Abstract
Credneramides A (1) and B (2), two vinyl chloride-containing metabolites, were isolated from a Papua New Guinea collection of cf. Trichodesmium sp. nov and expand a recently described class of vinyl chloride-containing natural products. The precursor fatty acid, credneric acid (3), was isolated from both the aqueous and organic fractions of the parent fraction as well as from another geographically and phylogenetically distinct cyanobacterial collection (Panama). Credneramides A and B inhibited spontaneous calcium oscillations in murine cerebrocortical neurons at low micromolar concentrations (1, IC50 4.0 μM; 2, IC50 3.8 μM).
It has been exceptionally productive to search for structurally novel and biologically active natural products in marine cyanobacteria, especially among filamentous types.1, 2 For example, the vinyl chloride-containing jamaicamides A–C and the structurally diverse malyngamides A–X have been found from marine cyanobacteria or opisthobranch mollusks that feed on these photosynthetic prokaryotes.1,3 However, it is important to note that field-collected cyanobacteria are rich substrates for heterotrophic bacterial growth,4 and thus it is conceivable that the latter organisms are the source for some of these natural products.5 In a few cases, a microbial symbiont has been shown rigorously to be the actual producer of a compound obtained from a macroorganism.6–8 Thus, the biosynthetic machinery of the bacterial symbiont Endobugula sertula has been shown to produce the anticancer bryostatin natural products that were first reported from the host bryozoan Bugula neritina.7 Similarly, the unicellular symbiotic cyanobacterium Prochloron sp. has been shown to be the biosynthetic source of the patellamides, originally reported from the host tunicate Lissoclinum patella.8 Thus, in many cases the metabolic origin of marine secondary metabolites remains largely speculative at present.6
Interestingly, the jamaicamides and malynamides have a common biosynthetic theme in that they possess a PKS-derived long lipid chain and a NRPS-derived peptidic moiety.9 Furthermore, all three jamaicamides and 22 of the 29 reported malyngamides contain the unusual and highly distinctive vinyl chloride functional group.1,3 Because the vinyl chloride unit is predicted to reside on C-1 of the glycine-derived unit, the sp2 carbon bearing the chloride is predicted to derive from the C-2 carbon of another pendant acetate unit.10 Likewise, in the jamaicamides,11 the vinyl chloride carbon has been shown to be derived from C-2 of acetate via a complex biosynthetic process involving an HMGCoA synthase catalytic unit, and to be highly similar to that responsible for cyclopropyl ring formation in curacin A.3,12
Among cyanobacterial natural products, particularly Lyngbya majuscula, the phenethylamine moiety has been reported in several natural products, including grenadamide (lyngbyamide A), hermitamide A, and lyngbyamide B, which harbors a p-hydroxy group on the phenethylamine group.13–15 Phenethylamines (PEA) are well known for their psychotropic effects and have been isolated from a variety of organisms, including octocorals, the edible mushroom Laetiporus sulphureus, Streptomyces sp., Enterococcus sp., and other species of cyanobacteria.16–20 In accordance with the neurotropic activity of PEAs, grenadamide exhibits cannabinoid receptor binding activity (Ki 4.7 μM)13 and brine shrimp toxicity (LD50 5 μg/ml), whereas hermitamide A is cytotoxic to neuro-2A murine neuroblastoma cells (IC50 2.2 μM) and brine shrimp (LD50 5 μM).13,14 Lyngbyamide B was reported to be cytotoxic to brine shrimp (LD50= 6.5 ppm).15 For each of the cyanobacterial compounds containing a phenethylamine moiety, the “precursor” fatty acid was also isolated along with the elaborated natural product.13,14 As seen with the malyngamides, the precursor fatty acid is a chemical scaffold to which the cyanobacterium adds further functional groups, and this appears to enhance to their biological activity.21 This theme of chemical scaffolding to functionalize and enhance bioactivity is also observed with the credneramides. Herein, we report the isolation and structure elucidation of credneramides A (1) and B (2) and credneric acid (3), neuromodulatory cyanobacterial compounds isolated from a Papua New Guinea collection of cf. Trichodesmium sp. nov. Additionally, credneric acid (3) was also isolated from a strain of cf. Oscillatoria sp. obtained from the Eastern Pacific (Panama). Because credneric acid was also reported recently from a field collection of the bacterium Rhodopirellula baltica,22 several interesting questions emerge as to the origin of this compound class, the phylogenetic relationships of the producing organisms, and the evolutionary history of the putative biosynthetic pathway.
RESULTS AND DISCUSSION
Natural Product Isolation and Structure Elucidation
Credneramides A (1) and B (2) were isolated from the organic extract of the cyanobacterium cf. Trichodesmium sp. nov. collected near the Credner Islands, Papua New Guinea. Using silica gel vacuum-liquid chromatography, the extract was fractionated into nine subfractions (A–I) of increasing polarity, and fraction E was further purified using reversed-phase HPLC. The HRESIMS data ([M+H]+ m/z 320.1779) of credneramide A (1) established the molecular formula as C19H26ClNO with seven degrees of unsaturation; the presence of one chlorine atom was supported by the isotopic pattern. The 1H NMR spectrum was well dispersed and contained two distinctive resonances similar to that of the malyngamide series and diagnostic of a vinyl chloride functionality and a disubstituted olefin. In particular, these resonances were a sharp singlet at 5.77 ppm (δC 113.1 ppm) and two narrowly resolved multiplets at 5.37 and 5.45 ppm (δC 127.8, 131.2, respectively; Table 1). Three resonances in the aromatic region of the 1H NMR spectrum (δH 7.19, δC 128.7; δH 7.24, δC 126.5; δH 7.32, δC 128.7) were suggestive of the presence of a mono-substituted phenyl group, and a carbon resonance at 172.1 ppm indicated the presence of an ester or amide carbonyl.
Table 1.
