Abstract
Localized Gurken (Grk) translation specifies the anterior–posterior and dorsal–ventral axes of the developing Drosophila oocyte; spindle-class females lay ventralized eggs resulting from inefficient grk translation. This phenotype is thought to result from inhibition of the Vasa RNA helicase. In a screen for modifiers of the eggshell phenotype in spn-B flies, we identified a mutation in the lnk gene. We show that lnk mutations restore Grk expression but do not suppress the persistence of double-strand breaks nor other spn-B phenotypes. This suppression does not affect Egfr directly, but rather overcomes the translational block of grk messages seen in spindle mutants. Lnk was recently identified as a component of the insulin/insulin-like growth factor signaling (IIS) and TOR pathway. Interestingly, direct inhibition of TOR with rapamycin in spn-B or vas mutant mothers can also suppress the ventralized eggshell phenotype. When dietary protein is inadequate, reduced IIS–TOR activity inhibits cap-dependent translation by promoting the activity of the translation inhibitor eIF4E-binding protein (4EBP). We hypothesize that reduced TOR activity promotes grk translation independent of the canonical Vasa- and cap-dependent mechanism. This model might explain how flies can maintain the translation of developmentally important transcripts during periods of nutrient limitation when bulk cap-dependent translation is repressed.
Key words: IRES, TOR, Gurken, lnk, Translation
Introduction
Reproduction represents a substantial energy investment for an organism. Many studies have shown that ovarian physiology is exquisitely sensitive to nutritional status. Limitation of dietary protein intake results in a dramatic slowing of egg chamber maturation through developmental arrest, programmed cell death or loss of germline stem cells (Drummond-Barbosa and Spradling, 2001; Hsu et al., 2008; LaFever and Drummond-Barbosa, 2005). Several signaling pathways are integrated to bring about this response including 20-hydroxyecdysone, Juvenile Hormone (JH), and insulin/insulin-like signaling (IIS). IIS is stimulated by protein feeding and is required for oogenesis to progress. The IIS pathway integrates nutritional signals at two distinct points during oogenesis. The first is in region 2A of the germarium where developing germline cysts undergo apoptosis in the absence of a source of maternal dietary protein. The second point of nutritional control is at stage 8 of oogenesis during the onset of vitellogenesis. In the absence of food, egg chambers develop to stage 8, where they are arrested until a favorable food source is located. These two checkpoints represent points at which the energetically expensive process of oogenesis can be halted if insufficient resources are available (Hsu et al., 2008; LaFever and Drummond-Barbosa, 2005).
The IIS pathway elicits its effect on Drosophila physiology through several effector pathways, namely the Drosophila Foxo transcription factor and the kinase Target of Rapamycin (TOR) (Grewal, 2009; Teleman, 2010). IIS inhibits Foxo activity by promoting its phosphorylation by PKB (Akt) and subsequent exclusion from the nucleus. Starvation or mutations in the insulin pathway allow Foxo to translocate to the nucleus where it directs the transcription of genes that promote longevity, stress resistance, fat storage and growth attenuation (Hwangbo et al., 2004; Giannakou et al., 2004; Junger et al., 2003). TOR activity is stimulated by both IIS, through the Rheb GTPase, and by amino acids through Rag GTPases (Grewal, 2009; Gao and Pan, 2001; Kim et al., 2008; Sancak et al., 2008). When nutrients are plentiful, high TOR activity stimulates the translation of mRNA by phosphorylating S6K, which in turn phosphorylates eIF4B and promotes its interaction with eIF3 (Holz et al., 2005). These steps are crucial for recruiting the translation preinitiation complex (PIC) to the m7G cap at the 5′-end of the mRNA. Once bound, the PIC recruits the small ribosomal subunit and proceeds to scan the transcript for an initiating AUG codon. This process requires the activity of the eIF4A RNA helicase (Sonenberg and Hinnebusch, 2009). TOR also phosphorylates and inactivates the inhibitory eIF4E-binding protein (4EBP). Starvation inhibits cap-dependent translation through reduced TOR activity. When nutrients are limiting and TOR activity is low, eIF4B is not phosphorylated and can no longer participate in PIC assembly; furthermore, 4EBP inhibition is lifted and it proceeds to inhibit cap-recognition by eIF4E (Richter and Sonenberg, 2005). Both activities have the effect of strongly blocking cap-dependent translation initiation when nutrients are scarce. A select few transcripts escape this translational block by upregulating the utilization of an alternative mechanism that relies on an internal ribosomal entry site (IRES) that obviates the requirement for cap recognition and start codon scanning. The list of transcripts that contain IRES sequences is growing (Mokrejs et al., 2009) and includes numerous growth factors such as VEGFA (Huez et al., 2001), FGF2 (Arnaud et al., 1999), PDGF2 (Bernstein et al., 1997) and IGF2 (Pedersen et al., 2002). A prominent example of IRES-mediated nutritional adaptation is the Drosophila insulin receptor InR, the translation of which is upregulated in response to starvation as a way to sensitize the cell to insulin when nutrients become available (Marr et al., 2007).
Control of translation is vitally important to developmental patterning. The transcripts of many morphogens, including nanos, oskar and gurken, are co-transcriptionally packaged into silencing particles and transported in a translationally quiescent form (Tomancak et al., 1998; Besse and Ephrussi, 2008; Chekulaeva et al., 2006; Martin and Ephrussi, 2009; Delanoue et al., 2007; Norvell et al., 1999). Once localized, this repression is alleviated and translation proceeds in the developmentally appropriate locale. Gurken (Grk) is a TGF-α-related ligand for the Drosophila Egfr. Localized translation of the spatially restricted grk transcript results in signaling by germline-derived Grk to the Egfr in the overlying follicle cells. This signal is required to specify the posterior fate in early oogenesis and the dorsal fate during mid-oogenesis (Gonzalez-Reyes et al., 1995; Roth and Lynch, 2009). Mutations that reduce grk translation are female sterile, owing to their inability to correctly pattern the developing oocyte, and result in concomitant patterning defects in the embryo. grk translation requires the eIF4A-related DEAD-box helicase Vasa (encoded by vas). Mutations in vas are female sterile owing to a failure to specify dorsal structures in the eggshell or posterior structures in the embryo (Tomancak et al., 1998; Styhler et al., 1998; Lasko and Ashburner, 1988; Tinker et al., 1998; Schüpbach and Wieschaus, 1986).
spindle-class genes are responsible for repairing DNA double-strand breaks (DSBs), which are induced during homologous recombination in Drosophila oogenesis (Jang et al., 2003; McKim et al., 2002; Ghabrial et al., 1998; Staeva-Vieira et al., 2003). In wild-type females, DSBs are induced in germline cells entering pachytene in region 2A of the germarium. This process is initiated by the Spo11 homologue Mei-W68 and Mei-P22, a protein that aids in break site selection (Liu, H. et al., 2002; McKim and Hayashi-Hagihara, 1998). These breaks are then repaired by homologous recombination, a process that requires the RAD51 homologue spindle-B (spn-B). Mutations in spn-B result in an accumulation of unrepaired DSBs that lead to the activation of a meiotic checkpoint (Ghabrial et al., 1998; Ghabrial and Schüpbach, 1999; Jang et al., 2003). The checkpoint is comprised of the ATR homologue mei-41 and the downstream kinase chk2 (also known as lok) (Ghabrial et al., 1998; Ghabrial and Schüpbach, 1999; Abdu et al., 2002; Jang et al., 2003). Persistent DSBs in spn-BBU females activate the checkpoint that requires the Mei-41 and Chk2 kinases and leads to inefficient grk translation and ventralized eggshell phenotypes. Checkpoint activation also results in phosphorylation of Vasa, a modification that is thought to inhibit its function (Ghabrial and Schüpbach, 1999; Abdu et al., 2002). Early in oogenesis, the oocyte nucleus becomes arrested in pachytene and forms a compact structure called the karyosome. The formation of the karyosome is disrupted in spindle-class mutants where the chromatin appears fractured or ellipsoid (Ghabrial et al., 1998; Abdu et al., 2002). Weak grk translation and an inability to properly form the karyosome are both spindle phenotypes that are consistent with reduced Vasa activity (Tomancak et al., 1998).
In the current study, we identified the SH2B family adaptor gene lnk in a genetic screen for modifiers of the ventralized eggshell phenotype seen in spn-BBU mutant flies. SH2B proteins are known to regulate intracellular signaling by membrane-bound receptor tyrosine kinases (RTKs). SH2Bs can promote signaling by scaffolding downstream effectors to the RTK or mediate proteosomal receptor destruction by recruiting the Cbl ubiquitin ligase. Lnk was recently identified as a positive regulator of the IIS pathway that functions at the level of the insulin receptor substrate Chico (Slack et al., 2010; Werz et al., 2009). Here, we show that lnk mutations can promote grk translation and suppress the ventralized eggshell phenotype in a spn-BBU mutant background. This suppression is independent of Vasa activity and does not suppress the karyosome phenotype. We did not find any genetic interactions with a weak grk allele or downstream targets of Egfr, suggesting that lnk-mediated suppression of spindle phenotypes does not occur by directly modulating Egfr activity. Our data suggest that lnk mutations promote grk translation by inhibiting TOR activity as Rapamycin-feeding experiments can also suppress the eggshell phenotype of spn-B and vas mutant flies. We propose a model in which reduced IIS and/or TOR signaling inhibits cap-dependent translation and promotes utilization of an alternative translation initiation mechanism of the grk mRNA. This mechanism enables flies to faithfully pattern their oocytes when nutrients are scarce.
