Since the initial identification of pathogenic mutations in 1988, more than 100 point mutations and numerous large-scale deletions of human mtDNA have been associated with a wide range of clinical phenotypes (1). The development, by King and Attardi, of cells in culture called cybrids (cytoplasmic hybrids) provided an in vitro method to study mtDNA mutations against a uniform nuclear (nDNA) background (2). Although cybrid technology has been very useful in defining some pathogenic effects of mtDNA mutations, until this year, researchers have been hindered by the lack of animal models with mtDNA mutations. By contrast, manipulation of mouse embryonic stem (ES) cells has already produced a few lines of mice with defects of nDNA-encoded mitochondrial proteins (3, 4).
In a recent issue of PNAS, Sligh et al. describe how they have succeeded in producing transmitochondrial mice by transferring exogenous mtDNA into embryo (5). These are the first mice with a transmitochondrial mtDNA point mutation to express an abnormal phenotype, a mitochondrial disease. This exciting work and the recent report by Inoue et al. of a transmitochondrial mouse harboring a pathogenic mtDNA deletion (6) are likely to be the harbingers of a new wave of research with animal models of human mitochondrial disorders. Curiously, these developments mirror the sequence in which human mutations were reported: large-scale deletions first, followed shortly thereafter by a pathogenic point mutation.
Mitochondria are vital organelles that provide most of the cellular energy in the form of ATP. This process requires the transfer of reducing equivalents through enzyme complexes of the respiratory chain, thereby generating a proton gradient across the inner mitochondrial membrane, which, in turn, drives oxidative phosphorylation to produce ATP. Four of the five respiratory chain complexes are under dual genetic control because some of their structural subunits are encoded by mtDNA and others by nDNA. Thus, mitochondria are the products of two genomes, and mutations in either can impair energy metabolism that frequently manifests in patients as encephalomyopathies, because brain and muscle have high energy requirements.
These are the first mice with a transmitochondrial mtDNA point mutation to express an abnormal phenotype. . . . .
To understand inherited mitochondrial disorders, the unusual properties of mtDNA must be considered. Mitochondrial genetics differs from Mendelian genetics in two ways. First, mtDNA is exclusively maternally transmitted. Second, because hundreds to thousands of copies of mtDNA reside within each cell, the proportions of mutant genomes can vary considerably from individual to individual within a family, a concept known as heteroplasmy. Typically, mutations of mtDNA are heteroplasmic and show variable tissue distribution; both factors, along with the distinct bioenergetic requirements of different tissues, contribute to the diversity of the clinical phenotype. Transmitochondrial mice offer the exciting possibility of studying the effects of segregation of mtDNA mutations at multiple levels, from individual cells to the whole organism.
Other types of transmitochondrial mice have already provided intriguing information about mtDNA. More than 20 years ago, mouse teratoma stem cells harboring a mtDNA mutation conferring chloramphenicol resistance (CAPR) were microinjected into blastocysts of a different mouse strain to produce mice with mosaic nDNA. However, it remained unclear whether this mtDNA mutation was transferred (7). In the laboratory of Eric Shoubridge, Jenuth et al. successfully transferred enucleated cells (cytoplasts) derived from one embryo into the embryo of a different mouse strain to produce heteroplasmic animals (8). By tracking the distribution of these neutral mtDNA polymorphisms, Jenuth et al. identified a low number of segregating units, about 200, in the female germline and, surprisingly, demonstrated variable tissue-specific and age-related segregation patterns depending on the particular mtDNA genotype (8, 9).
Last year, two groups produced chimeric transmitochondrial mice harboring mutant CAPR mtDNA by transfecting ES cells (10, 11). Marchington et al. fused enucleated cells harboring high levels of the CAPR mutation (A2379T) with female ES cells, which were injected into donor blastocysts. The resulting chimeric mice harbored variably low levels of the CAPR mutation in tissues (up to 6%) (10). By contrast, Levy et al. fused cytoplasts with ES cells depleted of mitochondria by exposure to the lipophilic dye rhodamine-6-G and produced chimeric mice with up to 50% of the T2433C CAPR mutation in tissues (11).
