A WDxR motif–containing protein required for gametogenesis and seed development was identified and found to interact with the CUL4 interactor protein DDB1.
Abstract
CUL4-RING ubiquitin E3 ligases (CRL4s) were recently shown to exert their specificity through the binding of various substrate receptors, which bind the CUL4 interactor DNA DAMAGED BINDING PROTEIN1 (DDB1) through a WDxR motif. In a segregation-based mutagenesis screen, we identified a WDxR motif–containing protein (WDR55) required for male and female gametogenesis and seed development. We demonstrate that WDR55 physically interacts with Arabidopsis thaliana DDB1A in planta, suggesting that WDR55 may be a novel substrate recruiter of CRL4 complexes. Examination of mutants revealed a failure in the fusion of the polar cells in embryo sac development, in addition to embryo and endosperm developmental arrest at various stages ranging from the zygote stage to the globular stage. wdr55-2 embryos suggest a defect in the transition to bilateral symmetry in the apical embryo domain, further supported by aberrant apical embryo localization of DORNROESCHEN, a direct target of the auxin response factor protein MONOPTEROS. Moreover, the auxin response pattern, as determined using the synthetic auxin-responsive reporter ProDR5:GREEN FLUORESCENT PROTEIN, was shifted in the basal embryo and suspensor but does not support a strong direct link to auxin response. Interestingly, the observed embryo and endosperm phenotype is reminiscent of CUL4 or DDB1A/B loss of function and thus may support a regulatory role of a putative CRL4WDR55 E3 ligase complex.
INTRODUCTION
The making of a plant seed is a coordinated effort of transcriptional programs in the different organisms that constitute a seed (Berger et al., 2006). Seed development also requires tight developmental control of male and female gametophytes and their products to secure double fertilization and establish parent-of-origin–specific cues and expression states (Nowack et al., 2010). In Arabidopsis thaliana, our understanding of the transcriptional networks that shape and orchestrate plant reproduction has been greatly enhanced in the postgenome era and involves different levels of transcriptionally based regulation, including the various roles of chromatin, and the distribution of hormone gradients (reviewed in Nawy et al., 2008; Berger and Chaudhury, 2009; Sundberg and Østergaard, 2009; De Smet et al., 2010; Ge et al., 2010). However, the levels of regulatory proteins are also regulated by proteolysis, and, like in other eukaryotes, a major route is through the UBIQUITIN-26S proteasome (UPS)-dependent pathway (Vierstra, 2009). More than 5% of the Arabidopsis proteome consists of components of the UPS pathway (Smalle and Vierstra, 2004), and accumulating data suggest its targets to be major regulators in processes ranging from photomorphogenesis to hormone signaling and pathogen response (Vierstra, 2009). With growing evidence of proteolytic and nonproteolytic roles of ubiquitylation, this regulation system thus rivals the role of transcription as the foremost regulatory mechanism (Smalle and Vierstra, 2004; Dreher and Callis, 2007).
The ubiquitylation machinery consists of three enzymes: a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin-protein ligase (E3) (Hershko and Ciechanover, 1998). The ubiquitin-protein ligase is responsible for the specificity of protein targeting, and all eukaryotes have a large number of E3 ubiquitin ligases, belonging to two major families: the HECT family and the RING family (Jackson and Xiong, 2009). In Arabidopsis, more than 1400 genes potentially encode components of E3 ubiquitin ligases (Vierstra, 2009). The RING family of E3 ligases can be subdivided into either monomeric RING E3s or multimeric RING E3s that scaffold on cullin proteins and are called cullin-RING E3 ligases (CRLs) (Petroski and Deshaies, 2005; Jackson and Xiong, 2009). Among the different CRLs, the composition of CUL4-based E3 ligases (CRL4s) was only recently identified (Higa and Zhang, 2007).
Orthologs of CUL4 are found in fission yeast, worms, flies, plants, and mammals (Chen et al., 2006). As its orthologs, Arabidopsis CUL4 interacts with the RING E3 ligase RING-BOX 1/REGULATOR OF CULLINS 1 (RBX1/ROC1) and the adaptor protein DNA DAMAGED BINDING PROTEIN1 (DDB1) (Bernhardt et al., 2006; reviewed in Biedermann and Hellmann, 2011) and is involved in important processes, such as light-dependent growth regulation, modulation of chromosomal structure, and DNA repair.
Mutation of Arabidopsis CUL4 affects embryo and endosperm development and leads to recessive seed abortion as well as a small decrease in male and female gametophyte transmission, indicating that CUL4 is essential for seed development (Dumbliauskas et al., 2011). Arabidopsis has two DDB1 paralogs sharing more than 90% protein identity: DDB1A and DDB1B, both interacting with CUL4 (Schroeder et al., 2002). ddb1a ddb1b double mutants have shown a critical requirement for DDB1 function both in embryo development as well as gametophyte development or function (Bernhardt et al., 2010; Dumbliauskas et al., 2011). DDB1 proteins are adaptors that bridge CUL4 to the substrate receptors referred to as DDB1-CUL4 associated factor (DCAF) proteins (Angers et al., 2006; He et al., 2006; Higa et al., 2006; Lee et al., 2008). Many DCAFs consist of WD40 proteins carrying a WDxR signature that is necessary for efficient DDB1 binding (He et al., 2006). Interestingly, two DCAFs, DDB1-CUL4 ASSOCIATED FACTOR1 (DCAF1) and MULTICOPY-SUPRESSOR OF IRA1, which have both been shown to interact with DDB1, have been recently reported to play crucial roles in embryogenesis and the latter also in gametogenesis (Lee et al., 2008; Zhang et al., 2008; Dumbliauskas et al., 2011).
Here, we provide data supporting that WDR55 is required for gametophyte development and function as well as embryo and endosperm development. WDR55 carries a signature WDxR motif and was shown to interact with DDB1A proteins in plant cells. Our results suggest that a WDR55-DDB1 complex is involved in the establishment of bilateral symmetry in the Arabidopsis embryo and may support a first link to CRL4-mediated regulation of this process.
RESULTS
Isolation of Mutant Alleles
A screen based on the distorted segregation of a transposon-borne NPTII marker conferring resistance to kanamycin was used to identify mutants affected in male and female gametophyte development. The mutant line dsg3127 showed reduced transmission through the male and female gametophyte and during self-fertilization (Table 1) and was thus selected for further studies. Using a Vectorette PCR (see Methods), the 5′ and 3′ ends of the DsG transposon were found to reside in the second exon of the gene At2g34260 (Figure 1A; see Supplemental Table 1 online). This gene encodes a WD40 repeat protein, and we termed it WDR55 due to its high similarity to the WD repeat protein 55 family (WDR55, IPR017422; see Supplemental Figure 1A online). The mutant line dsg3127 was accordingly named wdr55-1. The insertion leads to a truncated protein with two predicted stop codons immediately after the insertion site (Figure 1B; see Supplemental Table 1 online). Two splice variants, WDR55.1 and WDR55.2, were verified by cloning and sequencing (data not shown), and the DsG element insertion site in wdr55-1 corresponds to the N-terminal sequence of the longer splice variant WDR55.1, whereas the coding sequence of the shorter splice form WDR55.2 is unaffected by wdr55-1 insertion (Figures 1B and 1C).
Table 1.
Transmission of the Mutant Alleles wdr55-1 and wdr55-2
| Genotype of Cross | Ecotype | Mutant Line | SMa | %SMR (N)b | sd | Tc | TEd | Δ%e |
| wdr55-1 +/− self | Ler | dsg3127 | Km | 55.6 (7412) | 5.18 | 0.74 | 0.00 | 74.85 |
| wdr55-1 +/− self | Col | dsg3127 | Km | 50.2 (998) | 2.59 | 0.67 | 0.02 | 73.87 |
| wdr55-2 +/− self | Col | WiscDslox430F06 | BASTA | 57.7 (1103) | 5.00 | 0.77 | 0.01 | 73.89 |
| Wild type × wdr55-1 +/− | Ler | dsg3127 | Km | 29.5 (1000) | 2.96 | 0.59 | 0.42 | 29.08 |
| Wild type × wdr55-1 +/− | Col | dsg3127 | Km | 21.1 (1167) | 5.83 | 0.42 | 0.27 | 36.64 |
| Wild type × wdr55-2 +/− | Col | WiscDslox430F06 | BASTA | 38.9 (848) | 12.05 | 0.78 | 0.64 | 18.15 |
| wdr55-1 +/− × wild type | Ler | dsg3127 | Km | 39.2 (1624) | 3.57 | 0.78 | 0.64 | 17.81 |
| wdr55-1 +/− × wild type | Col | dsg3127 | Km | 44.9 (958) | 3.83 | 0.90 | 0.81 | 9.28 |
| wdr55-2 +/− × wild type | Col | WiscDslox430F06 | BASTA | 44.7 (1555) | 4.75 | 0.89 | 0.81 | 9.59 |
All crosses are female × male.
Selection marker (SM), kanamycin (Km), and BASTA.
Percentage of SM-resistant (SMR) plants in progeny. N is the number of individuals counted.
Transmission of the mutant allele (T) calculated as T(observed)/T(expected).
Transmission efficiency.
Frequency of defective male or female gametophytes (Δ%) as expected from transmission efficiency (TE). D = 0.5(1 − TE) × 100. TE was calculated according to Howden et al. (1998).
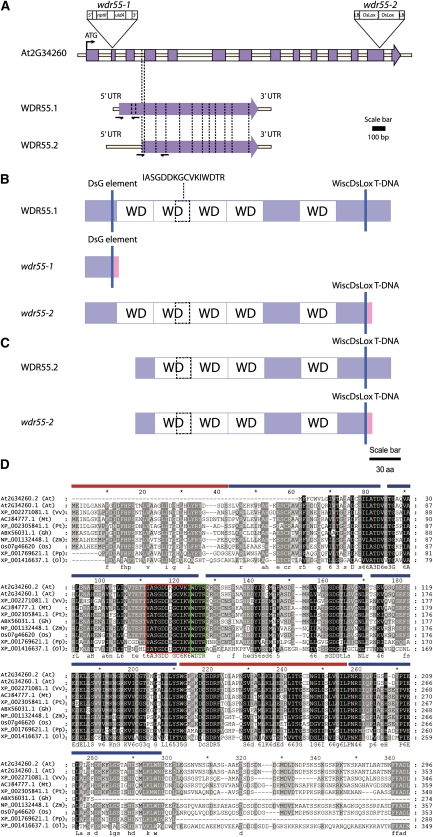
Figure 1.
WDR55 Is a WD Repeat–Containing Protein Conserved in Other Plants.
(A) to (D) Diagrams of WDR55 with insertional mutants, wdr55-1 and wdr55-2 (A), diagrams of the splice forms WDR55.1 (B) and WDR55.2 (C), indicating truncated proteins resulting from T-DNA insertion, and alignment of WDR55 plant orthologs (D).
(A) An 8-bp target site duplication occurred at the insertion site of the DsG element in the DsG line wdr55-1. Two copies of the T-DNA were inserted in reverse orientation into the same loci in wdr55-2. The vertical dashed lines highlight the difference between the two splice forms. Intron 3 in the genomic sequence is part of the 5′ untranslated region (UTR) of WDR55.2. The WDR55.2 transcriptional start site includes 22 bp of the 3′ end of WDR55.1 intron 3. Purple boxes, exons; white boxes, introns; yellow line, WDR55 transcribed region.
(B) WDR55.1 contains five annotated WD40 motifs and is affected by both wdr55-1 and wdr55-2 insertion. wdr55-1 results in a protein that is truncated to 34 amino acids. The wdr55-2 mutant results in a protein that is truncated to 306 amino acids, missing the last 47 amino acids. White boxes with WD indicate annotated WD40 motifs; purple boxes, not annotated.
(C) WDR55.2 contains four annotated WD40 motifs and is only affected by wdr55-2. The insertion results in a truncated protein of 247 amino acids that is missing the last 47 amino acids. Blue vertical lines indicate the position of the DsG element for the wdr55-1 mutant and the T-DNA for the wdr55-2 mutant. Dashed box indicates position of the partial DWD motif.
(D) WDR55 plant homologs were identified using WU-BLAST software and aligned using CulstalW2 software. Conservation is marked in black, and the conserved amino acid is indicated in capital letters below the alignment. Residues conserved across most species are indicated in lowercase letters and numbers. Light gray marks the less conserved residues. Gaps are indicated with dashed lines. WD-repeats identified by SMART are indicated by blue lines. Red lines indicate putative additional WD-repeats identified by PyMOL. The DWD motif is marked by red and a green-boxed area, where the green-boxed area is the WDxR motif. At, Arabidopsis; Vv, Vitis vinifera; Mt, Medicago truncatula; Pt, Populus trichocarpa; Gh, Gossypium hirsutum; Zm, Zea mays; Os, O. sativa; Pp, Physcomitrella patens; Ol, Ostreococcus lucimarinus. aa, amino acids.
We further analyzed putative alleles of wdr55-1 from the sequence indexed SALK T-DNA insertion database (see Supplemental Table 1 online). In the T-DNA line WISCDSLOX430F06, both T-DNA borders were found to reside in exon 13 (of 14 total) in WDR55.1, corresponding to exon 10 (of 11 total) in WDR55.2 (Figure 1; see Supplemental Table 1 online). We named this allele wdr55-2. A stop codon (TGA) is formed in the T-DNA, two amino acids downstream of the insertion site of the left border sequence of the T-DNA. For both splice variants, this predicts a truncated protein missing the last 47 amino acids (Figures 1B and 1C; see Supplemental Table 1 online).
WDR55 Encodes a WD40 Repeat Protein
WDR55 encodes a WD40 repeat protein (WDR). The best-characterized protein in this family, and the first to have its structure elucidated, is the Gβ subunit of the heterotrimeric G proteins. The Gβ subunit adopts a β-propeller fold, which is a symmetrical structure made up of repeats each consisting of a small four-stranded antiparallel β-sheet. Each propeller blade contains the first three strands of one repeat and the last strand in the next (Smith et al., 1999). The WD repeat–containing domains and their implied propeller structure are predicted to act as coordinators for reversible multidomain complex assemblies by supplying a rigid platform or scaffold where a surface for various protein–protein interactions has evolved (Yu et al., 2000).