Results
lnk mutations suppress dorsal–ventral patterning defects in spn-B mutants
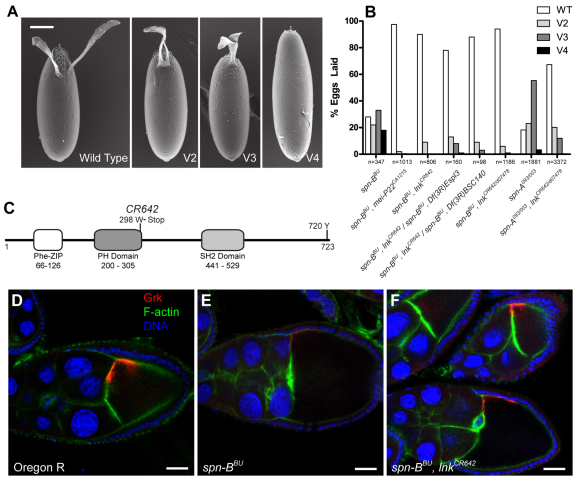
To identify factors that regulate the interaction between the meiotic checkpoint and grk translation, we conducted an ethyl methanesulfonate (EMS) screen on the third chromosome for mutations that could suppress the ventralized eggshell phenotype seen in spn-BBU females (Fig. 1A). Two lines, CA1215 and CR642, yielded strong suppression and produced a majority of wild-type eggs despite being mutant for spn-BBU (Fig. 1B). The mutations were positionally cloned and the molecular lesions were identified. CA1215 is an allele of the mei-P22 gene. mei-P22 is required to initiate DSB formation in pachytene, and mutations in mei-P22 suppress ovarian phenotypes associated with spn-BBU mutations (Ghabrial and Schüpbach, 1999; Liu, H. et al., 2002). In agreement with previous studies, mei-P22CA1215, spn-BBU mutants laid eggs with wild-type dorsal–ventral polarity, normal Grk expression and no indication of DSB formation in the germarium (Fig. 1B and Fig. 5D). This result validated our experimental approach and illustrated that we were able to isolate alleles of genes known to be involved in the meiotic recombination process.
Fig. 1.
lnk mutations suppress dorsal–ventral patterning defects in eggs laid by spn-B flies. Mutations in spindle-class genes result in female sterility and variable ventralization of the eggs laid by homozygous females. (A) Scanning electron microscopy images of eggs laid by spn-BBU flies demonstrating a range of ventralization phenotypes. Dorsal–ventral polarity was classified as wild type if two distinct dorsal appendages were evident. Fusion at the base of the appendages was classified as mild class 2 ventralization (V2), whereas fusion past the anterior aspect of the egg when viewed dorsally was classified as moderate class 3 ventralization (V3). Severely ventralized eggs lacking all appendage material were designated as V4. Scale bar: 100 μm. (B) Genetic suppression of the spn-BBU ventralized eggshell phenotype by mutations in mei-P22CA1215 and several allelic combinations with lnkCR642. Mutations in lnk are also able to suppress dorsal–ventral patterning defects in eggs laid by spn-A093/003 mutant females. (C) A domain map of the Lnk protein illustrating the location of the CR642 mutation and conserved tyrosine phosphorylation site. (D–F) The eggshell suppression reflects a rescue of Grk protein expression in the ovary. Grk is stained green; F-actin is shown in red. Scale bars: 25 μm.
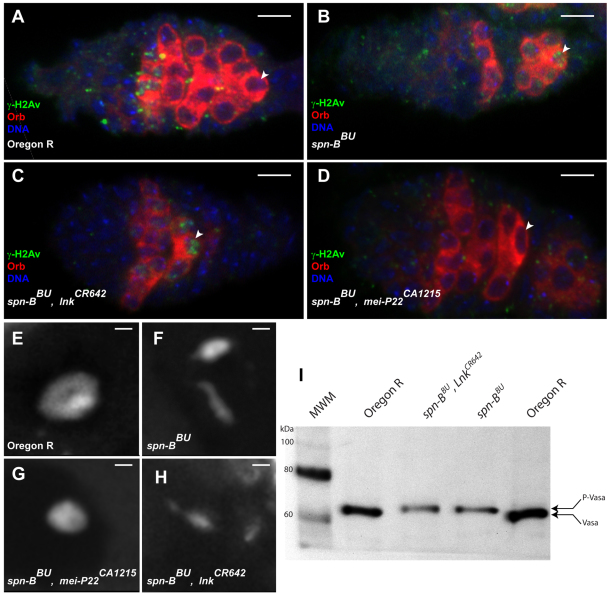
Fig. 5.
lnk suppression does not affect DNA repair or vas phenotypes. (A–D) Immunostaining of γ-H2Av foci (green) in germaria. Orb (red) stains individual cysts in the germarium and accumulates in the oocyte in region 2B. DNA is shown in blue. γ-H2Av foci appear in region 2A of wild-type germaria and are resolved in oocytes in region 2B and 3 (arrowheads) whereas DSBs are evident in oocytes between region 2B and stage 4 in spn-BBU ovarioles. Although spn-BBU, mei-P22CA1215 and spn-BBU, lnkCR642 flies both lay wild-type eggs, only mei-P22CA1215 does so by blocking the formation of DSBs. spn-BBU, lnkCR642 germaria have persistent DSBs in region 3 to a similar extent as those in spn-BBU. (E–H) A single confocal section of the karyosome from mid-stage egg chambers stained with Hoechst 33342 reveals that the karyosome fails to properly condense in spn-BBU, lnkCR642 oocytes and resembles the phenotype seen in spn-BBU. (I) Western blot of Vasa protein from ovarian extracts reveals that the electrophoretic mobility of Vasa from spn-BBU, lnkCR642 retains the modification seen in spn-BBU. Scale bars: 5 μm (A–D), 1 μm (E–H).
CR642, a second strong suppressor mutation, was mapped to the lnk locus and found to be a G to A transition mutation at nucleotide 894 of the lnk open reading frame (Fig. 1C). Lnk is a member of the SH2B adapter family that is known to modulate RTK signaling. Three functional domains have been described, including a phenylalanine zipper dimerization domain, a plextrin homology domain and an SH2 domain. There is also a conserved C-terminal tyrosine (Y720) that is required for binding to the Cbl ubiquitin ligase in mammalian homologues (Hu and Hubbard, 2005). The lnkCR642 allele results in a nonsense codon after serine 297 that truncates the protein leaving the dimerization domain and most of the PH domain intact. The lnkCR642 allele was tested in trans to the Df(3R)Espl3 and Df(3R)BSC140 deficiencies as well as with transposon integration alleles. The lnkf02642 and lnkf05062 alleles are PBac{WH} insertions in the first intron of the coding sequence, whereas the lnkd07478 allele is a P{XP} insertion in the 5′-untranslated region (5′-UTR) (Thibault et al., 2004). Homozygous and hemizygous allelic combinations yielded similar levels of suppression with greater than 80% wild-type eggs being laid by spn-B, lnk double mutants. Notably, heteroallelic spn-BBU, lnkCR642/lnkd07478 females showed particularly strong suppression and laid 96% wild-type eggs in contrast to spn-BBU flies that only laid 28% wild-type eggs (Fig. 1B). lnkCR642/lnkd07478 also suppressed the spindle-A mutation, indicating that this effect is not specific to spn-B. The suppression of spn-BBU reflects restored Grk protein levels in mid-oogenesis (Fig. 1D–F). Genomic lnk rescue constructs were able to reverse the suppression phenotype and produced a ventralization profile similar to that seen in spn-BBU flies (Fig 2E). Although loss of lnk activity was able to suppress the ventralized eggshell phenotype, eggs laid by spn-BBU, lnkCR642 females did not hatch. We have also observed that eggs laid by females homozygous mutant for lnkCR642 but wild-type for spn-B were patterned correctly, however 18% did not hatch.
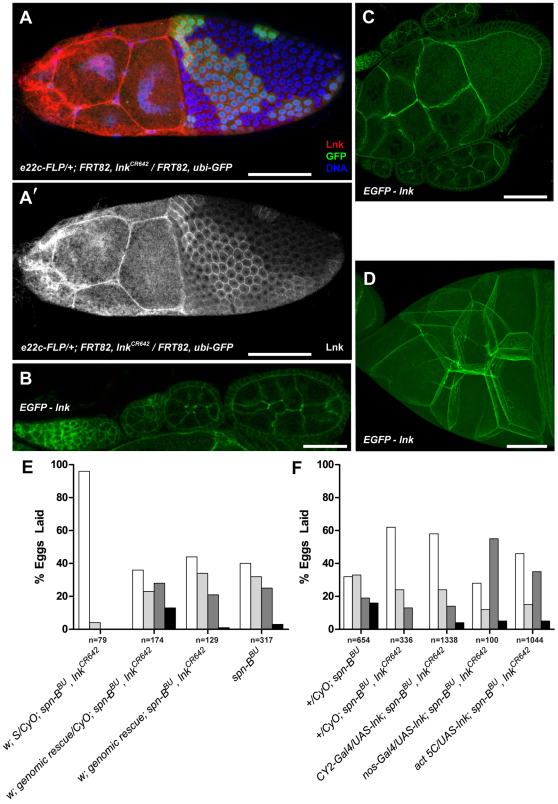
Fig. 2.
