It has been an interesting decade in human evolutionary genetics. We have gone from hope to confidence and from confidence to despair. A series of recent studies, two of which appear in this and a recent issue of PNAS (1, 2), gives new grounds for optimism.
The initial hope to which I refer was a product of the first large-scale studies of variation in human mitochondrial DNA (3). For years, population geneticists had been studying genetic variants that were either the same or different. There was no meaningful measure of the magnitude of difference between two alleles. The only clock-like process we had to work with was genetic drift, and this clock does not keep accurate time. It ticks at a rate that varies as the population grows or shrinks. Knowing little about past population sizes, we were left with no useful measure of time.
All this changed with the first large-scale surveys of variation in mitochondrial DNA. With these data, we could measure the differences between alleles, and there was good reason to think that these differences accumulated at a fairly constant rate. Genetics began immediately to play a more important role in the study of human history. Various authors argued early on that the human population must have passed through a bottleneck—a period of small population size—sometime during the late Pleistocene (4–8).
The next few years saw improvements both in the quantity of mitochondrial data available and in the statistical methods available for dealing with those data. By the mid-1990s, we were able not only to reject the hypothesis of stationary population size but also to place an upper bound on the preexpansion population size, a lower bound on the postexpansion size, and both bounds on the time of the expansion (9–11). The evidence indicated that, sometime between 30,000 and 130,000 years ago, our ancestors expanded to fill the globe from an initial population of roughly 10,000 breeding individuals. Then, just as this story seemed to be gaining momentum, the bottom fell out from under it.
The problem is that natural selection and population growth can be hard to tell apart.
In the late 1990s, as DNA sequencing got cheaper, people began assembling large data sets from the human nuclear genome. The trouble was that each genetic locus seemed to tell a different story (12). The mitochondrial story received support from data from the Y chromosome and from extensive sets of short tandem repeat (STR) loci distributed throughout the genome (13–16). But other genes seemed to imply a long history of constant population size (17), and still others suggested the action of balancing selection or of geographic population structure (18). How can a single species have genes with such disparate histories? Presumably because natural selection has affected different loci in different ways (19, 20). And if the pattern we see is telling us mainly about the history of selection, then it is unlikely ever to tell us much about the history of population size.
This was not, of course, the first time that anyone had suggested a role for natural selection in the evolution of human mitochondria (21–24). The problem is that selection and population growth can be hard to tell apart. A favorable mutation may sweep through the population under the influence of natural selection. If we focus on the carriers of this favorable mutation, the process looks just like population growth: the number of carriers is small at first, then increases, and then levels off. For practical purposes, the two processes have identical effects on genetic variation. There is still no clean way of distinguishing them except by comparing DNA from different genetic loci. Population growth should affect every locus in the same way, whereas selection should affect different loci in different ways. The disparate results that we see from different loci suggest that human genetic variation is influenced strongly by natural selection. If this view is correct, genetics may have little to tell us about population history.
This view, however, may be unnecessarily gloomy. There are two aspects of the emerging pattern that are puzzling under the view that it is all a product of natural selection (25): First, the genetic loci that show the signature of a selective sweep are precisely the ones that, on a priori grounds, seem most likely to be neutral—the sweeps all seem to have occurred in DNA that does not code for protein. It is coding only regions that seem clearly consistent with selective neutrality and constant population size. This pattern is not as implausible as it may sound: Because of linkage, the signature of a selective sweep may extend from the coding region on which selection has acted into the noncoding regions that surround it. Nonetheless, it is surprising that the effects of selection should be most clearly visible in those portions of the genome that do not code for protein. It is also hard to imagine that selection at linked loci was responsible for the pattern seen in the STRs, because these are distributed widely throughout the genome.
There is another puzzling observation, which has to do with the timing of the presumptive selective sweeps. It has been possible, for several data sets, to estimate the time at which the (presumptive) selective sweep occurred, and in each case the sweep appears to have occurred at roughly the same time, in the late Pleistocene (25). This rough simultaneity would be expected under the hypothesis of a population expansion, but it is surprising under the selection hypothesis. In view of all this, two sets of authors have proposed a hypothesis that may seem far-fetched: that a population expansion has indeed occurred, but that its effects have been obscured by pervasive balancing selection within the coding portions of the nuclear genome (20, 25). This hypothesis seems far-fetched because, for the past 30 years, most geneticists have downplayed the importance of balancing selection as a cause of genetic variation. Although Harpending and I (25) argued that the case is at least plausible, we were far from convinced that it is true. We have therefore watched with great interest as, during the past 12 months, data from several additional noncoding genetic systems have become available.
