to calculate distance estimates (the Jones Taylor Thornton matrix model of evolution) for a neighbor-joining tree with the PHYLIP software package (Felsenstein et al., 2004). To provide statistical support for each node in the tree, a consensus tree was generated from 1000 bootstrap data sets.
Ectopic Expression of Ruby in Tobacco
The coding sequence of the Ruby cDNA was amplified with primers PMC-GWF and PMC-GWR and cloned in a pBin19-derived binary vector, previously equipped with a double CaMV 35S promoter, the CaMV terminator with attR recombination sites in between, using Gateway recombination technology (Invitrogen). The resulting plasmid was transferred to Agrobacterium tumefaciens strain LBA4404 and used to transform tobacco (Nicotiana tabacum cv Samsun).
Protoplast Transfection Assays
Tobacco protoplasts were isolated from 3- to 5-week-old leaves of N. tabacum cv Samsun following the procedure described by Negrutiu et al. (1987). For each transfection, 10 μg of plasmid DNA containing the flavanone-3-hydroxylase or dihydroflavonol-4-reductase promoters from snapdragon (Antirrhinum majus) fused to the β-glucuronidase (GUS) reporter gene were used. To measure expression from the promoters, plasmid DNA containing the cDNA sequences coding for the transcriptional activators Am Rosea1 (4 μg; Schwinn et al., 2006) and Cs MYC2 (5.5 μg) under the control of the double CaMV 35S promoter were used. Different amounts of an empty plasmid containing the double CaMV 35S promoter were used to ensure that equal amounts of total DNA and viral promoter were used for each transfection. After incubation for 40 h, protoplasts were collected by centrifugation, and GUS activity in the cell lysate was determined according to Jefferson (1987) and expressed as nanomoles of methylumbelliferone per milligram of protein per minute. All transfections were performed in triplicate, and GUS activity was measured in duplicate for each transfection.
DNA Gel Blots
Citrus leaves were ground in liquid nitrogen, and DNA was extracted using caesium chloride density gradient purification. DNA (10 μg per sample) was digested with AseI (and numerous other restriction enzymes for mapping) for 5 h and then separated by electrophoresis. Denatured DNA was transferred to nitrocellulose membrane filters. Filters were hybridized with randomly primed 32P-labeled probes overnight at 60°C and washed in 0.1× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate), 0.5% SDS at 60°C for 2 h before exposure to x-ray film (Fuji RX-100).
Isolation of Ruby Promoters
The upstream regions of Ruby from Moro and Cadenera (CRA, Sicily) were isolated by chromosome walking using the GenomeWalker kit (Clontech) and gene-specific primers PMC-47 and PMC-109.
Isolation of Tcs1, Tcs2, and Ruby Deletion Allele
A Tcs1 fragment was initially obtained from Tarocco and Moro (Reeds Nursery). DNA from leaves was digested with BsrGI and self-ligated. The Tcs1 fragment was isolated by inverse PCR using primers PMC-CF and PMC-NR1. Full-length Tcs1 was obtained by conventional PCR using primers PMC-G3 and PMC-U.
A Tcs2 fragment was initially obtained from Jingxian DNA, digested with BstYI, self-ligated, and amplified by inverse PCR using primers PMCi4 and PMCiD. Full-length Tcs2 was obtained by conventional PCR using primers PMC-G2 and PMC-S5. The sequence of the Ruby deletion allele (r) was obtained from OTA7 DNA using the same inverse PCR procedure and primers PMCi4 and PMCiD.
Expression Analysis of Ruby and Tcs1
Total RNA was extracted from 3 mL of juice of Moro, Tarocco, Navelina, and Valencia fruit (all from CRA) using a modified protocol described by Ancillo et al. (2007). DNase-treated total RNA was further purified using the RNA Cleanup protocol (Qiagen) and retrotranscribed into cDNA using a High-Capacity cDNA Archive kit (Applied Biosystems). Quantitative real-time PCR was performed in optical 96-well plates with an ABI Prism 7000 sequence detection system (Applied Biosystems). The PCR mixture (final volume 25 μL) contained 15 μL Power SYBR Green mix, 0.2 μM of each gene-specific forward and reverse primer, and 100 ng of cDNA sample, using the protocol for the Power SYBR Green PCR Master Mix (Applied Biosystems). The following standard thermal profile was used for all PCRs: 50°C for 2 min, 95°C for 10 min, 40 cycles of 95°C for 15 s, and 60°C for 1 min. Three replicates were assayed, and a no-template negative control (water control) was performed. The analyses used the relative quantification standard curve method.
