Transmembrane LRR-RLKs are a major class of plant proteins. This study investigates the functional contributions of multiple FLS2 protein domains and modifications to provide insight into structure-function relationships of LRR-RLK proteins in general.
Abstract
FLAGELLIN SENSING2 (FLS2) is a transmembrane receptor kinase that activates antimicrobial defense responses upon binding of bacterial flagellin or the flagellin-derived peptide flg22. We find that some Arabidopsis thaliana FLS2 is present in FLS2-FLS2 complexes before and after plant exposure to flg22. flg22 binding capability is not required for FLS2-FLS2 association. Cys pairs flank the extracellular leucine rich repeat (LRR) domain in FLS2 and many other LRR receptors, and we find that the Cys pair N-terminal to the FLS2 LRR is required for normal processing, stability, and function, possibly due to undescribed endoplasmic reticulum quality control mechanisms. By contrast, disruption of the membrane-proximal Cys pair does not block FLS2 function, instead increasing responsiveness to flg22, as indicated by a stronger oxidative burst. There was no evidence for intermolecular FLS2-FLS2 disulfide bridges. Truncated FLS2 containing only the intracellular domain associates with full-length FLS2 and exerts a dominant-negative effect on wild-type FLS2 function that is dependent on expression level but independent of the protein kinase capacity of the truncated protein. FLS2 is insensitive to disruption of multiple N-glycosylation sites, in contrast with the related receptor EF-Tu RECEPTOR that can be rendered nonfunctional by disruption of single glycosylation sites. These and additional findings more precisely define the molecular mechanisms of FLS2 receptor function.
INTRODUCTION
Arabidopsis thaliana FLAGELLIN SENSING2 (FLS2) is a transmembrane leucine-rich repeat (LRR) receptor kinase (Gomez-Gomez and Boller, 2000; Torii, 2004; Boller and Felix, 2009). FLS2 is the pattern recognition receptor that mediates plant basal defenses triggered by bacterial flagellin, a microbe-associated molecular pattern (MAMP) (Gómez-Gómez and Boller, 2000). The flg22 region within the conserved N terminus of bacterial flagellins carries the elicitation determinant of bacterial flagellin that is recognized by many plants (Felix et al., 1999; Sun et al., 2006; Robatzek et al., 2007; Takai et al., 2008). FLS2 directly binds flg22 (Chinchilla et al., 2007). Site-directed mutagenesis together with structural modeling implicates the conserved cluster of residues across the β-strand/β-turn region of repeats 9 to 14 of the FLS2 LRR as a likely binding region for flg22 peptide (Dunning et al., 2007).
Multiple elements have been identified that participate with FLS2 to accomplish defense signaling in Arabidopsis. For instance, FLS2 associates with the cytoplasmic protein kinase BOTRYTIS-INDUCED KINASE1 (BIK1) in the absence of flagellin or flg peptides, and FLS2 associates with the transmembrane kinases BRI1-ASSOCIATED RECEPTOR KINASE1 (BAK1) and SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE4 (SERK4; also called BKK1) almost immediately after exposure to flagellin or flg22 (Chinchilla et al., 2007; Heese et al., 2007; Lu et al., 2010; Schulze et al., 2010; Zhang et al., 2010; Roux et al., 2011). Upon flg22 treatment, BIK1, BAK1, and FLS2 gain phosphorylation and then BIK1 dissociates from the FLS2 complex (Chinchilla et al., 2007; Heese et al., 2007; Lu et al., 2010; Schulze et al., 2010; Zhang et al., 2010). The LRR-kinase BAK1-INTERACTING RECEPTOR-LIKE KINASE1 negatively regulates multiple defense signaling pathways, including FLS2 pathways (Gao et al., 2009), while overexpression of KINASE ASSOCIATED PROTEIN PHOSPHATASE suppresses both flagellin-induced signaling and flg22 binding (Gómez-Gómez et al., 2001). FLS2 undergoes ligand-induced endocytosis following flg22 treatment (Robatzek et al., 2006; Chinchilla et al., 2007). Early downstream signaling events for FLS2 also include a Ca2+-associated membrane depolarization, RESPIRATORY BURST OXIDASE HOMOLOG D–mediated oxidative burst, and activation of calcium-dependent protein kinases and mitogen-activated protein kinase cascades (Felix et al., 1999; Peck et al., 2001; Asai et al., 2002; Zhang et al., 2007; Schwessinger and Zipfel, 2008; Boller and Felix, 2009; Boudsocq et al., 2010; Jeworutzki et al., 2010). Specific effector proteins from bacterial pathogens have been identified that can suppress plant defenses by interaction with FLS2, BAK1, or BIK1 (Shan et al., 2008; Xiang et al., 2008; Zhang et al., 2010; Xiang et al., 2011). Despite this extensive information, it remains unclear how FLS2 itself becomes physically activated so that the extracellular flagellin binding event is transduced to activate the FLS2 kinase domain and/or FLS2 partner proteins to initiate defense signaling. Similar structure/function questions remain for many plant receptor-like kinases (RLKs) (Torii, 2004; Morillo and Tax, 2006; Boller and Felix, 2009).
Activity after ligand-induced dimerization is a common theme for mammalian transmembrane receptor Tyr kinases (Ward et al., 2007; De Meyts, 2008). However, preligand receptor associations are also well known, for example, in the TNF family of receptors (Zhang, 2004). Signaling mediated by plasma membrane–spanning Toll-like receptors (TLRs; which are animal MAMP receptors) can involve receptor homodimers or heterodimers and an adaptor complex. For example, TLR2 can form heteromeric receptors with TLR6 or TLR1 that differ in their ligand specificity (Triantafilou et al., 2006). In many cases, ligand binding promotes dimerization or oligomerization of TLRs, including TLR3, TLR4, TLR5, and TLR9. Receptor dimerization promotes conformational changes in the ectodomains and facilitates stable protein–protein interaction (Weber et al., 2005; Bell et al., 2006; Gay and Gangloff, 2007; Latz et al., 2007).
Knowledge of receptor dimerization or oligomerization in plants is relatively scarce, but an emerging theme is that these associations often exist prior to ligand exposure. Arabidopsis SERK1 can form homooligomers, and S-locus receptor kinases in Brassica form homodimers in the absence of ligand (Shah et al., 2001; Naithani et al., 2007). The best-characterized RLK, the brassinolide hormone receptor BRI1, also is thought to form homodimers independent of ligand binding (Wang et al., 2005; Gendron and Wang, 2007; see also Hothorn et al., 2011). BAK1 and SERK1 were also found to exist in the BRI1 multimeric complex (Karlova et al., 2006). The findings that FLS2 forms heteromeric complexes with BIK1 prior to flagellin exposure and with BAK1 after flg22 exposure are noteworthy not only in showing the roles of these proteins in multiple cellular signaling pathways, but also in demonstrating ligand-dependent association of a plant receptor kinase with a signaling partner (Chinchilla et al., 2007; Heese et al., 2007; Lu et al., 2010). These studies did not explore FLS2-FLS2 associations. Based upon failure to detect signal in bimolecular fluorescence complementation and fluorescence resonance energy transfer assays, Ali et al. (2007) suggested that FLS2 exists in monomeric form before and after exposure to flg22, at least when constitutively overexpressed in protoplasts. However, the homomeric and heteromeric interactions of FLS2 merit further investigation.
Like many other plant and animal LRR-containing RLKs, TLRs, and receptor-like proteins (Diévart and Clark, 2003; Gay and Gangloff, 2007), FLS2 has highly conserved Cys pairs immediately flanking the N- and C-terminal ends of the LRR (in FLS2, C61-C68 and C783-C792). These Cys pair domains are often called LRRNT and LRRCT (van der Hoorn et al., 2005; Gay and Gangloff, 2007). In human TLRs, the conserved Cys pairs form disulfide bonds and are crucial for the formation of capping structures at both ends of the LRR (Kajava, 1998; Kim et al., 2007). In plants, the bri1-5 product, which harbors a Cys69Tyr mutation in the LRRNT domain, is a functional BR receptor, although it is mainly retained in the endoplasmic reticulum (ER; Hong et al., 2008). Other work with tomato (Solanum lycopersicum) Cf-9 transiently expressed in tobacco (Nicotiana tabacum) did show impacts on protein function despite protein accumulation (van der Hoorn et al., 2005; Kolade et al., 2006). However, the influences of the conserved Cys pairs on FLS2 stability, processing, and function have not been reported.
Another significant feature of LRR receptors and other plant and animal plasma membrane receptors is that the extracellular domains are often glycosylated at multiple sites, or where this has not been explicitly determined, they often carry a large number of putative N-linked glycosylation sites (PGSs; N in NxS/T motifs) (van der Hoorn et al., 2005; Ohtsubo and Marth, 2006). FLS2 contains 20 PGSs in its LRR domain and one more in the LRRCT region. Glycosylation of extracellular domains can facilitate protein folding and transport to the cell surface, as well as appropriate ligand binding, signaling, and receptor stability (Ohtsubo and Marth, 2006). In tomato Cf-9, all PGSs except PGS18 were demonstrated to be N-glycosylated, and functional analyses of PGS mutants showed that all of the glycosylation sites were important for Cf-9 activity (van der Hoorn et al., 2005). Multiple studies on the importance of glycosylation for Arabidopsis EF-Tu RECEPTOR (EFR) function have recently been published, some of which also touch upon FLS2 glycosylation (Li et al., 2009; Nekrasov et al., 2009; Saijo et al., 2009; Häweker et al., 2010).
In this study, we investigate the functional contributions of multiple FLS2 protein domains and modifications. The majority of tests use stable transformation of Arabidopsis to study modified FLS2 proteins in their natural environment and, for any single experimental treatment, report the behavior of many independent T1 transformants. Signaling output assays are coupled with immunoprecipitation assays to discover multiple previously unidentified relationships between FLS2 structure and function.
RESULTS
FLS2-FLS2 Associations Are Present before and after Ligand Binding
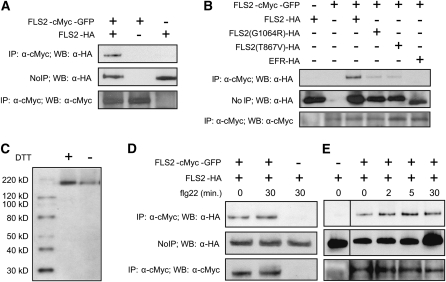
To test if FLS2 associates with FLS2 in vivo, we coexpressed, from separate constructs, two differently tagged FLS2 molecules in the same seedlings. Tissue was homogenized in the presence of 0.5% Triton X-100 to solubilize membranes and then proteins carrying one of the tags were immunoprecipitated, followed by a test for the second tag in the immunoprecipitate. The basic result is shown in Figure 1A: When hemagglutinin A (HA)-tagged FLS2 and FLS2-cMyc-green fluorescent protein (GFP) are coexpressed under the control of the native FLS2 promoter in Wassilewskija-0 (Ws-0) plants (naturally fls2− due to a premature stop codon in the FLS2 gene), FLS2-HA copurifies with FLS2-cMyc-GFP in the immune complex precipitated by anti-cMyc antibody. Both epitope-tagged FLS2 alleles have previously been studied and shown to function like wild-type FLS2 (Robatzek et al., 2006; Dunning et al., 2007; this study). These and all other experiments in this study were repeated at least one other time with similar results unless specifically noted. Control experiments were performed to exclude nonspecific association of the two proteins in these assays, as might occur for example by incomplete solubilization of the membranes. EFR, the transmembrane LRR kinase receptor for the bacterial MAMP EF-Tu (Zipfel et al., 2006), did not associate with FLS2 (Figure 1B). In tag-switch experiments, again with FLS2 transgenes expressed under the control of the native FLS2 promoter in stable transgenic plants, FLS2-FLS2 association is observed using FLS2-FLAG in place of FLS2-myc-GFP and is also observed in a Columbia-0 (Col-0) fls2-101 genetic background in addition to Ws-0 (see Supplemental Figure 1A online; Figure 2B). Supplemental Figure 1A online also shows the relative signal when FLS2 coimmunoprecipitation (co-IP) of FLS2 is compared with FLS2 co-IP of BAK1. FLS2-HA was detected in FLS2-cMyc-GFP immune complexes precipitated by either anti-myc antibody or by anti-GFP polyclonal antibody and not by anti-FLAG antibody of the same IgG1 immunoglobulin subtype (see Supplemental Figure 1B online). To document concentration-dependent detection and the relative sensitivity of detection of in vivo FLS2-FLS2 interaction, co-IP experiments were performed with different amounts of total protein extract (see Supplemental Figure 1C online).