Lnk functions in both the germline and follicle cells. (A) Immunostaining of Lnk protein (red in A and white in A′) in follicle cell mosaics generated in e22c-FLP/+; FRT82, lnkCR642/FRT82, ubi-GFP flies. Clones are marked by the absence of EGFP and DNA was stained blue with Hoechst 33342. Images are a maximum intensity projection of multiple confocal sections. (B–D) Localization of an N-terminal EGFP–Lnk fusion construct expressed from the genomic promoter. B is a single confocal section through the germarium and early stage cysts, C is a section through a stage 10A cyst, and D shows a maximum intensity projection of multiple sections through the nurse cell cluster of a stage 10B egg chamber. (E) Expression of a genomic lnk rescue construct can restore a ventralized eggshell phenotype to spn-BBU, lnkCR642 flies. (F) nos-Gal4 or CY2-Gal4 were used to drive expression of a UAS-lnk construct in the germline or follicle cells respectively of spn-BBU, lnkCR642 flies. Act 5C-Gal4 was used to drive expression in both tissues. Germline expression of lnk resulted in a significant increase in the number of ventralized eggs whereas follicle cell expression did not. Eggshell classes are as described in Fig. 1. Scale bars: 50 μm (A,C,D), 25 μm (B).
Lnk is expressed in both germline and follicle cells
To determine where Lnk protein is expressed, we generated a rabbit polyclonal antibody against amino acids 56–542 of Lnk. Whole-mount immunostaining of lnkCR642 follicle cell clones revealed a membrane localization in heterozygous and twin spot follicle cells but a nearly undetectable level of expression in lnkCR642 clones (Fig. 2A,A′). Lnk protein was also strongly expressed in the nurse cells and accumulated at the membrane. Genomic EGFP-lnk fusion constructs recapitulated the localization seen by immunofluorescence (Fig. 2B–D). The level of expression in the germline was somewhat more robust than in follicle cells, especially in regions 1 and 2A of the germarium where nutritional control of germline cells is most pronounced. The localization of Lnk throughout the ovary prompted us to assess the functional requirements for Lnk expression. We generated a UAS-lnk expression construct and crossed it with the follicle cell driver CY2-Gal4 or the germline driver nos-Gal4 to determine which tissue mediated the spn-BBU suppression. Consistent with a role in the regulation of the germline morphogen Grk, nos-Gal4 driven UAS-lnk rescued the suppression seen in spn-BBU, lnkCR642 flies, whereas expression in the somatic follicle cells did not (Fig. 2F).
The rate of division of developing cysts in the germarium drops rapidly in flies with a protein-poor diet (Drummond-Barbosa and Spradling, 2001). These results are consistent with the role of SH2B proteins as signal transduction scaffolds that interface with membrane bound RTKs. If Lnk attenuates Egfr signaling, loss of this activity could potentiate signaling and counteract the inefficient grk translation seen in spn-BBU flies. Alternatively, the suppression phenotype might be related to the ability of lnk to regulate signaling by InR which modulates ovarian development in response to nutritional availability (Slack et al., 2010; Werz et al., 2009).
lnk does not directly modulate Egfr signaling in the ovary
In mid-oogenesis Grk is translated in the future dorsal anterior of the oocyte and interacts with Egfr to specify the dorsal fate in the follicle cell epithelium whose default fate is ventral (reviewed by Roth and Lynch, 2009). It has been shown that there is crosstalk between the IIS pathway and the Egfr in the developing eye (McNeill et al., 2008). Furthermore, mammalian SH2B proteins have been shown to recruit the Cbl ubiquitin ligase and promote clathrin-mediated endocytosis and recycling of ligand-bound receptors as a negative-feedback mechanism (Yokouchi et al., 1999; Ahmed and Pillay, 2001; Thien and Langdon, 2005). Mutations in Drosophila cbl lead to activation of the Egfr even in lateral and ventral follicle cells that are exposed to the lowest levels of the Grk gradient (Pai et al., 2000; Pai et al., 2006; Chang et al., 2008). Therefore we tested the hypothesis that Lnk can directly regulate the activity of the Egfr in ovarian follicle cells.
If lnk mutations were to sensitize the Egfr to Grk signaling, reduced Grk levels caused by spn-BBU mutations should have the same effect as a hypomorphic allele of the grk gene from the perspective of the follicle cells. To test this hypothesis, we made flies that were double-mutant for lnkCR642 and grkED22. The grkED22 allele is a missense mutation in the EGF domain that affects mRNA levels and might also reduce Egfr binding. grkED22 females lay eggs with a single appendage due to inefficient activation of the Egfr (Clifford and Schüpbach, 1989; Neuman-Silberberg and Schüpbach, 1993; Thio et al., 2000). Surprisingly, the eggs laid by grkED22; lnkCR642 flies are indistinguishable from those produced by grkED22 single-mutants and all have a single appendage (data not shown).
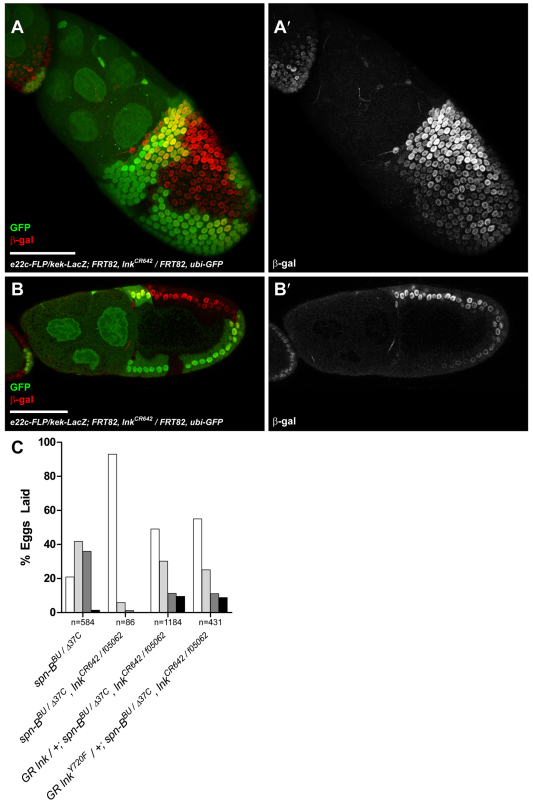
Mutations that promote Egfr signaling result in ectopic dorsal fates in follicle cell clones. For example, follicle cell clones of cbl mutations cause dorsalization of lateral domains of the eggshell due to hyperactivation of Egfr (Pai et al., 2000). If Lnk attenuates Egfr signaling by promoting Cbl-dependent Egfr endocytosis, mutations in lnk should also result in ectopic Egfr activation. Unlike cbl mutant clones, eggs laid by lnkCR642 mutant flies are indistinguishable from wild type (data not shown). This result was supported by looking directly at a LacZ enhancer trap for the Egfr transcriptional target kekkon (kek-LacZ) in lnkCR642 follicle cell clones. The intensity of kek-LacZ expression is proportional to the level of Egfr activity. In contrast to cbl mutant clones (Pai et al., 2000) no difference in kek-LacZ activity was observed across the border of lnk homozygous mutant, heterozygous or wild-type twin spot clones (Fig. 3A,B).
Fig. 3.
lnk mutations do not affect Egfr activity in the follicle cells. (A,B) Immunostaining of follicle cell clones in stage 10 e22c-FLP/kek-LacZ; FRT82, lnkCR642/FRT82, ubi-GFP ovaries. β-galactosidase (red) expression from the kek-LacZ enhancer trap reflects the gradient of Egfr activity in the follicle cells. Homozygous lnkCR642 cells are marked by the absence of EGFP. A is a maximum intensity projection of multiple confocal sections encompassing the peak of Egfr activity at the future dorsal anterior that declines towards the ventral and posterior axes; B is a single confocal section with the dorsal anterior on the top left. kek-LacZ expression is unaffected by the lnk genotype. Scale bars: 50 μm. (C) Cbl binds to the conserved tyrosine residue at 720 in SH2-B family members. Mutation of Lnk tyrosine 720 to a non-phosphorylatable phenylalanine residue in a genomic rescue construct does not affect the ability of the transgene to restore a ventralized eggshell phenotype in spn-BBU/spn-BΔ37C, lnkCR642/lnkf05062 flies. Eggshell classes are as described in Fig. 1.