1H and 13C NMR Data for Credneramide A (1) and Credneramide B (2).
| Credneramide A (1) | Credneramide B (2) | |||||
|---|---|---|---|---|---|---|
| carbon | δC, mult.a | δH, mult. (J in Hz)b | HMBCb | δC, mult.c | δH, mult. (J in Hz)d | HMBCd |
| 1 | 172.1, qC | 172.1, qC | ||||
| 2 | 36.4, CH2 | 2.19, t (7.0) | 1, 3, 4 | 36.4, CH2 | 2.22, t (6.9) | 1, 3, 4 |
| 3 | 28.4, CH2 | 2.32, dt (7.0, 6.6) | 1, 2, 4, 5 | 28.5, CH2 | 2.35, dt (6.9, 6.1) | 1, 2, 4, 5 |
| 4 | 131.2, CH | 5.43, dt (15.3, 6.6) | 2, 5, 6 | 131.3, CH | 5.48, dt (14.9, 6.1) | 2, 3, 5, 6 |
| 5 | 127.8, CH | 5.47, dt (15.3, 6.7) | 3, 4 | 127.8, CH | 5.41, dt (14.9, 6.1) | 3, 4, 6, 7 |
| 6 | 37.9, CH2 | 2.70, d (6.7) | 4, 5, 7, 8, 11 | 37.9, CH2 | 2.72, d (6.1) | 4, 5, 7, 8, 11 |
| 7 | 141.5, qC | 141.5, qC | ||||
| 8 | 32.3, CH2 | 2.15, t (6.7) | 6, 7, 9, 10, 11 | 32.2, CH2 | 2.15, t (7.6) | 6, 7, 9, 10, 11 |
| 9 | 20.2, CH2 | 1.43 h (7.5) | 7, 8, 10 | 20.2, CH2 | 1.43, h (7.6) | 7, 8, 10 |
| 10 | 13.9, CH3 | 0.91 t (7.5) | 8, 9 | 13.9, CH3 | 0.91, m | 8, 9 |
| 11 | 113.1, CH | 5.77, s | 6, 7, 8 | 113.1, CH | 5.78, s | 6, 7, 8, 9 |
| NH | 5.42 d (6.2) | 5.39 | ||||
| 1′ | 40.5, CH2 | 3.53 (dt (6.9, 6.2) | 1, 2′, 3′ | 37.8, CH2 | 3.26, dt (7.2, 6.5) | 1, 2′, 3′ |
| 2′ | 35.7, CH2 | 2.81, t (6.9) | 1′, 3′, 4′ | 38.5, CH2 | 1.36, dt (7.5, 7.2) | 1′, 3′, 4′, 5′ |
| 3′ | 138.9, qC | 25.9, CH | 1.60, m | 2′, 4′, 5′ | ||
| 4′ | 128.7, CH | 7.19, d (7.2) | 2′, 5′, 6′, 7′ | 22.4, CH3 | 0.90, m | 3′, 5′ |
| 5′ | 128.7, CH | 7.32, dd (7.2) | 3′, 4′ | 22.4, CH3 | 0.90, m | 3′, 4′ |
| 6′ | 128.5, CH | 7.24, t (7.2) | 4′ | |||
| 7′ | 128.7, CH | 7.32, t (7.2) | 3′, 4′ | |||
| 8′ | 128.7, CH | 7.19, d (7.2) | 2′, 5′, 6′, 7′ | |||
75 MHz in CDCl3.
500 MHz in CDCl3.
150 MHz CDCl3.
600 MHz in CDCl3.
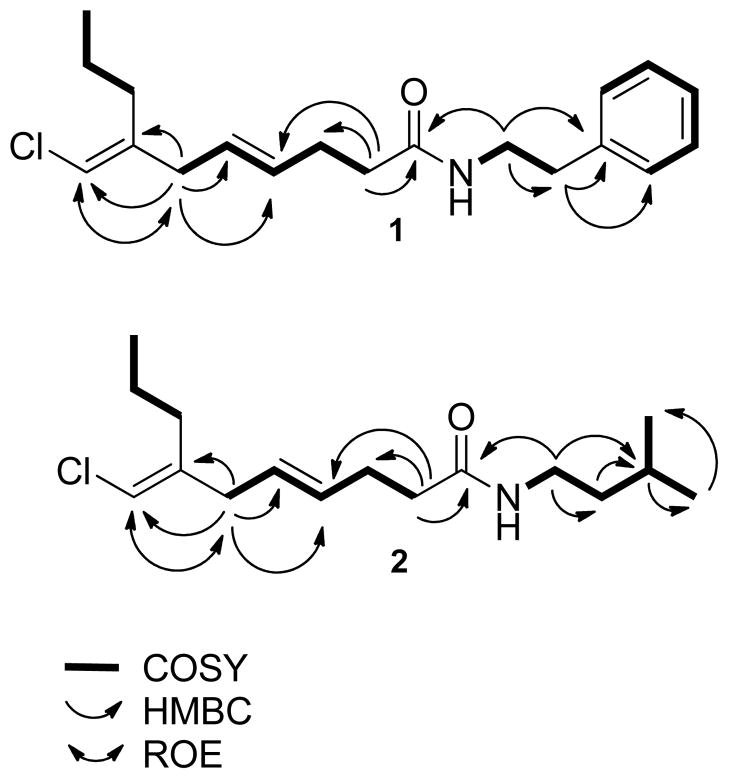
A combination of 2D COSY, HSQC, and HMBC correlations was used subsequently to complete the planar structure of credneramide A (1) (Figure 1). COSY correlations were used to assign consecutively two methylenes (δ1.43 and 2.15) adjacent to the terminal methyl at C-10 as well as to relate the C-3 and C-6 methylenes (δH 2.32 and 2.70) with each methine (δH 5.37 and 5.45) of the C-4/C-5 olefin. A HMBC correlation between H-2 and C-3 permitted connection of the C-2/C-3 methylenes, and a correlation between H-2 and C-1 allowed placement of the ester/amide carbonyl group adjacent to the C-2 methylene. Additional HMBC correlations between H-6 and C-11/C-7 permitted attachment of the vinyl chloride to this carbon chain. Integrated 1H NMR and COSY data supported the presence of a phenethyl moiety with five aromatic protons and coupling of the C-1′/C-2′ methylenes at 3.53 and 2.81 ppm, respectively (Figure 1). The complement of 1H NMR and 13C NMR chemical shifts for H-1′/C-1′ (δH 3.53; δC 40.5) supported its direct attachment to a nitrogen atom. HMBC correlations between H-1′ and C-2′/C-3′ and multiple correlations between the protons and carbons of the aromatic ring confirmed the presence of a phenethylamine moiety (Table 1). Lastly, a key HMBC correlation between H-1′ and C-1 permitted connection of the phenethylamine through an amide linkage to the vinyl chloride-containing fatty acid portion, thus completing the planar structure of credneramide A (1).