Bioinformatic Analysis of LTR Retrotransposons in Citrus
To estimate the proportion of the orange genome made up of full-length Tcs1-like elements, the genome sequence of sweet orange was scanned for Tcs1 sequences using the BLASTN program and the 5413-bp Tcs1 transposon as the query sequence. Only hits with an e-value < e−50 and sequence identity to the Tcs1 sequence >80% were used and filter masking was turned off.
The number of complete transposons identified was 78 where the BLAST high-scoring segment pair covered >70% of the length of the Tcs1 sequence. A further 47 LTR regions were identified that covered >80% of the Tcs1 LTR region. Many of these hits to the LTR region had flanking hits to other regions of the transposon and were assumed to be full-length transposons. Therefore, the estimated number of full length transposons is 125, and the proportion of the haploid genome sequence (296 megabases; derived from the file called Csinensis_154.fa [July, 2010] from the Phytozome website) occupied by these Tcs1-like elements is 0.23%.
To estimate the number of potentially active transposons, a Perl script was written to retrieve and translate the DNA sequence in the region of the open reading frame of the 78 complete transposons identified above. None of these translated regions were found to be without an internal stop codon; therefore, the corresponding elements are likely to be inactive.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: JN402329, Ruby gene (Navelina); JN402330, LTR+Ruby gene (Moro); JN402331, deleted allele (OTA7); JN402332, Tcs1 (Tarocco); JN402333, Tcs2 (Jingxian); JN402334, Ruby mRNA (Moro); and JN402335, Ruby mRNA (Jingxian).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. “Aranci, Bergamotti, Cedri, Limoni e Lumie” (1715), Oil on Canvas by Bartolomeo Bimbi, Villa Medicea di Poggio a Caiano.
Supplemental Figure 2. The Production of Anthocyanin Pigment as a Result of 35S:Ruby Expression in Tobacco Is Particularly Strong in Older Senescent Leaves.
Supplemental Figure 3. Cs MYC2 Regulates Anthocyanin Biosynthetic Gene Expression.
Supplemental Figure 4. Expression Analysis of Ruby.
Supplemental Table 1. Primers Used in This Study.
Supplemental Data Set 1. Text File of Alignment Used for Phylogenetic Analysis in Figure 2.
Acknowledgments
E.B., C.L., S.M., G.R.-R., and C.M. were supported by the European Union FP6 FLORA project (FOOD-CT-01730), and E.B., S.M., and C.M. were supported by the European Union FP7 ATHENA collaborative project (Grant Agreement 245121). Y.Z. is supported by a Rotation studentship from the John Innes Foundation. C.M., S.M., P.B., and E.B. are also supported by the core strategic grant of the Biological and Biotechnological Science Research Council to the John Innes Centre. C.L. and G.R.-R. were also supported by the Agronanotech Project MIPAF. We thank David Hopwood for his encouragement and invaluable comments on the manuscript.
AUTHOR CONTRIBUTIONS
E.B. cloned the Ruby locus, performed the molecular analysis, expressed Ruby in tobacco, and contributed to the planning and writing of the article. C.L. cloned part of the Ruby locus and performed the expression analysis by qRT-PCR and the transfection assays in tobacco protoplasts. Y.Z. and J.L. provided samples of Jingxian orange and performed analysis. S.M. prepared DNA from samples of Citrus leaves. P.B. performed bioinformatic analyses of retroelements and Myb transcription factors in Citrus. G.R.-R. developed the OMO and OTA hybrids by performing interspecific crosses. C.M. performed DNA gel blot analyses, crossed and analyzed transgenic tobacco plants, and contributed to the planning and writing of the article.