To rule out further that the eggshell suppression was due to effects on Cbl, we generated lnk mutants at locations that are known to block interactions with Cbl in mammalian Lnk orthologues. Phosphorylation of the intracellular domain of tyrosine kinase receptors induces the association of APS family proteins. The human insulin receptor recruits APS (SH2B2) following insulin binding and subsequently phosphorylates APS at a C-terminal tyrosine residue. Phosphorylation of this tyrosine residue in APS is strictly required to recruit the tyrosine-kinase-binding domain of Cbl to the activated insulin receptor (Liu, J. et al., 2002). The context of a conserved tyrosine residue at position 720 in Drosophila Lnk conforms to the consensus binding site for Cbl and is identical (RAVxNQYS) to human APS (Hu and Hubbard, 2005). To determine whether this tyrosine residue is required to rescue lnk suppression of spn-BBU, we performed a rescue experiment with a wild-type lnk transgene or a lnkY720F allele driven by the genomic promoter. These rescue constructs were integrated into the attP40-docking site at 25C7 using the ΦC31 site-specific integrase system to control for position effects on transgene expression (Markstein et al., 2008; Venken and Bellen, 2007). Both alleles were able to rescue the suppression of spn-BBU/spn-BΔ37C, lnkCR642/lnkf05062 to a similar extent, indicating that phosphorylation of this residue is not crucial for recruiting Cbl activity and thus downregulation of Egfr in the follicle cells (Fig. 3C). Taken together, these data suggest that Lnk does not directly shape the gradient of Egfr activity in the follicle cells.
lnkCR642 reduces IIS and slows the rate of oogenesis
Lnk is a positive regulator of IIS in Drosophila (Werz et al., 2009; Slack et al., 2010). On the basis of on epistatic analysis, Lnk has been shown to function upstream of phosphoinositide 3-kinase (PI3K) at the level of the Chico insulin receptor substrate. chico1/chico2, lnk4Q3/lnk6S2 double-mutants exhibit synthetic lethality that can be suppressed by heterozygosity of the pten phosphatidylinositol-(3,4,5)-trisphosphate [PtdIns(3,4,5)P3] phosphatase (Werz et al., 2009). Reduction of IIS either by mutation or dietary restriction results in atrophic ovaries with a dearth of egg chambers older than stage 8. This nutritional control point coincides with the onset of the energetically expensive process of vitellogenesis and arrests development until adequate resources are available. We find that, although the P-element insertion lnkd07478 allele produces a classical IIS ovarian atrophy with minimal yolk production (Fig. 4B,E), lnkCR642 ovaries are highly vitellogenic and morphologically indistinguishable from wild type (Fig. 4C,F).
Fig. 4.
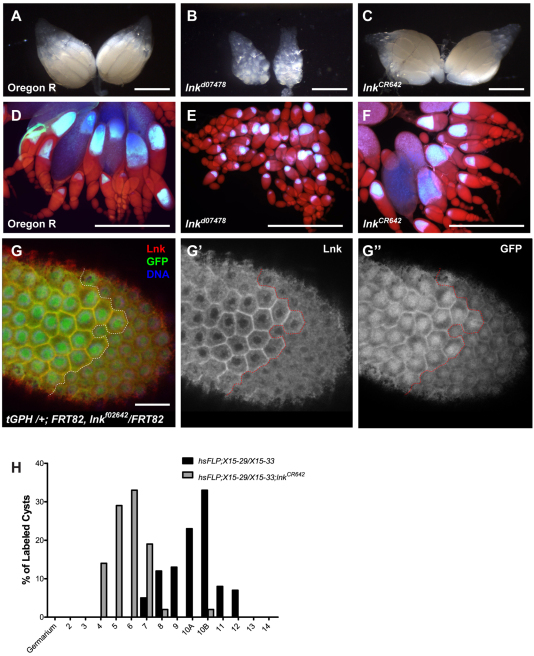
lnk mutations affect IIS and slow the rate of oogenesis. Whole ovaries were imaged from Oregon R (A,D), lnkd07478 (B,E) and lnkCR642 (C,F) flies. Ovaries in D–F were stained with Draq5 and imaged with UV epi-illumination. Following the onset of vitellogenesis, the oocyte autofluoresces blue. Note the strong arrest in early vitellogenesis in lnkd07478 ovaries, whereas many late stage egg chambers are evident in lnkCR642 ovaries. (G) Membrane localization of the tGPH reporter is sensitive to PtdIns(3,4,5)P3 levels. A follicle cell clone from tGPH /+; FRT82, lnkf02642/FRT82 ovaries stained with anti-Lnk antibodies (red, and G′) illustrating the reduced membrane localization of tGPH (green, G″) in cells lacking Lnk activity. The clone border is illustrated by the dotted line with heterozygous tissue on the left and lnkf02642 homozygous cells on the right. (H) Linage tracing in wild-type and lnkCR642 ovaries using the hsFLP X15 system. Clones were induced in the germarium by transient heat shock and allowed to develop on plates with live yeast paste for 4 days. The latest stage egg chamber with multiply marked nurse cell nuclei in each ovariole was recorded for each genotype. lnkCR642 egg chambers developed more slowly than wild type. Scale bars: 500 μm (A–F), 10 μm (G).
In light of the vitellogenic phenotype of lnkCR642 ovaries, we wanted to determine the effect of lnk mutations on PI3K signaling in the ovary. The tGPH reporter is sensitive to the levels of PtdIns(3,4,5)P3 in the cell and exhibits strong membrane localization when PI3K activity is high (Britton et al., 2002). Follicle cell clones of the strong lnkf02642 allele reduced the membrane localization of the tGPH reporter (Fig. 4G). This is consistent with results obtained by Wertz et al. in the larval fat body (Werz et al., 2009).
To determine whether lnkCR642 causes a developmental delay in oogenesis similar to that seen under conditions of moderate nutritional deprivation we monitored the rate of oocyte maturation by lineage tracing (Drummond-Barbosa and Spradling, 2001). The X-15 marker system was used to induce lacZ+ clones in dividing germline cells in the germarium (Harrison and Perrimon, 1993). At 4 days following heat shock Flp induction, the extent of egg chamber maturation was determined by counting the most mature cyst in each ovariole with multiple β-galactosidase-positive nuclei. The data show distinctly different rates of development, with wild type ovaries typically supporting development to stage 10B, whereas lnkCR642 egg chambers were most frequently observed at developmental stage 6 (Fig. 4G). These data suggest that lnkCR642 brings about a modest reduction in IIS that slows oogenesis but is not severe enough to completely arrest development in the same manner as the more severe lnkd07478 allele.
lnk does not affect DNA repair or vas phenotypes
In region 2A of the germarium Mei-W68 and Mei-P22 generate DSBs as an obligate step in meiotic recombination, and these can be detected by immunostaining of phosphorylated His2Av (γ-H2Av) (Mehrotra and McKim, 2006). In wild-type flies, these breaks are initiated in both nurse cells and pro-oocytes; however, they are repaired in region 2B and by the time cysts reach region 3 and bud off into the vitellarium no γ-H2Av foci are evident (McKim et al., 2002). Mutations in the spindle-class genes delay the formation of breaks until region three of the germarium and preclude the repair of these DSBs, resulting in γ-H2Av foci that are evident until stage 4 in the vitellarium (Staeva-Vieira et al., 2003; Jang et al., 2003). It is unclear whether the disappearance of γ-H2Av in spindle mutants indicates that other repair pathways such as non-homologous end joining (NHEJ) are compensating for a lack of homologous recombination or whether γ-H2Av is simply dissipating while the DBSs remain. Because lnkCR642 slows the rate of oogenesis, it is possible that this delay provides additional time to repair the breaks that persist in spn-BBU mutants. If compensatory repair were occurring, one would predict that all of the ovarian phenotypes seen in spn-BBU flies, such as karyosome malformation and Vasa phosphorylation, would also be suppressed.
To determine the prevalence of DSBs, we stained ovaries for γ-H2Av. Consistent with previous results, spn-BBU ovaries had breaks in region 3, whereas spn-BBU, mei-P22CA1215 ovaries did not exhibit any staining owing to a defect in the generation of DSBs (Fig. 5B,D). Despite the rate of oogenesis being slower in spn-BBU, lnkCR642 mutants, γ-H2Av foci were still prevalent in region 3, indicating that DNA repair activity is not enhanced in lnk mutants (Fig. 5C). Consistent with previous findings for spn-BBU (Jang et al., 2003), the γ-H2Av foci in spn-BBU, lnkCR642 ovaries persisted until stage 4 after which they disappeared (data not shown). We also examined the morphology of the karyosome in mid-oogenesis in these mutant backgrounds. In a spn-BBU background, mutation of mei-P22 was able to suppress the fragmented karyosome (Fig. 5G); however, similar to the γ-H2Av result, lnkCR642 was not able to rescue the karyosome malformation (Fig. 5H).
The fragmented karyosome phenotype seen in spindle mutant flies is shared with flies carrying alleles of the vas RNA helicase. Although this has yet to be shown directly, Vasa phosphorylation resulting from Mei-41 and Chk2 checkpoint activation is believed to inhibit Vasa activity and leads to both karyosome malformation and reduced Grk translation. We examined the phosphorylation state of Vasa in spn-BBU, lnkCR642 ovaries by western blotting. The mobility of ovarian Vasa from spn-BBU, lnkCR642 ovaries was slower than for wild-type ovaries and identical to Vasa from spn-BBU ovaries, indicating that Vasa is still phosphorylated in this background (Fig. 5I). These results were somewhat surprising given that the primary etiology of the ventralized eggshell phenotype in spindle mutants is inefficient grk translation, a process that is thought to require the activity of Vasa.
spn-B and vas mutants can be suppressed by rapamycin
The observation that robust grk translation can be supported despite continued phosphorylation of Vasa in spn-BBU, lnkCR642 mutants suggests that the cap-dependent mode of translation initiation that typically governs grk translation is being bypassed (Clouse et al., 2008). IIS is known to regulate cap-dependent translation by controlling the activity of the 4EBP. Low levels of IIS and TOR activity, such as occurs during starvation or rapamycin feeding, allow 4EBP to interact with eIF4E and inhibit its cap-binding activity and subsequent translation initiation. When IIS activity is high, it promotes the activity of TOR, which in turn phosphorylates 4EBP. This inhibitory phosphorylation blocks the interaction between 4EBP and eIF4E and permits eIF4E to participate in cap-dependent translation initiation. (Ruggero and Sonenberg, 2005). Recently, it was shown that the Drosophila InR transcript uses an alternative translational initiation mechanism to respond to nutrient availability. In times of nutrient limitation when IIS signaling is low and 4EBP activity is high, the InR is robustly translated at an IRES. The IRES in the 5′-UTR of the InR mRNA allows translation to bypass the 4EBP block of bulk cap-dependent translation (Marr et al., 2007).