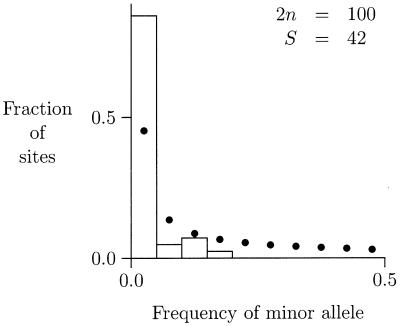
One of these data sets, that of Alonso and Armour (1), appears in this issue of PNAS. It is from a noncoding region within a functional gene (an intron). To explain its import, I will concentrate on what is known as the “site frequency spectrum.” A polymorphic nucleotide site is ordinarily present in only two states within a sample; let us call the rarer of these the “minor allele.” The site frequency spectrum is a histogram showing the fraction of polymorphic sites at which the minor allele is present in one copy, in two copies, and so on. Fig. 1 shows the site frequency spectrum of Alonso and Armour (1). It summarizes the same data as their figure 2, but without separating the data by continent. The rectangles in my figure show the observed spectrum, and the bold dots show the spectrum that is expected under a model of selective neutrality and constant population size. There is a clear excess, compared with the neutral model, of sites at which the minor allele is rare.
Figure 1.
Site frequency spectrum of Alonso and Armour (1). Open rectangles show the observed spectrum, with the horizontal axis aggregated into 10 bins of width 0.05. The bold dots show the spectrum expected under the infinite sites model with no selection and constant population size. 2n is the number of chromosomes sampled, S is the number of segregating sites, and the horizontal axis is the frequency of the minor (i.e., rarest) allele at a nucleotide site.
This excess is typical of populations that have undergone an expansion. It reflects the fact that small populations maintain little genetic diversity. If our species were small before, say, 50,000 years ago, then most of its genetic diversity would have arisen after this time. Because most variants would then be recent, few of them would yet have drifted to high frequency within the population. Consequently, most variants would be rare, and we would see a pattern like that in Fig. 1.
Alonso and Armour (1) attack this problem by using various methods and leave little doubt that something has happened at this locus. But was that something a population expansion or a selective sweep? Alonso and Armour argue in favor of a population expansion, pointing out that the DNA they sequenced is flanked by regions of high recombination and is therefore unlikely to be tightly linked to the coding regions (exons) upstream and downstream. Another pair of authors, however, reach opposite conclusions in the face of very similar data. Nachman and Crowell (26) study DNA sequences from two introns within the human Duchenne muscular dystrophy gene, which is another region of high recombination. In one intron, there is an excess of rare alleles, like that seen in Fig. 1; in the other, there is no such excess. Nachman and Crowell suggest that the first intron reflects a selective sweep, the second a long history of constant population size. But how can we be sure that the first intron does not reflect an expansion of population size and the second a long history of balancing selection?
In view of these ambiguities, it is reassuring that the signature of population growth has shown up recently in other studies of noncoding DNA. Three new studies of microsatellites have produced evidence of a population expansion (27–29). Similar results also emerge from studies of DNA sequence data. Zhao et al. (2) find a strong excess of low-frequency variants in sequence data from a 10-kb noncoding region on chromosome 22. Yu et al. (30) obtain the same result from a 10-kb noncoding region on chromosome 1. Of the noncoding regions that have been studied to date, all of those outside of functional genes show evidence of a population expansion, and so do several of those from introns. Within coding regions, on the other hand, no such evidence is consistently found. In short, the pattern that we noticed last year seems to be holding up.
The consistent pattern seen in noncoding DNA argues that we should take seriously once again the evidence for an expansion of human population size during the late Pleistocene. The consistent failure of coding DNA to reflect this pattern argues that we should take seriously the possibility that geneticists from the 1930s through the 1960s may have been right after all about the importance of balancing selection (31).
Acknowledgments
I thank Elizabeth Cashdan, Henry Harpending, Mark Jeffries, Lynn Jorde, Scott Watkins, and Stephen Wooding for comments; I thank Ning Yu and Wen-Hsiung Li for permission to mention a paper while it was in press.
