Abstract
Laminin-binding integrins (α3β1, α6β1, α6β4, α7β1) are almost always expressed together with tetraspanin CD151. In every coexpressing cell analyzed to date, CD151 makes a fundamental contribution to integrin-dependent motility, invasion, morphology, adhesion and/or signaling. However, there has been minimal mechanistic insight into how CD151 affects integrin functions. In MDA-MB-231 mammary cells, tetraspanin CD151 knockdown impairs α6 integrin clustering and functions without decreasing α6 integrin expression or activation. Furthermore, CD151 knockdown minimally affects the magnitude of α6 integrin diffusion, as measured using single particle tracking. Instead, CD151 knockdown has a novel and unexpected dysregulating effect on the mode of α6 integrin diffusion. In control cells α6 integrin shows mostly random-confined diffusion (RCD) and some directed motion (DMO). In sharp contrast, in CD151-knockdown cells α6 integrin shows mostly DMO. In control cells α6 diffusion mode is sensitive to actin disruption, talin knockdown and phorbol ester stimulation. By contrast, CD151 knockdown cell α6 integrin is sensitive to actin disruption but desensitized to talin knockdown or phorbol ester stimulation, indicating dysregulation. Both phorbol ester and EGF stimulate cell spreading and promote α6 RCD in control cells. By contrast, CD151-ablated cells retain EGF effects but lose phorbol-ester-stimulated spreading and α6 RCD. For α6 integrins, physical association with CD151 promotes α6 RCD, in support of α6-mediated cable formation and adhesion. By comparison, for integrins not associated with CD151 (e.g. αv integrins), CD151 affects neither diffusion mode nor αv function. Hence, CD151 support of α6 RCD is specific and functionally relevant, and probably underlies diverse CD151 functions in skin, kidney and cancer cells.
Key words: Integrin, Tetraspanin, CD151, Single particle tracking, Laminin
Introduction
Among the 24 different αβ heterodimers in the integrin family, the laminin-binding integrins (α3β1, α6β1, α6β4, α7β1) are a distinct subgroup, based on functional and structural similarities (Belkin and Stepp, 2000) and their close association with cell surface proteins in the tetraspanin family (Sterk et al., 2002; Stipp et al., 2003b). Among the tetraspanin proteins, CD151 shows the most robust association with laminin-binding integrins, in terms of stability and stoichiometry. CD151 association with α3 and α6 integrins occurs through direct protein–protein interaction, occurs early in biosynthesis, and can affect integrin glycosylation (Baldwin et al., 2008; Berditchevski et al., 2001; Kazarov et al., 2002; Yauch et al., 1998; Yauch et al., 2000).
CD151 expression on cancer cells correlates with poor clinical outcome and/or high grade in non-small cell lung (Tokuhara et al., 2001), prostate (Ang et al., 2004), hepatocellular (Liu et al., 2007), breast (Novitskaya et al., 2010; Sadej et al., 2009; Yang et al., 2008) and other cancers (Romanska and Berditchevski, 2011). Furthermore, CD151 is enriched on prostate-tumor-initiating cells (Rajasekhar et al., 2011), contributes functionally to tumor cell metastasis (Kohno et al., 2002; Zijlstra et al., 2008), supports breast cancer cell resistance to ErbB2 antagonists (Yang et al., 2010) and accelerates primary breast cancer growth in a human-mouse xenograft model (Sadej et al., 2009; Yang et al., 2008). Consistent with the multiple roles for CD151 in cancer, anti-CD151 antibodies that can inhibit tumor growth and metastasis are being evaluated for potential clinical application (Haeuw et al., 2011).
CD151 appears to function largely through its effects on laminin-binding integrins. A human CD151 mutation has been found to be associated with end-stage kidney failure, regional skin blistering and other defects in two individuals (Kagan et al., 1988; Karamatic Crew et al., 2004). Consistent with this, laminin-binding integrin α3, α6, and β4 subunits also contribute to skin and kidney development (Belkin and Stepp, 2000). As further evidence for CD151 working through laminin-binding integrins, ablation or mutation of CD151 markedly disrupts integrin-dependent effects on cell migration, proliferation, cable formation, morphology and signaling (Berditchevski et al., 2002; Johnson et al., 2009; Kazarov et al., 2002; Lammerding et al., 2003; Novitskaya et al., 2010; Sadej et al., 2009; Stipp et al., 2003a; Winterwood et al., 2006; Yang et al., 2002; Zevian et al., 2011; Zhang et al., 2002; Zuo et al., 2010).