Figure 1.
Intermolecular FLS2-FLS2 Association in Vivo.
(A) FLS2-HA expressed from FLS2 promoter is present in the FLS2-cMyc-GFP complex immunoprecipitated with an anti-cMyc antibody. Top: Immunoblot of immunoprecipitation products from seedling extracts, pulled down using anti-cMyc antibodies, probed with anti-HA antibody. Middle: Immunoblot of crude plant extract supernatant (no immunoprecipitation) probed with anti-HA antibody. Bottom: Immunoblot of immunoprecipitation products from seedling extracts pulled down using anti-cMyc, probed with anti-cMyc antibody. Vertically aligned lanes from all three panels derived from the same initial extracts of pooled Arabidopsis Ws-0 T1 seedlings transformed, as noted at the top of the figure, with FLS2-myc-GFP and/or FLS2-HA.
(B) The kinase-inactive mutants FLS2G1064R-HA and FLS2T867V-HA partially lose the FLS2-FLS2 interaction and Arabidopsis EFR-HA, another LRR-kinase, fails to coimmunoprecipitate with FLS2-cMyc-GFP. Panels are as in (A) except as noted.
(C) No apparent molecular weight shift for FLS2 from Arabidopsis protein samples treated with or without the reducing regent (DTT) prior to electrophoresis. Immunoblot detection used anti-HA antibody.
(D) FLS2-FLS2 association occurs in the presence or absence of flg22 ligand. Panels are as in (A), except with or without 30-min exposure to 10 μM flg22 as noted.
(E) Time course showing increase in FLS2-FLS2 association after flg22 treatment. Panels are as in (D) except as noted; all lanes in (E) are from same gel and blot.
Within each panel of this and all other figures, all sample lanes were loaded with equal amounts of total plant protein, and all co-IP experiments were repeated at least twice with similar results, unless specifically noted. For this figure, all FLS2 constructs except in (B) were expressed from FLS2 promoters.
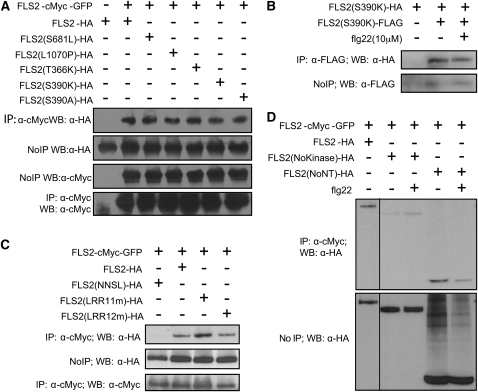
Figure 2.
In Vivo Association of FLS2 with Mutant FLS2 Proteins.
co-IP experiments were performed and labeled as in Figure 1. All experiments were repeated at least twice with similar results.
(A) Complex formation in vivo between FLS2-myc-GFP and HA-tagged FLS2T366K and FLS2S390K (which lack flg22 binding activity; Dunning et al., 2007) as well as FLS2S390A (which retains flg22 binding [Dunning et al., 2007], as well as two other mutants, FLS2S681L and FLS2L1070P; these mutations also in FLS2NNSL below).
(B) Complex formation in vivo between FLS2S390K-HA and FLS2S390K-FLAG.
(C) FLS2T342A/H344A and FLS2T363A/T366A (double mutants in the 11th and 12th LRR repeats, respectively) form FLS2-FLS2 multimers with FLS2-myc-GFP, while the FLS2NNSL (FLS2N179D, N388D, S681L, L1070P) quadruple mutant does not.
(D) FLS2 can associate with the truncated FLS2 proteins FLS2NoKinase and FLS2NoNT in vivo and associates with or without flg22 treatment. FLS2NoKinase-HA and FLS2NoNT-HA expressed from 35S promoter. All lanes are from same gel and blot and were loaded with equivalent amounts of total plant protein. Schematic of protein constructs is shown in Figure 5A.
The impact of the function-blocking FLS2T867V and FLS2G1064R intracellular domain mutations (Gómez-Gómez et al., 2001; Robatzek et al., 2006) on FLS2-FLS2 association was investigated. Reduced co-IP of these mutant FLS2 proteins was observed (Figure 1B). This suggests that the mutated proteins have a reduced capacity to undergo FLS2-FLS2 associations, although in the case of FLS2T867V-HA and FLS2G1064R-HA, the somewhat reduced abundance of these proteins compared with FLS2WT-HA may also contribute to this effect.
To test the possibility that FLS2 molecules interact with each other through intermolecular disulfide bonds, we performed immunoblot analyses on protein samples in the presence or absence of the reducing reagent DTT. Addition of the reducing agent did not cause molecular weight shift, suggesting that FLS2 apparently does not form intermolecular disulfide bonds with adjacent FLS2 molecules (Figure 1C).
To test if flg22 treatment alters FLS2-FLS2 association, seedlings were incubated with 10 μM flg22. Relative to the untreated control, the intensity of the FLS2-FLS2 association co-IP product remained similar or was slightly increased after 30 min of exposure to flg22 (for example, see Figure 1D). Previous studies demonstrated that early FLS2-mediated responses to flg peptides, such as FLS2 association with and phosphorylation of BAK1, are observable less than a minute after flg22 treatment (Felix et al., 1999; Chinchilla et al., 2006; Heese et al., 2007; Lu et al., 2010; Schulze et al., 2010; Zhang et al., 2010). Therefore, the effect of flg22 on FLS2-FLS2 association was also examined at earlier time points. Slightly elevated amounts of FLS2-HA were present in the anti-cMyc antibody pulldown complex at 2 and 5 min after flg22 addition (Figure 1E). Similar results were obtained in repeat experiments in Col-0 and Ws-0 genetic backgrounds (for example, see Supplemental Figure 1D online). Some variability in the amount of FLS2 protein was observed over the time course of flg22 exposure (lanes were loaded with equal amounts of total plant protein). The data indicate that in vivo, FLS2 is present at least partially in FLS2-FLS2 complexes whose abundance relative to total FLS2 undergoes a small increase in the presence of flg22.
FLS2-FLS2 Association Is Separable from Ability to Bind flg22
co-IP experiments were conducted to directly investigate if the flg22 binding activity of FLS2 is necessary for its ability to form FLS2-FLS2 associations. We previously identified solvent-exposed residues along the proposed concave face of repeats #9-15 of the FLS2 LRR that play an essential role in flg22 perception (Dunning et al., 2007). Several point mutations in this region, including FLS2T366K and FLS2S390K, but not FLS2S390A, cause FLS2 to lose flg22 binding activity (Dunning et al., 2007). FLS2T366K and FLS2S390K still interacted with FLS2WT (Figure 2A) even though they have lost flg22 binding activity. Likewise, the alleles FLS2T342A/H344A (LRR11m) and FLS2T363A/T366A (LRR12m), which have significantly lost flg22 responsiveness (Dunning et al., 2007), retained the ability to interact with FLS2-cMyc-GFP (Figure 2C). FLS2S390A, which retains flg22 binding, also interacts. FLS2-FLS2 association in fls2− lines coexpressing FLS2S390K-HA and FLS2S390K-FLAG demonstrated that a wild-type FLS2 is not required to drive this association (Figure 2B).
We also found FLS2 mutants that failed to form FLS2-FLS2 associations. For example, FLS2NNSL, a flg22-insensitive spontaneous PCR mutant with four point mutations encoding N179D, N388D, S681L, and L1070P, failed to coimmunoprecipitate with FLS2-cMyc-GFP, indicating that FLS2 proteins are not generically pulled down in this assay (Figure 2C; see also Figure 3). To follow up on the FLS2NNSL result, the single mutations S681L and L1070P were constructed. These proteins interact with FLS2-cMyc-GFP (Figure 2A), despite the fact that Col-0 fls2-101 plants expressing the FLS2L1070P allele lack FLS2 activity (lack a response to flg22 in seedling growth inhibition assays; data not shown).
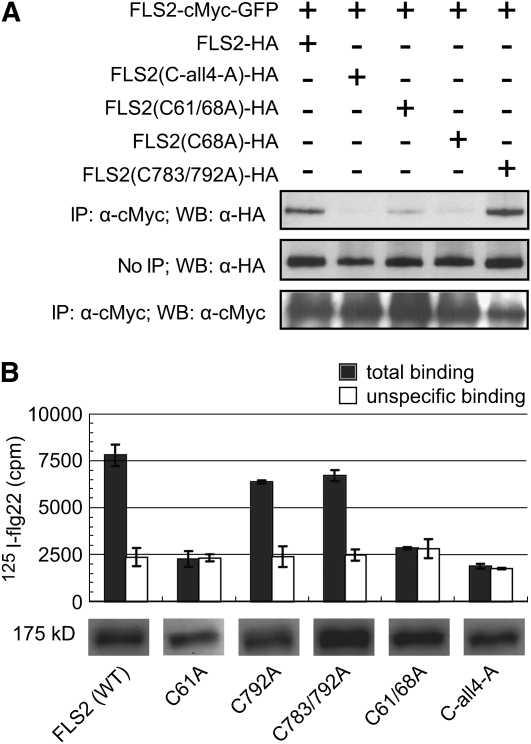
Figure 3.
Mutation of the N-Terminal Cys Pair of FLS2 Disrupts FLS2-FLS2 Association and flg22 Binding.
FLS2-HA is wild-type HA-tagged FLS2; C-all4-A carries Cys-to-Ala mutations at Cys-61, Cys-68, Cys-783, and Cys-792; other lanes carry single or double mutants as noted. FLS2-HA constructs were expressed from CaMV 35S promoters. Similar results were obtained in replicate experiments.
(A) co-IP experiment (performed and labeled as in Figure 1) using the HA-tagged alleles noted above each lane, together with FLS2-myc-GFP.
(B) Binding of 125I-flg22 by seedling extracts made from Col-0 fls2-101 plants expressing the designated FLS2-HA constructs driven by the 35S promoter. Data are mean ± se; specific binding is the difference between total binding (125I-flg22 added with no unlabeled flg22 competitor) and unspecific binding (125I-flg22 bound in presence of 1000-fold molar excess of unlabeled flg22). Aliquots of the plant extracts used in binding experiment were separated by SDS-PAGE, blotted, and probed with anti-HA antibody to detect abundance of FLS2 in each extract (total plant protein per lane was equal for all lanes; all images are from the same blot). cpm, counts per minute; WT, wild type.
FLS2-FLS2 Association Occurs through the Intracellular and Extracellular Domains
Truncated FLS2 proteins were used in co-IP experiments to investigate further the portions of FLS2 that mediate FLS2-FLS2 association, again using extracts from stably transformed Arabidopsis plants. Figure 2D shows that full-length FLS2 coimmunoprecipitated a truncated FLS2(NoNT) consisting only of the predicted intracellular portions, including the FLS2 protein kinase but lacking the LRRs. More weakly but also reproducibly, full-length FLS2 coimmunoprecipitated a truncated FLS2(NoKinase) that carries only the predicted extracellular domains, including the FLS2 LRRs but lacking the protein kinase region (Figure 2D). The results suggest that FLS2-FLS2 association is mediated both through intracellular domain interactions and extracellular domain interactions.
The Conserved LRR N-Terminal Cys Pair Impacts FLS2-FLS2 Association and flg22 Binding Activity, but the Membrane-Proximal Cys Pair Does Not
To study the role of the conserved Cys pairs in the LRR-capping domains, we constructed single, double, triple, and quadruple mutations to change the relevant LRRNT and LRRCT Cys codons of FLS2 to Ala codons. Constructs were then expressed by stable transformation of fls2− mutant plants (Col-0 fls2-101) as epitope-tagged FLS2-HA proteins, under the control of the cauliflower mosaic virus (CaMV) 35S promoter in the first set of experiments to ensure maximal expression. All constructs caused substantial accumulation of the corresponding protein (Figure 3A). The FLS2 LRRNT and LRRCT Cys pair mutants were then tested for interaction with wild-type FLS2-cMyc-GFP in vivo. As shown in Figure 3A, FLS2C68A, FLS2C61/68A, and FLS2C-all4-A exhibited reduced FLS2-FLS2 association, whereas FLS2C783/792A resembled FLS2WT-HA in its co-IP with wild-type FLS2-cMyc-GFP protein. The flg22 binding capacity of FLS2 Cys pair mutants was also tested. In contrast with wild-type FLS2-HA, plants expressing FLS2 with a disrupted Cys pair in the LRRNT lacked detectable flg22 binding activity (Figure 3B). Plants expressing FLS2 with a disrupted Cys pair in the LRRCT but an intact Cys pair in the LRRNT could still bind flg22. Thus, the conserved Cys pair at the FLS2 LRR N-terminal region is a prerequisite both for successful FLS2-FLS2 association and for flg22 binding, while the membrane-proximal Cys pair is not.