Figure 1.
Key COSY, HMBC, and ROE correlations of credneramides A (1) and B (2).
Assignment of the geometry of the two olefins in 1 was accomplished by 1D NOE, 2D ROE, and homonuclear coupling constants. A key NOE/ROE correlation between the vinyl proton and the H6 methylene established the C-7 olefin as E. A 3JH-H 15.3 Hz coupling constant between the vinyl protons at H-4 and H-5 protons permitted assignment of the C-4 olefin as E as well.
The second credneramide metabolite, credneramide B (2), was isolated in only about 60% of the yield of credneramide A (1), and showed by HRESIMS a [M+H]+ peak at m/z 286.1963 for a molecular formula of C16H28ClNO. A comparison of the 1H and 13C NMR chemical shifts of 1 and 2 revealed their close similarity with the notable absence of aromatic absorptions in the spectra for 2. Moreover, the 1H NMR of compound 2 lacked the distinctive triplet at δ 2.81 present in the spectrum of 1, and this was replaced with an additional methylene triplets at δ 1.36 and a methine multiplet at δ 1.60. Analysis of gHSQC data indicated the presence of two additional terminal methyl groups in the higher field region. COSY correlations defined the adjacent positions of the doublet of triplets at δ 1.36 to the C-1′ methylene at δ 3.26 and to the C-3′ methine at δ 1.60. A HMBC correlation (δH =0.90 ppm, δC =22.4 ppm) suggested a terminal gem-dimethyl grouping, and a correlation between the C-3′ methine and the two terminal methyls allowed the assignment of an isopentylamine moiety. Finally, a key HMBC correlation between the C-1′ methylene and the carbonyl (δC 172.0) permitted attachment of the isopentylamine to the rest of the molecule to complete the planar structure of 2. In similar fashion to credneramide A (1), a key NOE correlation between the vinyl proton and the H-6 methylene established the C-7 olefin as having E geometry in credneramide B (2), and a 3JHH 14.9 Hz coupling constant between the vinyl protons permitted assignment of the C-4 olefin as also being E.
The precursor vinyl chloride fatty acid, credneric acid (3), was isolated from both the aqueous and organic portions of the same parent extract. The initial analysis of 3 was done by 1H NMR spectroscopy which revealed that it was a short fatty acid. Analysis by COSY and HSQC revealed the presence of an n-propyl group. Further analysis by the same methods revealed another spin system consisting of two consecutive methylene groups, a di-substituted olefin, and another methylene group. A tri-substituted vinyl chloride functionality was postulated based on the two- and three-bond heteronuclear coupling information obtained by HMBC analysis. Finally, HMBC analysis also allowed connection of the three substructures to form the fatty acid portion of credneramides A and B. Again, the stereochemical assignment of the two olefins in 3 were made by the coupling constant between the two vicinal vinyl protons (5.42 ppm and 5.48 ppm; J = 15.4 Hz) and 1D ROESY correlations of key protons on and around the olefins, and these were in accordance with those of compounds 1 and 2.21
Credneramides A (1) and B (2), along with the recently reported grenadamides B and C, constitute a new class of vinyl chloride-containing fatty acids in cyanobacteria.23 A commonality shared among the credneramides is an apparent reductive decarboxylation of the amide head group. Reductive decarboxylation of phenylalanine (Phe) is expected to proceed via a phenylalanine decarboxylase or a tyrosine (Tyr) decarboxylase with promiscuity towards the related Phe residue.20 Similarly, the isopentylamine functionality is predicted to arise from reductive decarboxylation of leucine by a putative leucine decarboxylase. Vinyl chloride formation in the credneramides is predicted to proceed via a β-branch acetate extension as seen with the jamaicamides and most likely occurs in malyngamide biosynthesis.3,12,21 A non-heme Fe(II), α-ketoglutarate dependent halogenase similar to the Jam halogenase is expected to halogenate C-2 of acetate during β-branch formation. Following dehydration, a decarboxylase similar to Jam ECH2 must decarboxylate this pendant group in a regiospecific manner to yield the vinyl chloride.3,12
Evolution, Taxonomy, and Metabolic Origin of the Credneramides
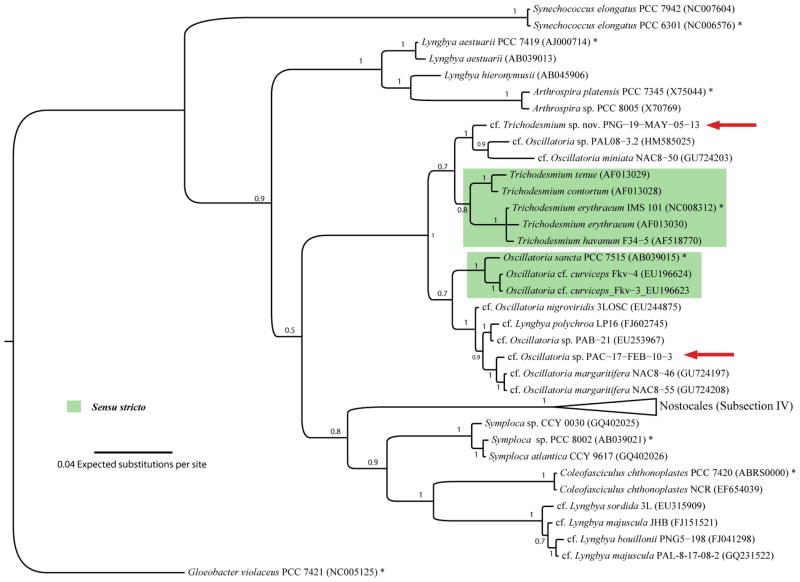
Morphologically, the credneramide-producing cyanobacterial specimen from Papua New Guinea (PNG-05/19/05-13; GenBank acc. Nr. 1452341) agreed well with the current definition of either Lyngbya or Oscillatoria (Figure 2, for morphological description, see Supporting Information). Marine forms of Lyngbya and Oscillatoria typically share their entire range of cellular types and are often misidentified.24,25 Phylogenetic inferences of the SSU (16S) rRNA gene from two credneramide-producing specimens was used to determine the taxonomic identity by inferring its evolutionarily relationship to relevant reference and type-strains. The credneramide-producing strain formed a clade with other benthic tropical marine cyanobacteria with similar morphology, including the strains PAL08-3.2 (GenBank acc. Nr. HM585025) from Palmyra atoll and NAC8-50 (GenBank acc. Nr. GU724203) from the Caribbean island of Curacao. This group is evolutionarily most closely related to the planktonic genus Trichodesmium (reference/type-strains = Trichodesmium erythraeum IMS 101T; p-distance 2.3%). The ecological and evolutionarily distinction suggests that the credneramide-producing strain, in fact, represents a new taxon. Technically, this new taxon could either be included in the genus Trichodesmium, which would require a change in the definition of this planktonic genus to also include benthic forms, or alternatively, this group needs to be erected as a separate generic entity. More careful taxonomic comparison between the two groups must be performed to establish firmly the placement of this new taxon. Presently, this strain is best defined as a cf. Trichodesmium sp. nov.