WDR55.1 has five WD40 repeats predicted by SMART (http://smart.embl-heidelberg.de/) (Figure 1B). We further compared WDR55 to the determined structure of the WD40 protein AGB1, which was chosen due to its sequence similarity to WDR55, using PyMOL (http://www.pymol.org), to check for the presence of other WD repeats than the ones predicted by SMART. The similarities between WDR55 and AGB1 were restricted to the WD40 repeats, which determine the propeller blades of AGB1, indicating the importance of conservation of these sequence domains for the three dimensional structure. We could identify seven possible blades, and, although blades one and seven were not complete in terms of number of β-sheets, a direct alignment of the WD repeats is indicative of a seven-repeat WDR structure (see Supplemental Figure 1B online).
A common feature among WDR proteins is that they have a proposed propeller structure and no enzymatic activity. Some WDR proteins have additional known functional domains, such as the DWD-box (DDB1 binding WD40 box). WDR55 contains a submotif of the DWD-box, named the WDxR motif (Zhang et al., 2008). Both the DWD box and the WDxR motif are signatures for potential substrate receptors of the CUL4 E3 ubiquitin ligases referred to as putative DCAFs (DDB1-CUL4 associated factor). WDxR facilitates assembly with DDB1 and CUL4 proteins, whereas other parts of DCAF bind to proteins to be targeted for ubiquitination (Angers et al., 2006; He et al., 2006).
Finally, WDR55.1 contains a signature WDR55 domain encompassing all seven predicted WD repeats. WDR55 is unique in Arabidopsis but is conserved throughout eukaryotes with highest sequence similarity to plant orthologs (Figure 1D; see Supplemental Figure 1A online). The closest metazoan homolog is found in Nematostella vectensis (UniprotID: A7RNE8, 42% identity).
Outside the regions of the protein occupied by the WD propellers, no functional domains or repeat structures could be identified. This fits well with the predictions from DISOPRED (http://bioinf.cs.ucl.ac.uk/disopred/) that a structured region occurs in the N-terminal part of the protein, whereas the C-terminal regions (after the WD repeats) of both WDR55.1 and WDR55.2 are unstructured (see Supplemental Figures 1C and 1D online). Disordered protein regions have no stable structures without their partner molecules, and due to this flexibility, they may be more adaptable for interaction with multiple partners with high specificity but low affinity (Dunker et al., 2001; Ishida and Kinoshita, 2008).
WDR55 Splice Variants Are Present throughout the Arabidopsis Life Cycle
Various public expression databases report that WDR55 is expressed throughout the Arabidopsis life cycle and also in most tissues (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi; www.genevestigator.com). To confirm these findings and investigate the abundance of the two splice variants, we performed real-time quantitative PCR with primers specific for the two splice variants. Both splice variants were present in all tissues examined, with highest abundance in roots (Figures 2A and 2B). Similarly, both splice forms could be detected throughout seed development (see Supplemental Figures 2A and 2B online).
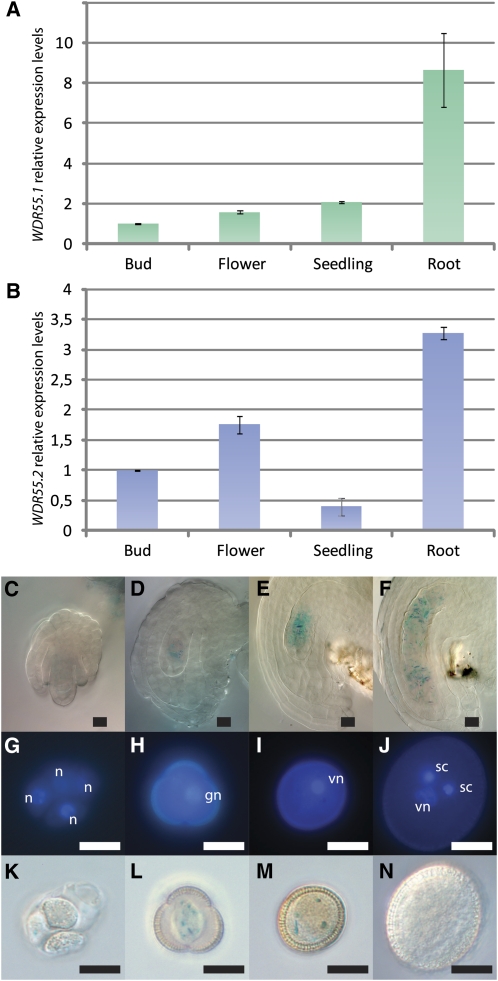
Figure 2.
WDR55 Is Expressed in Various Tissues and in the Female and Male Gametophyte.
(A) to (N) Expression of WDR55 in various tissues by means of real-time PCR of WDR55.1 (A) and WDR55.2 (B) and micrographs showing WDR55 expression using a GUS promoter reporter construct during female gametophyte development ([C] to [F]) and male gametophyte development ([G] to [N]).
(A) and (B) Relative mRNA levels of the splice variants WDR55.1 (A) and WDR55.2 (B) in buds, flowers, seedlings, and roots in wild-type Arabidopsis Col. The graph shows the average relative expression and sd calculated from the three independent biological replicas. WDR55 transcript levels were normalized to ACTIN11 levels.
(C) to (F) ProWDR55:GUS is expressed with increasing strength throughout embryo sac development from meiosis (C), one-nucleate embryo sac (D), four-nucleate embryo sac (E), and mature seven-celled embryo sac (F).
(G) to (J) The developmental stages of GUS-stained pollen visualized by DAPI. Development from after meiosis, showing a tetrad of haploid cells (G), which are released as uninucleate pollen (H). After asymmetric division to form the two-nucleate stage (I) with one vegetative cell and one generative cell, the generative cell then divides to form two sperm cells (J). gn, generative nucleus; n, nucleus; sc, sperm cell; vn, vegetative nucleus.
(K) to (N) The fusion construct ProWDR55:GUS demonstrated weak expression from meiosis (K) up to the two-nucleate stage (M) of pollen development. Note, no expression in mature pollen (N).
Bar = 5 μm.
Using a WDR55 promoter β-glucuronidase (GUS) fusion construct (ProWDR55:GUS), expression in most tissues could be verified using three independent single locus transgenic lines (see Supplemental Figures 2C to 2Q online). A thorough analysis of male and female gametophyte stages revealed prominent ProWDR55:GUS expression throughout embryo sac development (Figures 2C to 2F), and expression could also be found in the embryo proper until 2 d after pollination (DAP) (see Supplemental Figures 2C and 2D online). Only low levels of ProWDR55:GUS expression in the seed could be detected at later developmental stages, but a prominent GUS signal accumulated in the cotyledons of the mature embryo (see Supplemental Figure 2F online) and persisted in the cotyledons and roots after germination, and subsequently in leaves, buds, and flowers (see Supplemental Figures 2G to 2R online). Similarly, ProWDR55:GUS expression was detected during pollen development from meiosis up to the second pollen mitosis (Figures 2G to 2N). At mature (three nucleate) stages, there was no evident GUS signal (Figures 2J and 2N).
WDR Mutants Show Transmission Failure
Next, we performed a thorough analysis of wdr55 on the genetic and morphological level. Adult heterozygous wdr55 plants were indistinguishable from the wild type. Transmission of the wdr55-1 allele was analyzed in successive generations F3 to F10 and showed a stable segregation of 55.6% (n = 7412; Table 1) during self-fertilization. No lines homozygous for the transposon containing NPTII were detected among more than 14 sublines analyzed, and no homozygous plants were identified by genotyping using the DsG transposon and gene-specific primers. To determine which sex of the gametophyte generation was affected in wdr55-1, reciprocal crosses were performed. The transmission of the wdr55-1 allele was reduced through the male gametophyte (29.5%, n = 1000) and to a lesser extent through the female gametophyte (39.2%, n = 1624) (Table 1), thus suggesting that the wdr55-1 mutation impairs male and female development or function.
The wdr55-2 allele segregated 57.7% BASTA-resistant progeny in selfed plants (n = 1103; Table 1), suggesting similar effects as observed in wdr55-1. However, only a slight reduction in male and female transmission of the wdr55-2 allele could be observed in the progeny of reciprocal crosses (Table 1). To determine whether the gametophyte effect in wdr55-1 was ecotype specific (wdr55-1 is in Landsberg erecta [Ler], whereas wdr55-2 is in the Columbia [Col] accession), we introgressed wdr55-1 into Col (crossed six times). Col-introgressed wdr55-1 displayed even more reduced transmission through the male gametophyte, but segregation through the female gametophyte was slightly increased (Table 1).
wdr55-1 Affects Polar Nuclei Fusion
In wdr55-1, a weak effect on embryo sac development was expected based on transmission data (Table 1). Close inspection of unfertilized siliques 3 d after emasculation (DAE) revealed a modest frequency of unfused polar nuclei in wdr55-1 embryo sacs (29%, n = 101) compared with wild-type Ler (5%, n = 53; Figure 3; see Supplemental Table 2 online). The frequency of unfused polar nuclei decreased at 4 DAE, suggesting a delay in polar nuclei migration and karyogamy (16%, n = 55; Table 2). Upon hand-pollination of female wdr55-1 3 DAE, the frequency of unfused polar nuclei at 2 DAP decreased further compared with Ler controls (7%, n = 92 and 4%, n = 57, respectively; Table 2, Figure 3). Finally, no unfused polar nuclei were found in 4 DAE pollinated siliques at 2 DAP (see Supplemental Table 2 online), suggesting that the fusion of polar nuclei to form the diploid central cell nucleus is delayed in the wdr55-1 mutant in the Ler background. A similar delay was not observed in the Col-introgressed wdr55-1 allele (see Supplemental Table 2 online) or in the wdr55-2 allele, suggesting that ecotype-specific modifiers influence the penetrance of the wdr55-1 phenotype. This supports the observation that all crosses involving a maternal Ler partner showed a slight delay in polar nuclei fusion compared with Col (see Supplemental Table 2 online). Furthermore, these observations are in line with the observed increased maternal transmission of the wdr55-1 allele in the Col background (Table 1).
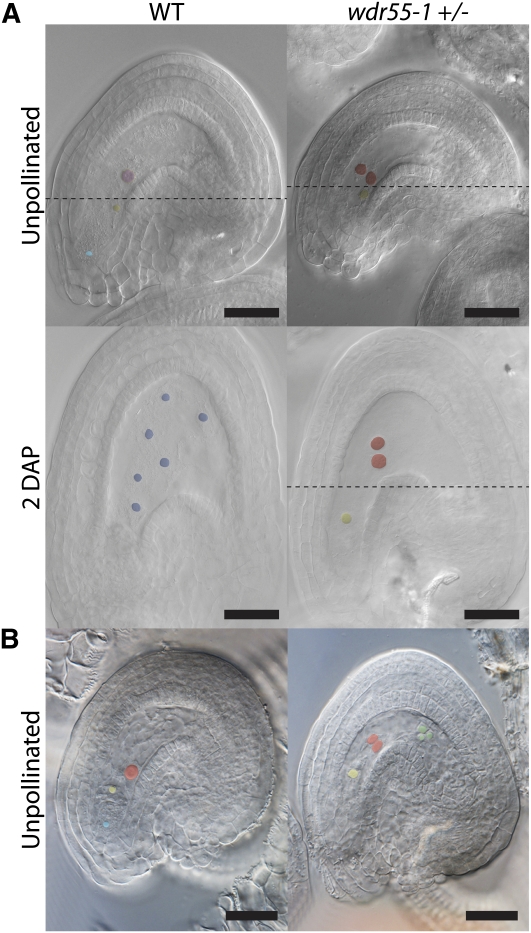
Figure 3.
Polar Nuclei Fusion Is Delayed in the wdr55-1 Embryo Sac.
(A) and (B) Light micrographs showing the embryo sac phenotype in unpollinated wild-type (WT) Ler and wdr55-1 ovules and ovules 2 DAP in Ler wild type and in wdr55-1 crossed to wild-type Ler. The light micrograph with a dotted line in (A) represents two focal planes that were assembled using Adobe Photoshop. Pink coloration indicates central cell nuclei, purple color shows developed endosperm nuclei, red color marks unfused polar nuclei, antipodal cells are marked green, and the egg cell nucleus is shown in yellow. Bars = 20 μm.
(A) Fused polar nuclei (central cell nucleus) in the wild type (left) compared with wdr55-1 ovules showing unfused polar nuclei in unpollinated ovules 2 DAE and also at a lower percentage in pollinated ovules 2 DAP. Unpollinated ovules were examined and crossed 3 DAE.
(B) Wild-type female gametophyte (left) with fused polar cells (central cell nucleus) and degenerated antipodal cells and wdr55-1 female gametophyte (right) displaying unfused polar cells and visible antipodal cells.
Table 2.
Embryo Development Arrests in wdr55-1 and wdr55-2 Alleles
| Genotype of Cross | Total | Wild Type | Developed Seed Late Arrest | Developed Seed Early Arrest | Collapsed Undeveloped |
| Self | |||||
| wdr55-1 3–14 DAP | 784 | 73.9% | 0.0% | 13.9% | 12.2% |
| wdr55-2 8–12 DAP | 548 | 77.2% | 17.9% | 0.6% | 4.4% |
| Col 8–12DAP | 340 | 98.5% | 0.3% | 0.6% | 0.6% |
| Ler 14 DAP | 131 | 96.2% | 0.8% | 3.1% | 0.0% |
| Cross to the wild type | |||||
| wdr55-1 × Ler 7–14 DAP | 187 | 87.2% | 0.0% | 4.8% | 8.0% |
| wdr55-2 × Col 7 DAP | 83 | 96.4% | 0.0% | 0.0% | 3.6% |
| Complementation cross | |||||
| wdr55-1 × wdr55-2 8–14 DAP | 286 | 80.8% | 6.6% | 1.8% | 10.8% |
| wdr55-2 × wdr55-1 8–14 DAP | 167 | 83.8% | 11.4% | 0.6% | 4.2% |
Embryo phenotype count for wdr55-1 and wdr55-2 showing percentage of total counts in four classes: wild-type development, developed seed late arrest, developed seed early arrest, and undeveloped seeds that have not been fertilized. All crosses are female × male.