The Conserved LRRNT Cys Pair Is Essential for FLS2 Processing, Stability, and Function; the LRRCT Cys Pair Is Not
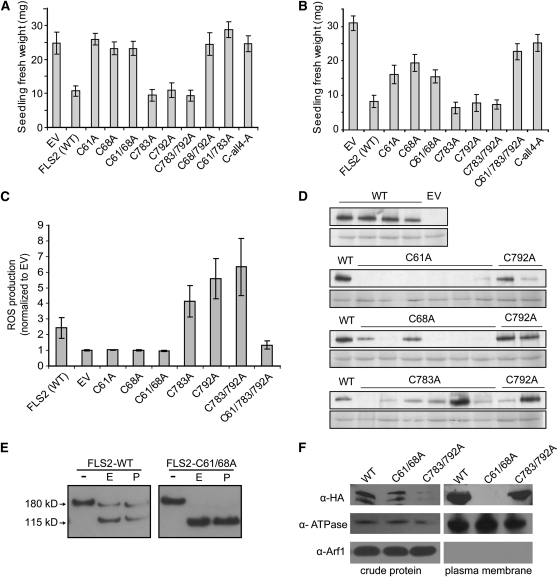
To further investigate the impact of the LRR-flanking Cys pairs on FLS2 function, transformed Col-0 fls2-101 T1 plants carrying the various FLS2 alleles with LRRNT and LRRCT mutations were tested for FLS2-mediated responses using the standard assay for seedling growth inhibition in response to flg22 (e.g., Gómez-Gómez et al., 1999; Chinchilla et al., 2006; Dunning et al., 2007; Heese et al., 2007). As previously reported (Dunning et al., 2007), the full-length FLS2WT construct with its HA-tag was functional and caused strongly reduced seedling fresh weight in the presence of flg22, in comparison to the empty-vector (EV) control (Figure 4A). However, all of the FLS2 constructs encoding the C61A and/or the C68A mutation in the LRRNT domain lost flg22 responsiveness. The constructs containing only the C783A and/or the C792A mutations in the LRRCT domain retained strong FLS2 function. Although variable levels of expression between independent transgenic plants carrying the same construct were evident, as is commonly observed, immunoblot analyses (see Supplemental Figure 2A online) showed that HA-tagged FLS2 was readily detectable for all of the 35S-FLS2 mutant alleles used to generate Figure 4A. The data demonstrate that the conserved Cys pair at the LRRNT region is functionally important, whereas the Cys pair at the LRRCT (membrane-proximal) region is not essential for FLS2 function.
Figure 4.
The Conserved Cys Pair at the FLS2 LRR N Terminus Impacts FLS2 Production, Processing, and/or Stability, in Addition to Its Function, but the Membrane-Proximal Cys Pair Does Not.
EV, no FLS2; FLS2 (WT), wild-type FLS2; C61A, Cys-61 mutated to Ala (similar nomenclature for other single and double mutants); C-all4-A, FLS2 quadruple mutant encoding C61A, C68A, C783A, and C792A. All FLS2 alleles encoded an HA-tagged protein under control of the 35S promoter.
(A) Functional test of FLS2 Cys-to-Ala alleles driven by 35S promoter in transgenic T1 seedlings of Arabidopsis Col-0 fls2-101, transformed with the specified FLS2 allele and subjected to seedling growth inhibition assay in the presence of 2.5 μM flg22. Mean ± se is shown for multiple independent transformants (usually 10) for each allele. WT, wild type.
(B) Functional test of FLS2 Cys-to-Ala alleles driven by native FLS2 promoter, tested in transgenic T1 seedlings of Arabidopsis Ws-0 accession (Ws-0 naturally lacks a functional FLS2). Seedling growth inhibition assay in the presence of 2.5 μM flg22; mean ± se are shown. All FLS2 alleles were expressed as cMyc-GFP–tagged proteins.
(C) Production of ROS induced by 1 μM flg22 in leaf samples from plants described in (B), detected using luminol reagent. Area under the curve for 30 min. oxidative burst (see Supplemental Figure 2B online) was calculated for each sample and then normalized to mean value for EV transgenics tested within same experiment. Data shown are combined from three independent experiments and depict mean ± se; n ≥ 8 independent T1 lines for each construct.
(D) FLS2 expression driven by the native FLS2 promoter, detected by immunoblot analysis using an anti-cMyc antibody, for randomly chosen individual transgenic T1 seedlings from the experiment in (B). Bottom: Ponceau S staining of same blot to detect total protein.
(E) Glycosylation state of FLS2 and an FLS2 mutant lacking the LRRNT Cys pair, detected by immunoblot after the treatment of Endo H and PNGase F. FLS2 expression driven by 35S promoter in Col-0 fls2-101. -, No treatment; E, Endo H treatment; P, PNGase treatment.
(F) Presence of FLS2 proteins in plasma membrane–enriched fraction. Two-phase partitioning experiment was performed using transgenic plants with HA-tagged FLS2-WT, FLS2-C61/68A, or FLS2-C783/792A expressed from FLS2 promoter. α-Arf1 detects the cytosolic ADP-ribosylation factor 1 protein; α-ATPase detects the plasma membrane H+-ATPase.
We also studied FLS2 LRRNT and LRRCT mutants expressed from a native FLS2 promoter. Seedling growth inhibition assays showed, as for the 35S constructs, that LRRCT Cys mutants conferred a flg22 response similar to wild-type FLS2, while the LRRNT Cys mutants had a clearly diminished flg responsiveness (Figure 4B). Additionally, transgenic T1 plants expressing FLS2 C61A, C68A, or C61/68A (LRRNT) mutants did not produce an oxidative burst after flg22 treatment, whereas transgenic plants with FLS2 C783A, C792A, or C783/792A (LRRCT) mutations reproducibly responded to flg22 treatment with a stronger oxidative burst than the wild type, as long as the FLS2 in question did not also carry an LRRNT mutation (Figure 4C; see Supplemental Figure 2B online). In callose deposition assays with the native promoter constructs in Ws-0, flg22 treatment induced callose deposition in only a small proportion of seedlings carrying an FLS2 transgene encoding C61A or C68A mutations, whereas new callose was prominent in the majority of seedlings expressing FLS2WT, or C783A or C792A mutations (data not shown). Protein abundance experiments were consistent with these functional results. In transgenic Ws-0 ecotype (naturally fls2−) carrying the FLS2-HA transgene under the control of the native FLS2 promoter, immunoblot analyses with randomly chosen T1 seedlings showed that only 25% of seedlings (three out of 12) had detectable FLS2 expression when C61 or C68 were mutated in FLS2. By contrast, most of the seedlings (11 out of 12) expressing the FLS2C783A or FLS2C792A alleles had detectable FLS2 protein level (Figure 4D).
The possibility of incomplete/incorrect processing of newly synthesized FLS2 was then investigated using standard Endoglycosidase H (Endo H) and N-glycosidase F (PNGase F) tests. These enzymes cleave off glycans from ER-localized immature glycosylated proteins that are in the early stages of maturation before processing through the Golgi, while mature membrane proteins that have been successfully delivered after processing through the Golgi are largely insensitive to Endo H or PNGase F (Nekrasov et al., 2009). Treatment of T1 seedling extracts revealed that C68 and C61/68 mutations eliminate the Endo H–insensitive or PNGase-insensitive pool of FLS2 (Figure 4E), suggesting little or no presence of mature protein, whereas the C783/792 mutant retains the Endo H–insensitive fraction expected for mature receptor proteins (see Supplemental Figure 2C online). Two-phase partitioning experiments further confirmed that FLS2 with C61/68A mutations no longer localizes to the plasma membrane fraction, unlike wild-type FLS2 and FLS2 with C783A/C792A mutations (Figure 4F). The results in Figures 4D to 4F indicate that the LRRNT Cys pair is required for correct processing and stability of FLS2. Together with the FLS2-FLS2 association and flg22 binding data, and with the data that FLS2-HA LRRNT mutant proteins confer only partial FLS2 function and do so only in a minority of transgenic lines, these protein abundance and glycosidase experiments indicate that the LRRNT Cys pair is very important but not absolutely required at multiple stages: for FLS2 processing, stability, and function.
Truncated FLS2NoNT Protein Interferes with the Function of Endogenous FLS2
To further dissect the structure and function of FLS2, seven specifically chosen FLS2 mutant alleles were expressed with a C-terminal HA tag under the control of the 35S promoter (Figure 5A). The FLS2G1064R mutation is found in fls2-17 plants, a widely used mutant background for FLS2 studies in which flg22 binding and FLS2 signaling are nonfunctional (Gómez-Gómez et al., 2001). FLS2T867V, mutated in a predicted phosphorylation site of the intracellular juxtamembrane domain, was previously shown to exhibit normal flg22 binding but loss of FLS2 signaling and of FLS2 protein endocytosis after flg22 stimulation (Robatzek et al., 2006). FLS2NNSL, as noted above, is a spontaneous PCR mutant with four point mutations encoding N179D, N388D, S681L, and L1070P. Four other newly constructed alleles encode truncated FLS2 gene constructs FLS2NoKinase (FLS2 amino acids 1 to 869), FLS2NoNT (amino acids 777 to end), FLS2NoLRR (amino acids 1 to 42 and 799 to end; deletion of amino acids 43 to 798), and FLS2NoCT (amino acids 1 to 1153), as is depicted in Figure 5A. These FLS2 mutant alleles failed to restore flg22 responsiveness when they were expressed from a 35S promoter in the fls2-101 background (Figure 5B). Expression of the protein products except FLS2NoCT was readily detectable by immunoblot analyses, although it was weak for the FLS2G1064R allele (see Supplemental Figure 3A online). Previous studies reported that the FLS2G1064R allele, carrying a mutation in the kinase domain, does not confer FLS2 function and produces an unstable FLS2 protein when expressed under its native promoter (Gómez-Gómez et al., 2001; Robatzek et al., 2006). Our results show that the FLS2G1064R and FLS2T867V mutants are nonfunctional even when protein abundance is restored by expression from a strong promoter. Immunoblot analyses (see Supplemental Figure 3 online; additional replicates not shown) also demonstrated that the FLS2NoCT variant consistently had a low abundance compared with wild-type FLS2, suggesting that the C-terminal tail in FLS2 has an important role in FLS2 stability.
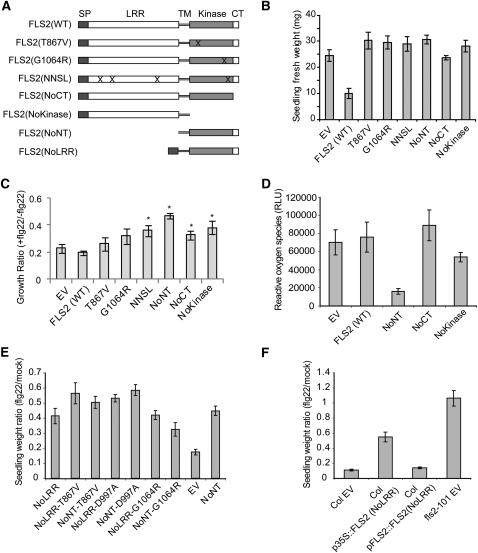
Figure 5.
FLS2NoNT and FLS2NoLRR Have a Dominant-Negative Effect on FLS2 Function, and the Dominant-Negative Effect Is Dependent on Expression Level but Independent of Kinase Activity.