References
- Ancillo G., Gadea J., Forment J., Guerri J., Navarro L. (2007). Class prediction of closely related plant varieties using gene expression profiling. J. Exp. Bot. 58: 1927–1933 [DOI] [PubMed] [Google Scholar]
- Bailey P.C., Dicks J., Wang T.L., Martin C. (2008). IT3F: A web-based tool for functional analysis of transcription factors in plants. Phytochemistry 69: 2417–2425 [DOI] [PubMed] [Google Scholar]
- Barrett H.C., Rhodes A.M. (1976). A numerical taxonomic study of affinity relationships in cultivated Citrus and its close relatives. Syst. Bot. 1: 105–136 [Google Scholar]
- Bernardi J., Licciardello C., Russo M.P., Luisa Chiusano M., Carletti G., Recupero G.R., Marocco A. (2010). Use of a custom array to study differentially expressed genes during blood orange (Citrus sinensis L. Osbeck) ripening. J. Plant Physiol. 167: 301–310 [DOI] [PubMed] [Google Scholar]
- Bonina F.P., Leotta C., Scalia G., Puglia C., Trombetta D., Tringali G., Roccazzello A.M., Rapisarda P., Saija A. (2002). Evaluation of oxidative stress in diabetic patients after supplementation with a standardised red orange extract. Diabetes Nutr. Metab. 15: 14–19 [PubMed] [Google Scholar]
- Butelli E., Titta L., Giorgio M., Mock H.P., Matros A., Peterek S., Schijlen E.G.W.M., Hall R.D., Bovy A.G., Luo J., Martin C. (2008). Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 26: 1301–1308 [DOI] [PubMed] [Google Scholar]
- Chapot H. (1963). Quelques oranges sanguines. Cah. Rech. Agron. (Rabat) 18: 61–87 [Google Scholar]
- Cultrone A., Controneo P.S., Reforgiato-Recupero G. (2010). Cloning and molecular characterisation of R2R3-MYB and bHLH-MYC transcription factors from Citrus sinensis. Tree Genet. Genomes 6: 101–112 [Google Scholar]
- Crifò T., Puglisi I., Petrone G., Recupero G.R., Lo Piero A.R. (2011). Expression analysis in response to low temperature stress in blood oranges: implication of the flavonoid biosynthetic pathway. Gene 476: 1–9 [DOI] [PubMed] [Google Scholar]
- De Felice B., Wilson R.R., Argenziano C., Kafantaris I., Conicella C. (2009). A transcriptionally active copia-like retroelement in Citrus limon. Cell. Mol. Biol. Lett. 14: 289–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pascual-Teresa S., Moreno D.A., García-Viguera C. (2010). Flavanols and anthocyanins in cardiovascular health: A review of current evidence. Int. J. Mol. Sci. 11: 1679–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies K.M. (2007). Genetic modification of plant metabolism for human health benefits. Mutat. Res. 622: 122–137 [DOI] [PubMed] [Google Scholar]
- Espley R.V., Hellens R.P., Putterill J., Stevenson D.E., Kutty-Amma S., Allan A.C. (2007). Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 49: 414–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. (2004). PHYLIP (Phylogeny Inference Package) Version 3.6. (Seattle, WA: University of Washington; ). [Google Scholar]
- Ferrari G.B. (1646). Hesperides Sive Malorum Aureorum Cultura et Usu. (Romae, sumptibus Hermanni Scheus; ). [Google Scholar]
- Feschotte C., Jiang N., Wessler S.R. (2002). Plant transposable elements: where genetics meets genomics. Nat. Rev. Genet. 3: 329–341 [DOI] [PubMed] [Google Scholar]
- Fournier-Level A., Lacombe T., Le Cunff L., Boursiquot J.M., This P. (2010). Evolution of the VvMybA gene family, the major determinant of berry colour in cultivated grapevine (Vitis vinifera L.). Heredity (Edinb) 104: 351–362 [DOI] [PubMed] [Google Scholar]
- Geekiyanage S., Takase T., Ogura Y., Kiyosue T. (2007). Anthocyanin production by over-expression of grape transcription factor gene VlmybA2 in transgenic tobacco and Arabidopsis. Plant Biotech. Rep. 1: 11–18 [Google Scholar]
- Guarnieri S., Riso P., Porrini M. (2007). Orange juice vs vitamin C: Effect on hydrogen peroxide-induced DNA damage in mononuclear blood cells. Br. J. Nutr. 97: 639–643 [DOI] [PubMed] [Google Scholar]
- Heim M.A., Jakoby M., Werber M., Martin C., Weisshaar B., Bailey P.C. (2003). The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20: 735–747 [DOI] [PubMed] [Google Scholar]