The antipodal cells undergo programmed cell death (PCD), and the timing of this event has been suggested to be in part regulated by the presence of a functional central cell (Gross-Hardt et al., 2007; Kägi et al., 2010). We therefore inspected PCD of wdr55-1 antipodal cells by means of whole-mount microscopy. We found no strict correlation between polar cell fusion and antipodal degeneration in wdr55-1 (Figure 3B). In 2 DAE embryo sacs, when approximately half of the mutant polar nuclei were unfused, compared with most being fused in the wild-type control (59 and 3% unfused, n = 100 and 39, respectively), a relatively similar frequency of antipodal cells was observed in the mutant and in the wild type (11 and 18%, n = 100 and 39, respectively), suggesting that antipodal PCD in wdr55-1 is similar to the wild type. Notably, all antipodals observed in the mutant were correlated with unfused polar nuclei.
A simple hypothesis derived from these experiments would be that if polar nuclei were given the time to fuse properly, the wdr55-1 mutation in Ler would segregate as the wild type (i.e., in a 1:1 ratio). To investigate this, wdr55-1 siliques were pollinated with Ler 4 DAE and the progeny seeds were selected for the transmission of the mutation. Surprisingly, the transmission was not restored to the hypothetical 1:1 ratio (50%) but actually decreased (26.6%, n = 143) compared with the wdr55-1 siliques pollinated 3 DAE (39.2%, n = 1624; Table 1). This may suggest that wdr55 is also involved in other processes affecting the timing or specification of the egg apparatus.
We further examined the role of WDR55 in the female gametophyte by ectopically expressing WDR55 splice variants under the control of the promoter of the ubiquitously expressed cell cycle regulator CDKA;1 (Nowack et al., 2006; Dissmeyer et al., 2007). More than half of the recovered pCDKA;1 overexpression lines displayed a variable albeit high frequency of undeveloped seeds at 2 DAP (see Supplemental Figure 3A online). Closer inspection of representative ProCDK:WDR55.1 lines at the wild-type octant embryo stage (see Supplemental Figures 3A and 3B online) revealed uniform arrest of 49 to 80% of the female gametophytes before the first mitotic division (66% ± 22%, n = 122; see Supplemental Figure 3C online). Similarly, ProCDK:WDR55.2 lines inspected at the early globular stage of embryogenesis (see Supplemental Figure 3D online) displayed arrest at the one-nucleate embryo sac stage, in a few cases allowing up to two mitotic divisions (45% ± 13%, n = 84; see Supplemental Figure 3F online). Moreover, ProCDK:WDR55.2 lines also resulted in a variable frequency of aberrant seeds with underdeveloped endosperm (see Supplemental Figure 3E online), suggesting that ProCDK:WDR55.2 is less detrimental to embryo sac development than the longer WDR55.1 splice form.
The WDR55 Level Is Crucial for Pollen Function
The transmission efficiency of wdr55-1 in pollen (Table 1) and WDR55 expression at early stages of male gametogenesis (Figures 2K to 2N) suggest that WDR55 is required for male gametophyte development or function. However, upon inspection of whole-mount 4′,6-diamidino-2-phenylindole (DAPI)–stained pollen of wdr55-1 heterozygous plants, no obvious phenotype could be detected, as all mature pollen contained a vegetative cell and two sperm cells (see Supplemental Figure 4A online). Furthermore, wdr55-1 was crossed to quartet, which is deficient in the separation of microspores after meiosis and thus allows tetrad analysis (Preuss et al., 1994), without revealing any observable phenotype (see Supplemental Figures 4B and 4C online). All pollen could be shown to be viable using Alexander’s Stain (Alexander, 1969; see Supplemental Figure 4D online; n = 342). Finally, to investigate if sperm cell fate is disturbed in wdr55-1 pollen, we introduced the germ line markers ProH3.3:H3.3-RFP (for red fluorescent protein) and ProDUO:DUO-RFP (Rotman et al., 2005; Ingouff et al., 2007; Brownfield et al., 2009). Both markers were uniformly expressed in sperm cells of pollen from heterozygous wdr55-1 plants (see Supplemental Figures 4E to 4H online), and mature wdr55-1 pollen were thus by our criteria indistinguishable from those of the wild type. Considering the reduced transmission of the mutant allele, WDR55 is therefore suggested to be required for male gametophyte function, such as pollen tube growth, guidance, or fertilization. We therefore compared wild-type and wdr55-1 pollen tube germination in vitro (see Supplemental Figures 4I and 4J online). The overall germination rate in the mutant was moderately lower than in the wild type (71%, n = 402; and 80%, n = 406, respectively). We further compared the frequency of germinating pollen with short tubes (tube span less than one pollen diameter) between the wild type and mutant. The frequency of germinating pollen with short tubes in the mutant was reminiscent of the wild type frequency, and no obvious difference in pollen tube morphology could be observed (see Supplemental Figure 5K online). Albeit not fully explaining the wdr55-1 male transmission deficiency, our results hint at a requirement for WDR55 in pollen tube germination.
In our expression analysis, ProWDR55:GUS displayed a modest level of expression during the early microspore stages but could not be detected in the mature pollen grain (Figure 2). This may suggest that fine-tuning of the WDR55 level is crucial for pollen function. This hypothesis was supported by analyzing pollen development in ProCDK:WDR55.1 and ProCDK:WDR55.2 lines. Two out of six ProCDK:WDR55.1 lines examined and four out of nine ProCDK:WDR55.2 lines checked displayed high frequencies of aborted pollen (see Supplemental Figures 5A to 5H online). In ProCDK:WDR55.1 lines, meiosis appeared to be affected, producing early-stage abortions in microspores (see Supplemental Figures 5I to 5L online). No meiotic defects were detected in ProCDK:WDR55.2 lines, but microsporogenesis appeared to be hampered already in the first microspore mitosis (see Supplemental Figures 5M to 5P online).
WDR55 Is Required for Embryo and Endosperm Development
Since the gametophyte lethality in wdr55 alleles is not fully penetrant, we expected homozygous wdr55-1/wdr55-1 plants to segregate from a self-cross. However, no homozygous plants could be identified by genotyping experiments for either allele. Since the reduced transmission rates of the mutant alleles cannot be explained by gametophyte failure alone, the genetic analyses of both alleles therefore suggest that WDR55 is also required post fertilization (Table 1).
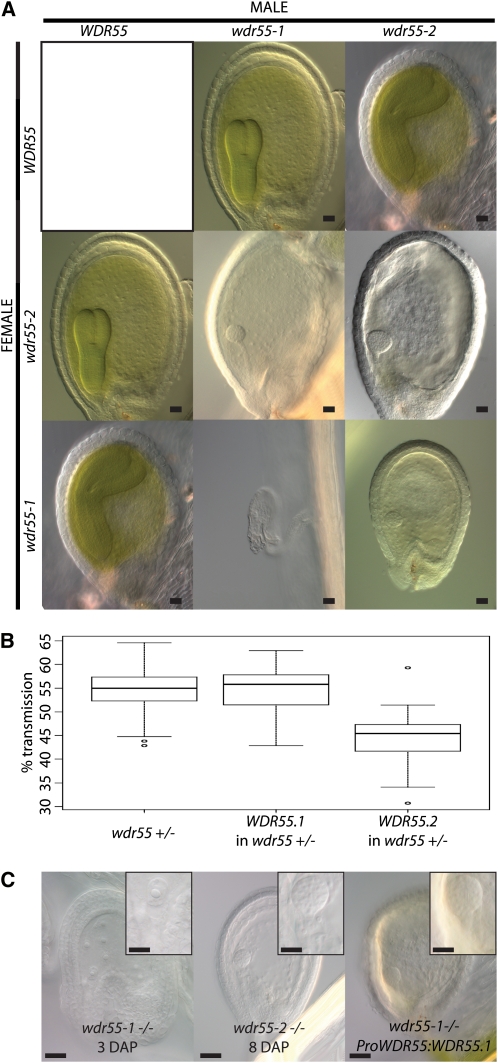
We investigated the requirement of WDR55 in the developing embryo and endosperm by inspecting cleared whole-mount preparations of heterozygous hand-pollinated wdr55-1 and wdr55-2 compared with the wild type at 1 to 12 DAP. In the wild type, the embryo develops through a series of invariant cell divisions, resulting in a fully differentiated bent-cotyledon embryo (Figure 4). The wild-type endosperm proliferates through syncytial nuclear divisions in parallel with the developing embryo and is gradually consumed by it (Figure 4). In siliques from heterozygous wdr55-1 and wdr55-2, however, a significant fraction of the seeds were arrested in development (14 and 18%, respectively; Table 2). To rule out gametophyte maternal effects, wdr55-1 and wdr55-2 were crossed as females to wild-type plants. No significant seed abortion was observed in these crosses (Table 2), in agreement with our genetic data (Table 1), suggesting aborted seeds to be homozygous wdr55.
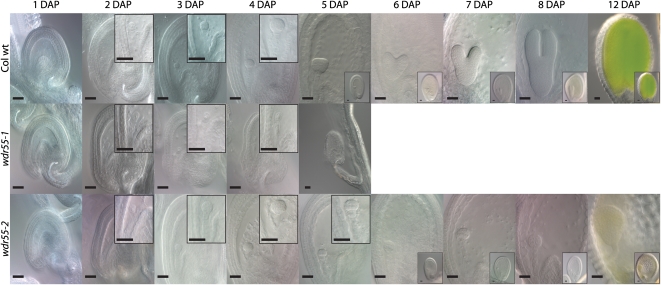
Figure 4.
wdr55-1 and wdr55-2 Have Arrested Embryo Development.
Light micrographs showing the embryo and endosperm phenotype for wdr55-1 (middle panel) and wdr55-2 (bottom panel) compared with wild-type (wt) Col (top panel) at 1 to 8 and 12 DAP. Insets at 2 to 5 DAP show embryo detail. Insets at 5 to 12 DAP show whole seed. Top panel: The wild-type Col embryo and endosperm develops from the zygote (1 to 2 DAP), to the early globular stage (3 to 4 DAP), to the heart stage (5 to 7 DAP), to the torpedo stage (8 DAP), and then through to the bent cotyledon stage (12 DAP). Middle panel: The wdr55-1 −/− embryo and endosperm appear to develop normally up to 2 DAP with three to four endosperm nuclear divisions and the initial asymmetric division of the zygote, but then arrests at around 3 DAP, after which the seed starts to collapse (4 to 5 DAP). Bottom panel: The wdr55-2 −/− embryo and endosperm develop normally up to the mid-globular stage (4 DAP), but the embryo is then delayed in development and does not initiate the cotyledon primordial throughout the stages investigated (5 to 8 and 12 DAP). The endosperm is also delayed and does not appear to have cellularized by 8 DAP. Bars = 20 μm.
[See online article for color version of this figure.]
In wdr55-1, the first three to four endosperm nuclear divisions as well as the initial asymmetric division of the zygote appeared to occur normally up to 2 DAP (Figure 4). However, subsequent development of the proembryo and the embryo suspensor was not observed in the affected class after this stage. Similarly, the endosperm did not progress beyond the fourth syncytial division and seemed to start to degenerate in the syncytial state 4 DAP (Figure 4). Concurrent with the embryo and endosperm arrest, no further development or elongation of the ovule integuments were observed (Figure 4).
In wdr55-2, we observed wild-type progression in embryo and endosperm development up to the embryo mid-globular stage 4 DAP (Figure 4). In wild-type globular embryos, the cotyledon primordia initiate at 5 DAP and are clearly recognizable in the 6 DAP heart-stage embryo (Figure 4). Syncytial endosperm divisions continue, and endosperm domains are visible at 5 DAP, with peripheral endosperm nuclei lining the endothelium of the inner integuments. wdr55-2 embryos, however, remained in the globular state up to 8 DAP (Figure 4). In contrast with the wild type, no initiation of cotyledon primordia could be observed, from 5 throughout 8 DAP (Figure 4). Endosperm development was also delayed, and syncytial endosperm nuclei were enlarged and reduced in number and appeared disorganized compared with the wild type (Figure 4). Cellularization was not observed up to 8 DAP, and the endosperm did not appear to degenerate. Most wdr55-2 seeds still appeared to be viable at 12 DAP, and these embryos were comparable in size with 6 DAP wild-type heart stage embryos but entirely lacked the organized cell division pattern and a distinguished bilateral symmetry in the apical region (Figure 4). However, upon silique ripening, further development of the mutant seeds was not supported.
Taken together, WDR55 is required for the early stages of embryo and endosperm development, as evidenced by the arrest of the wdr55-1 proembryo at 2 DAP, as well as the involvement of WDR55 in the establishment of bilateral symmetry in the transition from the globular to the heart embryo stage.
Both wdr55 Splice Forms Are Required for Embryo Development
Due to the phenotypic divergence between the wdr55 alleles, we performed reciprocal genetic complementation crosses and inspected the resulting siliques directly. As expected, wdr55-1 and wdr55-2 did not complement each other and are therefore allelic (Table 2). Interestingly, closer inspection using whole-mount cleared preparations revealed that the reciprocal crosses of the two alleles gave rise to the same embryo phenotype of wdr55-2. This finding further supports the zygotic requirement of WDR55, since one copy of the weaker allele wdr55-2 is sufficient to sustain embryo development up to the globular stage, regardless of its parental contribution (Table 2, Figure 5).
Figure 5.
Complementation Cross between wdr55-1 and wdr55-2 and Splice Variant Complementation of wdr55-1.
Light micrographs of phenotypes resulting from complementation crosses between wdr55-1 and wdr55-2 (A), box plot representation of the effect of splice variant-specific (WDR55.1 and WDR55.2) complementation on wdr55-1 segregation (B), and light micrographs of WDR55.1 splice variant complementation of the wdr55-1 phenotype (C). Reciprocal crosses between wdr55-1 and wdr55-2 give a wdr55-2 phenotype both when used as mother and father (A). When using the cDNA sequence of the two splice variants, WDR55.1 and WDR55.2, under the control of their endogenous promoter to separately complement the wdr55-1 segregation phenotype on kanamycin selection plates, neither rescues the phenotype completely and WDR55.2 seems to have a negative effect on the transmission (B). The box plot values indicated are as follows: the smallest observation (sample minimum), lower quartile, median (in bold) upper quartile, and the largest observation (sample maximum). Observations considered outliers are indicated by small circles. For the box plot, 26 observations of segregating wdr55-1 lines were used, and a total of 6280 seedlings were counted on kanamycin selection. For the WDR55.1 splice variant in the wdr55-1 background, 31 observations from individual transformants were used and a total of 5964 seedlings were counted on kanamycin selection. For the WDR55.2 splice variant in the wdr55-1 background, 26 observations from individual transformants were used and a total of 4066 seedlings were counted on kanamycin selection. Complementation of wdr55-1 by the splice variant WDR55.1 resulted in a wdr55-2 phenotype in the wdr55-1 background (C). Insets are magnified versions of the embryo region from the respective seed shown. Bar = 20 μm. Inset bars = 10 μm.