(A) Schematic of the mutated and truncated FLS2 genes tested. CT, C terminus (C-terminal 20 amino acids); Kinase, protein kinase domain; NNSL, N179D, N388D, S681L, and L1070P quadruple mutant; SP, native N-terminal export signal amino acids; TM, transmembrane domain; WT, wild type. “X” marks indicate approximate location of amino acid changes; all constructs were made in pGWB14 (see Methods).
(B) Functional test of the mutated FLS2 proteins in an fls2− genetic background, determined by seedling growth inhibition assay. All FLS2 variants (see [A]) were placed under control of the CaMV 35S promoter and transformed into Col-0 fls2-101 plants. Data for each allele are for multiple independent T1 transgenic seedlings grown in the presence of 2.5 μM flg22.
(C) Functional test of the mutated FLS2 proteins in a Col-0 (FLS2+) genetic background, determined by seedling growth inhibition assay with 2.5 μM flg22. The constructs in (A) were transformed into wild-type Col-0 plants. Similar results were obtained in three repeat experiments.
(D) Functional test of the mutated FLS2 proteins in a Col-0 (FLS2+) genetic background, determined by oxidative burst measurements. The area under the oxidative burst curve (relative luminescence units × time) was determined for each transgenic seedling; histogram shows mean ± se for eight T1 seedlings. Averaged ROS traces (luminescence over time) are shown in Supplemental Figure 3D online.
(E) Functional test of mutated FLS2 constructs transformed into Col-0 (FLS2+) genetic background, determined by seedling growth inhibition assay of T1 seedlings in 2.5 μM flg22.
(F) Functional test of FLS2NoLRR proteins with expression driven by 35S promoter or FLS2 native promoter, in a Col-0 (FLS2+) genetic background, determined by seedling growth inhibition assay of T1 seedlings in 2.5 μM flg22.
In (B) to (F), mean ± se are shown.
The FLS2 constructs of Figure 5A were also expressed under the control of the 35S promoter in the Col-0 ecotype, which carries a wild-type form of FLS2 and therefore responds to flg22. The mutant FLS2 proteins were all produced in Col-0, although FLS2NoKinase and FLS2NoCT accumulated to lower levels than FLS2WT (see Supplemental Figures 3B and 3C online). Assays with multiple independent T1 seedlings showed that Col-0 plants transformed with EV displayed the typical seedling growth inhibition in response to flg22, resulting in a very low seedling fresh weight as in previous studies (Figure 5C). However, the FLS2 kinase construct FLS2NoNT strongly and reproducibly reduced flg22-induced growth inhibition, indicating a dominant-negative effect on wild-type FLS2 function. Overexpression of some of the other 35S-FLS2 constructs, including FLS2WT, seemed to partially reduce overall FLS2 activity in this set of experiments, possibly due to weak dominant-negative activity or to occasional cosuppression of both the transgene and the endogenous FLS2 gene. However, when the flg22-induced oxidative burst was monitored in experiments with other transgenic plants, a strong dominant-negative action was observed only for FLS2NoNT (Figure 5D). 35S-driven expression of the FLS2NoNT construct in transgenic Col-0 T1 and T2 plants blocked all detectable oxidative burst induced by flg22 treatment, whereas reactive oxygen species (ROS) were still generated after flg22 treatment in transgenic Col-0 plants expressing the FLS2NoCT, FLS2NoKinase, and FLS2WT constructs (T1 plants in Figure 5D and Supplemental Figure 3D online; T2 plants in Supplemental Figure 3E online). Consistently, flg22-induced ethylene production, mitogen-activated protein kinase cascade activation (MPK3 and MPK6 phosphorylation), and restriction of the growth of Pseudomonas syringae pv tomato strain DC3000 also were inhibited in FLS2NoNT transgenic Col-0 plants (see Supplemental Figures 3G to 3I online). Overexpression of the FLS2NoNT construct did not reduce the levels of epitope-tagged wild-type FLS2 in Ws-0 plants (see Supplemental Figure 3F online).
The dominant-negative activity of the FLS2 intracellular kinase domain (FLS2NoNT) was further dissected using a kinase-dead allele in which a D997A mutation disrupts the core Asp that is central to the catalytic activity of this type of protein kinase (Knighton et al., 1991). Full-length FLS2D997A fails to confer flg22 responsiveness (see Supplemental Figure 3J online). The FLS2NoNT/D997A construct with the D997A mutation still retained dominant-negative activity (Figure 5E). The same was true of the similar FLS2NoLRR/D997A construct. FLS2NoNT and FLS2NoLRR constructs with T867V or G1064R mutations also retained dominant-negative activity over wild-type FLS2 when overexpressed under a CaMV 35S promoter (Figure 5E). The quantitative rather than qualitative nature of this dominant-negative activity was further documented by the finding that function of native FLS2 (in wild-type Col-0) was retained if expression of the FLS2NoNT transgene was under the control of a native FLS2 promoter, or if the 35S-FLS2NoNT construct was transformed into a Ws line that strongly expresses a full-length wild-type FLS2-myc-GFP transgene (Figure 5F; data not shown).
To test if the dominant-negative effect of FLS2NoNT is due to nonproductive occupation of shared downstream signaling partners, such as BAK1 or BIK1, which are also used in other signaling pathways, we performed seedling growth inhibition assays to test for disruption of the EFR-mediated response to elf18 treatment. The response was similar with or without FLS2NoNT overexpression, at each of three concentrations of elf18 (see Supplemental Figure 3K online). Furthermore, co-IP assays that detected the previously documented flg22-dependent interaction of BAK1 with FLS2 did not detect an interaction between BAK1 and FLS2NoNT (see Supplemental Figure 3L online).
Unlike EFR, FLS2 Is Relatively Insensitive to Mutation of Putative N-Glycosylation Sites
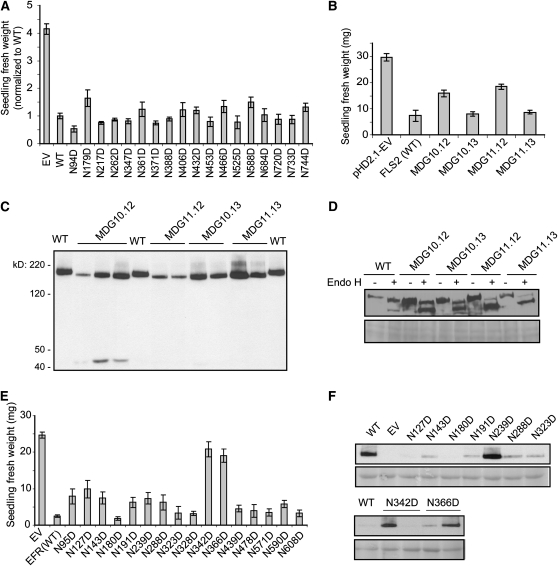
FLS2 appears to be a glycosylated protein, based on its unexpectedly slow migration in SDS-PAGE and the 21 PGSs in the FLS2 extracellular domain. In vitro removal of carbohydrate from the FLS2 protein backbone using the endoglycosidases PNGase F and Endo H (Figure 4E) corroborated the previous observation that FLS2 is a glycosylated protein (Chinchilla et al., 2006). To determine the functional significance of N-linked glycosylation in FLS2 extracellular domain, we used site-directed mutagenesis to replace the Asn codon with an Asp codon in the PGSs in FLS2 constructs driven by the native FLS2 promoter. For each allele, multiple independent T1 transgenic seedlings were generated and tested for responsiveness to flg22. Repeated seedling growth inhibition assays showed that none of the single-PGS mutations caused a reproducible difference from the wild-type FLS2 in responsiveness to flg22 (Figure 6A). These results were confirmed in assays for flg22-elicited ethylene production, which was not significantly different between fls2-101 plants transformed with FLS2WT or single PGS mutation alleles for all 15 of the PGS alleles that were tested (data not shown). In subsequent experiments, FLS2 alleles with double, quadruple, sextuple, and octuple PGS replacements were made and expressed downstream of native FLS2 promoter in stable transgenic plants. Among these, none showed a major loss of FLS2 function, and only two mutants, both with octuple PGS mutations, partially lost flg22 responsiveness (Figure 6B; data not shown). Immunoblot analyses showed a detectable molecular weight reduction for all of the octuple PGS mutants, suggesting a depletion of glycosylation (Figure 6C). The four octuple PGS mutants were further investigated using Endo H. Although all of these mutants still generate the Endo H–insensitive form expected of protein that has been delivered from endomembrane system to plasma membrane, MDG11.12 and 11.13 produced much less (Figure 6D). Immunoblots also detected, in separate experiments, smaller C-terminal HA-tagged bands in plants expressing the MDG10.12 and 11.12 proteins, apparently due to proteolytic release from what was originally full-length FLS2 protein (for example, see Figure 6C).
Figure 6.
FLS2 Is Apparently Less Sensitive to Disruption of PGSs Than EFR.
(A) Replacement of single PGSs in FLS2 does not detectably alter response to flg22. Multiple independent transgenic T1 seedlings of Col-0 fls2-101 transformed with mutant FLS2-HA constructs as noted (all expressed from native FLS2 promoter) were assayed by seedling growth inhibition in 10 μM flg22. Data shown are mean ± se. Each seedling weight was normalized to average weight of Col-0 fls2-101 transformed with wild-type (WT) FLS2-HA, from the same experiment, to allow comparison of data from separate experiments.
(B) The FLS2MDG10.12 and FLS2MDG11.12 mutants that each carry eight PGS mutations partially lose responsiveness to flg22, while the other two octuple PGS mutants respond normally to flg22, as tested by seedling growth inhibition assays in 2.5 μM flg22. Data shown are mean ± se. pHD2.1-EV expresses FLS2 lacking the LRR region (controlled by native FLS2 promoter); same vector with wild-type or mutant LRR-encoding region used in other samples. The octuple PGS mutants are as follows: MDG10.12: N94D, N179D, N217D, N262D, N388D, N406D, N432D, and N525D; MDG11.12: N262D, N347D, N361D, N371D, N388D, N406D, N432D, and N525D; MDG10.13: N94D, N179D, N217D, N262D, N588D, N684D, N733D, and N744D; MDG11.13: N262D, N347D, N361D, N371D, N588D, N684D, N733D, and N744D.
(C) Immunoblot with anti-HA antibody used for detection, showing that mutation of PGS leads to a reduction in the size (apparent molecular weight) of FLS2, presumably due to reduced glycosylation. WT, wild-type FLS2-HA (loaded three times); octuple-PGS FLS2-HA products are described in (B).
(D) Immunoblot analysis with anti-HA antibody used for detection of FLS2 MDG10.12, MDG10.13, MDG11.12, and MDG11.13 after treatment with or without Endo H. Bottom: Same blot after Ponceau staining for total protein.
(E) EFRN342D and EFRN366D mutants with single PGS mutations lose responsiveness to 1 μM elf18, and a partially reduced response was also observed with some other EFR single PGS mutants. Data shown are mean ± se.
(F) Detectable levels of EFR-HA protein in randomly selected transgenic T1 seedlings are highly variable but similar for PGS mutants and wild-type EFR. Immunoblot with anti-HA antibody used for detection. Bottom: Same blot after Ponceau S staining for total protein.
EFR is another plasma membrane receptor for a bacterial MAMP, EF-Tu, and EFR has 16 PGSs in the extracellular LRR domain (Zipfel et al., 2006). We made a set of single PGS mutations in EFR and tested for elf18 responsiveness. Two single PGS mutants (EFRN342D and EFRN366D) completely lost elf18-induced seedling growth inhibition, and at least six other single PGS mutants exhibited partial loss of elf18 responsiveness (Figure 6E). Like FLS2 and many other transgenically expressed proteins, EFR levels were variable between transgenic lines, but in the nonfunctional N342D and N366D mutants, EFR protein was not reproducibly absent (Figure 6F). Impacts of disrupted glycosylation on EFR function were then also reported by others (Li et al., 2009; Nekrasov et al., 2009; Saijo et al., 2009; Häweker et al., 2010). In summary, although N-linked glycosylation is a common feature of plant LRR RLK receptors and FLS2 carries this glycosylation, and although for two of four octuple PGS mutants tested a subpool of FLS2 did exhibit some perturbations, FLS2 function (unlike EFR) was remarkably unperturbed by mutation of multiple PGSs.