- Hodgson R.W. (1967). History, world distribution, botany and varieties. In The Citrus Industry, Vol. I, Reuther W., Webber H.J., Batchelor L.D., eds (Berkeley, CA: University of California; ), pp. 431–591 [Google Scholar]
- Holmes E.M. (1924). The origin of the Maltese blood orange. Pharm. J. Citrus 614–615 [Google Scholar]
- Jayaprakasha G.K., Patl B.S. (2007). In vitro evaluation of the antioxidant activities in fruit extracts from citron and blood orange. Food Chem. 101: 410–418 [Google Scholar]
- Jefferson R.A. (1987). Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol. Biol. Rep. 5: 387–405 [Google Scholar]
- Kelebek H., Canbas A., Selli S. (2008). Determination of phenolic composition and antioxidant capacity of blood orange juices obtained from cvs Moro and Sanguinelli (Citrus sinensis (L) Osbeck) grown in Turkey. Food Chem. 107: 1710–1716 [Google Scholar]
- Kim J.M., Vanguri S., Boeke J.D., Gabriel A., Voytas D.F. (1998). Transposable elements and genome organization: A comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 8: 464–478 [DOI] [PubMed] [Google Scholar]
- Kobayashi S., Goto-Yamamoto N., Hirochika H. (2004). Retrotransposon-induced mutations in grape skin color. Science 304: 982. [DOI] [PubMed] [Google Scholar]
- Latado R.R., Tognato P.C., Silva-Stenico M.E., do Nascimento L.M., dos Santos P.C. (2008). Anthocyanin accumulation and physical and chemical characteristics of blood orange fruits during cold storage. Rev. Bras. Frutic. 30: 604–610 [Google Scholar]
- Li X., Xie R., Lu Z., Zhou Z. (2010). The origin of cultivated Citrus as inferred from internal transcribed spacer and chloroplast DNA sequence and amplified fragment length polymorphism fingerprints. J. Am. Soc. Hortic. Sci. 135: 341–350 [Google Scholar]
- Licciardello C., Russo M.P., Vale’ G., Recupero-Reforiato G. (2008). Identification of differentially expressed genes in the flesh of blood and common oranges. Tree Genet. Genomes 4: 315–331 [Google Scholar]
- Lin-Wang K., Bolitho K., Grafton K., Kortstee A., Karunairetnam S., McGhie T.K., Espley R.V., Hellens R.P., Allan A.C. (2010). An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 10: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löytynoja A., Goldman N. (2008). Phylogeny-aware gap placement prevents errors in sequence alignment and evolutionary analysis. Science 320: 1632–1635 [DOI] [PubMed] [Google Scholar]
- Mabberley D.J. (1997). A classification of edible Citrus (Rutaceae). Telopea (Syd.) 7: 167–172 [Google Scholar]
- McClintock B. (1984). The significance of responses of the genome to challenge. Science 226: 792–801 [DOI] [PubMed] [Google Scholar]
- Moore G.A. (2001). Oranges and lemons: Clues to the taxonomy of Citrus from molecular markers. Trends Genet. 17: 536–540 [DOI] [PubMed] [Google Scholar]
- Negrutiu I., Shillito R., Potrykus I., Biasini G., Sala F. (1987). Hybrid genes in the analysis of transformation conditions. I. Setting up a simple method for direct gene transfer into plant protoplasts. Plant Mol. Biol. 8: 363–373 [DOI] [PubMed] [Google Scholar]
- Nicolosi E., Deng Z.N., Gentile A., La Malfa S., Continella G., Tribulato E. (2000). Citrus phylogeny and genetic origin of important species as investigated by molecular markers. Theor. Appl. Genet. 100: 1155–1166 [Google Scholar]
- Paredes-López O., Cervantes-Ceja M.L., Vigna-Pérez M., Hernández-Pérez T. (2010). Berries: Improving human health and healthy aging, and promoting quality life—a review. Plant Foods Hum. Nutr. 65: 299–308 [DOI] [PubMed] [Google Scholar]
- Parisod C., Alix K., Just J., Petit M., Sarilar V., Mhiri C., Ainouche M., Chalhoub B., Grandbastien M.A. (2010). Impact of transposable elements on the organization and function of allopolyploid genomes. New Phytol. 186: 37–45 [DOI] [PubMed] [Google Scholar]
- Pelsy F. (2010). Molecular and cellular mechanisms of diversity within grapevine varieties. Heredity (Edinb.) 104: 331–340 [DOI] [PubMed] [Google Scholar]