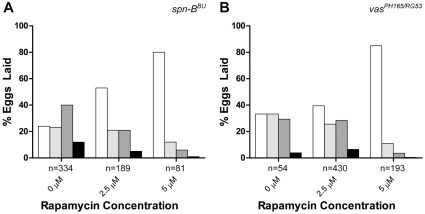
To determine whether grk translation can overcome a block in the cap-dependent pathway induced by spn-BBU or vasPH165/vasRG53 mutations, we attempted to suppress the ventralized eggshell phenotype of these flies by promoting 4EBP activity with rapamycin. Through interactions with FK506-binding proteins, rapamycin inhibits TOR activity, thereby alleviating the repression of 4EBP. Rapamycin was added to yeast paste and grape juice agar and fed to the flies for 7 days. The eggshell phenotypes were scored on days 4–6. Surprisingly, rapamycin promoted a greater proportion of wild-type eggs in spn-BBU flies up to a concentration of 5 μM, whereas 10 μM caused completely arrested oogenesis (Fig. 6A). This indicates that moderate inhibition of TOR activity can phenocopy the lnkCR642 mutation and promote grk translation. This result suggests that, even in situations where the translation of most cellular mRNA is repressed, grk translation can persist.
Fig. 6.
Suppression of spn-B and vas eggshell phenotypes by rapamycin. (A) spn-BBU and (B) vasPH165/RG53 flies were fed yeast paste on grape juice agar containing the indicated concentration of rapamycin. Eggs from days 4–7 were scored for eggshell phenotypes. In both cases, TOR inhibition results in strong suppression of eggshell ventralization. At 10 μM rapamycin completely inhibited egg deposition in spn-BBU flies. Eggshell classes are as described in Fig. 1.
Finally, we looked directly at the phenotype of vas mutants that are unable to translate grk (Fig. 6B). Strong vas mutants still accumulate grk mRNA in the oocyte, however Grk protein is not translated and strongly ventralized eggs are laid (Styhler et al., 1998). We hypothesize that in these flies, cap-dependent translation of grk is blocked due to an inability to navigate secondary structure that is encountered during scanning of the 5′ UTR and/or to promote joining of the 60S ribosomal subunit through interactions with eIF5B (Johnstone and Lasko, 2004). Although the null vasPH165 allele did not produce many eggs, the weaker heteroallelic vasPH165/vasRG53 combination resulted in increased fecundity and an eggshell distribution similar to spn-BBU flies. When these flies were fed rapamycin, strong suppression of the ventralized eggshell phenotype was evident and greater than 80% of the eggs laid by the 5 μM rapamycin cohort were wild type (Fig. 6B).
Discussion
In the current study we demonstrate a new interaction between a meiotic checkpoint, the IIS pathway and translation of gurken mRNA in Drosophila oogenesis. Mutations in meiotic DNA repair enzymes such as spn-B result in persistent DSBs in early oogenesis that activate an ATR-Chk2-dependent meiotic checkpoint. Checkpoint activation results in phosphorylation of the eIF4A-like RNA helicase Vasa, the activity of which is important for grk translation. In these mutants, low levels of Grk protein are synthesized, which is insufficient to pattern the eggshell correctly and results in ventralized eggs. Using forward genetics, we isolated an allele of the insulin receptor adapter, lnk. This mutation can suppress the weak grk translation phenotype and restore normal patterning to eggs laid by spn-BBU flies. We have shown through clonal analysis that lnk mutations reduce IIS in a cell-autonomous manner in the ovary. As in mammals, Drosophila IIS controls the rate of cap-dependent translation initiation in the cell by regulating the activity of TOR. Rapamycin inhibits TOR activity and we have shown that feeding rapamycin can suppress the ventralized eggshell phenotype not only in spn-BBU females but also in vasPH165/vasRG53 flies. These data suggest an alternative translation initiation mechanism for the grk mRNA by which flies can maintain dorsal–ventral axis patterning in times of moderate nutrient limitation.
Inhibition of TOR activity leads to activation of grk translation
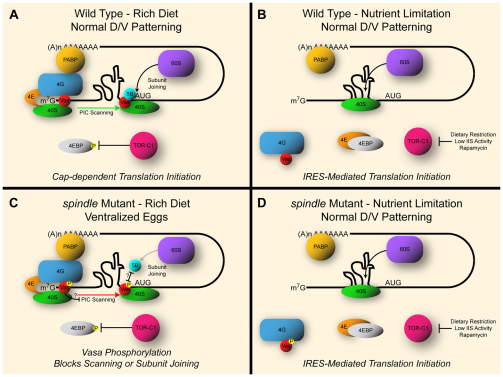
The discovery that mutations in lnk, a positive regulator of IIS, can suppress the patterning defects in spn-B flies was initially surprising. The eggshell phenotypes of the different genotypes were assessed after keeping the flies on apple or grape juice agar plates on which abundant amounts of yeast paste had been added, thus allowing the females to eat a protein-rich diet (Wieschaus and Nusslein-Volhard, 1998). A protein-rich diet stimulates the activity of the kinase TOR though two mechanisms. Drosophila insulin-like peptides (dilps) are secreted into the hemolymph by neuroendocrine cells in response to nutrient availability (Ikeya et al., 2002). This in turn activates the IIS cascade comprised of Chico–Lnk, PI3K, Akt, Tsc1 and 2 and Rheb, which promotes TOR-C1 activity (Grewal, 2009). The second mechanism acts more directly through the levels of intracellular amino acids that are imported in part by the slimfast and pathetic transporters. Both of these mechanisms stimulate TOR-C1 activity, which has been shown to promote cap-dependent translation by inhibiting 4EBP-mediated sequestration of eIF4E (Holz et al., 2005; Sonenberg and Hinnebusch, 2009; Teleman, 2010). Therefore, reducing TOR activity, either by a mutation in lnk or by addition of rapamycin, would be expected to interfere with cap-dependent translation and therefore further enhance the mutant phenotype. However, in spn-B mutant flies, cap-dependent translation is already inhibited by the activity of the checkpoint, presumably acting through Vasa modification. The fact that we observed a suppression of the ventralized phenotype in lnk mutants indicates that the reduction in TOR signaling must activate a second mode of translation that allows Grk protein to be produced independently of the block in cap-dependent translation.
A model of Vasa-independent grk translation
Several ovarian phenotypes are shared between mutations in spindle genes and vas mutants, including failure to form a compact karyosome, very weak grk translation, and ventralized eggs. Combined with the reproducible phosphorylation of Vasa protein in spindle-class mutants, these phenotypes are consistent with a defect in Vasa activity. Although the specific effect of this phosphorylation is unknown, Vasa has several functions in cap-dependent translation initiation of grk mRNA. Vasa has been shown to interact with eIF5B, and mutations that interfere with this interaction inhibit grk translation (Johnstone and Lasko, 2004). This interaction is thought to facilitate assembly of the 60S ribosomal subunit at the AUG start codon. Furthermore, as a DEAD-box RNA helicase, Vasa might permit the PIC to scan the 5′-UTR of grk and negotiate secondary structures that might impede the progress of this complex (Liang et al., 1994). IRES sequences adopt strong secondary structures in the 5′-UTR of RNAs that they regulate (Kanamori and Nakashima, 2001). If it can be demonstrated in the future that grk possess an IRES sequence, this might explain the requirement for Vasa helicase activity to unwind this structure when translation is initiated from the 5′-cap during conditions of adequate nutrient availability. Whether the checkpoint-dependent phosphorylation of Vasa affects its stability, RNA helicase activity or its eIF5B interaction, the expected result is a block in cap-dependent translation initiation of grk mRNA and concomitant dorsal–ventral patterning defects. Our observation that grk translation can be induced spn-BBU and in vasPH165/vasRG53 flies indicates that an alternative mechanism for supporting translation initiation is taking place. Because reduced IIS and TOR activity both block bulk cap-dependent translation initiation through sequestration of eIF4E by 4EBP, yet stimulate IRES activity, we propose that the latter might provide an explanation for our results.
Proposed mechanism of spindle phenotype suppression by IIS and TOR
Grk plays a central role in shaping the development of the egg and subsequent embryo. Mutations that disrupt Grk and/or Egfr signaling during oogenesis result in female sterility (Schüpbach, 1987). Blocking the translation of this essential morphogen in spindle-class mutants that are unable to repair DNA damage is an effective mechanism to prevent the transmission of mutations to the progeny (Abdu et al., 2002; Ghabrial and Schüpbach, 1999). This reproductive checkpoint is effective when nutrients are abundant, however as we have demonstrated, the strategy breaks down when IIS and/or TOR activity is low. Under these conditions, grk can be translated and result in eggs that are patterned correctly (Fig. 1), even though the DNA damage and karyosome malformation phenotypes persist (Fig. 5). We propose that this difference occurs because the DNA damage checkpoint can only impinge on one of the two mechanisms by which grk translation can be initiated.
One mechanism by which suppression of the dorsal–ventral patterning defects of spn-BBU might occur is through the effects of the additional time that lnkCR642 egg chambers spend completing oogenesis. Although Grk production is reduced in spn-BBU flies, it is not completely blocked and some Grk protein is made. If the reduced rate of Grk production is integrated over the extended time spent during mid oogenesis, sufficient Grk levels could accumulate and support normal dorsal–ventral patterning. However, this model is inconsistent with the inability of lnkCR642 to suppress the ventralized eggs laid by grkED22 females. These flies do retain some Grk activity, as is
Fig. 7.