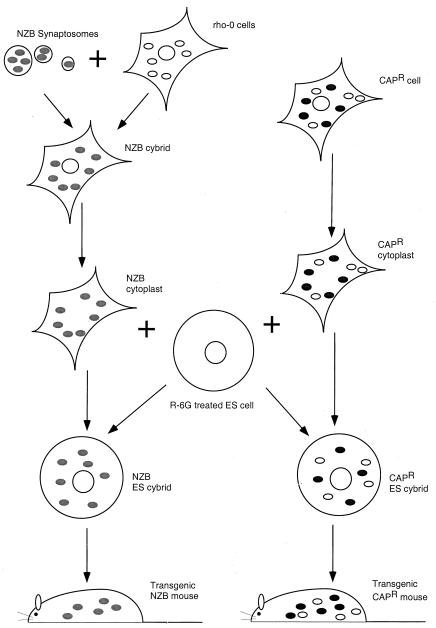
There are important differences between these pioneering attempts to obtain transmitochondrial mice and the present work by Sligh et al. (5). Starting with transgenic chimeric female mice, Sligh et al. have been able to transmit mtDNA mutations to succeeding generations and, even more notably, have produced heteroplasmic mice with sufficiently high levels of the CAPR mutation to express phenotypic abnormalities (Fig. 1). In addition, they have trapped mtDNA from synaptosomes of NZB mice by using ρ0 cells to produce cybrids that were enucleated and fused with the rhodamine-6-G cells to produce another transmitochondrial mouse (Fig. 1). A similar strategy was used by Inoue et al. to produce still another transmitochondrial mouse to capture synaptosome mtDNA in ρ0 cells (6).
Figure 1.
Experimental strategy used by Sligh et al. (5) to produce two types of transmitochondrial mice. To produce transmitochondrial mice with NZB mtDNA (indicated by gray ovals), synaptosomes from NZB mice were fused with ρ0 cells (mitochondria lacking DNA; indicated by empty ovals) to generate cybrids harboring NZB mitochondria. These cybrids were enucleated and fused with ES cells depleted of mitochondria with rhodamine-6G (R-6G). The resulting ES cybrids containing NZB mitochondria were injected into C57BL/6J blastocysts to produce chimeric and ultimately transmitochondrial mice (NZB mtDNA with C57BL/6J nDNA). To produce mice with heteroplasmic mtDNA mutation conferring CAPR (mitochondria with mutation; indicated by black ovals), cybrids harboring the mutation were enucleated and fused with R-6G-treated ES cells.
The CAPR chimeric mice showed congenital cataracts, retinal abnormalities, and optic nerve hamartomas, whereas their heteroplasmic and homoplasmic CAPR mutant progeny were more severely affected, dying in utero or within 11 days of birth. Histopathological studies of the CAPR mutant mice revealed dilated cardiomyopathy and degenerative changes with abnormal mitochondria in skeletal and cardiac muscle. Importantly, these CAPR mice prove the principle that transmitochondrial mice harboring pathogenic mtDNA point mutations can be produced and transmitted through germlines with sufficiently high levels of mutation to express a clinical phenotype. Nevertheless, as with any transgenic animal models, relevance of the transmitochondrial mice to human diseases must be assessed. For example, correlations between pathological changes and levels of heteroplasmy in various organs may demonstrate higher levels of mutation in severely affected tissues, as typically observed in human mitochondrial disorders.
The CAPR T2433C mutation is in the 3′-end of the16S rRNA gene. The human mtDNA mutation that most closely resembles the mouse mutation is the A1555G mutation in the 3′-end of the 12S rRNA gene, which is associated with aminoglycoside-induced and nonsyndromic deafness (12). The A1555G mutation is located at the base of a stem–loop structure; the homologous structure in bacteria is part of the aminoacyl site, where mRNAs are decoded (12). The human mutation appears to cause a protein translation defect that can be exacerbated by aminoglycosides (13, 14). Although both human and mouse mutations affect mitochondrial protein synthesis, and consequently the respiratory chain, the pathomechanisms for the translational defect probably differ and the clinical phenotypes are dramatically divergent, with one exception. A single family harboring heteroplasmic A1555G mtDNA mutation showed maternally inherited cardiomyopathy, which may be analogous to the cardiomyopathy seen in the CAPR mice (15). Clearly, additional studies of the CAPR mice must be performed to demonstrate defects in mitochondrial protein synthesis and respiratory chain functions.
In contrast to the CAPR mice, the transmitochondrial mice produced by Inoue and colleagues harbor large-scale rearrangements of mtDNA, which is directly analogous to the mutations typically associated with Kearns–Sayre syndrome, sporadic chronic progressive external ophthalmoplegia, and Pearson syndrome (6, 16–18). Most mice harboring high proportions of the mtDNA deletion died of kidney failure, a surprising finding because renal dysfunction is atypical in human mitochondrial diseases (6, 19). Nevertheless, the mice did reveal a mosaic pattern of cytochrome c oxidase-deficient and ragged-red fibers in muscle and lactic acidosis, indicative of mitochondrial dysfunction (6).