[See online article for color version of this figure.]
Next, we performed molecular complementation using the genomic sequence of WDR55 and 1283 bp of the upstream promoter region (see Methods for details). This construct was transformed directly into heterozygous wdr55-1 and selected for the presence of both the rescue construct and the wdr55-1 allele. In the T2 generation, several lines were found to be wdr55-1 homozygous, indicating full rescue of both the seed and the gametophyte deficiency in wdr55-1 plants (see Supplemental Table 3 online). We further crossed a line homozygous for wdr55-1 and the rescue transgene to a heterozygous wdr55-2 plant. wdr55-1 wdr55-2 seeds are lethal without the rescue transgene (Figure 5), but trans-homozygous plants were identified in the resulting progeny by genotyping (five of 16 plants investigated), indicating that both wdr55-2 and wdr55-1 are complemented by the WDR55 transgene.
To investigate whether the two WDR55 splice forms have specific roles in gametophyte or embryo development, we designed splice variant-specific rescue constructs containing the 1326-bp WDR55 promoter fused to the WDR55 long or short splice form cDNA sequence. The constructs were transformed separately into heterozygous wdr55-1 and selected for the presence of a splice form rescue construct and the wdr55-1 allele. No homozygous wdr55-1 plants were obtained in subsequent generations, indicating that both splice forms or regulatory sequences not present in our transgenes are required for embryo and endosperm development.
We further compared the effect of the single splice variant rescue constructs on the transmission of wdr55-1. The overall effect of all WDR55.1 rescue construct lines on wdr55-1 transmission was not significantly different from that of the wdr55-1 controls (Figure 5B). However, six transgenic lines with elevated transmission were rechecked in the T3 generation and seemed to have a stable higher transmission of ~64.9% (n = 1279; sd ± 0,8), indicating that WDR55.1 can partially rescue the gametophyte transmission defect in wdr55 -1. Furthermore, the homozygous embryo phenotype in the same outlier lines was partially rescued and arrested at the globular stage, similar to the phenotype observed in wdr55-2 (Figure 5C; 19% [n = 1423; sd ± 4.3] out of a total of 24.7% defective seeds [n = 1423; sd ± 4]). This may suggest that WDR55.1 is sufficient for embryo development up to the globular stage. However, both WDR55 splice forms appear to be required to complete seed development. Surprisingly, the shorter splice variant, WDR55.2, led to a decrease in wdr55-1 transmission (Figure 5B). We did not further inspect the possible phenotypic basis of this effect, but since the WDR55.2 transcript is not physically interrupted by the wdr55-1 mutation, and we have seen that overexpression of WDR55.2 induces gametophyte failure (see Supplemental Figures 4 and 5 online), one could speculate that a strict requirement for correct levels of WDR55.2 may be needed.
Embryo Patterning Is Disturbed in wdr55 Mutants
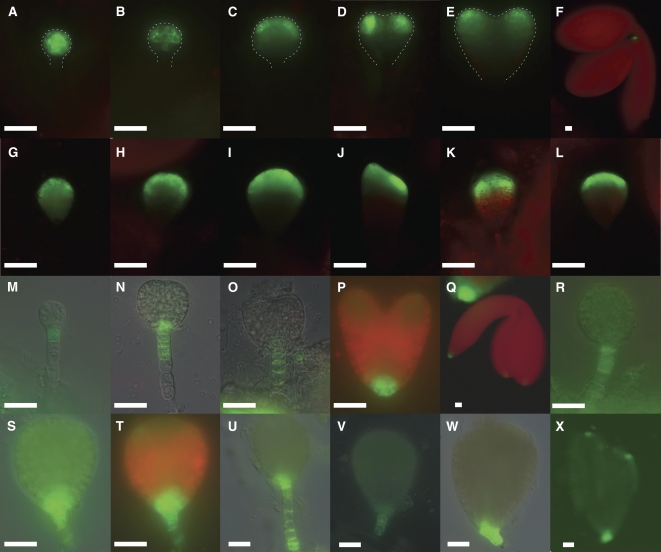
The observed phenotype in wdr55-2 embryos suggests a defect in the transition to bilateral symmetry in the apical embryo domain (i.e., cotyledon specification). Defects in the breakage of radial symmetry are associated with transcription factors like DORNRÖSCHEN (DRN)/DORNRÖSCHEN-LIKE (Chandler et al., 2007) or genes relating to auxin signaling or transport (Friml et al., 2003). The transcriptional auxin responses required for axialization are mediated by the auxin response factor (ARF) protein MONOPTEROS (MP) and the AUXIN/INDOLE-3-ACETIC ACID (AUX/IAA) protein IAA12/BODENLOS (Jenik et al., 2007). DRN has been shown to be directly regulated by MP in response to auxin, in cotyledon tips from the embryo heart stage and onwards, and, in early heart stage mp embryos, DRN is not downregulated in the shoot apical meristem (SAM) region (Cole et al., 2009).
We investigated the expression pattern of a ProDRN:GFP (for green fluorescent protein) transgene in developing wdr55-2 embryos. In the wild type, DRN:GFP accumulates in the cotyledon initials in the apical part of the globular embryo and remains symmetrically localized throughout the embryo heart stage. A weak signal can be observed at the bent cotyledon stage in addition to accumulation in the shoot meristem (Figures 6A to 6F; Cole et al., 2009). However, ProDRN:GFP expression in globular stage wdr55-2−/− embryos is present in the entire apical domain and appears to be weaker than in wild-type embryos (Figure 6G), similar to the situation at early wild-type stages before polarized localization (Cole et al., 2009). In subsequent stages, DRN:GFP localizes to the upper tier of the apical domain of the mutant embryos (Figures 6H and 6I), and occasionally nonsymmetric localization to one of the cotyledon initials could be observed (Figure 6J). At later stages, localization to the cotyledon initials was observed sporadically (Figure 6K), but the majority of mutant embryos failed to downregulate ProDRN:GFP expression in the SAM domain and possessed uniform localization in the entire uppermost tier (Figure 6L). This strongly supports our findings that WDR55 is required to break radial symmetry and establish bilateral symmetry in the apical embryo domain.
Figure 6.
ProDRN:GFP and ProDR5:GFP Expression in the wdr55-2 −/− Embryo.
(A) to (X) Fluorescence micrograph of ProDRN:GFP and ProDR5:GFP expression in wild-type ([A] to [F] and [M] to [Q]) and wdr55-2 ([G] to [L] and [R] to [X]) embryos. ProDRN:GFP ([A] to [L]) and ProDR5:GFP ([M] to [X]). Bars = 20 μm.
(A) to (C) In the wild-type preglobular stage, DRN is localized to the entire embryo proper (A) and then shifts apically (B) to be localized to the upper apical region where it marks the precotyledons (C).
(D) and (E) At the wild-type heart stage, expression is localized to the cotyledons.
(F) Wild-type bent cotyledon stage with DRN localized in the SAM.
(G) Expression in globular stage wdr55-2−/− embryos appears as in early wild-type stages before DRN is localized to the precotyledons (Cole et al., 2009), although expression is weak in the mutant.
(H) and (I) Expression is reduced to the upper layer of the wdr55-2−/− embryo but does not mark precotyledons.
(J) and (K) wdr55-2 −/− embryos showing occasional asymmetric localization (J) and symmetric localization (K) of DNR.
(L) Strong DRN expression also in the apical central domain in the wdr55-2 −/− embryo.
(M) to (O) ProDR5:GFP expression in the wild-type marks basal auxin maxima restricted to the upper suspensor and hypophysis in the embryo proper.
(P) A strong signal in the hypophyseal region is maintained at the heart stage and at the bent cotyledon stage marks the cotyledon tips and the root tip (Q).
(R) to (U) In wdr55-2, ProDR5:GFP expression expands toward the basal end of the suspensor. Expression in the hypophyseal region is weak, absent, or partly absent ([R], [V], and [W]) or in other cases strongly expressed ([S] and [T]).
(X) In rare cases, a moderately strong ProDR5:GFP signal is found in the apical region. wdr55-2 mutant embryos were inspected in stages corresponding to 9 to 12 DAP.
We therefore inspected the auxin response pattern in wdr55-2 embryos using the synthetic auxin-responsive reporter ProDR5:GFP (Ulmasov et al., 1997; Ottenschläger et al., 2003). In the wild type, the basal auxin maxima are restricted to the upper suspensor and hypophysis in the embryo proper (Figures 6M to 6Q)(Friml et al., 2003; Ottenschläger et al., 2003). ProDR5:GFP expression in the mutant largely resembles that in the wild type but appears somewhat more variable and is basally expanded in the suspensor and variably reduced in the hypophysis (Figures 6R to 6X). Whereas auxin accumulation was only rarely observed in basal suspensor cells in the wild type, this was the most frequent situation in the mutant (Figures 6R, 6S, and 6U). Auxin maxima in the hypophyseal region were in most cases weak (Figures 6R and 6V) but ranged from virtually absent (Figures 6R and 6V) to a prominent signal (Figures 6S and 6T). These results do not support a strong link to the auxin response. However, an indirect relationship between WDR55 action and auxin response cannot be ruled out.
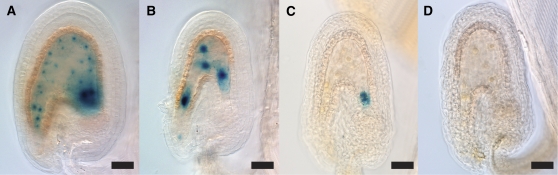
We further investigated endosperm development using specific marker lines. ProFIS2:FIS2-GUS (Luo et al., 1999) was used as an early endosperm marker in wdr55-1 and wdr55-2. Arrested endosperm nuclei in wdr55-1/wdr55-1 seeds expressed the marker (Figures 7A and 7B); however, a fraction of the mutant seeds did not show uniform expression as found in the wild type (Figure 7A), but expression was confined to the chalazal endosperm region (Figure 7C) or even appeared to be lost (Figure 7D; 20% of the cases as opposed to 6% in the wild type [n = 86 and 82, respectively]). Mutant seeds in heterozygous wdr55-2 siliques also displayed a similar ProFIS2:FIS2-GUS expression pattern (see Supplemental Figure 6 online). ProFIS2:FIS2-GUS expression in micropylar endosperm and peripheral endosperm nuclei was lost after four to five syncytial divisions and remained in the chalazal endosperm region, as also seen in the wild type (Luo et al., 1999), thus suggesting a functional chalazal endosperm in the wdr55-2 endosperm. Similar results were also observed using a ProKS117:GFP marker (Sørensen et al., 2001). ProKS117:GFP marks the early stages of syncytial endosperm development and is regarded as a marker of endosperm identity (Ungru et al., 2008). Although delayed compared with the wild-type endosperm, a uniform ProKS117:GFP expression pattern could be observed in the wdr55-2 endosperm at both the early and late stages of embryo development (see Supplemental Figure 6 online).
Figure 7.
Endosperm Marker Has Ectopic Expression in wdr55-1.
Light micrographs of the early endosperm marker ProFIS2:GUS in the wdr55-1 -/+ background. The expression of ProFIS2:GUS is shown in wild-type ovules 2 DAP (A) and in wdr55-1 -/+ ovules 2 DAP (B) and 3 DAP ([C] and [D]). Bars = 20 μm.
(A) In the wild type, ProFIS2:GUS is expressed in the endosperm from fertilization until about five rounds of nuclear divisions.
(B) to (D) ProFIS2:GUS is expressed in the wdr55-1 −/− endosperm after fertilization (B), but ectopic expression (C) or absence of expression in visible endosperm (D) was also observed.
[See online article for color version of this figure.]
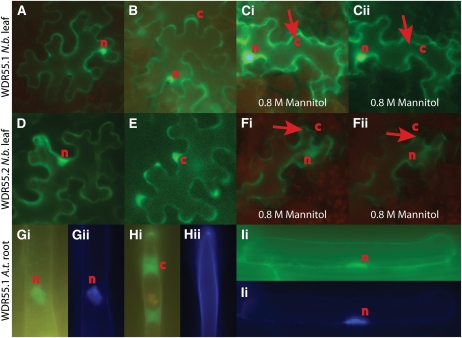
WDR55 Localizes to the Nucleus and the Cytoplasm
The lack of bilateral symmetry and the abnormal expression of a direct target of the ARF MP suggest that WDR55 may function in the nucleus, possibly by targeting components of the auxin regulatory pathway (see Discussion). However, WDR55 from Medaka was recently reported to be localized to the nucleolus and to the cytoplasm (Iwanami et al., 2008). To clarify the subcellular localization of Arabidopsis WDR55, C-terminal GFP fusion constructs of both WDR55 splice forms under the control of the cauliflower mosaic virus 35S promoter (Pro35S) were transformed into Agrobacterium tumefaciens for infiltration of tobacco (Nicotiana tabacum) leaves. Inspection of leaves 2 to 4 d after infiltration revealed a strong nuclear signal for both the long and the short splice forms (Figures 8A and 8D). Although weaker than the nuclear signal, both fusion proteins also appeared to be localized to the cytoplasm (Figures 8B and 8E). To distinguish true cytoplasmic localization, we incubated small pieces of the infiltrated leaves in 0.8 M mannitol for 2 h (Gagne and Clark, 2010). Upon treatment, neither the Pro35S:WDR55.1-GFP nor the Pro35S:WDR55.2-GFP signals were present in the extracellular space. The observed signals rather followed the cell wall–detached vacuoles, creating cytoplasmic strands marked by the GFP signal (Figures 8C and 8F). We also designed constructs for overexpression of Pro35S:WDR55-GFP in Arabidopsis. Upon inspection of seedling roots from transformed plants, Pro35S:WDR55.1-GFP was observed in both the nucleus and the cytoplasm (Figures 8G to 8I). Thus, in infiltrated tobacco leaves as well as in Arabidopsis roots, the nuclear localization of the fusion proteins appeared to mark the entire nucleus in contrast with nucleolar localization of its Medaka homolog (Iwanami et al., 2008).
Figure 8.
WDR55 Is Localized to the Nucleus and the Cytoplasm.