A model of Vasa-independent grk translation. (A) When nutrients are abundant, cap-dependent translation predominates. Vasa helicase activity facilitates 43S PIC scanning of the 5′-UTR, allowing it to navigate secondary structures. Once an AUG codon is identified, Vasa–eIF5B interactions promote joining of the 60S subunit to form the 80S ribosome. (B) When nutrients are low, IIS is compromised or flies are fed the TOR-C1 inhibitor rapamycin, TOR activity falls and 4EBP is free to inhibit the cap-binding protein eIF4E. The resulting increase in free ribosomes favors IRES translation initiation of the localized grk transcript allowing oocytes to be patterned correctly, thereby preserving their viability for when nutrients are available again. (C) spindle-class mutants such as spn-BBU inhibit grk translation by phosphorylating Vasa in a checkpoint-dependent manner. This disrupts scanning of the 43S PIC and/or interactions with eIF5B (5B) and therefore subunit joining at the initiation AUG codon. In these conditions, grk cannot be translated and ventralized eggs result. (D) Reduced TOR activity in a spn-B mutant background permits grk translation independent of Vasa activity. Despite the inhibition of cap-dependent translation initiation resulting from Vasa phosphorylation and increased 4EBP activity, grk translation persists. We hypothesize that this is due to alternative translation initiation at an IRES in the grk 5′-UTR.
evident by the single appendage that is specified; however, if the mechanism of suppression were through accumulation, then grkED22 should be suppressed by lnk mutations. Therefore, we favor the IRES-dependent model proposed herein.
The selective pressure that may have driven the evolution of this bi-modal translation mechanism for grk can be best understood by considering that in wild populations of Drosophila, females feed and oviposit at locations where yeast is abundant (Good and Tatar, 2001). This behavior ensures adequate nutrition to support oogenesis in the female as well as for the developing larvae (Drummond-Barbosa and Spradling, 2001; LaFever and Drummond-Barbosa, 2005). If, however, nutrients become scarce, females adjust the rate of oogenesis to match nutrient availability. In response to complete starvation, egg chambers undergo apoptosis and are reabsorbed; however, moderate reductions in IIS slow the rate of oogenesis until an abundant protein source is found. The conserved response to dietary restriction is to repress cap-dependent translation of most cellular transcripts while a select population of RNAs that are essential for survival escape this repression by utilizing a cap-independent IRES mechanism. We speculate that grk may be one such transcript. Oocytes that are in mid-development when nutrients are scarce must still be patterned appropriately so that the resulting eggs are fertile. IRES activity might facilitate Grk expression in order to maintain normal dorsal–ventral patterning when food is scarce, whereas, when nutrients are abundant, cap-dependent translation predominates.
Materials and Methods
Eggshell phenotype scoring
Five female flies of the indicated genotypes were placed in egg-laying blocks with an equal number of sibling males of any genotype (Wieschaus and Nusslein-Volhard, 1998). The blocks were inverted on apple juice or grape juice agar plates, onto which active yeast paste had been spotted, and kept at 25°C. The plates were changed daily before the flies had consumed the available yeast to guard against dietary restriction. Eggshell phenotypes were scored on days 4–7 as previously described (Ghabrial et al., 1998). When indicated, rapamycin (LC Laboratories) dissolved in DMSO was added to both the agar substrate as well as the yeast paste. The final concentration of DMSO in the medium was 0.1%.
EMS mutagenesis screen
A total of 75–100 st, spn-BBU, sr, e/TM6B, Hu, e adult males were starved for 2 hours, then fed a 1% sucrose solution containing 0.25% EMS for 24 hours, and cleaned by transferring to fresh food four times. The mutagenized males were crossed to ras, st, e/TM8, DTS, Sb, st, e, th virgin females at 18°C. The males were discarded after 5–7 days. In the F1 generation single st, spn-BBU, sr, e */TM8, DTS, Sb, st, e, th (where * indicates that the chromosome was present during the mutagenesis) virgin females were crossed to P[w+, ovoD]/TM8, DTS, Sb, st, e, th males at 18°C. The F2 generation was crossed inter se at 29°C for 7 days and the cross was moved to 25°C for the remainder of development. In the F3 generation, 5 homozygous st, spn-BBU, sr, e * females and 3–5 males of any genotype were placed in laying chambers on yeasted apple juice plates at room temperature. Fresh yeasted plates were applied approximately every 24 hours. Eggs laid on days 5–7 were examined. Lines that showed a noticeable decrease in the percentage of ventralized eggs laid relative to spn-BBU were selected for further analysis.
Fly stocks
The spn-BBU allele is a G107E missense mutation and has been previously described (Ghabrial et al., 1998). The spn-BΔ37C allele was generated by imprecise excision of the P{GSV6}GS13042 P-element resulting in deletion of the entire spn-B open reading frame and 9 bp of the 3′-UTR; 633 bp of the P-element remained in the deficiency. mei-P22CA1215 was identified in this work and consists of an EMS-generated missense mutation at codon R31C and a 36-bp deletion that removes amino acids 67–78. lnkCR642 was identified in this work and consists of an EMS-generated nonsense mutation at codon 298. lnkd07478 is a P{XP} insertion in the 5′-UTR of the lnk locus, 701 bp upstream of the start codon; the lnkf02642 and lnkf05062 alleles are PBac{WH} insertions in the second intron (Thibault et al., 2004). The vasPH165 allele is a deletion generated by P-element excision (Styhler et al., 1998) and the vasRG53 allele is an undefined EMS-generated allele (Schüpbach and Wieschaus, 1991). P[w+ lac-Z]BB142 is an enhancer trap of the kekkon gene (Schüpbach and Roth, 1994). We generated follicle cell clones in e22c-Gal4, UAS-FLP/+; FRT82, lnkCR642/FRT82, ubi-GFP flies (Xu and Rubin, 1993; Duffy et al., 1998). To assess Egfr activity we generated clones in flies with the genotype e22c-Gal4, UAS-FLP/P[w+ lac-Z]BB142; FRT82, lnkCR642/FRT82, ubi-GFP. PI3K activity was compared in clones generated in ovaries with the genotype e22c-Gal4, UAS-FLP/tGPH; FRT82, lnkf02642/FRT82, ubi-GFP (Britton et al., 2002). UAS constructs were expressed using the CY2-Gal4 or nos-Gal4 drivers (Queenan et al., 1997; Van Doren et al., 1998). Lineage tracing experiments were conducted as previously described with flies of the genotypes hs-FLP; X15-29/X15-33; lnkCR642 (Harrison and Perrimon, 1993; Drummond-Barbosa and Spradling, 2001).
Mapping of CR642
The lnkCR642 suppressor mutation was positionally cloned by meiotic recombination. Rough meiotic mapping with visible markers placed the suppressor between ebony and claret. Fine-scale SNP mapping was conducted by recombination with the P{EPgy2}EY09907 insertion at 98B1 (Venken and Bellen, 2005). It was necessary to select sr, e, w+ recombinants to maintain spn-BBU in the background as no phenotype was evident in flies carrying only the CR642 mutation. Intergenic SNPs were identified that allowed us to define the region between 96F3 and 96F9. lnk was identified as one of four genes with ovarian expressed-sequence tags (ESTs) and the CR642 allele was confirmed by sequence comparison with the spn-BBU parental line.
Transgenic constructs
The lnk genomic rescue construct (pSF10) was amplified from BAC22N13 with primers 5′-ATGCGGTACCGAATTCGCTTTAATGTCTCCAATCATGCAG-3′ and 5′-ATGCGCGGCCGCTGAAAGAAGACCGACGTTGG-3′ and cloned into p[ACMAN}-attB-Amp as an EcoRI-NotI fragment (Venken et al., 2006). This construct contains 903 bp of sequence upstream of the start of transcription and the entire 3′-UTR. The lnkY720F rescue construct (pSF11) was generated by site-directed mutagenesis with primers 5′-GACGGGCCGTCGATAATCAGTTCAGCTTCACCTAAGTCCG-3′ and 5′-CGGACTTAGGTGAAGCTGAACTGATTATCGACGGCCCGTC-3′. The EGFP-tagged Lnk construct (pSF12) contains all of the sequence elements in pSF10; however, the EGFP sequence is cloned in frame with the 5′-end of the lnk open reading frame. These constructs were integrated into the attP16- and attP40-docking sites using the ΦC31 system (Markstein et al., 2008). A UAS-lnk expression construct (pSF44) was amplified from cDNA clone LD10453 with primers 5′-ACTGGGTACCATGGGTGGCAATAGCACAGG-3′ and 5′-ACTGGAATTCTTAGGTGAAGCTGTACTGATTATCG-3′ and cloned as a KpnI/EcoRI fragment into a derivative of pUASP called pTIGER. Transgenic flies were generated by standard P-element transgenesis (Genetic Services).