Figure 2.
Evolutionary tree of the credneramide-producing strain PNG-05/19/05-13. The phylogenetic proximity with the genus Trichodesmium (reference/type-strains = Trichodesmium erythraeum IMS) suggests that this strain represents a new taxa within this genus, which would also include the related and morphologically-similar strains PAL08-3.2 (GenBank acc. Nr. HM585025) and NAC8-50 (GenBank acc. Nr. GU724203) previously putatively identified as cf. Oscillatoria sp. Representative reference-strains obtained from Bergey’s Manual are highlighted with asterisks (*) and the sensu stricto for the genera Trichodesmium and Oscillatoria are highlighted with green boxes. In addition to the strain PNG-05/19/05-13, credneric acid (3) was also isolated from the Panamanian strain cf. Oscillatoria sp. PAC-17-FEB-10-3 (both credneric acid-producing strains are highlighted with red arrows). The cladogram is based on SSU (16S) rRNA gene sequences using the bayesian (MrBayes) method and the support values are indicated as posterior probability at the nodes. The specimens are indicated as species, strain, and access number in brackets. The scale bar is indicated at 0.04 expected nucleotide substitutions per site.
The predicted precursor of the credneramides, credneric acid (3), was also isolated from a filamentous cyanobacterial specimen (Oscillatoria sp., PAC-17-FEB-10-3; GenBank acc. Nr. 1452343) obtained from the Pacific coast of Panama. Phylogenetic comparison between the strain from Papua New Guinea and the Panamanian strain revealed that these two credneric acid-producing strains are unrelated and belong to two different genera (Figure 2). Strain PNG-05/19/05-13 is most closely related evolutionarily to the genus Trichodesmium whereas strain PAC-17-FEB-10-3 belongs to a clade of tropical marine Oscillatoria. To determine if this secondary metabolite is the result of a more ancient biosynthetic pathway, several related specimens, including the strains NAC8-50, 3LOSC (GenBank acc. Nr. EU244875), PAB-21 (EU253967), NAC8-46 (GU724197), and NAC8-55 (GU724208) were screened by ESI-LC-MS for a molecular weight corresponding to credneric acid. The fact that this secondary metabolite was not present in any other of these specimens suggests that its biosynthetic pathway has not been inherited vertically, at least within these groups of cyanobacteria.
Another possibility for this disjunct taxonomic distribution of credneric acid in these cyanobacteria is suggested by the recent isolation of credneric acid (3) from the epiphytic heterotrophic bacterium Rhodopirellula baltica.22 Since this bacterium is known to grow in association with other marine organisms, there is the possibility that credneric acid isolated from our cyanobacterial specimens is actually produced by R. baltica living in association with the cyanobacteria. To determine whether R. baltica was present in our collections and, therefore, the likely producer of credneric acid, an attempt was made to amplify the 16S rRNA gene sequence of R. baltica from the Panamanian and Papua New Guinean strains using specifically designed primers and to compare that data with the 16S rRNA sequence for R. baltica (H. S. Lee, personal communication). However, sequencing of several clones from libraries generated using these specific primers resulted in gene sequences of heterotrophic bacteria other than R. baltica. Thus, because the biosynthetic machinery for production of the vinyl chloride functional group has been carefully and quite thoroughly described in other marine cyanobacteria,3,12 and the fact that another well-known marine cyanobacterial metabolite (malyngic acid)26 was also isolated from the field collection of R. baltica,22 we believe that these metabolites derive from cyanobacterial metabolism. Their occurrence in the field-collected R. baltica therefore may have resulted from small filaments of Trichodesmium or Oscillatoria contaminating this environmental sample.
Biological Activity and Ecological Significance of Credneramides
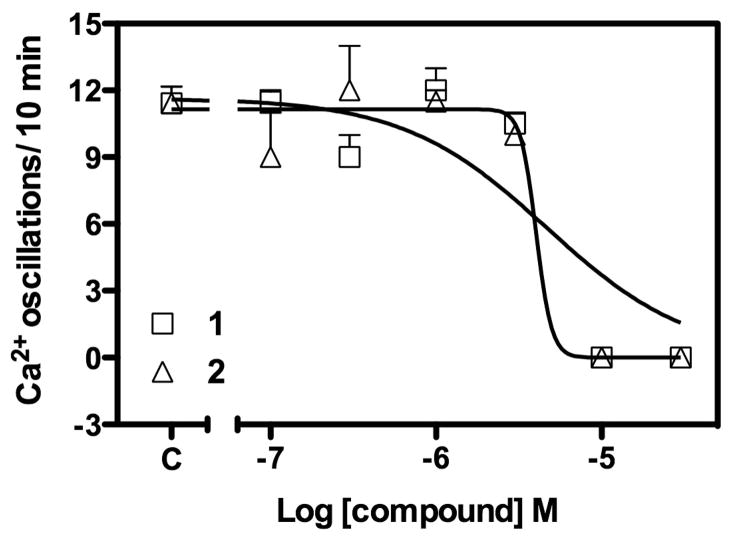
Credneramides A (1) and B (2), and credneric acid (3), were subjected to a suite of bioassays to explore their activities. Although the parent extract possessed sodium-channel blocking activity, the activity of 1 and 2 in this assay was very modest. Their most potent activity was inhibition of calcium oscillations in cerebrocortical mouse neurons (1, IC50 4.0 μM, 2, IC50 3.8 μM). Although 3 exhibited modest calcium oscillation inhibition (IC50 8.2 μM), the Hill slope of its concentration response curve suggested nonspecific activity or 1:1 ligand-receptor binding (data not shown). In contrast, the steep Hill slopes of the concentration curves for the 1 and 2 suggest positive cooperativity (Figure 3). Given that the credneric acid demonstrates a nonspecific response, and 1 and 2 confer more specific calcium inhibition, it is logical that the PEA and isopentylamine moieties may be responsible for the enhanced activity of each compound.21
Figure 3.