Fluorescent micrograph showing subcellular localization of WDR55.1-YFP and WDR55.2-YFP fusion protein in transfected tobacco (N.b.) leaf ([A] to [Fii]) and of WDR55.1-GFP and WDR55.2-GFP in transgenic Arabidopsis (A.t.) root ([Gi] to [Iii]). c, cytoplasm; n, nucleus.
(A) to (C) WDR55.1-YFP fusion protein in tobacco leaf. WDR55.1 is shown to be localized to the nucleus (A) and to the cytoplasm (B). When treated with 0.8 M mannitol for 2 h, the subcellular localization to the cytoplasm is demonstrated as strings marked by arrows that move as the vacuoles shrink upon UV exposure ([Ci] and [Cii]).
(D) to (F) WDR55.2-YFP fusion protein in tobacco leaf. WDR55.2 is shown to be localized also to the nucleus (D) and the cytoplasm (E) in the tobacco leaf. Upon 0.8 M mannitol treatment, the vacuoles shrink and cytoplasm localization is confirmed as GFP expression contracts with the plasma membrane ([Fi] and [Fii]; arrows).
(G) to (I) WDR55.1-GFP fusion protein in Arabidopsis root hair ([G] and [H]), while (I) shows WDR55.1-GFP fusion protein in an Arabidopsis root cell. Also, nuclear localization can be observed in Arabidopsis root hairs and root cells by GFP expression ([Gi] and [Ii]) with DAPI staining to confirm nuclear identity ([Gii] and [Iii]). Cytoplasmic localization can also be observed in root hair by the GFP around vacuoles (Hi), and the DAPI staining shows that there is no visible nucleus in this hair (Hii).
WDR55 Interacts with Arabidopsis DDB1A in Plant Cells
The presence of a signature WDxR motif suggests that WDR55 is a potential substrate receptor of the CUL4 E3 ubiquitin ligase complex. To test this possibility, we investigated the interaction between DDB1A/DDB1B and WDR55.1/WDR55.2 in reciprocal yeast two-hybrid assays, using both DDB1 and WDR55 as prey and bait. No interaction was detected upon repeated testing (see Supplemental Figure 7 online). Nevertheless, it is noteworthy that protein interactions between DCAFs and DDB1-related protein in yeast are often weak and sometimes difficult to detect (Molinier et al., 2008; Dumbliauskas et al., 2011). Therefore, we performed bimolecular fluorescence complementation (BiFC) experiments to investigate a physical interaction between WDR55.1/WDR55.2 and DDB1A in plant cells. Plasmids YC-WDR55.1/YC-WDR55.2 and YN-DDB1A were cobombarded into etiolated mustard (Sinapis alba) hypocotyls. A strong yellow fluorescent protein (YFP) signal was observed in the nucleus of cells (72.5%, n = 40) coexpressing YC-WDR55.1 and YN-DDB1A and of cells (80%, n = 42) coexpressing YC-WDR55.2 and YN-DDB1A (Figure 9). These data are similar to those obtained with cells transformed with the positive control YC-DDB2 + YN-DDB1A as previously reported (Figure 9A) (Molinier et al., 2008). Only a weak fluorescence signal was observed in <30% of the cells after bombardment with the negative control plasmid combinations YC-WDR55.1 + YN-empty vector (Figure 9A) and YC-WDR55.2 + YN-empty vector (29%, n = 55; and 22%, n = 55, respectively; Figure 9A).
Figure 9.
WDR55 Interacts with DDB1A in the Nuclei of Plant Cells.
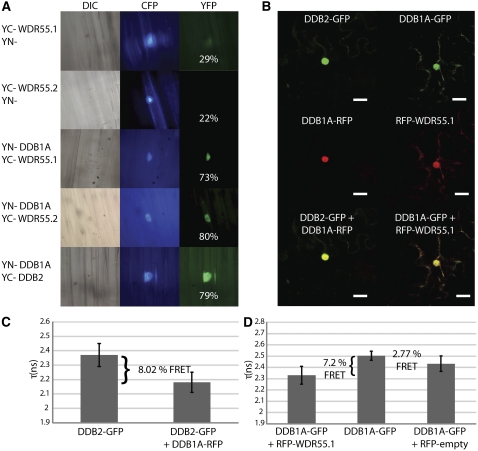
(A) BiFC of YN-DDB1A/YC-WDR55. Different combinations of plasmids expressing the indicated YN- and YC-fusion proteins were bombarded into hypocotyls of dark-grown S. alba seedlings. A transfection control CPRF2 expressing a fused CFP targeted to the nucleus was systematically included to identify transformed cells (middle panels). Images were recorded 20 h after bombardment via CFP- (middle panels) and YFP-specific (right panels) filters. Differential interference contrast (DIC) images are shown (left panels). Reconstitution of functional YFP as detected by YFP fluorescence occurs only in the nucleus with WDR55.1, WDR55.2, and DDB2 (used here as a positive control). Percentages in the YFP panel indicate the frequency of cells observed with YFP signal.
(B) Confocal images showing the subcellular localization of DDB2-GFP, DDB1A-GFP, and RFP-WDR55.1 protein fusions transiently expressed by the cauliflower mosaic virus 35S promoter in transformed tobacco cells. N. benthamiana plants were infiltrated with Agrobacteria harboring the different plasmids. The two fusion proteins colocalize mainly in the nucleus. Bars =10 μm.
(C) and (D) FRET analysis showing that DDB1A-GFP interacts with RFP-WDR55.1 in the nuclei of N. benthamiana cells in agroinfiltrated leaves. Bars represent mean fluorescence lifetime (τ, in nanoseconds [ns] ± se) of DDB1A-GFP alone or together with RFP-WDR55.1, or DDB2-GFP alone or together with DDB1A-RFP as positive control. Mean FRET values are indicated above the bars. The percentages indicate the relative change in fluorescence lifetime mediated by GFP-RFP fusion protein interactions compared with a single fluorescent protein.
To further support the interaction between WDR55.1 and DDB1A, we performed fluorescence resonance energy transfer (FRET) assays (Levitt et al., 2009). Visual inspection of DDB1A-GFP and RFP-WDR55.1 proteins transiently expressed in Nicotiana benthamiana plants showed colocalization of the proteins in the nuclear domain (Figure 9B). Cotransformed DDB1A-GFP and RFP-WDR55.1 revealed a mean FRET value of 7.2%, which was significantly higher than for the negative control using RFP alone (Figure 9C). As a positive control, we used DDB2, which is known to be a strong DDB1A interactor (Molinier et al., 2008), showing a similar level of interaction as between DDB1A-GFP and RFP-WDR55.1. These results indicate that WDR55 interacts with DDB1A and suggest that this protein might act as a CRL4 substrate recruiter. However, our attempts to immunoprecipitate CUL4 together with WDR55.1 remained unsuccessful; thus, further experimental approaches are required to determine whether WDR55.1 acts in the frame of a CRL4 complex.
DISCUSSION
WDR55 Is a WD40 Protein with a Conserved WDxR Motif and Interacts with DDB1
WD40 proteins in Arabidopsis have been classified into 143 distinct families, and WDR55 was designated into its own class with no paralogs but with orthologs in other species like Drosophila melanogaster (CG14722) or Homo sapiens (WDR55) (van Nocker and Ludwig, 2003). Some WD40 proteins contain a conserved 16–amino acid DDB1 binding WD40 (DWD) motif, and 85 Arabidopsis and 78 rice (Oryza sativa) proteins containing this motif have been identified in silico (Lee et al., 2008). Rice WDR55 has a perfect match with the 16–amino acid DWD motif, where the corresponding sequence in Arabidopsis WDR55 differs in position seven, since a Lys residue has substituted an Asp/Glu residue conserved in other WDR55 homologs. Site-directed mutagenesis of the DWD box at position seven in the DCAF DDB2 results in a substantially reduced DDB2-CUL4A association (He et al., 2006). The Arabidopsis Information Resource ecotype polymorphism search (http://www.Arabidopsis.org/) does not report any polymorphisms within the WDR55 potential DWD, and Arabidopsis lyrata also has the Lys in position seven, suggesting that this position is conserved within the Brassicaceae. The conserved amino acid WDxR stretch of the DWD box may thus be sufficient for DDB1 physical interaction and in silico identification of putative CUL4 E3 ligase substrate modules, rendering the substrate receptor family of Arabidopsis CRL4 protein much broader than previously anticipated (Zhang et al., 2008). Indeed, the previously reported DCAF1 carries only the WDxR motif in the WDR region and interacts with DDB1 both in humans and Arabidopsis (Angers et al., 2006; Zhang et al., 2008). Mutation of the WDxR motif of Arabidopsis DCAF MSI4 decreases its interaction with DDB1 and affects the function of the protein in the regulation of flowering time (Pazhouhandeh et al., 2011). Moreover, a point mutation in the WDxR motif of human DDB2, a DCAF involved in nucleotide excision repair, leads to the genetic disorder xeroderma pigmentosum, complementation group E (Chu and Chang, 1988), while in Arabidopsis, the WDxR is also important for the function of the protein in DNA repair (Castells et al., 2011). Our finding that WDR55 interacts with DDB1 in planta further suggests that the WDxR motif is most likely sufficient for DDB1 interaction. This is the first report showing that a member of the WDR55 protein family may act as part of a CRL4 complex and given the conservation of the WDxR motif in WDR55 metazoan orthologs, a similar function may be assigned to WDR55 orthologs.
WDR55 Is Required for Gametogenesis
Previous reports have attributed various biological functions to WDR55 proteins in eukaryotes. Mice homozygous for wdr55tm1Matn are embryonic lethal and die before embryonic day (E) 9.5 but can be maintained as heterozygotes without any obvious defects (Youngren et al., 2005). The Medaka (Oryzias latipes) WDR55 ortholog was identified in a screen for defective thymus development and is required for development of multiple organs (Iwanami et al., 2008). Defects in WDR55 cause the accumulation of incompletely processed rRNA precursors and may be a nucleolar modulator of rRNA synthesis and cell cycle progression in Medaka (Iwanami et al., 2008). Zebra fish null mutants are also thymus defective and have reduced organ size (Amsterdam et al., 2004; Iwanami et al., 2008). Interestingly, the medaka WDR55 mRNA appears to be maternally provided in the early zygote (Iwanami et al., 2009).
We show that Arabidopsis WDR55 is essential for seed development and is involved in both male and female gametogenesis. In line with our findings, WDR55 was expressed throughout embryo sac development, at early stages of pollen maturation, and in early stages of embryo development. As found for several other gametophytic mutations (Chen and McCormick, 1996; Park et al., 1998; Grini et al., 1999; Colombo et al., 2008), wdr55-1 is only partially penetrant. However, the segregation distortion observed for the allele wdr55-1 indicates a significant defect in both gametophytes, causing a reduction in the transfer of the mutation to the progeny. Interestingly, the wrd55-2 allele truncates only the unstructured region protein after the last WD40 repeat, and this may explain some of the differences in allelic strength of wdr55-1 and wdr55-2.
No obvious pollen phenotype could be observed upon direct inspection or using various cell fate markers. However, we did observe a moderate reduction in pollen tube germination in the mutant, suggesting a requirement of WDR55 in pollen tube formation. It is unlikely that WDR55 is required for the final events of fertilization, such as release of sperm cells or karyogamy. In such a case, a phenotype in the female gametophyte or in seed development would result, and this was not the case. Overexpression of either WDR55 splice forms, however, resulted in strong defects both in progamic pollen development and in female gametophyte development, suggesting that WDR55 plays an important role in gametogenesis and that the protein level may be regulated. However, effects of titration of the DDB1 proteins cannot be excluded, and further experiments will be needed to elucidate WDR55 function in the male gametophyte.
A modest frequency of unfused polar cells was observed in wdr55-1 embryo sacs. Several genes required for central cell fusion have previously been identified (Pagnussat et al., 2005; Portereiko et al., 2006b), including a heterodimer pair of MADS box AGAMOUS-LIKE (AGL) transcription factors, AGL61/DIANA and AGL80 (Portereiko et al., 2006a; Bemer et al., 2008; Steffen et al., 2008), the splice factors LACHESIS (LIS) and CLOTHO (Gross-Hardt et al., 2007), and genes with mitochondrial function (León et al., 2007; Kägi et al., 2010; Tan et al., 2010). Interestingly, LIS encodes a seven-repeat WDR protein predicted to be a CRL4 substrate receptor based on the presence of two WDxR motifs (Zhang et al., 2008). LIS is involved in preventing accessory cells from adopting a gametic cell fate, and its homolog in yeast, PRP4, is associated with the U4/U6 spliceosome complex (Gross-Hardt et al., 2007). In this respect, mutant WDR55 central cells support PCD of accessory antipodal cells and may therefore reflect a general delay in polar cell fusion as previously observed in SLOW WALKER mutants (Shi et al., 2005; Li et al., 2009)
DDB1A and DDB1B have been shown by double mutant analyses to be required for successful transmission through the male and female gametophyte (Bernhardt et al., 2010; Dumbliauskas et al., 2011), consistent with a possible function of the DDB1A binding WDR55 as a CRL4 substrate receptor. Interestingly, in reciprocal crosses using a homozygous mutant background of either ddb1a or ddb1b, a male requirement was found for DDB1A, whereas DDB1B was required for passage through the female gametophyte (Dumbliauskas et al., 2011). CUL4 mutants, however, show only a weak male and female gametophyte transmission failure (Dumbliauskas et al., 2011). One explanation for this could be carryover of CUL4 protein and that sufficient CUL4 is present after meiosis for gametogenesis to occur. An alternative scenario would be that, in gametogenesis, Arabidopsis WDR55 acts in a DDB1-dependent but CUL4-independent manner, as has recently been proposed for the mammalian WD40 protein Cdh1 (Lv et al., 2010). However, it is noteworthy that mice homozygous mutant for Cul4A are male infertile and defective in meiosis (Kopanja et al., 2011; Yin et al., 2011). Thus, further work will clarify whether Arabidopsis CUL4 is required for WDR55 function in gametogenesis.
WDR55 Is Required for Seed and Endosperm Development
Analysis of both wdr55-1 and wdr55-2 mutant lines suggested that WDR55 is required for embryo and endosperm development from 2 DAP and is involved in setting up apical bilateral symmetry in the transition from the globular to the heart stage embryo. We observed arrested embryo development, which often accompanied by a reduced number of endosperm nuclei, as early as in the zygote stage. No embryo stages later than the globular stage could be observed in wdr55-2 mutant seeds. The embryo undergoes an invariant pattern of cell differentiation; however, no major difference in the cell division pattern could be observed in mutant embryos, although they were severely delayed. Interestingly, both cul4 and ddb1 mutants display a globular arrest phenotype (Bernhardt et al., 2010; Dumbliauskas et al., 2011). Thus, WDR55 loss of function in embryogenesis, at least in part, phenocopies the CUL4- and DDB1A/B-deficient embryo phenotype and may suggest that a CUL4WDR55 complex is involved in embryo development.