Antibodies and immunohistochemistry
Grk staining was performed with monoclonal mouse anti-Grk ID12 at 1:10 as previously described (Queenan et al., 1999). Mouse anti-βGal was used at 1:1000 (Promega). Rabbit anti-γ-H2Av antibody (a gift of Kim McKim) was used at 1:500 (McKim et al., 2009; Mehrotra and McKim, 2006). Rabbit anti-Vasa antibody was generated by immunization with a KLH-conjugated peptide CGDGVGGSGGEGGGY and used at 1:3000 (Epitomics). Rabbit anti-Lnk antibody was used at 1:500. The antigen was expressed as a recombinant GST–Lnk-56–542 fusion protein and was purified on a GSTrap FF column (GE Healthcare) according to the manufacturer's instructions. This antigen contains sequences that are predicted to remain in protein made by the lnkCR642 (this work), lnkf02642 and lnkf05062 alleles (Thibault et al., 2004); however, the existence and/or stability of these truncations could not be determined. Attempts to detect Lnk protein by western blotting were unsuccessful. Secondary Alexa-Fluor-488 or -546-conjugated anti-mouse and/or anti-rabbit antibodies and Alexa-Fluor-546–phalloidin were used at 1:1000 (Invitrogen). Hoechst 33342 was used at 1 μg/mL. Horseradish peroxidase (HRP)-conjugated donkey anti-rabbit was used at 1:5000 (Jackson ImmunoResearch).
Acknowledgements
We thank Kim McKim, Spyros Artavanis-Tsakonas, the Developmental Studies Hybridoma Bank, and the Bloomington and Kyoto stock centers for providing flies and antibodies. Thank you to Gail Barcelo for technical support and Uri Abdu for assistance with the screen. We also thank the Schüpbach, Wieschaus, and Ferguson labs for useful discussions. We are grateful for microscopy assistance from Joe Goodhouse at the Confocal and Electron Microscopy Core Facility Laboratory at Princeton University, Wade Sigurdson at the Confocal Microscope and Flow Cytometry Facility in the School of Medicine and Biomedical Sciences, University at Buffalo, and Alan Siegel at The University at Buffalo North Campus Imaging Facility supported by Major Research Instrumentation [grant number DBI 0923133].
Footnotes
Funding
This work was supported by the Howard Hughes Medical Institute; US Public Health Service Grants [grant numbers PO1 CA41086, RO1 GM077620]; and Startup Funds and Summer Research Seed Grants from The State University of New York at Fredonia. Deposited in PMC for release after 6 months.
References
- Abdu U., Brodsky M., Schüpbach T. (2002). Activation of a meiotic checkpoint during Drosophila oogenesis regulates the translation of Gurken through Chk2/Mnk. Curr. Biol. 12, 1645-1651 [DOI] [PubMed] [Google Scholar]
- Ahmed Z., Pillay T. (2001). Functional effects of APS and SH2-B on insulin receptor signaling. Biochem. Soc. Trans. 29, 529-534 [DOI] [PubMed] [Google Scholar]
- Arnaud E., Touriol C., Boutonnet C., Gensac M., Vagner S., Prats H., Prats A. (1999). A new 34-kilodalton isoform of human fibroblast growth factor 2 is cap dependently synthesized by using a non-AUG start codon and behaves as a survival factor. Mol. Cell. Biol. 19, 505-514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein J., Sella O., Le S. Y., Elroy-Stein O. (1997). PDGF2/c-sis mRNA leader contains a differentiation-linked internal ribosomal entry site (D-IRES). J. Biol. Chem. 272, 9356-9362 [DOI] [PubMed] [Google Scholar]
- Besse F., Ephrussi A. (2008). Translational control of localized mRNAs: restricting protein synthesis in space and time. Nat. Rev. Mol. Cell Biol. 9, 971-980 [DOI] [PubMed] [Google Scholar]
- Britton J. S., Lockwood W. K., Li L., Cohen S. M., Edgar B. A. (2002). Drosophila's insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev. Cell 2, 239-249 [DOI] [PubMed] [Google Scholar]
- Chang W. L., Liou W., Pen H. C., Chou H. Y., Chang Y. W., Li W. H., Chiang W., Pai L. M. (2008). The gradient of Gurken, a long-range morphogen, is directly regulated by Cbl-mediated endocytosis. Development 135, 1923-1933 [DOI] [PubMed] [Google Scholar]
- Chekulaeva M., Hentze M., Ephrussi A. (2006). Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell 124, 521-533 [DOI] [PubMed] [Google Scholar]
- Clifford R. J., Schüpbach T. (1989). Coordinately and differentially mutable activities of torpedo, the Drosophila melanogaster homolog of the vertebrate EGF receptor gene. Genetics 123, 771-787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse K., Ferguson S., Schüpbach T. (2008). Squid, Cup, and PABP55B function together to regulate gurken translation in Drosophila. Dev. Biol. 313, 713-724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delanoue R., Herpers B., Soetaert J., Davis I., Rabouille C. (2007). Drosophila Squid/hnRNP helps dynein switch from a gurken mRNA transport motor to an ultrastructural static anchor in sponge bodies. Dev. Cel. 13, 523-538 [DOI] [PubMed] [Google Scholar]
- Drummond-Barbosa D., Spradling A. (2001). Stem cells and their progeny respond to nutritional changes during Drosophila oogenesis. Dev. Biol. 231, 265-278 [DOI] [PubMed] [Google Scholar]
- Duffy J., Harrison D., Perrimon N. (1998). Identifying loci required for follicular patterning using directed mosaics. Development.125, 2263-2271 [DOI] [PubMed] [Google Scholar]
- Gao X., Pan D. (2001). TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth. Genes Dev. 15, 1383-1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghabrial A., Schüpbach T. (1999). Activation of a meiotic checkpoint regulates translation of Gurken during Drosophila oogenesis. Nat. Cell Biol. 1, 354-357 [DOI] [PubMed] [Google Scholar]
- Ghabrial A., Ray R., Schüpbach T. (1998). okra and spindle-B encode components of the RAD52 DNA repair pathway and affect meiosis and patterning in Drosophila oogenesis. Genes Dev. 12, 2711-2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakou M. E., Goss M., Jünger M. A., Hafen E., Leevers S. J., Partridge L. (2004). Long-lived Drosophila with overexpressed dFOXO in adult fat body. Science 305, 361 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Reyes A., Elliott H., St Johnston D. (1995). Polarization of both major body axes in Drosophila by gurken-torpedo signalling. Nature 375, 654-658 [DOI] [PubMed] [Google Scholar]
- Good T., Tatar M. (2001). Age-specific mortality and reproduction respond to adult dietary restriction in Drosophila melanogaster. J. Insect Physiol. 47, 1467-1473 [DOI] [PubMed] [Google Scholar]
- Grewal S. (2009). Insulin/TOR signaling in growth and homeostasis: a view from the fly world. Int. J. Biochem. Cell Biol. 41, 1006-1010 [DOI] [PubMed] [Google Scholar]
- Harrison D., Perrimon N. (1993). Simple and efficient generation of marked clones in Drosophila. Curr. Biol. 3, 424-433 [DOI] [PubMed] [Google Scholar]
- Holz M. K., Ballif B. A., Gygi S. P., Blenis J. (2005). mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell 123, 569-580 [DOI] [PubMed] [Google Scholar]
- Hsu H., LaFever L., Drummond-Barbosa D. (2008). Diet controls normal and tumorous germline stem cells via insulin-dependent and -independent mechanisms in Drosophila. Dev. Biol. 313, 700-712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Hubbard S. R. (2005). Structural characterization of a novel Cbl phosphotyrosine recognition motif in the APS family of adapter proteins. J. Biol. Chem. 28018943-18949 [DOI] [PubMed] [Google Scholar]
- Huez I., Bornes S., Bresson D., Creancier L., Prats H. (2001). New vascular endothelial growth factor isoform generated by internal ribosome entry site-driven CUG translation initiation. Mol. Endocrinol. 15, 2197-2210 [DOI] [PubMed] [Google Scholar]
- Hwangbo D. S., Gersham B., Tu M., Palmer M., Tatar M. (2004). Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature 429, 562-566 [DOI] [PubMed] [Google Scholar]
- Ikeya T., Galic M., Belawat P., Nairz K., Hafen E. (2002). Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Curr. Biol. 12, 1293-1300 [DOI] [PubMed] [Google Scholar]
- Jang J., Sherizen D., Bhagat R., Manheim E., McKim K. (2003). Relationship of DNA double-strand breaks to synapsis in Drosophila. J. Cell Sci. 116, 3069-3077 [DOI] [PubMed] [Google Scholar]
- Johnstone O., Lasko P. (2004). Interaction with eIF5B is essential for Vasa function during development. Development 131, 4167-4178 [DOI] [PubMed] [Google Scholar]
- Junger M., Rintelen F., Stocker H., Wasserman J., Vegh M., Radimerski T., Greenberg M., Hafen E. (2003). The Drosophila Forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J. Biol. 2, 20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamori Y., Nakashima N. (2001). A tertiary structure model of the internal ribosome entry site (IRES) for methionine-independent initiation of translation. RNA 7, 266-274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E., Goraksha-Hicks P., Li L., Neufeld T. P., Guan K. (2008). Regulation of TORC1 by Rag GTPases in nutrient response. Nat. Cell Biol. 10, 935-945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFever L., Drummond-Barbosa D. (2005). Direct control of germline stem cell division and cyst growth by neural insulin in Drosophila. Science 309, 1071-1073 [DOI] [PubMed] [Google Scholar]
- Lasko P., Ashburner M. (1988). The product of the Drosophila gene vasa is very similar to eukaryotic initiation factor-4A. Nature 335, 611-617 [DOI] [PubMed] [Google Scholar]