DISCUSSION
Transmembrane LRR-RLKs are a major class of plant proteins, accounting for ~1% of the genes in the genomes of Arabidopsis, rice (Oryza sativa), and poplar (Populus spp; Shiu et al., 2004; Tuskan et al., 2006). The broader goal of this study was to provide insight into structure-function relationships within LRR-RLK proteins, using FLS2 as a particularly well-studied example in which models for receptor mode of action are becoming increasingly refined.
FLS2-FLS2 Association
We observed, in a variety of experiments, that a significant portion of FLS2 is present in FLS2-FLS2 associations prior to ligand exposure. This is consistent with previous observations for many types of transmembrane receptors, including mammalian Tyr kinases such as G-protein–coupled receptors or insulin receptors, and plant receptors, including S-receptor kinase and BRI1 (Zhang, 2004; Bulenger et al., 2005; Wang et al., 2005; Naithani et al., 2007; De Meyts, 2008; Harding and Hancock, 2008; Nakasako et al., 2008). The simplest and most likely model is that FLS2 is present as a dimer. However, we use the term “FLS2-FLS2 associations” because FLS2 may also be present in larger multiprotein complexes, such as receptor matrices or nanoclusters (Agnati et al., 2005; Harding and Hancock, 2008). Models cannot be eliminated in which FLS2-FLS2 association occurs through one or more intermediary bridging molecules (such as BIK1) between the FLS2 proteins. Such models seem less likely in light of our finding that in planta, truncated FLS2 proteins can form FLS2-FLS2 association through the intracellular domain or the extracellular domain. However, the reported failure of Ali et al. (2007) to observe FLS2-FLS2 association by fluorescence resonance energy transfer and bimolecular fluorescence complementation methods may also speak to this latter model. Their data could have arisen if constitutive overexpression of epitope-tagged FLS2 in protoplasts did not mimic the natural FLS2 context (other research groups have also encountered difficulty in obtaining validated results with FLS2 in protoplast systems) or if the paired fluorescent tag proteins did not orient appropriately despite some level of proximity. Alternatively, the negative data of Ali et al. (2007) may suggest that an intermediary bridging molecule does occupy a position between associated FLS2 monomers.
FLS2 is present in FLS2-FLS2 associations both prior to and after exposure to flg22. There are at least two possible explanations for this. First, the FLS2-FLS2 associations may be functionally important in flg22 signaling but recruit BAK1 and release BIK1 without FLS2-FLS2 dissociation. For example, FLS2 may remain dimerized before and after exposure to flg22 or flagellin but change conformation upon ligand binding to allow the dimer to dissociate from BIK1 and gain interacting BAK1 molecules as FLS2-mediated signaling becomes activated. There are examples of animal cell surface receptors that bind ligand and coreceptor proteins as receptor-dimers or multimers (Zhang, 2004; Bulenger et al., 2005; De Meyts, 2008). Signaling may initiate at the cell surface and/or ensue during endocytosis (Geldner and Robatzek, 2008). In an alternative model, the FLS2-FLS2 associations may be silent in signaling and a separate pool of unassociated FLS2 may function in signaling without forming FLS2-FLS2 associations. Although it is not conclusively eliminated, a third model (ligand-induced dissociation of FLS2-FLS2 associations to allow interaction with BAK1) seems less likely in light of our results. We observed, if anything, increased recruitment of FLS2 to FLS2-FLS2 associations at 2 and 5 min after exposure to flg22.
A limited correlation was observed between formation of FLS2-FLS2 associations and capacity for defense signaling. FLS2 proteins lacking one or both of the N-terminal Cys pair (Cys-61 or Cys-68) were defective for FLS2 binding/signaling, and for formation of FLS2-FLS2 associations, even when the mutant proteins were present at levels known to otherwise be adequate for FLS2 signaling (Figure 4; see also Dunning et al. [2007] regarding the very low levels of FLS2 needed for signaling). Subsequent work showed that most Cys-61 or Cys-68 mutant proteins are not at the plasma membrane. However, a partial reduction of FLS2-FLS2 association was also observed with the signaling-inactive FLS2T867V-HA and FLS2G1064R-HA alleles that are mutated in the predicted FLS2 intracellular domain. This result, together with prior data on FLS2T867V and FLS2G1064R (Gómez-Gómez et al., 2001; Robatzek et al., 2006), may suggest that FLS2 needs to be appropriately phosphorylated to remain in FLS2-FLS2 associations. As a third example, the FLS2NNSL mutant protein was present but also did not form FLS2-FLS2 associations and lacked FLS2 function. A reverse correlation (that nonfunctional FLS2 will not form FLS2-FLS2 associations) was not observed. The FLS2S390K and FLS2T366K proteins, which carry single amino acid changes on the predicted LRR surface that prevent binding of flg22 (Dunning et al., 2007), did still form FLS2-FLS2 associations. This latter result indicates that FLS2-FLS2 association alone is not sufficient for signaling function and is the expected result for receptors that exist as a dimer prior to ligand exposure. However, where capacity to form FLS2-FLS2 associations was reduced, defense signaling capacity was also impacted.
Cys Pairs
The Cys pairs that cap the N terminus and C terminus of extracellular LRR domains are very common in both plants and animals (Kajava, 1998; Diévart and Clark, 2003; van der Hoorn et al., 2005; Gay and Gangloff, 2007; Kim et al., 2007). One likely role for these caps is to stabilize overall protein structure by covering the hydrophobic core of the LRR that might otherwise be exposed at the ends of the LRR solenoid, but other contributions are possible. For example, presence/absence of disulfide bonds could serve as a molecular switch for conformation changes. Some LRR-containing receptors, such as the mammalian insulin receptor, undergo covalent intermolecular association via Cys linkages (Ward et al., 2007). A similar arrangement was proposed for the Arabidopsis receptor CLV1 (Trotochaud et al., 1999). When we compared plant extracts with and without exposure to high levels of DTT, we did not find any evidence that disulfide linkages couple FLS2 to other proteins.
The N-terminal Cys pair is particularly well conserved and is present in >100 different Arabidopsis LRR-RLK proteins (Diévart and Clark, 2003). We found that mutation of the FLS2 N-terminal Cys pair disrupted FLS2 signaling. However, FLS2 proteins carrying LRRNT mutations did retain a slight residual capacity to activate defense, manifested as a wide distribution of flg22 response phenotypes among replicates (most plants gave no detectable response, but a few plants with FLS2C61A and/or FLS2C68A expression driven by the native FLS2 promoter gave a partial or, rarely, a full response). Hence, FLS2 mutated at Cys-61 and/or Cys-68 can be a functionally competent receptor under some circumstances, possibly like the bri1-5 product (Hong et al., 2008).
When FLS2 transgenes with the LRRNT C61A and/or C68A mutations were expressed from the native FLS2 promoter, FLS2 protein abundance was substantially reduced, indicating a significant role for this Cys pair in FLS2 stability. When the FLS2 transgenes with C61A and/or C68A mutations were overexpressed from 35S promoter, overall FLS2 abundance was similar to that seen for wild-type FLS2 expressed from the native FLS2 promoter, yet most of these 35S-driven versions carrying LRRNT mutations still exhibited almost no detectable FLS2-FLS2 association or flg22 binding and still exhibited loss of FLS2 activity. Endo H assays indicated substantial retention of most LRRNT-mutated FLS2 in the ER, and two-phase partitioning confirmed absence from the plasma membrane fraction. We hypothesize that the folding of FLS2 lacking this N-terminal Cys pair is sufficiently unstable that the protein does not participate consistently in the protein processing and localization interactions that are characteristic of wild-type FLS2. A likely mechanism for this is that FLS2 with LRRNT Cys mutations is retained in the ER and is degraded via ER quality control (ERQC) mechanisms, such as by a proteasome-independent ER-associated degradation. For the BRI1-5 protein that has a Cys69Tyr mutation, it was demonstrated that the nearby free thiol group at Cys-62 in the LRRNT region is essential for a thiol-mediated ER retention mechanism (Hong et al., 2008). However, in our assays of FLS2 function, stability, and ER processing, double C61/C68A mutants behaved very similarly to C61A or C68A single mutants that would have the free thiol, suggesting that thiol-mediated ER retention during ERQC is not the primary mechanism of depleted abundance of the FLS2 LRRNT mutant proteins. Another notable difference between Cys mutation in FLS2 and BRI1 is that overexpression of BRI1-5 protein significantly suppressed the bri1-5 dwarf phenotype (Hong et al., 2008), whereas FLS2 Cys mutations disrupted FLS2 function even after overexpression.
Recent studies have shown that ERQC in plants, similar to yeast and mammals, relies on at least three different mechanisms, including the thiol-dependent retention process, a calnexin calreticulin cycle that is specific for glycoproteins, and an Hsp40/ERdj3B/Bip chaperone complex (Sitia and Braakman, 2003; Hong et al., 2008; Nekrasov et al., 2009; Saijo et al., 2009). Studies involving FLS2 have revealed that CRT3, UGGT, and STT3A acting in concert in an ER-resident N-glycosylation pathway, and the ER complex SDF2/ERdj3B/BiP, are dispensable for the biogenesis and function of FLS2 function and its proper accumulation (Nekrasov et al., 2009; Saijo et al., 2009). However, our Endo H experiments with MDG (glycosylation-defective) mutant FLS2 proteins showed that N-glycosylation does play a readily detectable role in ER retention, albeit not enough of a role in most cases to negatively impact overall FLS2 function (Figure 6). Our findings, together with those of Nekrasov et al. (2009) and Saijo et al. (2009), suggest that a presently undescribed fourth retention/degradation mechanism may be involved in the ERQC of cargo proteins such as FLS2. As one possibility, a thiol-independent ER retention mediated by ERp44 has been observed in mammals for formylglycine-generating enzyme, monomeric immunoglobulin K and J, and mutant μ chains (Anelli et al., 2002, 2003; Mariappan et al., 2008). Most recently, it was demonstrated that Arabidopsis reticulon-like RTNLB1 and RTNLB2 regulate the transport of newly synthesized FLS2 to plasma membrane (Lee et al., 2011). In the future, it may be of interest to use FLS2 N-glycosylation mutants (further discussed below) and FLS2 LRRNT mutants to help identify new mechanisms for ERQC.
At the other end of the LRR, the occurrence of a membrane-proximal Cys pair is common but is less universal than the N-terminal Cys pair in extracellular LRR proteins (Diévart and Clark, 2003). It is intriguing that mutation of the membrane-proximal LRRCT Cys pair C783/C792 not only failed to eliminate FLS2 function, but instead caused partially elevated FLS2 signaling (as long as LRRNT mutations were not also present). The seedling growth inhibition mediated by LRRCT mutant proteins was similar to the wild type, but elevated ROS production was consistently observed after flg22 stimulation (for example, see Figure 4C and Supplemental Figure 2B online). No basal activity in the absence of flg22 was detected for the FLS2 LRRCT mutants, in seedling growth inhibition, or ROS assays (data not shown). The elevated oxidative burst might arise if there is more LRRCT-mutated FLS2 in signaling-proficient locations prior to ligand exposure, or more rapid association/dissociation with phosphorylation substrates, or if FLS2 with LRRCT mutations remains in a signaling configuration for a longer period after exposure to ligand because of less efficient receptor recycling. We consider it unlikely that the effect has to do with a higher affinity for flg22 since our experiments were conducted with saturating flg22 concentrations.
Dominant-Negative Effects
It is also intriguing that the FLS2 kinase lacking an LRR (NoNT and NoLRR constructs; Figure 5) exhibited a dominant-negative effect on flg22-elicited signaling when overexpressed in plants that are wild-type for the endogenous FLS2. Dominant-negative impacts have been reported for a range of mutation types in other plant RLKs (Diévart and Clark, 2003; Morillo and Tax, 2006), but not for FLS2. Relatively high expression levels were required for dominant-negative activity, but experiments with D997A mutants indicate that kinase activity of FLS2NoLRR is not needed for the protein to exert dominant-negative activity. The dominant-negative action of FLS2NoLRR or FLS2NoNT may arise when they associate with wild-type FLS2 kinase (Figure 2D), if this disrupts normal FLS2-FLS2 association. Alternatively, although overexpressed FLS2NoNT did not detectably interact with BAK1 and did not block EFR-mediated signaling, the NoNT and NoLRR proteins may titrate out other signaling partners. The above findings suggest multiple avenues for future exploration of these and other hypotheses about FLS2 function.