In wild-type seeds, the endosperm undergoes three rounds of nuclear division before the first division of the zygote (Berger, 1999). In most wdr55 seeds, however, only a few endosperm division rounds could be observed. Interestingly, cul4 null mutants also display underdeveloped endosperm with large nuclei and nucleoli reminiscent of those in the wdr55 endosperm (Dumbliauskas et al., 2011). The underdeveloped wdr55 endosperm may also contribute to the observed embryo arrest phenotype as a consequence of an endosperm defect or failure in the crosstalk between embryo and endosperm (Nowack et al., 2010). Endosperm identity in wdr55 seeds appeared to be preserved, as evidenced by ProKS117:GFP and ProFIS2:FIS2-GUS marker expression, although ProFIS2:FIS2-GUS was deregulated or even absent in a fraction of wdr55-1 endosperm. Notably, FIS2 is imprinted and only expressed from the maternal genome due to DNA METHYLTRANFERASE1–mediated paternal silencing and activation of the maternal allele by removal of DNA methylation by the DNA glycosilase DEMETER (Jullien et al., 2006). A role for metazoan CRL4 substrate receptors in DNA methylation has been demonstrated previously (Xu et al., 2010), and FIS2 is part of the Polycomb Repressive Complex 2 (PRC2). Thus, its deregulation also indirectly influences chromatin modification by H3K27 methylation (Hennig and Derkacheva, 2009).
At present, we can only speculate about how such a putative CUL4-DDB1WDR55 ubiquitin ligase complex may regulate seed development and embryo bilateral symmetry. An alternative hypothesis would be that a WDR55-DDB1 complex acts independently of CUL4, as recently described for a mammalian WD40 protein interacting with DDB1 (Lv et al., 2010), or that WDR55 can even function in both CUL4-dependent and -independent mechanisms. CRL4s act mainly at the chromatin level in fungi, metazoans, and Arabidopsis, regulating DNA damage responses and heterochromatin silencing and mediating Polycomb histone methylation (Jia et al., 2005; Wysocka et al., 2005; Higa et al., 2006; Molinier et al., 2008; Jackson and Xiong, 2009; Xu et al., 2010; Dumbliauskas et al., 2011; Pazhouhandeh et al., 2011). Some of these functions require nonproteolytic ubiquitination by CRL4s. For instance, CUL4DDB2 modifies histones H2A, H3, and H4 during UV-damaged DNA repair (Kapetanaki et al., 2006; Wang et al., 2006). Recent work in Arabidopsis also revealed that CRL4s functionally interact with PRC2 to participate in epigenetic regulation by histone tail modification (Dumbliauskas et al., 2011; Pazhouhandeh et al., 2011). Thus, the mode of action of WDR55 together with DDB1 and eventually CUL4 in the regulation of embryo and endosperm development may not necessarily require proteolytic events.
Auxin is a major player in the polarization of the embryo and is transported toward the incipient cotyledon primordium in a basal to apical direction in the lateral protoderm (De Smet et al., 2010). DRN is a member of the AP2-type transcription factor family, which contributes to auxin transport and perception in the Arabidopsis embryo. After the division of the zygote, the protein is associated with apical cell fate and marks the position of incipient leaf primordia (Chandler et al., 2008, 2011; Cole et al., 2009). In wdr55-2, polarization and accordingly initiation of leaf primordia appears to be blocked and, consequently, DRN:GFP primordial localization also failed.
DRN is a direct target of MP, an ARF regulated by CRL1-mediated UPS degradation of the AUX/IAA BODENLOS (Chandler et al., 2008; Cole et al., 2009; De Smet et al., 2010). An involvement of CRL4s in the auxin regulation pathway has not been demonstrated until now, and our data do not support a role of WDR55 in this process. However, another putative DCAF, LRS1, might play such a role, as the lrs1-1 mutant is defective in lateral root development and displays auxin accumulation during lateral root primordium development and lateral root meristem emergence (Lee et al., 2010). Further investigations on WDR55 and its interacting DDB1 partner will be crucial to clarify its function in seed and gametophyte development.
METHODS
Plant Material and General Procedures
The wdr55-1 allele was isolated as line dsg3127 in a segregation distortion screen for mutants affected in male or female gametophyte transmission from populations of Dissociator (Ds)-transposed insertion lines in Arabidopsis thaliana in the Ler ecotype. The other plant lines used, SALK034787, SALK063054, and wdr55-2 (WiscDsLox430F06), were in the Col-0 ecotype and are T-DNA insertion mutants that were obtained from the Nottingham Arabidopsis Stock Centre (Alonso et al., 2003). The insertions in these lines were verified by PCR using gene-specific primers and primers against the insert (T-DNA or DsG element). Primer sequences used can be found in Supplemental Table 4 online.
Seeds were surface-sterilized using ethanol, bleach, and Tween 20 before germination on Murashige and Skoog media (Murashige and Skoog, 1962) supplemented with 2% Suc (MS-2). To select for wdr55-1 plants, 50 μg/L kanamycin was added to the media, and for wdr55-2, 100 μM BASTA was used. All plants were stratified on MS-2 plates at 4°C overnight before being incubated at 18°C for ~12 d until germination. The seedlings were then transferred to soil and grown at 18°C under long-day conditions (16 h). Constructs made during this study were transformed into Col, Ler, or the wdr55-1 mutant using Agrobacterium tumefaciens strain C58-pGV2260 or GV3101-pMP90-RK by the floral dip method (Clough and Bent, 1998), and transformants were selected for by the appropriate resistance encoded in the inserted T-DNA.
Molecular Cloning and DNA Work
To isolate genomic DNA from plants for genotyping, a quick protocol was followed, where a buffer (50 mL of 1 M Tris HCl, pH 7.2, 60 mL of 5 M NaCl, and 100 g Suc in 1 liter double-deionized water) was added to a tissue sample (e.g., a leaf) and homogenized. Then, 1.5 μL of this material was used in a 50-μL PCR reaction. For other purposes, such as amplifying genomic sequences for cloning, the Aqua Pure Genomic DNA isolation kit from Bio-Rad was used according to the manufacturer’s instruction manual.
For the Vectorette PCR, genomic DNA from the DsG line wdr55-1 was isolated and digested using restriction enzymes that have BamHI-compatible sites (i.e., BclI [Promega] and Bg1II [TaKara]). Vectorettes were made to ligate onto the ends of the fragments after the restriction digest by mixing the TopL strand and the restriction enzyme–specific bottom strand (see Supplemental Table 4B online for sequence) (Devon et al., 1995). The vectorettes were then ligated onto the end of the fragments from the restriction digest from the DsG line using T4 ligase. Two rounds of nested PCR was performed using the vectorette primers vec-1 and vec-2 and the 5′ and 3′ end nested primers of the DsG element: Ds 5-1 and Ds 5-2, and Ds 3-1 and Ds 3-2. The PCR products were isolated from the gel using the Wizard SV Gel and PCR Clean-Up System Kit from Promega. The isolated DNA fragments were cloned using TOPO TA cloning technology (Invitrogen) and sequenced.
The TOPO TA cloning technology was used according to the manufacturer’s instructions. The vectors used for cloning fragments were pCR-4TOPO or pCR-2.1-TOPO. In addition, the TOPO Gateway vector pCR8 was used to clone a genomic fragment later to be used as an entry clone in a Gateway reaction.
Constructs were made using Gateway technology (Invitrogen) according to the Invitrogen Gateway Technology Manual Version E (2003). The DNA fragments were amplified using primers with att sites (i.e., attB1 and attB2) (see Supplemental Table 4B online). The donor vector used in all construct design was pDONR/Zeo by Invitrogen. Primers used for creating donor vectors for WDR55.1 and WDR55.2 were attB1 SP/WDR55.1 and attB2 ASP/WDR55.1, and attB1 SP/WDR55.2 and attB2 ASP/WDR55.2, respectively, where cDNA from wild-type Col was used as template. These donor vectors were recombined with pGBKT7 and pADN (for yeast two-hybrid screen, verification, and direct mating), pK7WG2, pK7FWG2, pK7WGF2 (for transformation and subcellular localization in Arabidopsis), and pEarlygate101 (for transfection and subcellular localization in tobacco [Nicotiana tabacum]), MN27 (for overexpression in Arabidopsis), and a modified MN27 vector where the CDKA;1 promoter was replaced with the WDR55 promoter (for splice variant-specific rescue). For GUS expression studies, the WDR55 promoter region up to the next gene (1283 bp) was amplified using the primers SP At2g34260 Promoter Gateway and ASP At2g34260 Promoter Gateway, and the resulting entry clone was recombined with the destination vector pMDC162 (see Supplemental Table 4B online).
The genomic sequence of WDR55 and the corresponding 1283-bp upstream promoter region were amplified using the primers TOPO SP/At2g34260 rescue and TOPO ASP/At2g34260 rescue, and the fragment was cloned into the TOPO Gateway vector pCR8 by the TOPO TA cloning technology. The resulting entry clone was recombined with the destination vector pMDC123 by Gateway technology (for complementation of the wdr55-1 mutant).
To test the interaction of DDB1A and DDB1B with WDR55, they were cloned into both pGBKT7 and pADN. A Gateway vector was obtained for DDB1A (CG104978) from the ABRC, while a pda clone (pda20340) was obtained from the RIKEN Bio Research Centre for DDB1B. The primers attB1 SP/DDB1B and attB2 ASP/DDB1B were used to create the entry clone.
Subcellular Localization and Overexpression Constructs
Subcellular localization was performed by making both N- and C-terminal GFP fusion constructs with both splice variants WDR55.1 and WDR55.2 and transforming these into Arabidopsis Col ecotype. The GFP vectors used were pK7FWG2 and pK7WGF2. GFP was inspected in leaf and root cells under a Zeiss Axioplan Imaging2 microscope system.
To verify fusion, tissues from the subcellular expression lines were harvested and RNA was extracted using a Spectrum Plant Total RNA kit (Sigma-Aldrich). RT-PCR was performed using primers situated in the gene and in the GFP sequence to verify fusion after transcription. The controls were cDNA from wild-type Col. For WDR55.2 N-GFP, primers EGFP 625F and Trans 400sp were used. For WDR55.1 and WDR55.2, Egfp-pK7FWG2 and Trans 200asp primers were used. For the control, ASP At2g34260 RT PCR 5 and SP At2g34260 RT-PCR 5 primers were used. Primer sequences are presented in Supplemental Table 4B online.
For subcellular localization using the tobacco infiltration protocol, the short and long splice variant cDNA was recombined with the vector pEarlyGate101, which creates a C-terminal GFP fusion construct. These were then transformed into Agrobacterium (C58), mixed with Agrobacterium harboring the helper construct P19, and infiltrated into Nicotiana benthamiana leaves. The leaves were inspected under UV light after 2 to 4 d. Some small pieces of the infiltrated leaves were also incubated in 0.8 M mannitol for 2 h. This treatment results in the expulsion of water from the cell, which causes the plasma membrane to detach from the cell wall and the vacuoles to shrink, allowing for detection of extracellular proteins.
Complementation Analysis
A complementation construct was made to rescue the phenotype in the DsG line wdr55-1, which contained the genomic sequence of WDR55, its promoter region and the 3′ untranslated region. This product is 3902 bp and ranged from position 35031 to 38932 on the BAC F13P17 (GI: 201971539). A splice variant-specific complementation construct was made to investigate the role of the two splice variants. The constructs were made by excising the CDKA;1 promoter from the Gateway vector MN27 with the restriction enzymes AscI and XhoI and replacing it with the promoter region of WDR55 (SP pAt2g34260-AscI-2 and ASP pAt2g34260-XhoI-2). Ligation of DNA using T4 ligase was performed according to the manufacturer’s instructions and took place at 4 to 14°C overnight. cDNA from the two splice variants of WDR55 in the pDONR/Zeo vector was then recombined into the modified MN27 vector containing the WDR55 promoter by Gateway recombination.
Total RNA Isolation and cDNA Synthesis
RNA was isolated from various tissues from wild-type Arabidopsis ecotype Col plants using the Spectrum Plant Total RNA kit according to the manufacturer’s instructions, except all centrifugation was performed at 4°C and RNA was eluted in 50 μL. The tissue was harvested in liquid nitrogen in tubes prefilled with ceramic beads (MagNA Lyser Green Beads) and then homogenized with a MagNA Lyser (Roche Applied Science). The isolated RNA was treated with DNase I (Sigma-Aldrich) before cDNA synthesis using oligo(dT) and the reverse transcriptase enzyme SuperScript III, in the SuperScript III first-strand synthesis system for RT-PCR (Invitrogen), according to the manufacturer’s instructions. The resulting cDNA was then purified using the QIAquick PCR purification kit (Qiagen) and eluted in 30 μL. The cDNA concentration was measured on a NanoDrop 1000 spectrophotometer.
Real-Time Quantitative PCR Analysis
Real-time quantitative PCR (qPCR) was performed on cDNA from wild-type Col to determine the expression level of the gene of interest in different tissues relative to a reference tissue, using expression of ACTIN11 as a control. Three plants were harvested as one biological replica, and three biological replicas were used in total unless otherwise specified. Equipment required was the LightCycler 480 instrument and LightCycler Multiwell Plate 96 available from Roche Applied Science. The primers (see Supplemental Table 4B online) used to detect WDR55.1 were RP_At2g34260_54# and LP_At2g34260_54#, which give an amplicon of 78 bp. The primers used to detect WDR55.2 were ASP RT PCR WDR55.2 and SP RT PCR WDR55.2, giving a 166-bp product. Primers for ACTIN11 were ACT11-77-LP and ACT11-77-RP, giving a product of 100 bp. The primers spanned introns to exclude amplification of genomic sequence, and the product for WDR55.2 was verified to give one band by running the real-time qPCR product on a 1% agarose gel. Primer concentration was between 0.2 and 1 μM. Setup of the master mix and cDNA dilution series and running of the real-time qPCR were performed according to the instructions in the SYBR Premix Ex Taq (Perfect Real Time) manual (RR041A v.050614) from TaKaRa Bio. To determine efficiency, both biological and technical replicas were used. Technical replicas were produced by making a dilution series with concentrations of 100, 50, 20, and 5 ng. The relative expression ratios were calculated according to a model described by Pfaffl (2001).