Footnotes
References
- 1.Alonso S, Armour J A L. Proc Natl Acad Sci USA. 2001;98:864–869. doi: 10.1073/pnas.98.3.864. . (First Published December 19, 2000; 1.1073/pnas.011244998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao Z, Jin L, Fu Y-X, Ramsay M, Jenkins T, Leskinen E, Pamilo P, Trexler M, Patthy L, Jorde L B, et al. Proc Natl Acad Sci USA. 2000;97:11354–11358. doi: 10.1073/pnas.200348197. . (First Published September 26, 2000; 10.1073/pnas.200348197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cann R L, Stoneking M, Wilson A C. Nature (London) 1987;325:31–36. doi: 10.1038/325031a0. [DOI] [PubMed] [Google Scholar]
- 4.Brown W M. Proc Natl Acad Sci USA. 1980;77:3605–3609. doi: 10.1073/pnas.77.6.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haigh J, Maynard Smith J. Genet Res. 1972;19:73–89. doi: 10.1017/s0016672300014282. [DOI] [PubMed] [Google Scholar]
- 6.Jones J S, Rouhani S. Nature (London) 1986;319:449–450. doi: 10.1038/319449b0. [DOI] [PubMed] [Google Scholar]
- 7.Maynard Smith J. Nature (London) 1990;344:591–592. [Google Scholar]
- 8.Wills C. Nature (London) 1990;348:398. doi: 10.1038/348398a0. [DOI] [PubMed] [Google Scholar]
- 9.Rogers A R, Harpending H C. Mol Biol Evol. 1992;9:552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- 10.Harpending H C, Sherry S T, Rogers A R, Stoneking M. Curr Anthropol. 1993;34:483–496. [Google Scholar]
- 11.Rogers A R. Evolution (Lawrence, KS) 1995;49:608–615. doi: 10.1111/j.1558-5646.1995.tb02297.x. [DOI] [PubMed] [Google Scholar]
- 12.Hey J. Mol Biol Evol. 1997;14:166–172. doi: 10.1093/oxfordjournals.molbev.a025749. [DOI] [PubMed] [Google Scholar]
- 13.Di Rienzo A, Donnelly P, Toomajian C, Sisk B, Hill A, Petzl-Erler M L, Haines G K, Barch D H. Genetics. 1998;148:1269–1284. doi: 10.1093/genetics/148.3.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimmel M, Chakraborty R, King J P, Bamshad M, Watkins W S, Jorde L B. Genetics. 1998;148:1921–1930. doi: 10.1093/genetics/148.4.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reich D E, Goldstein D B. Proc Natl Acad Sci USA. 1998;95:8119–8123. doi: 10.1073/pnas.95.14.8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Underhill P A, Jin L, Lin A A, Mehdi S Q, Jenkins T, Vollrath D, Davis R W, Cavalli-Sforza L L, Oefner P J. Genome Res. 1997;7:996–1005. doi: 10.1101/gr.7.10.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harding R M, Fullerton S M, Griffths R C, Bond J, Cox M J, Schneider J A, Moalin D S, Clegg J B. Am J Hum Genet. 1997;60:722–789. [PMC free article] [PubMed] [Google Scholar]
- 18.Harris E E, Hey J. Proc Natl Acad Sci USA. 1999;96:3320–3324. doi: 10.1073/pnas.96.6.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Przeworski M, Hudson R R, Di Rienzo A. Trends Genet. 2000;16:296–302. doi: 10.1016/s0168-9525(00)02030-8. [DOI] [PubMed] [Google Scholar]
- 20.Wall J D, Przeworski M. Genetics. 2000;155:1865–1874. doi: 10.1093/genetics/155.4.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Excoffer L. J Mol Evol. 1990;30:125–139. doi: 10.1007/BF02099939. [DOI] [PubMed] [Google Scholar]
- 22.Merriwether D A, Clark A G, Ballinger S W, Schurr T G, Soodyall H, Jenkins T, Sherry S T, Wallace D C. J Mol Evol. 1991;33:543–555. doi: 10.1007/BF02102807. [DOI] [PubMed] [Google Scholar]
- 23.Templeton A R. Am Anthropol. 1993;95:51–72. [Google Scholar]
- 24.Wise C, Sraml M, Easteal S. Genetics. 1998;148:409–421. doi: 10.1093/genetics/148.1.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harpending H C, Rogers A R. Annu Rev Genom Hum Genet. 2000;1:361–385. doi: 10.1146/annurev.genom.1.1.361. [DOI] [PubMed] [Google Scholar]
- 26.Nachman M W, Crowell S L. Genetics. 2000;155:1855–1864. doi: 10.1093/genetics/155.4.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gonser R, Donnelly P, Nicholson G, Di Rienzo A. Genetics. 2000;154:1793–1807. doi: 10.1093/genetics/154.4.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhivotovsky L A, Bennett L, Bowcock A M, Feldman M W. Mol Biol Evol. 2000;17:757–767. doi: 10.1093/oxfordjournals.molbev.a026354. [DOI] [PubMed] [Google Scholar]
- 29.Pritchard J K, Seielstad M T, Perez-Lezuan A. Mol Biol Evol. 1999;16:1791–1798. doi: 10.1093/oxfordjournals.molbev.a026091. [DOI] [PubMed] [Google Scholar]
- 30.Yu, N., Zhao, Z., Fu, Y.-X., Sambuughin, N., Ramsay, M., Jenkins, T., Leskinen, E., Patthy, L., Jorde, L. B. & Li, W.-H. (2001) Mol. Biol. Evol, in press. [DOI] [PubMed]
- 31.Ford E. Ecological Genetics. London: Metheun; 1964. [Google Scholar]