Glycosylation
The function of glycosylation varies among cell-surface receptor proteins; it can contribute to protein folding, processing and secretion, to stability, and to interactions with ligands and other proteins (Hawtin et al., 2001; Yan et al., 2008; reviewed in Ohtsubo and Marth, 2006). In the example of the human innate immunity receptor TLR3, there are multiple glycosylations but none on a particular lateral face of the LRR solenoid, consistent with the observed homodimerization of this receptor along the nonglycosylated face (Liu et al., 2008). This glycosylation pattern may also help to guide appropriate positioning of TLR3 with respect to its large nucleic acid ligand prior to the final high-affinity docking that tightly sandwiches nucleic acid polymer between antiparallel TLR3 proteins (Liu et al., 2008).
N-glycosylation sites are widely predicted among plant extracellular receptors with roles in disease resistance, but tomato Cf-9 offers one of the few examples where this glycosylation has been functionally investigated. Cf-9 is highly dependent on proper glycosylation of the extracellular LRRs (van der Hoorn et al., 2005). Hence, it was somewhat surprising to discover that N-linked glycosylation makes only subtle quantitative contributions to the functional capacity of FLS2. Impacts on defense signaling capacity were observed for only some octuple-PGS mutants, and then only a partial loss of activity, despite clear molecular weight reductions (more rapid migration in SDS-PAGE) in all of the octuple PGS mutant forms that were examined. FLS2 protein abundance in plants was not detectably reduced for these octuple PGS mutants. We did not investigate other types of glycosylation, which are much less commonly relevant to the biology of extracellular receptors, but the data suggest that FLS2 stability and functional interactions can occur relatively independently of glycosylation.
When we conducted a similar mutational study of putative glycosylation sites for EFR (another Arabidopsis LRR-RLK MAMP receptor) to pursue these observations further, a simple screen of single-site PGS mutant alleles was sufficient to identify mutations that largely abolish function. This was also the case for tomato Cf-9 (van der Hoorn et al., 2005). For both of the nonfunctional alleles EFRN342D and EFRN366D, the loss of EFR function was not attributable to a consistent reduction of EFR protein abundance. During the preparation of this article, additional studies reported similar phenomena (Li et al., 2009; Nekrasov et al., 2009; Saijo et al., 2009; Häweker et al., 2010). It was also found that EFRN143Q lacking a single conserved N-glycosylation site from the EFR ectodomain accumulated to reduced levels and lost the ability to bind its ligand and to mediate elf18-elicited oxidative burst (Häweker et al., 2010). We found that EFRN143D only partially lost flg22-mediated inhibition of seedling growth, suggesting quantitative differences between EFR N143Q and N143D proteins, but we also identified two novel single PGS mutants EFRN342D and EFRN366D that almost completely lost elf18 responsiveness. The glycans attached to different Asn residues can play different roles in ERQC of plasma membrane–bound RLKs, and the distinct effects of N-glycosylation on FLS2 and EFR function may be due to differences in the engagement of ERQC mechanisms. Alternatively or additionally, the different effects of N-glycosylation on FLS2 and EFR may be a result of differences in the signaling partners used by different subsets of plant PRRs. It will be of interest to elucidate how and why N-glycosylation has different effects on these structurally and functionally related plant immunity receptors.
In conclusion, FLS2-FLS2 complexes are constitutively present in planta. The functional significance of these complexes is suggested by, among other things, the dominant-negative effect of truncated FLS2 proteins that associate with full-length FLS2 proteins. Although glycosylation of the FLS2 extracellular domain occurs, it seems to be largely dispensable for function, in stark contrast with EFR and other extracellular receptors. The Cys pair at the FLS2 LRRNT plays an important role in FLS2 processing, stabilization, and overall function, but it is not absolutely required for FLS2 function. The Cys pair at the FLS2 LRRCT is not required for overall function and has a negative regulatory role in modulating the extent of the FLS2-mediated oxidative burst. Intracellular and extracellular domains of FLS2 can each participate in FLS2-FLS2 association. The C-terminal tail of FLS2 is required for sustained FLS2 abundance. This progress in defining the FLS2 features necessary for function has been paralleled by work in other labs to identify proteins that functionally associate with FLS2. However, discovery of the full roster of FLS2-associated proteins and the physical configuration of FLS2 and these other proteins prior to, during, and after signaling remains as a significant challenge for future research into the molecular mechanisms by which ligand binding is converted to signaling activation.
METHODS
Plant Materials
Arabidopsis thaliana ecotype Col-0 was used as an FLS2-containing wild type. Ecotype Ws-0 was used as a natural fls2 mutant. The pFLS2:FLS2-cMyc-GFP transgenic plant in Ws-0 background was kindly provided by Silke Robatzek (Robatzek et al., 2006). Arabidopsis Col-0 T-DNA insertion line fls2-101 (Pfund et al., 2004) was used for plant transformation unless otherwise specified. The homozygous Arabidopsis efr T-DNA insertion mutant SALK_068675C was also obtained from the ABRC.
Gene Cloning and Construction
The FLS2 gene was amplified by PCR with Pfu Turbo DNA polymerase (Stratagene) using Col-0 genomic DNA; the truncated FLS2 constructs, including NoKinase, NoNT, and NoCT, were amplified from an FLS2 cDNA mimic, which was generated from the genomic FLS2 clone by PCR-splice overlap extension to precisely delete the one FLS2 intron. The oligonucleotide primer sets used in this study are listed in Supplemental Table 1 online. The resultant DNA was gel purified and cloned into pENTR/D TOPO vector (Invitrogen). Alternatively, the FLS2 gene was released from pCAMBIA2300-pFLS2:FLS2-cMyc-GFP construct (kindly provided by Silke Robatzek) using BamHI and XbaI and religated into pSTBlue-1 (Novagen). For point mutations of the conserved Cys pairs and kinase domain of FLS2, mutant FLS2 was generated from pENTR/D TOPO-FLS2 (for 35S promoter) or from pSTBlue-1-FLS2 (for FLS2 native promoter). For point mutations on PGSs and on conserved LRR regions, mutant LRRs were generated from pTOPO:FLS2 LRR template (Dunning et al., 2007) using site-directed mutagenesis. All FLS2 mutated/truncated constructs were verified by DNA sequence determination. The pENTR/D TOPO-FLS2 wild-type and mutant constructs were then recombined into the pGWB14 binary destination vector (courtesy of T. Nakagawa, Shimane University, Matsue, Japan) using LR clonase II mix (Invitrogen). The resulting pGWB14-derived plasmids contain the 35S promoter to drive FLS2 expression and a HA tag at the C terminus. The mutated FLS2 genes in pSTBlue-1 were released using BamHI and XbaI and religated into pCAMBIA2300 to reconstitute PFLS2:FLS2-cMyc-GFP constructs, or the mutated FLS2 LRR DNA fragments in pTOPO:FLS2 LRR were cut out with AscI and PacI (NEB), gel purified, and then cloned into pHD3300, in which C-terminally tagged FLS2-HA is driven by the native FLS2 promoter (Dunning et al., 2007).
The EFR gene constructs (with EFR native promoter, without stop codon) were made by PCR in pENTR/D vector (for primer sequences, see Supplemental Table 1 online), then recombined into pGWB13 (no promoter, HA tag) using LR clonase II mix. Site-directed mutagenesis alleles were made using pENTR/D-EFR as a template.
Site-Directed Mutagenesis
Point mutations were generated by circular PCR as per the instructions of the QuikChange mutagenesis kit (Stratagene). Briefly, two synthetic complementary oligonucleotide primers containing the desired mutation(s) were extended during temperature cycling by PfuTurbo DNA polymerase (Stratagene). After cycling, DpnI was added into PCR products to specifically digest the methylated parental DNA template. The linear PCR products were then transformed into DH5α electroporation competent cells. The resultant mutated plasmid constructs were verified by sequencing.
Arabidopsis Transformation and Selection of Transformed Plants
Plasmid constructs were electroporated into Agrobacterium tumefaciens strain GV3101 (pMP90) for Arabidopsis transformation. For most experiments, FLS2 binary plasmid constructs in Agrobacterium were transformed into Arabidopsis Col-0 fls2-101/fls2-101 (Pfund et al., 2004) or homozygous transgenic Ws-0 expressing FLS2-cMyc-GFP under the control of the FLS2 promoter (Ws PFLS2:FLS2–3×myc-GFP; Robatzek et al., 2006) by floral dip transformation (Clough and Bent, 1998). Other experiments used wild-type Col-0, Ws-0, or the Col efr mutant as noted. To select transformed plants with resistance to Basta (pHD3300-FLS2 and derivatives) or kanamycin (pCAMBIA2300-FLS2-cMyc-GFP and derivatives), T1 seeds were surface sterilized and plated on 0.8% agar plates carrying 0.5× Murashige and Skoog (MS) basal medium and Gamborg’s vitamins (Sigma-Aldrich), 1% Suc, 200 mg/L cefotaxime, and 10 mg/L Basta (Liberty, AgrEvo) or 50 mg/L kanamycin. Plates were kept at 4°C for 2 d and then grown in 16-h light/day at 23°C for 1 week. Healthy green seedlings were then tested for flg22 responsiveness in the seedling growth inhibition assay or grown out for other studies. To select transformed plants with hygromycin resistance (pGWB13 and pGWB14 derivatives), T1 seeds were plated on 0.5× MS plates with cefotaxime and 20 mg/L hygromycin after sterilization. Plates were kept in the dark at room temperature for 4 d after cold treatment. Healthy etiolated seedlings were then grown under daily light/dark regimen for further experimentation.
Seedling Growth Inhibition Assays
Seedling growth inhibition assays for flg22-dependent FLS2 activity were performed as described by Pfund et al. (2004). Typically, 10 Basta-resistant, kanamycin-resistant, or hygromycin-resistant Arabidopsis T1 seedlings (representing 10 independent transformation events) were transferred to a 24-well plate (one seedling per well), with each well carrying 400 μL of 0.5× MS salts and 2.5 μM flg22 peptide. After 10 to 14 d of further growth, each seedling was briefly blotted dry and weighed.
Protein Extraction, Immunoblotting, and Immunoprecipitation
Total proteins were extracted from Arabidopsis T1 seedlings as described by Karlova et al. (2006). Briefly, plant material was ground in liquid nitrogen and thawed in extraction buffer containing 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% Triton X-100, and 1× plant protease inhibitor cocktail (Sigma-Aldrich). The samples were centrifuged at 400g (Eppendorf Centrifuge 5810R) at 4°C for 3 min after 30 min incubation on ice. Protein concentrations were determined using the BCA protein assay (Pierce). After boiling 40 μg of total protein for 5 min in 1× SDS-PAGE sample buffer, the proteins were separated on a 7.5% SDS-PAGE gel and electrotransferred to Hybond-P (GE Healthcare). Proteins were analyzed by immunoblotting with anti-HA, anti-cMyc antibodies (1000× dilution; Covance) or anti-FLAG antibody (1000× dilution; Sigma-Aldrich). Additional antibodies included anti-ARF1/At2g47170 and anti-plasma membrane H+ATPase (Agrisera).
For immunoprecipitation, we routinely used 12 to 15 2-week-old Arabidopsis T1 seedlings to prepare a protein extract, as described above, with flg22 peptide, where noted, added to the plant growth solution at indicated times as a 100× stock (e.g., 100 μM peptide stock to achieve final concentration 1 μM). One milliliter of total plant protein extract (2.5 mg/mL) was incubated for 1 h at 4°C with 25 μL of 50% (v/v) Protein A beads slurry (Amersham Biosciences) in 50 mM Tris-HCl and 150 mM NaCl, pH 7.5. After centrifugation at 200g for 2 min at 4°C, the supernatant was incubated with 7 μL of anti-cMyc, anti-GFP, or anti-FLAG antibodies. After incubation with gentle mixing for 1 h at 4°C, 25 μL of fresh 50% slurry of protein A beads was added, and incubation was continued for 4 h. Protein A beads were spun down by centrifugation at 200g for 1 min, and the supernatant was removed. The beads were washed five times with 1 mL of wash buffer (50 mM Tris-HCl, pH 7.5, and 150 mM NaCl). After the last centrifugation, the wash buffer was removed completely, and 100 μL 1× SDS-PAGE sample buffer was added. After boiling, the samples were subjected to SDS-PAGE and immunoblot analysis. The FLS2-HA band shown in many figures consistently migrated at ~175 kD in SDS-PAGE, as previously reported (Chinchilla et al., 2006; Dunning et al., 2007).