- Liang L., Diehl-Jones W., Lasko P. (1994). Localization of vasa protein to the Drosophila pole plasm is independent of its RNA-binding and helicase activities. Development 120, 1201-1211 [DOI] [PubMed] [Google Scholar]
- Liu H., Jang J., Kato N., McKim K. (2002). mei-P22 encodes a chromosome-associated protein required for the initiation of meiotic recombination in Drosophila melanogaster. Genetics 162, 245-258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Kimura A., Baumann C. A., Saltiel A. R. (2002). APS facilitates c-Cbl tyrosine phosphorylation and GLUT4 translocation in response to insulin in 3T3-L1 adipocytes. Mol. Cell. Biol. 22, 3599-3609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M., Pitsouli C., Villalta C., Celniker S. E., Perrimon N. (2008). Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 40, 476-483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr M., D'Alessio J., Puig O., Tjian R. (2007). IRES-mediated functional coupling of transcription and translation amplifies insulin receptor feedback. Genes Dev. 21, 175-183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K., Ephrussi A. (2009). mRNA localization: gene expression in the spatial dimension. Cell 136, 719-730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim K. S., Hayashi-Hagihara A. (1998). mei-W68 in Drosophila melanogaster encodes a Spo11 homolog: evidence that the mechanism for initiating meiotic recombination is conserved. Genes Dev. 12, 2932-2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim K., Jang J., Manheim E. (2002). Meiotic recombination and chromosome segregation in Drosophila females. Annu. Rev. Genet. 36, 205-232 [DOI] [PubMed] [Google Scholar]
- McKim K. S., Joyce E. F., Jang J. K. (2009). Cytological analysis of meiosis in fixed Drosophila ovaries. In Methods in Molecular Biology: Meiosis Volume 2 Cytological Methods (ed. Keeney S.), pp. 197-216 New York: Humana Press; [DOI] [PubMed] [Google Scholar]
- McNeill H., Craig G. M., Bateman J. M. (2008). Regulation of neurogenesis and Egfr signalling by the insulin receptor/Target of Rapamycin pathway in Drosophila. Genetics. 179, 843-853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra S., McKim K. S. (2006). Temporal analysis of meiotic DNA double-strand break formation and repair in Drosophila females. PLoS Genet. 2, e200, 1883-1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokrejs M., Masek T., Vopalensky V., Hlubucek P., Delbos P., Pospisek M. (2009). IRESite–a tool for the examination of viral and cellular internal ribosome entry sites. Nucleic Acids Res. 38, D131-D136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman-Silberberg F., Schüpbach T. (1993). The Drosophila dorsoventral patterning gene gurken produces a dorsally localized RNA and encodes a TGF alpha-like protein. Cell 75, 165-174 [PubMed] [Google Scholar]
- Norvell A., Kelley R., Wehr K., Schüpbach T. (1999). Specific isoforms of squid, a Drosophila hnRNP, perform distinct roles in Gurken localization during oogenesis. Genes Dev. 13, 864-876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai L., Barcelo G., Schüpbach T. (2000). D-cbl, a negative regulator of the Egfr pathway, is required for dorsoventral patterning in Drosophila oogenesis. Cell 103, 51-61 [DOI] [PubMed] [Google Scholar]
- Pai L., Wang P., Chen S., Barcelo G., Chang W., Nilson L., Schüpbach T. (2006). Differential effects of Cbl isoforms on Egfr signaling in Drosophila. Mech. Dev. 123, 450-462 [DOI] [PubMed] [Google Scholar]
- Pedersen S. K., Christiansen J., Hansen T. V. O., Larsen M. R., Nielsen F. C. (2002). Human insulin-like growth factor II leader 2 mediates internal initiation of translation. Biochem. J. 363, 37-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queenan A., Ghabrial A., Schüpbach T. (1997). Ectopic activation of torpedo/Egfr, a Drosophila receptor tyrosine kinase, dorsalizes both the eggshell and the embryo. Development 124, 3871-3880 [DOI] [PubMed] [Google Scholar]
- Queenan A., Barcelo G., Van Buskirk C., Schüpbach T. (1999). The transmembrane region of Gurken is not required for biological activity, but is necessary for transport to the oocyte membrane in Drosophila. Mech. Dev. 89, 35-42 [DOI] [PubMed] [Google Scholar]
- Richter J. D., Sonenberg N. (2005). Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 433, 477-480 [DOI] [PubMed] [Google Scholar]
- Roth S., Lynch J. A. (2009). Symmetry breaking during Drosophila oogenesis. Cold Spring Harb. Perspect. Biol. 1, a001891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D., Sonenberg N. (2005). The Akt of translational control. Oncogene 24, 7426-7434 [DOI] [PubMed] [Google Scholar]
- Sancak Y., Peterson T. R., Shaul Y. D., Lindquist R. A., Thoreen C. C., Bar-Peled L., Sabatini D. M. (2008). The Rag GTPases bind Raptor and mediate amino acid signaling to mTORC1. Science 320, 1496-1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüpbach T. (1987). Germ line and soma cooperate during oogenesis to establish the dorsoventral pattern of egg shell and embryo in Drosophila melanogaster. Cell 49, 699-707 [DOI] [PubMed] [Google Scholar]
- Schüpbach T., Wieschaus E. (1986). Maternal-effect mutations altering the anterior-posterior pattern of the Drosophila embryo. Rouxs Arch. Dev. Biol. 195, 302-317 [DOI] [PubMed] [Google Scholar]
- Schüpbach T., Wieschaus E. (1991). Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics 129, 1119-1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüpbach T., Roth S. (1994). Dorsoventral patterning in Drosophila oogenesis. Curr. Opin. Genet. Dev. 4, 502-507 [DOI] [PubMed] [Google Scholar]
- Slack C., Werz C., Wieser D., Alic N., Foley A., Stocker H., Withers D., Thornton J., Hafen E., Partridge L. (2010). Regulation of lifespan, metabolism, and stress responses by the Drosophila SH2B protein, Lnk. PLoS Genet. 6, 1-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N., Hinnebusch A. G. (2009). Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 136, 731-745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staeva-Vieira E., Yoo S., Lehmann R. (2003). An essential role of DmRad51/SpnA in DNA repair and meiotic checkpoint control. EMBO J. 22, 5863-5874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styhler S., Nakamura A., Swan A., Suter B., Lasko P. (1998). vasa is required for GURKEN accumulation in the oocyte, and is involved in oocyte differentiation and germline cyst development. Development 125, 1569-1578 [DOI] [PubMed] [Google Scholar]
- Teleman A. (2010). Molecular mechanisms of metabolic regulation by insulin in Drosophila. Biochem. J. 425, 13-26 [DOI] [PubMed] [Google Scholar]
- Thibault S. T., Singer M. A., Miyazaki W. Y., Milash B., Dompe N. A., Singh C. M., Buchholz R., Demsky M., Fawcett R., Francis-Lang H. L., et al. (2004). A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36, 283-287 [DOI] [PubMed] [Google Scholar]
- Thien C., Langdon W. (2005). c-Cbl and Cbl-b ubiquitin ligases: substrate diversity and the negative regulation of signalling responses. Biochem. J. 391, 153-166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thio G. L., Ray R. P., Barcelo G., Schüpbach T. (2000). Localization of gurken RNA in Drosophila oogenesis requires elements in the 5′ and 3′ regions of the transcript. Dev. Biol. 221, 435-446 [DOI] [PubMed] [Google Scholar]
- Tinker R., Silver D., Montell D. (1998). Requirement for the vasa RNA helicase in gurken mRNA localization. Dev. Biol. 199, 1-10 [DOI] [PubMed] [Google Scholar]
- Tomancak P., Guichet A., Zavorszky P., Ephrussi A. (1998). Oocyte polarity depends on regulation of gurken by Vasa. Development 125, 1723-1732 [DOI] [PubMed] [Google Scholar]
- Van Doren M., Williamson A. L., Lehmann R. (1998). Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr. Biol. 8, 243-246 [DOI] [PubMed] [Google Scholar]
- Venken K. J., Bellen H. J. (2005). Emerging technologies for gene manipulation in Drosophila melanogaster. Nat. Rev. Genet. 6, 167-178 [DOI] [PubMed] [Google Scholar]
- Venken K. J., Bellen H. J. (2007). Transgenesis upgrades for Drosophila melanogaster. Development 134, 3571-3584 [DOI] [PubMed] [Google Scholar]
- Venken K., He Y., Hoskins R., Bellen H. (2006). P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314, 1747-1751 [DOI] [PubMed] [Google Scholar]
- Werz C., Kohler K., Hafen E., Stocker H. (2009). The Drosophila SH2B family adaptor Lnk acts in parallel to chico in the insulin signaling pathway. PLoS Genet. 5, 1-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieschaus E., Nusslein-Volhard C. (1998). Looking at embryos. In Drosophila: A Practical Approach. Second Ed. (ed. Roberts D. B.), pp. 179-214 Washington D.C: IRL Press; [Google Scholar]
- Xu T., Rubin G. (1993). Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 117, 1223-1237 [DOI] [PubMed] [Google Scholar]
- Yokouchi M., Wakioka T., Sakamoto H., Yasukawa H., Ohtsuka S., Sasaki A., Ohtsubo M., Valius M., Inoue A., Komiya S., et al. (1999). APS, an adaptor protein containing PH and SH2 domains, is associated with the PDGF receptor and c-Cbl and inhibits PDGF-induced mitogenesis. Oncogene 18, 759-767 [DOI] [PubMed] [Google Scholar]