MicroRNAs (miRNAs) and other small interfering RNAs (siRNAs) play an important regulatory role in the growth and development of eukaryotes. miRNAs in plants are known to regulate the expression of a number of key developmental and stress-related genes. New insight into miRNA function was gained recently with the discovery that several miRNA families target genes encoding nucleotide binding site–leucine-rich repeat (NBS-LRR) plant innate immune receptors in legumes (Zhai et al., 2011) and in the Solanaceae (Li et al., 2012).
New work by Shivaprasad et al. (pages 859–874) complements and extends these findings, demonstrating that the miR482/2118 family of miRNAs targets numerous NBS-LRR mRNAs encoding disease resistance proteins in tomato (Solanum lycopersicum) and other members of the Solanaceae. This miRNA-mediated silencing of disease resistance mRNAs is shown to be lost in virus- and bacteria-infected plants, thereby resulting in a mechanism for pathogen-induced expression of NBS-LRR defense-related genes. The results suggest that the miR482/2118 superfamily may be a master regulator of diseases resistance in tomato.
Shivaprasad et al. found the miR482/2118 superfamily to be highly variable both in sequence and in expression level across a wide range of plant families and especially abundant in the Rutaceae, Solanaceae, and Fabaceae. The miRNAs in this family are largely 22 nucleotides in length, which is known to be associated with the ability to generate secondary trans-acting siRNAs (Cuperus et al., 2010). The authors demonstrate the generation of secondary siRNAs and the accumulation of cleaved target mRNAs in phase with miR482, providing direct evidence of a cascade of miR482-mediated regulation of NBS-LRR gene expression.
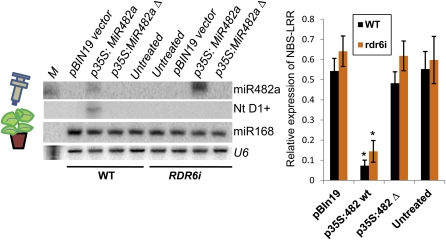
The need for defense against pathogens is considered a strong evolutionary force that gives rise to diversifying selection and high levels of variation in plant genes encoding key defense-related molecules, such as NBS-LRR defense-related genes. miRNA regulation of innate immunity adds a new weapon to the arsenal of defense mechanisms in plants. Shivaprasad et al. note that NBS-LRR proteins are normally associated with race-specific effector-triggered immunity, wherein the host NBS-LRR protein mediates recognition of a pathogenesis effector encoded by specific pathogen strains. Based on the effects of NBS-LRR protein overexpression (see figure), they propose that miRNA regulation of NBS-LRR gene expression might create a role for NBS-LRR proteins in non-race-specific immunity. In this scenario, pathogen-encoded suppressors of RNA silencing mechanisms in the plant might trigger the loss of miR482/2118-mediated suppression of NBS-LRR genes in a non-race-specific manner, resulting in the induction of multiple NBS-LRR defense proteins. This system also minimizes the cost to the plant for defense, as multiple NBS-LRR proteins can be maintained at a low level in the absence of a pathogen and rapidly induced under pathogen attack.
Transient overexpression of tomato miR482a (p35S:MiR482a, left panel) in Nicotiana benthamiana is associated with a decline in NBS-LRR mRNA carrying a miR482a target site (right panel). (Reprinted from Shivaprasad et al. [2012], Figure 6.) M, decade marker indicating a 20-nucleotide RNA; WT, wild type.
References
- Cuperus J.T., Carbonell A., Fahlgren N., Garcia-Ruiz H., Burke R.T., Takeda A., Sullivan C.M., Gilbert S.D., Montgomery T.A., Carrington J.C. (2010). Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat. Struct. Mol. Biol. 17: 997–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Pignatta D., Bendix C., Brunkard J.O., Cohn M.M., Tung J., Sun H., Kumar P., Baker B. (2012). MicroRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. USA 109: 1790–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad P.V., Chen H.-M., Patel K., Bond D.M., Santos B.A.C.M., Baulcombe D.C. (2012). A microRNA superfamily regulates nucleotide binding site–leucine-rich repeats and other mRNAs. Plant Cell 24: 859–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J.X., et al. (2011). MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 25: 2540–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]