Calcium oscillation inhibition in cerebrocortical mouse neurons by credneramides A (1) (IC50 4.0 μM) and B (2) (IC50 3.8 μM).
The calcium oscillations observed in primary and mature cerebrocortical neurons of mice are sensitive to AMPA receptor modulation as well as to group I metabotropic glutamate receptor (mgluR) agonism or antagonism.27 AMPA receptors regulate calcium oscillation frequency while mgluR mediate frequency and amplitude as well as release of intracellular calcium stores via phospholipase C (PLC). Thus, the bioactivity of the credneramides in this assay could be due to modulation of AMPA, mgluR, or PLC signaling or to TAAR1, dopamine, and GABA receptor activity; the latter three are known to be modulated by PEA.28–31 Unlike grenadamide A, a mixture of 1 and 2 did not possess cannabinoid receptor binding activity, and only marginal cytotoxic activity against the beet armyworm Spodoptera exigua was reported for grenadamides B and C, which are structurally most similar to the credneramides.23
Fatty acids with phenethylamine have been encountered increasingly among cyanobacterial metabolites. PEA derivatives have a well-recognized role in mammalian physiology as an endogenous neurotransmitter that has been shown to bind trace amine (TAAR), dopaminergic, and GABA receptors.28–31 By binding to these receptors, PEA derivatives modulate higher cognitive function via a capacity to act as hallucinogens, stimulants, and antidepressants.28,29 This class of compounds has also been implicated in the pathogenesis of several neurologic diseases including depression, attention deficit and hyperactivity disorder (ADHD), and schizophrenia.28,29,32 Given the substantial evidence of neuromodulation by phenethylamines, the neuromodulatory effects of credneramide A (1) are reasonable.
The ecologic and physiologic role of reductively decarboxylated fatty acid amides, particularly those containing PEA, in lower life forms is poorly understood. While they may serve in antipredatory or antifeedant roles, it is also possible that they serve a role similar to that of neurotransmitters in higher life forms, namely, to mediate intercellular signaling. Fatty acid amides with functionalized head groups similar to PEA mimic N-acyl homoserine lactones (AHL), which are quorum-sensing molecules in diverse bacteria.33 At threshold concentrations, AHLs bind “sensor peptides” that are transcriptional regulators and activate the expression of genes of varying functions such as those involved in antibiotic synthesis and motility.33 Malyngamide C and epi-malyngamide C are two other fatty acid amides isolated from cyanobacteria with demonstrated quorum sensing properties.34 Thus, it is possible that the credneramides also have roles in intercellular communication and quorum sensing.
CONCLUSIONS
Credneramides A (1) and B (2), neuromodulatory phenethylamine and isopentylamine derivatives of a vinyl chloride-containing fatty acid, were isolated from a Papua New Guinea collection of cf. Trichodesmium sp. nov. The precursor fatty acid, credneric acid (3), was also isolated from both the aqueous and organic fractions of the parent fraction, in addition, to a phylogenically distinct Panamanian collection. The true source of credneric acid production remains somewhat uncertain. However, the constellation of findings, the widespread and well characterized vinyl chloride functionality in other marine cyanobacterial metabolites, and the co-occurrence of another well known cyanobacterial metabolite in the report from the heterotrophic bacterium R. baltica, indicates that these filamentous cyanobacteria are the likely biosynthetic source. Credneric acid (3) appears to serve as a template for biochemical elaboration and enhancement of bioactivity.
EXPERIMENTAL SECTION
General Experimental Procedures
IR and UV spectra were obtained using a Nicolet IR-100 FT-IR and Beckman-Coulter DU800 spectrophotometer, respectively. NMR spectra were obtained using Varian Unity 300 and 500 MHz spectrometers and a Bruker 600 MHz spectrometer [CDCl3 (δH 7.26; δC 77.0) was used as an internal standard]. High-resolution mass spectra were obtained using an Agilent ESI-TOF mass spectrometer (TSRI, La Jolla, CA) and a Thermo Finnigan MAT900XL spectrometer (UCSD, La Jolla, CA). LC-ESI-MS profiling of extracts was performed with a Finnigan LCQ Advantage Max using a LiChrospher 100 RP18 column (5 μm, 125 × 4 mm) and elution gradients of CH3CN-MeOH (1:1) to 100% CH3CN over 15 to 30 min. Flash chromatography was performed using EM Science silica gel (230–400 mesh). VLC was performed using TLC grade silica gel. TLC was performed using EM Science pre-coated silica gel plates (Merck 60 F254). Extracts were processed using a HPLC Waters 515 pump, Waters 996 photodiode array detector, and Millenium software for acquisition and analysis of data.
Organism Collection
The cf. Trichodesmium sp. nov. sample (PNG-05/19/05-13) was collected in May 2005 by snorkel at a depth of 1–2 m near the Credner Islands of Papua New Guinea (4° 14.105’ S and 152° 25.605’ E). The Oscillatoria sp. sample (PAC-02/17/10-3) was collected in February 2010 in shallow waters near Isla Canales de Afuera of Panama (7° 41.723’ N and 81° 38.025’ W). Both specimens were stored in 70% EtOH in a 1 L container at −20 °C until extraction.