Histology and Imaging
Embryos and endosperms were viewed by directly mounting seeds in clearing solution (8:2:1 chloral hydrate:deionized water:glycerol). After ~2 h of incubation at room temperature, embryo and endosperm development could be viewed under the microscope.
To visualize pollen nuclei under UV radiation, an anther was mounted on a slide in 15 μl staining buffer (2.5 μg/mL DAPI solution, 50 mM NaPO4, pH 7.2, 5% DMSO, 0.01 Tween 20) and covered with a cover slide. The preparations were sealed with clear nail polish and stored at 4°C for 1 to 5 h before being visualized under the microscope by exciting with a 365/12-nm band-pass filter and recording emission using a 397-nm long-pass filter.
Alexander stain was made with 10 mL ethanol (95%), 1 mL malachite green (1% in 95% ethanol), 5 mL fuchsin acid (1% in water), 0.5 mL Orange G (1% in water), 5 g phenol, 5 g chloral hydrate, 2 mL glacial acetic acid, 25 mL glycerol, and 50 mL water. Anthers were mounted directly in the stain on a slide and covered with a cover slide. The slides were sealed with clear nail polish before being visualized under light microscopy. Alexander stain stains live pollen pink and dead pollen green or not at all.
GFP and YFP were visualized in various tissues by mounting the tissue in water on a slide with a cover slide and viewing directly under the microscope. For GFP, a 450- to 490-nm band-pass filter was used for excitation, and emission was recorded with a 515- to 565-nm band-pass filter. YFP was visualized using excitation with a 500/20-nm band-pass filter, and emission was measured with a 535/30-nm band-pass filter.
Histochemical GUS assays were performed by incubating the sampled tissue in substrate staining solution [0.05 M NaPO4, 0,1% Triton X-100, 2 mM K4Fe(CN)63H6O, 2 mM K3Fe(CN)6, and 2 mM X-Gluc] in the dark at 37°C, usually overnight. When staining the embryo sac, which has eight cell layers, the time of incubation was often longer or the X-Gluc concentration was increased. The substrate was then removed and 50% ethanol was added before slides were prepared with the materials mounted in a clearing solution (8:2:1 chloral hydrate:deionized water:glycerol) and incubated for 1 h at 4°C before inspection for GUS. Stained tissue could be stored at 4°C in 50% ethanol for later use.
For pollen tube germination assays, pollen grains (1 d after anthesis) were gently applied to plates containing pollen germination medium [18% Suc, 0.01% boric acid, 5 mM CaCl2, 5 mM Ca(NO3)2, 5 mM MgSO4, and 0.5% phytoagar]. The plates were covered, placed in a humidified plastic box, and kept at 22°C for up to 10 h before observation.
A Zeiss Axioplan Imaging2 microscope system equipped with Nomarski optics, epifluorescence attachment, and cooled liquid crystal display imaging facilities was used for examining GFP, YFP, and DAPI under UV and for differential interference contrast light microscopy. Some sporophytic tissues were examined using the stereo light microscope Leica WILD MZ8, and images were captured using a Nikon Coolpix 995 camera.
Protein Interaction Studies
The yeast two-hybrid system from Clontech was used to test full-length At2G34260 cDNA against a library of cDNA clones created from Col wild-type flowers and buds and recombined into pGADT7-Rec. To make a bait construct, full-length WDR55.1 cDNA was cloned in frame with the lexA DNA binding domain (vector pGBKT7) using Gateway technology. The bait construct was tested for autoactivation as described by the manufacturer (Clontech) and then transformed into the yeast strain Y187. To verify possible interacting partners by direct mating, the full-length sequence of the candidate genes was cloned into Gateway-compatible pGADT7 and transformed into yeast strain AH109. Mating was performed directly with the original bait construct and also with pGBKT7-WDR55.2 in Y187. The candidate genes were also cloned into pGBKT7 and WDR55 into pADN to test for interactions in both directions. The negative controls used were BD (binding domain) and Lam in pGBKT7. Media used were SD/-Leu/-Trp (-L/-W for short), SD/-Leu/-Trp/-His (triple dropout for short), and SD/-Leu/-Trp/-His/-Ade/ X-a-Gal (quadruple dropout for short).
For BiFC, the DDB1A, WDR55.1, and WDR55.2 coding sequences were cloned into the split YFP destination vectors by recombination (Invitrogen) to yield the YN-DDB1A, YC-WDR55.1, and YC-WDR55.2 constructs. The YC-DDB2 construct, which was used as a positive control, was described by Molinier et al. (2008). The YN-, YC-, and Pro35:CPRF2-CFP vectors and split YFP experiments were performed as described by Stolpe et al. (2005). Images were recorded 20 h after bombardment with a Nikon fluorescent stereomicroscope E800 equipped with a ×40 water immersion optic using CFP- and YFP-specific filters. Images were assembled using Adobe Photoshop and Adobe Illustrator software.
For fluorescence lifetime imaging microscopy, WDR55.1, DDB1A, and DDB2 coding sequences were cloned by recombination into destination vectors: WDR55.1 in pB7WGR2, DDB1A in pB7RWG2 and pB7FWG2, and DDB2 in pB7FWG2 (Ghent plasmids collection; http://bccm.belspo.be/index.php) to obtain RFP-WDR55.1, DDB1A-GFP, DDB1A-RFP, and DDB2-GFP constructs. The different constructs were transformed into Agrobacterium and infiltrated into N. benthamiana leaves together with the silencing suppressor P19. Two days after infiltration, the lifetime of the fluorescent fusion proteins was measured in the epidermal cells of infiltrated leaves either with DDB2-GFP or DDB1A-GFP alone or together with DDB1A-RFP or RFP-WDR55.1 using a LIFA frequency domain fluorescence lifetime imaging system (Lambert Instruments).
Sequencing and Bioinformatics
Sequencing was performed using an ABI 3730 high-throughput capillary electrophoresis sequencer by the ABI lab at the Departments of Biology and Molecular Biosciences at the University of Oslo. Sequences were processed using ContigExpress and Vector NTI AdvanceTM v.10 software (Invitrogen) and using the BLAST engine provided by the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/). Plant homologs were identified using WU-BLAST software (http://www.Arabidopsis.org/wublast/index2.jsp). Alignments were performed using ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html) and formatted using GeneDoc software (http://www.nrbsc.org/gfx/genedoc/). WD40 repeats were predicted using SMART (http://smart.embl-heidelberg.de/) and PyMOL (http://www.pymol.org).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: WDR55.1 (At2g34260.1, NP_565782.1), WDR55.2 (At2g34260.2, NP_973596.1), DDB1A (At4g05420, NP_192451.1), DDB1B (At4g21100, NP_193842.1), and CUL4 (At5g46210, NP_568658.1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Bioinformatics Analysis of WDR55.
Supplemental Figure 2. WDR55 Is Expressed during Early and Late Embryo Development and in Various Sporophytic Tissues.
Supplemental Figure 3. Overexpression of WDR55 Results in Arrest at the One-Nucleate Stage of Embryo Sac Development.
Supplemental Figure 4. Pollen Development and Pollen Tube Germination in wdr55-1.
Supplemental Figure 5. Overexpression of WDR55 Results in Various Developmental Defects in Pollen.
Supplemental Figure 6. FIS2:GUS and KS117:GFP in wdr55-2 Endosperm.
Supplemental Figure 7. Direct Mating of WDR55.1 with DDB1A and DDB1B in a Yeast Two-Hybrid Experiment Reveals No Interaction.
Supplemental Table 1. Insertion Details for Mutant Lines of WDR55.
Supplemental Table 2. Delayed Fusion of Polar Nuclei in wdr55-1 (Ler) Embryo Sacs.
Supplemental Table 3. Genomic Rescue of wdr55-1.
Supplemental Table 4. Primer List.
Acknowledgments
We thank Birgit Schwab and Gottfried Martin for the generation of the AcDs starter lines and Ueli Grossniklaus and Venkatesan Sundaresan for making the Ds launch pads available. We also thank Barbara M. Gloeckle for making the pollen germination assay work. We thank Roy Falleth, Charles Albin-Amiot, and Solveig Hauge Engebretsen for technical assistance and our colleagues Reza Shirzadi and Ellen Dehnes Andersen for careful reading and comments. This work was supported by a Norwegian Research Council FUGE Young Investigator Starting Grant to P.E.G.
AUTHOR CONTRIBUTIONS
K.N.B., S.J.-R., G.J., P.G., and P.E.G. designed the research. K.N.B., S.J.-R., and P.E.G. performed the research. K.N.B., S.J.-R., G.J., P.G., and P.E.G. analyzed the data. K.N.B., G.J., P.G., and P.E.G. wrote the article.
References
- Alexander M.P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44: 117–122 [DOI] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Amsterdam A., Nissen R.M., Sun Z., Swindell E.C., Farrington S., Hopkins N. (2004). Identification of 315 genes essential for early zebrafish development. Proc. Natl. Acad. Sci. USA 101: 12792–12797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angers S., Li T., Yi X., MacCoss M.J., Moon R.T., Zheng N. (2006). Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature 443: 590–593 [DOI] [PubMed] [Google Scholar]
- Bemer M., Wolters-Arts M., Grossniklaus U., Angenent G.C. (2008). The MADS domain protein DIANA acts together with AGAMOUS-LIKE80 to specify the central cell in Arabidopsis ovules. Plant Cell 20: 2088–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger F. (1999). Endosperm development. Curr. Opin. Plant Biol. 2: 28–32 [DOI] [PubMed] [Google Scholar]
- Berger F., Chaudhury A. (2009). Parental memories shape seeds. Trends Plant Sci. 14: 550–556 [DOI] [PubMed] [Google Scholar]
- Berger F., Grini P.E., Schnittger A. (2006). Endosperm: An integrator of seed growth and development. Curr. Opin. Plant Biol. 9: 664–670 [DOI] [PubMed] [Google Scholar]
- Bernhardt A., Lechner E., Hano P., Schade V., Dieterle M., Anders M., Dubin M.J., Benvenuto G., Bowler C., Genschik P., Hellmann H. (2006). CUL4 associates with DDB1 and DET1 and its downregulation affects diverse aspects of development in Arabidopsis thaliana. Plant J. 47: 591–603 [DOI] [PubMed] [Google Scholar]
- Bernhardt A., Mooney S., Hellmann H. (2010). Arabidopsis DDB1a and DDB1b are critical for embryo development. Planta 232: 555–566 [DOI] [PubMed] [Google Scholar]
- Biedermann S., Hellmann H. (2011). WD40 and CUL4-based E3 ligases: lubricating all aspects of life. Trends Plant Sci. 16: 38–46 [DOI] [PubMed] [Google Scholar]
- Brownfield L., Hafidh S., Borg M., Sidorova A., Mori T., Twell D. (2009). A plant germline-specific integrator of sperm specification and cell cycle progression. PLoS Genet. 5: e1000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castells E., Molinier J., Benvenuto G., Bourbousse C., Zabulon G., Zalc A., Cazzaniga S., Genschik P., Barneche F., Bowler C. (2011). The conserved factor DE-ETIOLATED 1 cooperates with CUL4-DDB1DDB2 to maintain genome integrity upon UV stress. EMBO J. 30: 1162–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler J.W., Cole M., Flier A., Grewe B., Werr W. (2007). The AP2 transcription factors DORNROSCHEN and DORNROSCHEN-LIKE redundantly control Arabidopsis embryo patterning via interaction with PHAVOLUTA. Development 134: 1653–1662 [DOI] [PubMed] [Google Scholar]
- Chandler J.W., Cole M., Jacobs B., Comelli P., Werr W. (2011). Genetic integration of DORNRÖSCHEN and DORNRÖSCHEN-LIKE reveals hierarchical interactions in auxin signalling and patterning of the Arabidopsis apical embryo. Plant Mol. Biol. 75: 223–236 [DOI] [PubMed] [Google Scholar]
- Chandler J.W., Cole M., Werr W. (2008). The role of DORNROESCHEN (DRN) and DRN-LIKE (DRNL) in Arabidopsis embryonic patterning. Plant Signal. Behav. 3: 49–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Shen Y., Tang X., Yu L., Wang J., Guo L., Zhang Y., Zhang H., Feng S., Strickland E., Zheng N., Deng X.W. (2006). Arabidopsis CULLIN4 forms an E3 ubiquitin ligase with RBX1 and the CDD complex in mediating light control of development. Plant Cell 18: 1991–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.C., McCormick S. (1996). sidecar pollen, an Arabidopsis thaliana male gametophytic mutant with aberrant cell divisions during pollen development. Development 122: 3243–3253 [DOI] [PubMed] [Google Scholar]
- Chu G., Chang E. (1988). Xeroderma pigmentosum group E cells lack a nuclear factor that binds to damaged DNA. Science 242: 564–567 [DOI] [PubMed] [Google Scholar]
- Clough S.,J., Bent A., F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cole M., Chandler J., Weijers D., Jacobs B., Comelli P., Werr W. (2009). DORNROSCHEN is a direct target of the auxin response factor MONOPTEROS in the Arabidopsis embryo. Development 136: 1643–1651 [DOI] [PubMed] [Google Scholar]
- Colombo M., Masiero S., Vanzulli S., Lardelli P., Kater M.M., Colombo L. (2008). AGL23, a type I MADS-box gene that controls female gametophyte and embryo development in Arabidopsis. Plant J. 54: 1037–1048 [DOI] [PubMed] [Google Scholar]
- De Smet I., Lau S., Mayer U., Jürgens G. (2010). Embryogenesis - The humble beginnings of plant life. Plant J. 61: 959–970 [DOI] [PubMed] [Google Scholar]
- Devon R.S., Porteous D.J., Brookes A.J. (1995). Splinkerettes—Improved vectorettes for greater efficiency in PCR walking. Nucleic Acids Res. 23: 1644–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissmeyer N., Nowack M.K., Pusch S., Stals H., Inzé D., Grini P.E., Schnittger A. (2007). T-loop phosphorylation of Arabidopsis CDKA;1 is required for its function and can be partially substituted by an aspartate residue. Plant Cell 19: 972–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher K., Callis J. (2007). Ubiquitin, hormones and biotic stress in plants. Ann. Bot. (Lond.) 99: 787–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumbliauskas E., Lechner E., Jaciubek M., Berr A., Pazhouhandeh M., Alioua M., Cognat V., Brukhin V., Koncz C., Grossniklaus U., Molinier J., Genschik P. (2011). The Arabidopsis CUL4-DDB1 complex interacts with MSI1 and is required to maintain MEDEA parental imprinting. EMBO J. 30: 731–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker A.K., et al. (2001). Intrinsically disordered protein. J. Mol. Graph. Model. 19: 26–59 [DOI] [PubMed] [Google Scholar]