For the deglycosylation experiments, the crude protein extract (~40 μg) was treated with PNGase F or Endo H (New England Biolabs) according to the manufacturer’s instructions. For disulfide linkage experiments (Figure 1C), one-fifth volume of 5× sample buffer (0.225 M Tris-HCl, pH 6.8, 50% glycerol, 5% SDS, and 0.05% bromophenol blue with or without 0.25 M DTT) was added to crude protein samples. The samples were boiled for 5 min and separated on a 7.5% Tris-HCl polyacrylamide gel. The protein was transferred to Hybond P membrane (GE Healthcare) and used for protein gel blotting with an anti-HA antibody.
Binding Assay
Binding of 125I-Tyr-flg22 to plant homogenates was done as described previously (Bauer et al., 2001). In brief, plant homogenates were incubated in binding buffer (25 mM MES-KOH, pH 6.0, 3 mM MgCl2, and 10 mM NaCl) in a total volume of 100 μL with 125I-Tyr-flg22 (60 fmol per sample) for 30 min either alone (total binding) or with an excess (10 μM) of unlabeled competing flg22 peptide (unspecific binding). Plant homogenates were collected by vacuum filtration on glass fiber filters (Macherey-Nagel MN-GF2; 2.5-cm diameter, preincubated with 1% BSA, 1% tryptone, and 1% peptone in binding buffer) and washed with 10 mL of ice-cold binding buffer. Radioactivity on filters was quantified by γ-counting.
Oxidative Burst Assay
The production of ROS was measured by a luminol-dependent assay (Kunze et al., 2004). Briefly, leaf punches from fully expanded leaves of 3- to 6-week-old Arabidopsis plants were floated on 50 μL 1% DMSO solution in the 96-well plate overnight. Forty microliters of water supplied with 1 μg luminol and 1 μg of horseradish peroxidase (Sigma-Aldrich) was added into each well and then 10 μL of 10 μM flg22 was added immediately before measurement. Luminescence was measured in a luminometer (Synergy HT plate reader; Bio-Tek) for 30 min after addition of flg22 peptide.
Ethylene Production Assay
Ethylene production in response to flg22 (Felix et al., 1999) was monitored with leaf strips from leaves of 4- to 8-week-old plants using a Shimadzu GC-14A gas chromatograph, C-R4A chromatopac, and aluminum oxide column as described (Dunning et al., 2007). Alternatively, one or two 2-week-old seedlings (grown in 24-well plates) were transferred into 2-mL vials with 0.5 mL 0.5× MS liquid medium, vials were left on the lab bench overnight, and the next morning flg22 was added immediately prior to gas-tight capping of tubes. Vials were rocked gently for 4.0 h and then the ethylene concentration in the airspace was determined.
Callose Deposition
Callose deposition was monitored as described (Gómez-Gómez et al., 1999). Approximately six Arabidopsis seedlings per treatment were selected from Basta or kanamycin selection medium and transferred to 24-well plates (one seedling per well) containing 400 μL of liquid media (no agar) with flg22. Twenty-four hours after treatment, seedlings were fixed overnight in 1% (v/v) glutaraldehyde, 5 mM citric acid, and 90 mM Na2HPO4, pH 7.4, and then cleared and dehydrated with 100% ethanol. After aniline blue staining, callose was visualized using UV epifluorescence microscopy.
MPK Phosphorylation
Leaves from 6-week-old Arabidopsis were treated with 5 μM flg22 or water (for control) by syringe infiltration. Five minutes later, infiltrated leaf tissues were ground in liquid nitrogen and crude proteins were extracted in 2× SDS loading buffer. After separation on 12% SDS-PAGE gels, samples were transferred to Hybond P membranes and phosphorylated MPK3 and MPK6 were detected by P44/P42 polyclonal antibody (Cell Signaling Technology).
Bacterial Growth Assay
Overnight cultures of Pseudomonas syringae pv tomato strain DC3000 or DC3000 ΔhrcC were resuspended in 10 mM MgCl2 solution. Plant rosettes (age 6 weeks, grown in potting mix at 22°C under 9 h of light per day) were dipped in 5 × 108 colony-forming units/mL (OD600 = 0.5) of bacteria with 0.02% Silwet L-77. Three days later, leaves were removed, surface sterilized in 70% ethanol for 10 s, and then rinsed in sterilized water. For each sample, leaf discs were then removed from four different leaves and combined and ground in 10 mM MgCl2, and then samples were serially diluted and plated on NYGA plates with 50 mM rifampicin.
Transient Expression in Arabidopsis Protoplasts
Transient expression Gateway vectors were made by ligating DNA fragments containing 35S promoter and Nos terminator amplified from pGWB14 and pGWB17 into prelinearized pUC19 digested by SmaI, resulting in Gateway plasmids pUC-GW14 and pUC-GW17, respectively. BAK1 and FLS2 were cloned into pUC-GW17 and pUC-GW14 by LR reaction using LR clonase II (Invitrogen), resulting in pUC-GW17-BAK1 and pUC-GW14-FLS2, respectively. Arabidopsis mesophyll protoplasts were isolated from 6-week-old transgenic plants and used according to the method described by Yoo et al. (2007). About 100 μg plasmids pUC-GW17-BAK1 and pUC-GW14-FLS2 were cotransformed into protoplasts from fls2-101 plants, and ~100 μg plasmid pUC-GW17-BAK1 was transformed into protoplasts from a Col-0 transgenic line expressing 35S-FLS2NoNT. Co-IPs were performed using the methods described above.
Two-Phase Partitioning Experiment
Eight to ten grams of fresh Arabidopsis seedlings (fls2-101 stably transformed to express FLS2-WT, FLS2-C61/68A, or FLS2-C783/792A) grown for 2 weeks under low light in liquid MS medium were homogenized in 10 mL homogenization buffer (50 mM Tris, pH 7.6, 100 mM KCl, 1 mM EDTA, 0.1 mM MgCl2, 8% Suc, and 1× Sigma-Aldrich plant protease inhibitor cocktail) on ice and filtered through Miracloth. The filtrate was centrifuged for 15 min at 10,000g at 4°C. Plasma membranes were purified according to the procedures described by Larsson et al. (1987). Finally, the purified plasma membranes were resuspended in 200 μL 1× SDS loading buffer. Crude protein and purified plasma membrane were used for immunoblots.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: FLS2 (At5g46330), EFR (At5g20480), BAK1 (At4g33430), MPK3 (At3g45640), MPK6 (At2g43790), MPK4 (At4g01370), Arf1 (At2g47170), and plasma membrane H+-ATPase (At2g18960).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Intermolecular FLS2-FLS2 Interaction in Vivo.
Supplemental Figure 2. The Conserved FLS2 LRRNT Cys Pair Is Essential for FLS2 Function but the Membrane-Proximal Cys Pair Is Not.
Supplemental Figure 3. FLS2-NoNT and FLS2-NoLRR Have a Dominant-Negative Effect on FLS2 Function.
Supplemental Table 1. PCR Primer Sets Used for Site-Directed Mutagenesis and Gene Cloning.
Acknowledgments
A.F.B. thanks T.B. for hosting his sabbatical research. We also thank Silke Robatzek (Sainsbury Laboratory) for the pFLS2:FLS2-cMyc-GFP construct and transgenic Ws-0 plant line, Tsuyoshi Nakagawa (Shimane University, Japan) for pGWB series plasmids, and Sebastian Bednarek, Marisa Otegui, Michael Havey, and Janejira Duangjit of the University of Wisconsin–Madison for use of reagents and equipment. The majority of this work was supported by the Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science, U.S. Department of Energy (Contract DE-FG02-02ER15342) to A.F.B. W.S. was also supported by the National Natural Science Foundation of China (30971893). Work in the Boller lab was supported by the Swiss National Science Foundation. K.J.L. received support from the University of Wisconsin–Madison Hilldale Fellowship program.
AUTHOR CONTRIBUTIONS
W.S. and Y.C. designed the research, performed the research, analyzed data, and wrote the article. K.J.L. and P.B. performed research, analyzed data, and edited the article. T.B. and A.F.B. designed the research, analyzed data, and wrote the article.
References
- Agnati L.F., Tarakanov A.O., Ferré S., Fuxe K., Guidolin D. (2005). Receptor-receptor interactions, receptor mosaics, and basic principles of molecular network organization: possible implications for drug development. J. Mol. Neurosci. 26: 193–208 [DOI] [PubMed] [Google Scholar]
- Ali G.S., Prasad K.V.S.K., Day I., Reddy A.S.N. (2007). Ligand-dependent reduction in the membrane mobility of FLAGELLIN SENSITIVE2, an Arabidopsis receptor-like kinase. Plant Cell Physiol. 48: 1601–1611 [DOI] [PubMed] [Google Scholar]
- Anelli T., Alessio M., Bachi A., Bergamelli L., Bertoli G., Camerini S., Mezghrani A., Ruffato E., Simmen T., Sitia R. (2003). Thiol-mediated protein retention in the endoplasmic reticulum: The role of ERp44. EMBO J. 22: 5015–5022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anelli T., Alessio M., Mezghrani A., Simmen T., Talamo F., Bachi A., Sitia R. (2002). ERp44, a novel endoplasmic reticulum folding assistant of the thioredoxin family. EMBO J. 21: 835–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.L., Gomez-Gomez L., Boller T., Ausubel F.M., Sheen J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Bauer Z., Gómez-Gómez L., Boller T., Felix G. (2001). Sensitivity of different ecotypes and mutants of Arabidopsis thaliana toward the bacterial elicitor flagellin correlates with the presence of receptor-binding sites. J. Biol. Chem. 276: 45669–45676 [DOI] [PubMed] [Google Scholar]
- Bell J.K., Askins J., Hall P.R., Davies D.R., Segal D.M. (2006). The dsRNA binding site of human Toll-like receptor 3. Proc. Natl. Acad. Sci. USA 103: 8792–8797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Boudsocq M., Willmann M.R., McCormack M., Lee H., Shan L., He P., Bush J., Cheng S.H., Sheen J. (2010). Differential innate immune signalling via Ca(2+) sensor protein kinases. Nature 464: 418–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulenger S., Marullo S., Bouvier M. (2005). Emerging role of homo- and heterodimerization in G-protein-coupled receptor biosynthesis and maturation. Trends Pharmacol. Sci. 26: 131–137 [DOI] [PubMed] [Google Scholar]
- Chinchilla D., Bauer Z., Regenass M., Boller T., Felix G. (2006). The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18: 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nürnberger T., Jones J.D., Felix G., Boller T. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- De Meyts P. (2008). The insulin receptor: A prototype for dimeric, allosteric membrane receptors? Trends Biochem. Sci. 33: 376–384 [DOI] [PubMed] [Google Scholar]
- Diévart A., Clark S.E. (2003). Using mutant alleles to determine the structure and function of leucine-rich repeat receptor-like kinases. Curr. Opin. Plant Biol. 6: 507–516 [DOI] [PubMed] [Google Scholar]
- Dunning F.M., Sun W., Jansen K.L., Helft L., Bent A.F. (2007). Identification and mutational analysis of Arabidopsis FLS2 leucine-rich repeat domain residues that contribute to flagellin perception. Plant Cell 19: 3297–3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix G., Duran J.D., Volko S., Boller T. (1999). Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18: 265–276 [DOI] [PubMed] [Google Scholar]
- Gao M., Wang X., Wang D., Xu F., Ding X., Zhang Z., Bi D., Cheng Y.T., Chen S., Li X., Zhang Y. (2009). Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell Host Microbe 6: 34–44 [DOI] [PubMed] [Google Scholar]
- Gay N.J., Gangloff M. (2007). Structure and function of Toll receptors and their ligands. Annu. Rev. Biochem. 76: 141–165 [DOI] [PubMed] [Google Scholar]
- Geldner N., Robatzek S. (2008). Plant receptors go endosomal: A moving view on signal transduction. Plant Physiol. 147: 1565–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron J.M., Wang Z.Y. (2007). Multiple mechanisms modulate brassinosteroid signaling. Curr. Opin. Plant Biol. 10: 436–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez L., Bauer Z., Boller T. (2001). Both the extracellular leucine-rich repeat domain and the kinase activity of FSL2 are required for flagellin binding and signaling in Arabidopsis. Plant Cell 13: 1155–1163 [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez L., Boller T. (2000). FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L., Felix G., Boller T. (1999). A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J. 18: 277–284 [DOI] [PubMed] [Google Scholar]