DNA Extraction, Amplification, and Sequencing
Cyanobacterial biomass (~50 mg) was partly cleaned under an Olympus VMZ dissecting microscope. Genomic DNA was extracted using the Wizard® Genomic DNA Purification Kit (Promega) following the manufacturer’s specifications. DNA concentration and purity were measured on a DU-800 spectrophotometer (Beckman Coulter). The 16S rRNA genes were PCR-amplified from isolated DNA using the modified lineage-specific primers, OT106F 5′-GGACGGGTGAGTAACGCGTGA-3′ and OT1445R 5′-AGTAATGACTTCGGGCGTG-3′. The PCR reaction volumes were 25 μL containing 0.5 μL (~50 ng) of DNA, 2.5 μL of 10 × PfuUltra IV reaction buffer, 0.5 μL (25 mM) of dNTP mix, 0.5 μL of each primer (10 μM), 0.5 μL of PfuUltra IV fusion HS DNA polymerase and 20.5 μL dH2O. The PCR reactions were performed in an Eppendorf Mastercycler gradient as follows: initial denaturation for 2 min at 95 °C, 25 cycles of amplification, followed by 20 sec at 95 °C, 20 sec at 55 °C and 1.5 min at 72 °C, and final elongation for 3 min at 72 °C. PCR products were purified using a MinElute PCR Purification Kit (Qiagen) before subcloning using the Zero Blunt TOPO PCR Cloning Kit (Invitrogen) following the manufacturer’s specifications. Plasmid DNA was isolated using the QIAprep Spin Miniprep Kit (Qiagen) and sequenced with M13 primers. The 16S rRNA gene sequences are available in the DDBJ/EMBL/GenBank databases under acc. No. 1452341 and 1452343.
Phylogenetic Inference
The 16S rRNA gene sequence of PAC-17-FEB-10-2 was aligned with evolutionary informative cyanobacteria using the L-INS-I algorithm in MAFFT 6.71735 and refined using the SSU secondary structures model for Escherichia coli J0169536 without data exclusion. The best-fitting nucleotide substitution model optimized by maximum likelihood was selected using corrected Akaike/Bayesian Information Criterion (AICC/BIC) in jModeltest 0.1.1.37 The evolutionary histories of the cyanobacterial genes were inferred using Maximum likelihood (ML) and Bayesian inference algorithms. The ML inference was performed using GARLI 1.038 for the GTR+I+G model assuming a heterogeneous substitution rates and gamma substitution of variable sites (proportion of invariable sites (pINV) = 0.494, shape parameter (α) = 0.485, number of rate categories = 4) with 1,000 bootstrap-replicates. Bayesian inference was conducted using MrBayes 3.139 with four Metropoliscoupled MCMC chains (one cold and three heated) ran for 3,000,000 generations. The first 25% were discarded as burn-in and the following data set were sampled with a frequency of every 100 generations. The MCMC convergence was detected by AWTY.40
Extraction and Isolation
The cf. Trichodesmium sp. nov. collection was extracted ten times with heat using CH2Cl2-MeOH (2:1) to yield a crude organic extract of mass 12.2 g. Approximately 3 g of this crude extract was processed further on silica gel using vacuum-liquid chromatography with a 100% hexanes-EtOAc-MeOH gradient to generate nine subfractions, A–I. Fraction E eluted with 60/40 EtOAc-hexanes and harbored the most potent sodium channel blocking activity in neuro-2A neuroblastoma cells. Fraction E (240.5 mg) was filtered subsequently using a Waters RP C18 SPE cartridge and processed further using reversed-phase HPLC (Phenomenex RP C18 Jupiter 250 × 10 mm, 10 μm; 50:50 CH3CN-H2O to 100 CH3CN in 30 min; 100 CH3CN 30–45 min), to yield 7.0 mg of a mixture of credneramides A (1) and B (2), and 9.6 mg of credneric acid (3). The mixture was processed further (C18 Kromasil 250 × 4.6 mm, 5 μm; 70:30 CH3CN-H2O isocratic), to yield 1.4 mg of pure credneramide A (1) and 0.8 mg of credneramide B (2). The remaining aqueous layers and the EtOH used for preservation of the alga were combined and the volatiles were removed under vacuum. The residues were resuspended in EtOH and the insolubles were removed by filtration. Filtration of this material over a pad of water-treated silica gel with EtOH was followed by silica gel solid-phase extraction (SPE) chromatography, sequentially eluting with 1:1 CH2Cl2-EtOAc, EtOAc, 1:9 MeOHCH2Cl2, and 3:7 MeOH-CH2Cl2). Four fractions (4 mg, 422 mg, 1.08 g, and 1.09 g, respectively) were obtained. Based on TLC characteristics, the third SPE fraction was subjected to flash column chromatography with silica gel with a gradient mixture of MeOH and CH2Cl2 as the mobile phase to obtain several fractions. One of the fractions contained a partially purified compound (7 mg) and it was further purified by semi-preperative reversed-phase HPLC (250 ×10.00 mm, Synergi 4 μm Hydro-RP 80A column, CH3CN/H2O gradient) to obtain an additional 4 mg of credneric acid (3) as a colorless oil.
The Oscillatoria sp. collection was extracted five times using CH2Cl2-MeOH (2:1) to yield a crude organic extract of mass 90.6 mg. This crude extract (82.9 mg) was processed further on silica gel using vacuum-liquid chromatography, as described above, to generate nine subfractions, A–I. Fraction E eluted with 60:40 EtOAc-hexanes (5.8 mg) and was purified further using preparative TLC [hexanes-EtOAc (1:1) + 0.1% AcOH] to yield 1.9 mg credneric acid (3).
Credneramide A (1): clear oil: [α]D +14 (c 0.1, MeOH); UV (MeOH) λmax (log ε) 206 (4.21), 257 (2.36) nm; IR (neat) vmax 3289, 3068, 3028, 2959, 2930, 2869, 1644, 1551, 1453, 970, 746 cm−1; 1H NMR (CDCl3, 500 MHz) and 13C NMR (CDCl3, 75 MHz) data, see Table 1; HRESI(+)MS m/z 320.1779 [M+H]+ (calcd for C19H27ClNO, 320.1781).
Credneramide B (2): clear oil: [α]D −2.5 (c 0.1, MeOH); UV (MeOH) λmax (log ε) 203 (3.98) nm; IR (neat) vmax 3305, 2958, 2926, 2857, 1709, 1644, 1554, 1461, 1376, 1172, 1071, 605 cm−1; 1H NMR (CDCl3, 500 MHz) and 13C NMR (CDCl3, 75 MHz) data, see Table 1; HRESI(+)MS m/z 286.1963 [M+H]+ (calcd for C16H29ClNO, 286.1938).