- Friml J., Vieten A., Sauer M., Weijers D., Schwarz H., Hamann T., Offringa R., Jürgens G. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Gagne J.M., Clark S.E. (2010). The Arabidopsis stem cell factor POLTERGEIST is membrane localized and phospholipid stimulated. Plant Cell 22: 729–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X., Chang F., Ma H. (2010). Signaling and transcriptional control of reproductive development in Arabidopsis. Curr. Biol. 20: R988–R997 [DOI] [PubMed] [Google Scholar]
- Grini P.E., Schnittger A., Schwarz H., Zimmermann I., Schwab B., Jürgens G., Hülskamp M. (1999). Isolation of ethyl methanesulfonate-induced gametophytic mutants in Arabidopsis thaliana by a segregation distortion assay using the multimarker chromosome 1. Genetics 151: 849–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross-Hardt R., Kägi C., Baumann N., Moore J.M., Baskar R., Gagliano W.B., Jürgens G., Grossniklaus U. (2007). LACHESIS restricts gametic cell fate in the female gametophyte of Arabidopsis. PLoS Biol. 5: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y.J., McCall C.M., Hu J., Zeng Y., Xiong Y. (2006). DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4-ROC1 ubiquitin ligases. Genes Dev. 20: 2949–2954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig L., Derkacheva M. (2009). Diversity of Polycomb group complexes in plants: same rules, different players? Trends Genet. 25: 414–423 [DOI] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67: 425–479 [DOI] [PubMed] [Google Scholar]
- Higa L.A., Zhang H. (2007). Stealing the spotlight: CUL4-DDB1 ubiquitin ligase docks WD40-repeat proteins to destroy. Cell Div. 2: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa L.A., Wu M., Ye T., Kobayashi R., Sun H., Zhang H. (2006). CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat. Cell Biol. 8: 1277–1283 [DOI] [PubMed] [Google Scholar]
- Howden R., Park S.K., Moore J.M., Orme J., Grossniklaus U., Twell D. (1998). Selection of T-DNA-tagged male and female gametophytic mutants by segregation distortion in Arabidopsis. Genetics 149: 621–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingouff M., Hamamura Y., Gourgues M., Higashiyama T., Berger F. (2007). Distinct dynamics of HISTONE3 variants between the two fertilization products in plants. Curr. Biol. 17: 1032–1037 [DOI] [PubMed] [Google Scholar]
- Ishida T., Kinoshita K. (2008). Prediction of disordered regions in proteins based on the meta approach. Bioinformatics 24: 1344–1348 [DOI] [PubMed] [Google Scholar]
- Iwanami N., et al. (2008). WDR55 is a nucleolar modulator of ribosomal RNA synthesis, cell cycle progression, and teleost organ development. PLoS Genet. 4: e1000171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwanami N., Okada M., Hoa V.Q., Seo Y., Mitani H., Sasaki T., Shimizu N., Kondoh H., Furutani-Seiki M., Takahama Y. (2009). Ethylnitrosourea-induced thymus-defective mutants identify roles of KIAA1440, TRRAP, and SKIV2L2 in teleost organ development. Eur. J. Immunol. 39: 2606–2616 [DOI] [PubMed] [Google Scholar]
- Jackson S., Xiong Y. (2009). CRL4s: The CUL4-RING E3 ubiquitin ligases. Trends Biochem. Sci. 34: 562–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenik P.D., Gillmor C.S., Lukowitz W. (2007). Embryonic patterning in Arabidopsis thaliana. Annu. Rev. Cell Dev. Biol. 23: 207–236 [DOI] [PubMed] [Google Scholar]
- Jia S., Kobayashi R., Grewal S.I. (2005). Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nat. Cell Biol. 7: 1007–1013 [DOI] [PubMed] [Google Scholar]
- Jullien P.E., Kinoshita T., Ohad N., Berger F. (2006). Maintenance of DNA methylation during the Arabidopsis life cycle is essential for parental imprinting. Plant Cell 18: 1360–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kägi C., Baumann N., Nielsen N., Stierhof Y.D., Gross-Hardt R. (2010). The gametic central cell of Arabidopsis determines the lifespan of adjacent accessory cells. Proc. Natl. Acad. Sci. USA 107: 22350–22355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapetanaki M.G., Guerrero-Santoro J., Bisi D.C., Hsieh C.L., Rapić-Otrin V., Levine A.S. (2006). The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc. Natl. Acad. Sci. USA 103: 2588–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopanja D., Roy N., Stoyanova T., Hess R.A., Bagchi S., Raychaudhuri P. (2011). Cul4A is essential for spermatogenesis and male fertility. Dev. Biol. 352: 278–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I., Ambaru B., Thakkar P., Marcotte E.M., Rhee S.Y. (2010). Rational association of genes with traits using a genome-scale gene network for Arabidopsis thaliana. Nat. Biotechnol. 28: 149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Terzaghi W., Gusmaroli G., Charron J.-B.F., Yoon H.-J., Chen H., He Y.J., Xiong Y., Deng X.W. (2008). Characterization of Arabidopsis and rice DWD proteins and their roles as substrate receptors for CUL4-RING E3 ubiquitin ligases. Plant Cell 20: 152–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- León G., Holuigue L., Jordana X. (2007). Mitochondrial complex II Is essential for gametophyte development in Arabidopsis. Plant Physiol. 143: 1534–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt J.A., Matthews D.R., Ameer-Beg S.M., Suhling K. (2009). Fluorescence lifetime and polarization-resolved imaging in cell biology. Curr. Opin. Biotechnol. 20: 28–36 [DOI] [PubMed] [Google Scholar]
- Li N., Yuan L., Liu N., Shi D., Li X., Tang Z., Liu J., Sundaresan V., Yang W.C. (2009). SLOW WALKER2, a NOC1/MAK21 homologue, is essential for coordinated cell cycle progression during female gametophyte development in Arabidopsis. Plant Physiol. 151: 1486–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Bilodeau P., Koltunow A., Dennis E.S., Peacock W.J., Chaudhury A.M. (1999). Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96: 296–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv X.B., Xie F., Hu K., Wu Y., Cao L.L., Han X., Sang Y., Zeng Y.X., Kang T. (2010). Damaged DNA-binding protein 1 (DDB1) interacts with Cdh1 and modulates the function of APC/CCdh1. J. Biol. Chem. 285: 18234–18240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinier J., Lechner E., Dumbliauskas E., Genschik P. (2008). Regulation and role of Arabidopsis CUL4-DDB1A-DDB2 in maintaining genome integrity upon UV stress. PLoS Genet. 4: e1000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T., Skoog T. (1962). A revised medium for rapid growth and bioassays with tobacco cultures. Physiol. Plant. 15: 473–497 [Google Scholar]
- Nawy T., Lukowitz W., Bayer M. (2008). Talk global, act local-patterning the Arabidopsis embryo. Curr. Opin. Plant Biol. 11: 28–33 [DOI] [PubMed] [Google Scholar]
- Nowack M.K., Grini P.E., Jakoby M.J., Lafos M., Koncz C., Schnittger A. (2006). A positive signal from the fertilization of the egg cell sets off endosperm proliferation in angiosperm embryogenesis. Nat. Genet. 38: 63–67 [DOI] [PubMed] [Google Scholar]
- Nowack M.K., Ungru A., Bjerkan K.N., Grini P.E., Schnittger A. (2010). Reproductive cross-talk: Seed development in flowering plants. Biochem. Soc. Trans. 38: 604–612 [DOI] [PubMed] [Google Scholar]
- Ottenschläger I., Wolff P., Wolverton C., Bhalerao R.P., Sandberg G., Ishikawa H., Evans M., Palme K. (2003). Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proc. Natl. Acad. Sci. USA 100: 2987–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat G.C., Yu H.J., Ngo Q.A., Rajani S., Mayalagu S., Johnson C.S., Capron A., Xie L.F., Ye D., Sundaresan V. (2005). Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132: 603–614 [DOI] [PubMed] [Google Scholar]
- Park S.K., Howden R., Twell D. (1998). The Arabidopsis thaliana gametophytic mutation gemini pollen1 disrupts microspore polarity, division asymmetry and pollen cell fate. Development 125: 3789–3799 [DOI] [PubMed] [Google Scholar]
- Pazhouhandeh M., Molinier J., Berr A., Genschik P. (2011). MSI4/FVE interacts with CUL4-DDB1 and a PRC2-like complex to control epigenetic regulation of flowering time in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 3430–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroski M.D., Deshaies R.J. (2005). Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6: 9–20 [DOI] [PubMed] [Google Scholar]
- Pfaffl M.W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portereiko M.F., Lloyd A., Steffen J.G., Punwani J.A., Otsuga D., Drews G.N. (2006a). AGL80 is required for central cell and endosperm development in Arabidopsis. Plant Cell 18: 1862–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portereiko M.F., Sandaklie-Nikolova L., Lloyd A., Dever C.A., Otsuga D., Drews G.N. (2006b). NUCLEAR FUSION DEFECTIVE1 encodes the Arabidopsis RPL21M protein and is required for karyogamy during female gametophyte development and fertilization. Plant Physiol. 141: 957–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss D., Rhee S.Y., Davis R.W. (1994). Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science 264: 1458–1460 [DOI] [PubMed] [Google Scholar]
- Rotman N., Durbarry A., Wardle A., Yang W.C., Chaboud A., Faure J.E., Berger F., Twell D. (2005). A novel class of MYB factors controls sperm-cell formation in plants. Curr. Biol. 15: 244–248 [DOI] [PubMed] [Google Scholar]
- Schroeder D.F., Gahrtz M., Maxwell B.B., Cook R.K., Kan J.M., Alonso J.M., Ecker J.R., Chory J. (2002). De-etiolated 1 and damaged DNA binding protein 1 interact to regulate Arabidopsis photomorphogenesis. Curr. Biol. 12: 1462–1472 [DOI] [PubMed] [Google Scholar]
- Shi D.Q., Liu J., Xiang Y.H., Ye D., Sundaresan V., Yang W.C. (2005). SLOW WALKER1, essential for gametogenesis in Arabidopsis, encodes a WD40 protein involved in 18S ribosomal RNA biogenesis. Plant Cell 17: 2340–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J., Vierstra R.D. (2004). The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55: 555–590 [DOI] [PubMed] [Google Scholar]
- Smith T.F., Gaitatzes C., Saxena K., Neer E.J. (1999). The WD repeat: A common architecture for diverse functions. Trends Biochem. Sci. 24: 181–185 [DOI] [PubMed] [Google Scholar]
- Sørensen M.B., Chaudhury A.M., Robert H., Bancharel E., Berger F. (2001). Polycomb group genes control pattern formation in plant seed. Curr. Biol. 11: 277–281 [DOI] [PubMed] [Google Scholar]
- Steffen J.G., Kang I.H., Portereiko M.F., Lloyd A., Drews G.N. (2008). AGL61 interacts with AGL80 and is required for central cell development in Arabidopsis. Plant Physiol. 148: 259–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolpe T., Süsslin C., Marrocco K., Nick P., Kretsch T., Kircher S. (2005). In planta analysis of protein-protein interactions related to light signaling by bimolecular fluorescence complementation. Protoplasma 226: 137–146 [DOI] [PubMed] [Google Scholar]
- Sundberg E., Østergaard L. (2009). Distinct and dynamic auxin activities during reproductive development. Cold Spring Harb. Perspect. Biol. 1: a001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X.Y., Liu X.L., Wang W., Jia D.J., Chen L.Q., Zhang X.Q., Ye D. (2010). Mutations in the Arabidopsis nuclear-encoded mitochondrial phage-type RNA polymerase gene RPOTm led to defects in pollen tube growth, female gametogenesis and embryogenesis. Plant Cell Physiol. 51: 635–649 [DOI] [PubMed] [Google Scholar]
- Ulmasov T., Murfett J., Hagen G., Guilfoyle T.J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungru A., Nowack M.K., Reymond M., Shirzadi R., Kumar M., Biewers S., Grini P.E., Schnittger A. (2008). Natural variation in the degree of autonomous endosperm formation reveals independence and constraints of embryo growth during seed development in Arabidopsis thaliana. Genetics 179: 829–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nocker S., Ludwig P. (2003). The WD-repeat protein superfamily in Arabidopsis: conservation and divergence in structure and function. BMC Genomics 4: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra R.D. (2009). The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Wang H., Zhai L., Xu J., Joo H.Y., Jackson S., Erdjument-Bromage H., Tempst P., Xiong Y., Zhang Y. (2006). Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol. Cell 22: 383–394 [DOI] [PubMed] [Google Scholar]
- Wysocka J., Swigut T., Milne T.A., Dou Y., Zhang X., Burlingame A.L., Roeder R.G., Brivanlou A.H., Allis C.D. (2005). WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 121: 859–872 [DOI] [PubMed] [Google Scholar]
- Youngren K.K., et al. (2005). The Ter mutation in the dead end gene causes germ cell loss and testicular germ cell tumours. Nature 435: 360–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Wang J., Hu Q., Quan Y., Chen H., Cao Y., Li C., Wang Y., He Q. (2010). DCAF26, an adaptor protein of Cul4-based E3, is essential for DNA methylation in Neurospora crassa. PLoS Genet. 6: e1001132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., et al. (2011). The E3 ubiquitin ligase Cullin 4A regulates meiotic progression in mouse spermatogenesis. Dev. Biol. 356: 51–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Gaitatzes C., Neer E., Smith T.F. (2000). Thirty-plus functional families from a single motif. Protein Sci. 9: 2470–2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Feng S., Chen F., Chen H., Wang J., McCall C., Xiong Y., Deng X.W. (2008). Arabidopsis DDB1-CUL4 ASSOCIATED FACTOR1 forms a nuclear E3 ubiquitin ligase with DDB1 and CUL4 that is involved in multiple plant developmental processes. Plant Cell 20: 1437–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]