- Harding A.S., Hancock J.F. (2008). Using plasma membrane nanoclusters to build better signaling circuits. Trends Cell Biol. 18: 364–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häweker H., Rips S., Koiwa H., Salomon S., Saijo Y., Chinchilla D., Robatzek S., von Schaewen A. (2010). Pattern recognition receptors require N-glycosylation to mediate plant immunity. J. Biol. Chem. 285: 4629–4636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawtin S.R., Davies A.R., Matthews G., Wheatley M. (2001). Identification of the glycosylation sites utilized on the V1a vasopressin receptor and assessment of their role in receptor signalling and expression. Biochem. J. 357: 73–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese A., Hann D.R., Gimenez-Ibanez S., Jones A.M., He K., Li J., Schroeder J.I., Peck S.C., Rathjen J.P. (2007). The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. USA 104: 12217–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z., Jin H., Tzfira T., Li J. (2008). Multiple mechanism-mediated retention of a defective brassinosteroid receptor in the endoplasmic reticulum of Arabidopsis. Plant Cell 20: 3418–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn M., Belkhadir Y., Dreux M., Dabi T., Noel J.P., Wilson I.A., Chory J. (2011). Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 474: 467–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeworutzki E., Roelfsema M.R., Anschütz U., Krol E., Elzenga J.T., Felix G., Boller T., Hedrich R., Becker D. (2010). Early signaling through the Arabidopsis pattern recognition receptors FLS2 and EFR involves Ca-associated opening of plasma membrane anion channels. Plant J. 62: 367–378 [DOI] [PubMed] [Google Scholar]
- Kajava A.V. (1998). Structural diversity of leucine-rich repeat proteins. J. Mol. Biol. 277: 519–527 [DOI] [PubMed] [Google Scholar]
- Karlova R., Boeren S., Russinova E., Aker J., Vervoort J., de Vries S. (2006). The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 protein complex includes BRASSINOSTEROID-INSENSITIVE1. Plant Cell 18: 626–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.M., Park B.S., Kim J.-I., Kim S.E., Lee J., Oh S.C., Enkhbayar P., Matsushima N., Lee H., Yoo O.J., Lee J.-O. (2007). Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist Eritoran. Cell 130: 906–917 [DOI] [PubMed] [Google Scholar]
- Knighton D.R., Zheng J.H., Ten Eyck L.F., Ashford V.A., Xuong N.H., Taylor S.S., Sowadski J.M. (1991). Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 253: 407–414 [DOI] [PubMed] [Google Scholar]
- Kolade O.O., Bamford V.A., Ancillo Anton G., Jones J.D.G., Vera P., Hemmings A.M. (2006). In vitro characterization of the cysteine-rich capping domains in a plant leucine rich repeat protein. Biochim. Biophys. Acta 1764: 1043–1053 [DOI] [PubMed] [Google Scholar]
- Kunze G., Zipfel C., Robatzek S., Niehaus K., Boller T., Felix G. (2004). The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16: 3496–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson C., Widell S., Kjelbom P. (1987). Preparation of high-purity plasma membranes. Meth. Enzymol. 148: 558–568 [Google Scholar]
- Latz E., Verma A., Visintin A., Gong M., Sirois C.M., Klein D.C.G., Monks B.G., McKnight C.J., Lamphier M.S., Duprex W.P., Espevik T., Golenbock D.T. (2007). Ligand-induced conformational changes allosterically activate Toll-like receptor 9. Nat. Immunol. 8: 772–779 [DOI] [PubMed] [Google Scholar]
- Lee H.Y., Bowen C.H., Popescu G.V., Kang H.G., Kato N., Ma S., Dinesh-Kumar S., Snyder M., Popescu S.C. (2011). Arabidopsis RTNLB1 and RTNLB2 Reticulon-like proteins regulate intracellular trafficking and activity of the FLS2 immune receptor. Plant Cell 23: 3374–3391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Zhao-Hui C., Batoux M., Nekrasov V., Roux M., Chinchilla D., Zipfel C., Jones J.D. (2009). Specific ER quality control components required for biogenesis of the plant innate immune receptor EFR. Proc. Natl. Acad. Sci. USA 106: 15973–15978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Botos I., Wang Y., Leonard J.N., Shiloach J., Segal D.M., Davies D.R. (2008). Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science 320: 379–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Wu S., Gao X., Zhang Y., Shan L., He P. (2010). A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. USA 107: 496–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariappan M.M., Shetty M., Sataranatarajan K., Choudhury G.G., Kasinath B.S. (2008). Glycogen synthase kinase 3beta is a novel regulator of high glucose- and high insulin-induced extracellular matrix protein synthesis in renal proximal tubular epithelial cells. J. Biol. Chem. 283: 30566–30575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillo S.A., Tax F.E. (2006). Functional analysis of receptor-like kinases in monocots and dicots. Curr. Opin. Plant Biol. 9: 460–469 [DOI] [PubMed] [Google Scholar]
- Naithani S., Chookajorn T., Ripoll D.R., Nasrallah J.B. (2007). Structural modules for receptor dimerization in the S-locus receptor kinase extracellular domain. Proc. Natl. Acad. Sci. USA 104: 12211–12216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakasako M., Zikihara K., Matsuoka D., Katsura H., Tokutomi S. (2008). Structural basis of the LOV1 dimerization of Arabidopsis phototropins 1 and 2. J. Mol. Biol. 381: 718–733 [DOI] [PubMed] [Google Scholar]
- Nekrasov V., et al. (2009). Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. EMBO J. 28: 3428–3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo K., Marth J.D. (2006). Glycosylation in cellular mechanisms of health and disease. Cell 126: 855–867 [DOI] [PubMed] [Google Scholar]
- Peck S.C., Nühse T.S., Hess D., Iglesias A., Meins F., Boller T. (2001). Directed proteomics identifies a plant-specific protein rapidly phosphorylated in response to bacterial and fungal elicitors. Plant Cell 13: 1467–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfund C., Tans-Kersten J., Dunning F.M., Alonso J.M., Ecker J.R., Allen C., Bent A.F. (2004). Flagellin is not a major defense elicitor in Ralstonia solanacearum cells or extracts applied to Arabidopsis thaliana. Mol. Plant Microbe Interact. 17: 696–706 [DOI] [PubMed] [Google Scholar]
- Robatzek S., Bittel P., Chinchilla D., Köchner P., Felix G., Shiu S.H., Boller T. (2007). Molecular identification and characterization of the tomato flagellin receptor LeFLS2, an orthologue of Arabidopsis FLS2 exhibiting characteristically different perception specificities. Plant Mol. Biol. 64: 539–547 [DOI] [PubMed] [Google Scholar]
- Robatzek S., Chinchilla D., Boller T. (2006). Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 20: 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux M., Schwessinger B., Albrecht C., Chinchilla D., Jones A., Holton N., Malinovsky F.G., Tör M., de Vries S., Zipfel C. (2011). The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23: 2440–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y., Tintor N., Lu X., Rauf P., Pajerowska-Mukhtar K., Häweker H., Dong X., Robatzek S., Schulze-Lefert P. (2009). Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO J. 28: 3439–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze B., Mentzel T., Jehle A.K., Mueller K., Beeler S., Boller T., Felix G., Chinchilla D. (2010). Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J. Biol. Chem. 285: 9444–9451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwessinger B., Zipfel C. (2008). News from the frontline: Recent insights into PAMP-triggered immunity in plants. Curr. Opin. Plant Biol. 11: 389–395 [DOI] [PubMed] [Google Scholar]
- Shah K., Gadella T.W., Jr, van Erp H., Hecht V., de Vries S.C. (2001). Subcellular localization and oligomerization of the Arabidopsis thaliana somatic embryogenesis receptor kinase 1 protein. J. Mol. Biol. 309: 641–655 [DOI] [PubMed] [Google Scholar]
- Shan L., He P., Li J., Heese A., Peck S.C., Nürnberger T., Martin G.B., Sheen J. (2008). Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4: 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu S.H., Karlowski W.M., Pan R., Tzeng Y.H., Mayer K.F., Li W.H. (2004). Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16: 1220–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitia R., Braakman I. (2003). Quality control in the endoplasmic reticulum protein factory. Nature 426: 891–894 [DOI] [PubMed] [Google Scholar]
- Sun W., Dunning F.M., Pfund C., Weingarten R., Bent A.F. (2006). Within-species flagellin polymorphism in Xanthomonas campestris pv campestris and its impact on elicitation of Arabidopsis FLAGELLIN SENSING2-dependent defenses. Plant Cell 18: 764–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai R., Isogai A., Takayama S., Che F.S. (2008). Analysis of flagellin perception mediated by flg22 receptor OsFLS2 in rice. Mol. Plant Microbe Interact. 21: 1635–1642 [DOI] [PubMed] [Google Scholar]
- Torii K.U. (2004). Leucine-rich repeat receptor kinases in plants: Structure, function, and signal transduction pathways. Int. Rev. Cytol. 234: 1–46 [DOI] [PubMed] [Google Scholar]
- Triantafilou M., Gamper F.G., Haston R.M., Mouratis M.A., Morath S., Hartung T., Triantafilou K. (2006). Membrane sorting of toll-like receptor (TLR)-2/6 and TLR2/1 heterodimers at the cell surface determines heterotypic associations with CD36 and intracellular targeting. J. Biol. Chem. 281: 31002–31011 [DOI] [PubMed] [Google Scholar]
- Trotochaud A.E., Hao T., Wu G., Yang Z., Clark S.E. (1999). The CLAVATA1 receptor-like kinase requires CLAVATA3 for its assembly into a signaling complex that includes KAPP and a Rho-related protein. Plant Cell 11: 393–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan G.A., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604 [DOI] [PubMed] [Google Scholar]
- van der Hoorn R.A., Wulff B.B., Rivas S., Durrant M.C., van der Ploeg A., de Wit P.J., Jones J.D. (2005). Structure-function analysis of cf-9, a receptor-like protein with extracytoplasmic leucine-rich repeats. Plant Cell 17: 1000–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Li X., Meisenhelder J., Hunter T., Yoshida S., Asami T., Chory J. (2005). Autoregulation and homodimerization are involved in the activation of the plant steroid receptor BRI1. Dev. Cell 8: 855–865 [DOI] [PubMed] [Google Scholar]
- Ward C.W., Lawrence M.C., Streltsov V.A., Adams T.E., McKern N.M. (2007). The insulin and EGF receptor structures: new insights into ligand-induced receptor activation. Trends Biochem. Sci. 32: 129–137 [DOI] [PubMed] [Google Scholar]
- Weber A.N.R., Moncrieffe M.C., Gangloff M., Imler J.-L., Gay N.J. (2005). Ligand-receptor and receptor-receptor interactions act in concert to activate signaling in the Drosophila toll pathway. J. Biol. Chem. 280: 22793–22799 [DOI] [PubMed] [Google Scholar]
- Xiang T., Zong N., Zhang J., Chen J., Chen M., Zhou J.M. (2011). BAK1 is not a target of the Pseudomonas syringae effector AvrPto. Mol. Plant Microbe Interact. 24: 100–107 [DOI] [PubMed] [Google Scholar]
- Xiang T., Zong N., Zou Y., Wu Y., Zhang J., Xing W., Li Y., Tang X., Zhu L., Chai J., Zhou J.-M. (2008). Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr. Biol. 18: 74–80 [DOI] [PubMed] [Google Scholar]
- Yan Y., Scott D.J., Wilkinson T.N., Ji J., Tregear G.W., Bathgate R.A. (2008). Identification of the N-linked glycosylation sites of the human relaxin receptor and effect of glycosylation on receptor function. Biochemistry 47: 6953–6968 [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhang G. (2004). Tumor necrosis factor family ligand-receptor binding. Curr. Opin. Struct. Biol. 14: 154–160 [DOI] [PubMed] [Google Scholar]
- Zhang J., et al. (2007). A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 1: 175–185 [DOI] [PubMed] [Google Scholar]
- Zhang J., et al. (2010). Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7: 290–301 [DOI] [PubMed] [Google Scholar]
- Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J.D., Boller T., Felix G. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]