- Prior R.L., Wu X.L. (2006). Anthocyanins: Structural characteristics that result in unique metabolic patterns and biological activities. Free Radic. Res. 40: 1014–1028 [DOI] [PubMed] [Google Scholar]
- Proteggente A.R., Saija A., De Pasquale A., Rice-Evans C.A. (2003). The compositional characterisation and antioxidant activity of fresh juices from sicilian sweet orange (Citrus sinensis L. Osbeck) varieties. Free Radic. Res. 37: 681–687 [DOI] [PubMed] [Google Scholar]
- Ramsay N.A., Glover B.J. (2005). MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 10: 63–70 [DOI] [PubMed] [Google Scholar]
- Rapisarda P., Fabroni S., Peterek S., Russo G., Mock H.P. (2009). Juice of new citrus hybrids (Citrus clementina Hort. ex Tan. x C.sinensis L. Osbeck) as a source of natural antioxidants. Food Chem. 117: 212–218 [Google Scholar]
- Rapisarda P., Tomaino A., Lo Cascio R., Bonina F., De Pasquale A., Saija A. (1999). Antioxidant effectiveness as influenced by phenolic content of fresh orange juices. J. Agric. Food Chem. 47: 4718–4723 [DOI] [PubMed] [Google Scholar]
- Rico-Cabanas L., Martínez-Izquierdo J.A. (2007). CIRE1, a novel transcriptionally active Ty1-copia retrotransposon from Citrus sinensis. Mol. Genet. Genomics 277: 365–377 [DOI] [PubMed] [Google Scholar]
- Riso P., Visioli F., Gardana C., Grande S., Brusamolino A., Galvano F., Galvano G., Porrini M. (2005). Effects of blood orange juice intake on antioxidant bioavailability and on different markers related to oxidative stress. J. Agric. Food Chem. 53: 941–947 [DOI] [PubMed] [Google Scholar]
- Schwinn K., Venail J., Shang Y.J., Mackay S., Alm V., Butelli E., Oyama R., Bailey P., Davies K., Martin C. (2006). A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell 18: 831–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scora R.W. (1975). On the history and origin of Citrus. Bull. Torrey Bot. Club 102: 369–375 [Google Scholar]
- Shapiro J.A. (2005). Retrotransposons and regulatory suites. Bioessays 27: 122–125 [DOI] [PubMed] [Google Scholar]
- Spiegel-Roy P. (1979). Chimeral nature of the shamouti orange. Euphytica 28: 361–365 [Google Scholar]
- Stracke R., Werber M., Weisshaar B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4: 447–456 [DOI] [PubMed] [Google Scholar]
- Takos A.M., Jaffé F.W., Jacob S.R., Bogs J., Robinson S.P., Walker A.R. (2006). Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 142: 1216–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao N.G., Xu J., Cheng Y.J., Hong L., Guo W.W., Yi H.L., Deng X.X. (2005). Isolation and characterization of Copia-like retrotransposons from 12 sweet orange (Citrus sinensis Osbeek) cultivars. J. Int. Biol. 47: 1507–1515 [Google Scholar]
- Titta L., et al. (2010). Blood orange juice inhibits fat accumulation in mice. Int J Obes (Lond) 34: 578–588 [DOI] [PubMed] [Google Scholar]
- Toufektsian M.C., et al. (2008). Chronic dietary intake of plant-derived anthocyanins protects the rat heart against ischemia-reperfusion injury. J. Nutr. 138: 747–752 [DOI] [PubMed] [Google Scholar]
- Walker A.R., Lee E., Bogs J., McDavid D.A.J., Thomas M.R., Robinson S.P. (2007). White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J. 49: 772–785 [DOI] [PubMed] [Google Scholar]
- Wessler S.R. (1996). Turned on by stress. Plant retrotransposons. Curr. Biol. 6: 959–961 [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B. (2001). Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126: 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan F., Long G., Deng Z. (2008). Jingxian blood orange: The only pigmented sweet orange cultivar originated in China. In Abstracts of the 11th International Citrus Congress, Proceedings of the International Society of Citriculture, Vol. 1 (Beijing: China Agriculture Press; ), pp 70–72 China [Google Scholar]
- Zarba A.S., Pulvirenti G. (2006). The consumption of Sicilian red oranges: Implications for firms involved in commercialization. J. Business Chemistry 3: 22–41 [Google Scholar]
- Zimmermann I.M., Heim M.A., Weisshaar B., Uhrig J.F. (2004). Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 40: 22–34 [DOI] [PubMed] [Google Scholar]