Credneric acid (3): clear oil: [α]D +7.4 (c 0.2, MeOH); UV (MeOH) λmax (log ε) 203 (3.77), 269 (2.81) nm; IR (neat) vmax 3428 (br), 2965, 2930, 2873, 1711, 1632, 1431, 1410, 1284, 1252, 1212, 1158, 968, 795 cm−1; 1H NMR (CDCl3, 500 MHz) δ 5.79 (1H, s, H-11), 5.48 (1H, dt, J = 15.4, 6.2, H-4), 5.42 (1H, dt, J = 15.4, 6.4, H-5), 2.73 (2H, d, J = 6.2, H2-6), 2.44 (2H, t, J = 7.4, H2-2), 2.35 (2H, dt, J = 7.0, 6.4, H2-3), 2.16 (2H, t, J = 7.7, H2-8), 1.44 (2H, h, J = 7.5, H2-9), 0.92 (3H, t, J = 7.3, H3-10); 13C NMR (CDCl3, 75 MHz) δ 178.9 (C, C-1), 141.4 (C, C-7), 130.5 (CH, C-4), 128.1 (CH, C-5), 113.1 (CH, C-11), 37.9 (CH2, C-6), 33.7 (CH2, C-2), 32.2 (CH2, C-8), 27.4 (CH2, C-3), 20.2 (CH2, C-9), 13.9 (CH3, C-10); HREIMS m/z 217.0995 [M+H]+ (calcd for C11H18O2Cl, 217.0990).
Biological Testing
A mixture containing 1 and 2 was screened for biological activity in an H460 cytotoxicity assay, neuro-2A sodium channel activation and blocking assays, and malaria, Chagas’ disease, and leishmania antiparasitic assays, and a cannabinoid receptor-binding assay. Compounds 1–3 were also submitted for testing in a neocortical neuron calcium oscillation assay. See reported references for the calcium oscillation assay,27 the H460 cytotoxicity assay,41 the neuro-2A assays,42 and the anti-parasitic assays used.43–46
Supplementary Material
Acknowledgments
We thank the governments of Papua New Guinea and Panama for permission to make these cyanobacterial collections, J. Wingerd for the H-460 cytotoxicity and Neuro-2A sodium assay screening, A. Ligresti and V. Di Marzo for cannabinomimetic assay screening, the H. S. Lee laboratory (Korea) for the R. baltica 16S RNA sequence, H. Choi for help with collection of analytical data, and The Scripps Research Institute and UCSD mass spectrometry facilities for their analytical services. This work was supported by NIH grants NS 053398 and ICBG FIC TW006634.
References
- 1.Gerwick WH, Tan LT, Sitachitta N. In: The Alkaloids. Cordell G, editor. Vol. 57. Academic Press; San Diego: 2001. pp. 75–184. Chapter 2. [DOI] [PubMed] [Google Scholar]
- 2.Tidgewell K, Clark BR, Gerwick WH. In: Comprehensive Natural Products Chemistry. Mander L, Lui H-W, editors. Vol. 2. Elsevier; Oxford, UK: 2010. pp. 141–188. Ser. [Google Scholar]
- 3.Edwards DJ, Marquez BL, Nogle LM, McPhail K, Goeger DE, Roberts MA, Gerwick WH. Chem Biol. 2004;11:817–833. doi: 10.1016/j.chembiol.2004.03.030. [DOI] [PubMed] [Google Scholar]
- 4.Berg KA, Lyra C, Sivonen K, Paulin L, Suomalainen S, Tuomi P, Rapala J. ISME J. 2009;3:314–325. doi: 10.1038/ismej.2008.110. [DOI] [PubMed] [Google Scholar]
- 5.Garson MJ. In: Molluscs from Chemo-ecological Study to Biotechnological Application. Cimino G, Gavagnin M, editors. Springer-Verlag; Berlin, Heidelberg: 1996. pp. 159–174. [Google Scholar]
- 6.Haygood MG, Schmidt EW, Davidson SK, Faulkner DJ. J Mol Microbiol Biotechnol. 1999;1:33–43. [PubMed] [Google Scholar]
- 7.Davidson SK, Allen SW, Lim GE, Anderson CM, Haygood MG. Appl Environ Microbiol. 2001;67:4531–4537. doi: 10.1128/AEM.67.10.4531-4537.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmidt EW, Nelson JT, Rasko DA, Sudek S, Eisen JA, Haygood MG, Ravel J. Proc Natl Acad Sci USA. 2005;102:7315–7320. doi: 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischbach MA, Walsh CT. Chem Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 10.Dewick PM. Medicinal Natural Products: A Biosynthetic Approach. 2. John Wiley & Sons Ltd; Chichester, UK: 2001. [Google Scholar]
- 11.Vaillancourt FH, Yeh E, Vosburg DA, Garneau-Tsodikova S, Walsh CT. Chem Rev. 2006;106:3364–3378. doi: 10.1021/cr050313i. [DOI] [PubMed] [Google Scholar]
- 12.Gu L, Wang B, Kulkarni A, Geders TW, Grindberg RV, Gerwick L, Hakansson K, Wipf P, Smith JL, Gerwick WH, Sherman DH. Nature. 2009;459:731–735. doi: 10.1038/nature07870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sitachitta N, Gerwick WH. J Nat Prod. 1998;61:681–684. doi: 10.1021/np970576a. [DOI] [PubMed] [Google Scholar]
- 14.Tan LT, Okino T, Gerwick WH. J Nat Prod. 2000;63:952–955. doi: 10.1021/np000037x. [DOI] [PubMed] [Google Scholar]
- 15.Nannini C. MS Thesis. Oregon State University; Corvallis, OR: 2002. Novel Secondary Metabolites from a Madagascar Collection of Lyngbya majuscula; pp. 25–58. [Google Scholar]
- 16.Shiono Y, Tamesada Y, Muravayev YD, Murayama T, Ikeda M. Nat Prod Res. 2005;19:363–366. doi: 10.1080/14786410412331280113. [DOI] [PubMed] [Google Scholar]
- 17.Zhao PJ, Wang HX, Li GH, Li HD, Liu J, Shen YM. Chem Biodiversity. 2007;4:899–904. doi: 10.1002/cbdv.200790078. [DOI] [PubMed] [Google Scholar]