The transmitochondrial mice produced by Sligh and colleagues and by Inoue and colleagues have admirably proven that, in principle, heteroplasmic mtDNA mutations can be transmitted through germlines and will produce phenotypic abnormalities (5, 6). These animal models offer the promise of a greater understanding of the basic biology of mtDNA in normal and disease states and will allow more effective testing of therapies. Certainly, transmitochondrial mice may provide new avenues of research and indicate that the answer to the question, “Mutations of mtDNA: Are we scraping the bottom of the barrel?” is a resounding “no” (1).
Acknowledgments
I thank Drs. Salvatore DiMauro, Eric A. Schon, and Eduardo Bonilla for their thoughtful insights. This work was supported by grants from the National Institutes of Health (R01-HD37529 and R01-HL596457) and the Muscular Dystrophy Association.
Footnotes
See companion article on page 14461 in issue 26 of volume 97.
References
- 1.DiMauro S, Andreu A. Brain Pathol. 2000;10:431–441. doi: 10.1111/j.1750-3639.2000.tb00275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.King M P, Attardi G. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- 3.Larsson N-G, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh G S, Clayton D A. Nat Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 4.Wallace D C. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 5.Sligh J E, Levy S E, Waymire K G, Allard P, Dillehay D L, Nusinowitz S, Heckenlively J R, MacGregor G R, Wallace D C. Proc Natl Acad Sci USA. 2000;97:14461–14466. doi: 10.1073/pnas.250491597. . (First Published December 5, 2000; 10.1073/pnas.250491597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inoue K, Nakada K, Ogura A, Isobe K, Goto Y, Nonaka I, Hayashi J I. Nat Genet. 2000;26:176–181. doi: 10.1038/82826. [DOI] [PubMed] [Google Scholar]
- 7.Watanabe T, Dewey M J, Mintz B. Proc Natl Acad Sci USA. 1978;75:5113–5117. doi: 10.1073/pnas.75.10.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jenuth J P, Peterson A C, Fu K, Shoubridge E A. Nat Genet. 1996;14:146–151. doi: 10.1038/ng1096-146. [DOI] [PubMed] [Google Scholar]
- 9.Jenuth J P, Peterson A C, Shoubridge E A. Nat Genet. 1997;16:93–95. doi: 10.1038/ng0597-93. [DOI] [PubMed] [Google Scholar]
- 10.Marchington D R, Barlow D, Poulton J. Nat Med. 1999;5:957–960. doi: 10.1038/11403. [DOI] [PubMed] [Google Scholar]
- 11.Levy S E, Waymire K G, Kim Y L, MacGregor G R, Wallace D C. Transgenic Res. 1999;8:137–145. doi: 10.1023/a:1008967412955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prezant T R, Agapian J V, Bohlman M C, Bu X, Oztas S, Qiu W-Q, Arnos K S, Cortopassi G A, Jaber L, Rotter J I, Shohat M, Fischel-Ghodsian N. Nat Genet. 1993;4:289–294. doi: 10.1038/ng0793-289. [DOI] [PubMed] [Google Scholar]
- 13.Guan M X, Fischel-Ghodsian N, Attardi G. Hum Mol Genet. 1996;5:963–971. doi: 10.1093/hmg/5.7.963. [DOI] [PubMed] [Google Scholar]
- 14.Inoue K, Takai D, Soejima A, Isobe K, Yamasoba T, Oka Y, Goto Y, Hayashi J. Biochem Biophys Res Commun. 1996;223:496–501. doi: 10.1006/bbrc.1996.0923. [DOI] [PubMed] [Google Scholar]
- 15.Santorelli F M, DiMauro S, Manta P, Tanji K, Casali C, Hays A P, Addonizio L C, Mancini D M, Hirano M. Am J Hum Genet. 1999;64:295–300. doi: 10.1086/302188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holt I J, Harding A E, Morgan Hughes J A. Nature (London) 1988;331:717–719. doi: 10.1038/331717a0. [DOI] [PubMed] [Google Scholar]
- 17.Zeviani M, Moraes C T, DiMauro S, Nakase H, Bonilla E, Schon E A, Rowland L P. Neurology. 1988;38:1339–1346. doi: 10.1212/wnl.38.9.1339. [DOI] [PubMed] [Google Scholar]
- 18.Rötig A, Colonna M, Bonnefont J P, Blanche S, Fischer A, Saudubray J M, Munnich A. Lancet. 1989;1:902–903. doi: 10.1016/s0140-6736(89)92897-3. [DOI] [PubMed] [Google Scholar]
- 19.Shoubridge E A. Nat Genet. 2000;26:132–134. doi: 10.1038/79832. [DOI] [PubMed] [Google Scholar]