- 18.Gutierrez M, Capson TL, Guzman HM, Gonzalez J, Ortega-Barria E, Quinoa E, Riguera R. J Nat Prod. 2006;69:1379–1383. doi: 10.1021/np060007f. [DOI] [PubMed] [Google Scholar]
- 19.Liyanage GK, Schmitz FJ. J Nat Prod. 1998;61:1180. doi: 10.1021/np980264n. [DOI] [PubMed] [Google Scholar]
- 20.Marcobal A, de las Rivas B, Muñoz R. FEMS Microbiol Lett. 2006;258:144–149. doi: 10.1111/j.1574-6968.2006.00206.x. [DOI] [PubMed] [Google Scholar]
- 21.Suyama TL. PhD Thesis. University of California; San Diego, La Jolla, CA: 2009. Organic Synthesis as an Effective Approach to Chemical, Pharmaceutical, and Biosynthetic Investigations of Natural Products; pp. 101–125. [Google Scholar]
- 22.Lee Y-J, Lee JW, Oh C, Heo S-J, Kang D-H, Shin HJ, Lee H-S. Bull Korean Chem Soc. 2010;31:3421–3422. [Google Scholar]
- 23.Jimenez JI, Vansach T, Yoshida WY, Sakamoto B, Pörzgen P, Horgen FD. J Nat Prod. 2009;72:1573–1578. doi: 10.1021/np900173d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castenholz RW, Rippka R, Herdman M. In: Bergey’s Manual of Systematic Bacteriology. Boone DR, Castenholz RW, editors. Vol. 1. Springer; New York: 2001. pp. 473–599. [Google Scholar]
- 25.Engene N, Choi H, Esquenazi E, Rottacker EC, Ellisman MH, Dorrestein PC, Gerwick WH. Environ Microbiol. 2011;13:1601–1610. doi: 10.1111/j.1462-2920.2011.02472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cardellina JH, Moore RE. Tetrahedron. 1980;36:993–996. [Google Scholar]
- 27.Dravid SM, Murray TF. Brain Res. 2004;1006:8–17. doi: 10.1016/j.brainres.2004.01.059. [DOI] [PubMed] [Google Scholar]
- 28.Sabelli H, Javaid J. J Neuropsych Clin N. 1995;7:6–14. doi: 10.1176/jnp.7.1.6. [DOI] [PubMed] [Google Scholar]
- 29.Kitamura T, Munakata M, Haginoya K, Tsuchiya S, Iinuma K. Tohoku J Exp Med. 2008;215:333–340. doi: 10.1620/tjem.215.333. [DOI] [PubMed] [Google Scholar]
- 30.Sotnikova TD, Caron MG, Gainetdinov RR. Mol Pharmacol. 2009;76:229–235. doi: 10.1124/mol.109.055970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borowsky B, Adham N, Jones KA, Raddatz R, Artymyshyn R, Ogozalek KL, Durkin MM, Lakhlani PP, Bonini JA, Pathirana S, Boyle N, Pu X, Kouranova E, Lichtblau H, Ochoa FY, Branchek TA, Gerald C. Proc Natl Acad Sci USA. 2001;98:8966–8971. doi: 10.1073/pnas.151105198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Myojin T, Taga C, Tsuji M. Psychiat Clin Neuros. 1989;43:171–176. doi: 10.1111/j.1440-1819.1989.tb02566.x. [DOI] [PubMed] [Google Scholar]
- 33.Uroz S, Dessaux Y, Oger P. ChemBioChem. 2009;10:205–216. doi: 10.1002/cbic.200800521. [DOI] [PubMed] [Google Scholar]
- 34.Kwan JC, Teplitski M, Gunasekera SP, Paul VJ, Luesch H. J Nat Prod. 2010;73:463–466. doi: 10.1021/np900614n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katoh K, Toh H. Brief Bioinf. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 36.Cannone JJ, Subramanin S, Schnare MN, Collett JR, D’Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, Pnde N, Schang Z, Yu N, Gutell RR. BMC Bioinformatics. 2002;3:1471–2105. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Posada D. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 38.Zwickl DJ. PhD Thesis. The University of Texas; Austin, TX: 2006. Genetic Algorithm Approaches for the Phylogenetic Analysis of Large Biological Sequence Datasets under the Maximum Likelihood Criterion; pp. 1–115. [Google Scholar]
- 39.Ronquist F, Huelsenbeck JP. Bioinformatics. 2003;12:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 40.Nylander JAA, Wilgenbusch JC, Warren DL, Swofford DL. Bioinformatics. 2008;15:581–583. doi: 10.1093/bioinformatics/btm388. [DOI] [PubMed] [Google Scholar]
- 41.Alley MC, Scudiero DA, Monks A, Hursey ML, Czerwinski MJ, Fine DL, Abbott BJ, Mayo JG, Shoemaker RH, Boyd MR. Cancer Res. 1988;48:589–601. [PubMed] [Google Scholar]
- 42.Manger RL, Leja LS, Lee SY, Hungerford JM, Hokama Y, Dickey RW, Granade HR, Lewis R, Yasumoto T, Wekell MM. J AOAC Int. 1995;78:521–527. [PubMed] [Google Scholar]
- 43.Corbett Y, Herrera L, Gonzalez J, Cubilla L, Capson TL, Coley PD, Kursar TA, Romero LI, Ortega-Barria E. Am J Trop Med Hyg. 2004;70:119–124. [PubMed] [Google Scholar]
- 44.Buckner FS, Verlinde CL, LaFlamme AC, Van Boris WC. Antimicrob Agents Chemother. 1996;40:2592–2597. doi: 10.1128/aac.40.11.2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gutierrez M, Capson TL, Guzman HM, Gonzalez J, Ortega-Barria E, Quinoa E, Riguera R. J Nat Prod. 2006;69:1379–1383. doi: 10.1021/np060007f. [DOI] [PubMed] [Google Scholar]
- 46.Calderón AI, Romero LI, Ortega-Barría E, Solís PN, Zacchino S, Gimenez A, Pinzón R, Cáceres A, Tamayo G, Guerra C, Espinosa A, Correa M, Gupta MP. Pharm Biol. 2010;48:545–553. doi: 10.3109/13880200903193344. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.