Although CD151 is known to modulate α6β1-, α6β4- and α3β1-integrin-dependent cell morphology, motility and neurite outgrowth (Ashman, 2002; Hemler, 2005), mechanistic details are lacking. Despite its close association, CD151 is not needed for α3 or α6 integrin expression (Sachs et al., 2006; Takeda et al., 2007; Wright et al., 2004). A suggested CD151 support of α3β1 integrin ‘activation’ neoepitopes (Nishiuchi et al., 2005) is counterbalanced by a report that α3β1 integrin is remarkably resistant to changes in neoepitopes, even when saturated with manganese (Bazzoni et al., 1998). CD151 also might affect integrin trafficking (Liu et al., 2007; Winterwood et al., 2006), and supports (Nishiuchi et al., 2005; Winterwood et al., 2006) or does not support (Berditchevski, 2001; Testa et al., 1999) integrin-mediated adhesion. In a case where CD151 did not influence initial cell binding to laminin-coated beads, it did affect subsequent adhesion strengthening (Lammerding et al., 2003). Laminin-binding integrins are linked, through CD151, to tetraspanin-enriched microdomains (Nydegger et al., 2006; Takeda et al., 2007; Yanez-Mo et al., 2009; Yang et al., 2008), which probably participate in the regulation of integrin functions.
Studies of CD151 in breast cancer (Sadej et al., 2009; Yang et al., 2008) have relied on mammary cell lines (MDA-MB-231 and/or MCF-10A) with basal-like properties (Neve et al., 2006). In these cells, CD151 affects α6-integrin-dependent migration, invasion, spreading and/or signaling in vitro, and both ectopic and orthotopic tumor growth in vivo. However, insights are needed into how CD151 fundamentally and specifically affects α6 integrin functions in basal-like mammary cell lines. Here, we show that CD151 ablation does not affect α6 integrin expression or activation. Instead, CD151 supports α6-integrin-dependent morphology, adhesion (especially at low laminin or integrin density) and antibody-induced clustering – events all suggestive of possible effects on integrin diffusion. To characterize α6 integrin diffusion, we utilized single particle tracking (SPT), a technique with suitable sensitivity for studying slowly diffusing receptors such as integrins (Cairo et al., 2006). Several studies have focused on the regulation of integrin diffusion magnitude (see Chen et al., 2007; Gaborski et al., 2008; Hirata et al., 2005; Kucik et al., 1996; Yauch et al., 1997). However, CD151 ablation only minimally affected α6 integrin diffusion magnitude. Instead, there was a substantial and unexpected effect on diffusion mode, as α6 shifted away from random-confined diffusion (RCD) and towards directed motion (DMO). The shift towards DMO was accompanied by loss of diffusion sensitivity to talin knockdown and phorbol ester stimulation, and by diminished phorbol-ester-stimulated cell spreading function. These results suggest that CD151 supports α6 integrin functions by preventing unregulated DMO while favoring RCD. Hence, α6 integrins can properly translate outside-in signals and extracellular laminin cues into appropriate cell adhesion, migration, morphology, signaling and other events.
Results
CD151 affects cell cable formation and adhesion
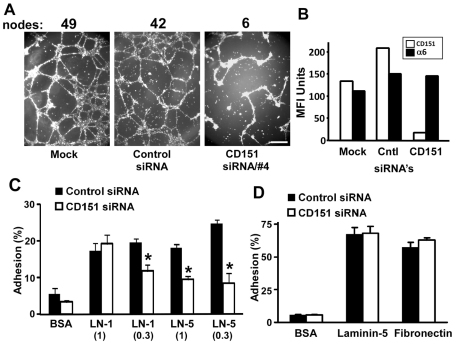
To gain new insights into functions of CD151 in MDA-MB-231 mammary cells, we first used a three-dimensional (3D) Matrigel ‘cable formation’ assay, which involves laminin adhesion and adhesion strengthening (Zhang et al., 2002) while modeling branching morphogenesis (Michaelson et al., 2005; Stahl et al., 1997). Alignment of MDA-MB-231 cells into a branching network of cellular cables was markedly diminished when CD151 was knocked down (Fig. 1A). CD151 surface expression was decreased by >80–90% upon siRNA treatment (Fig. 1B). Next we examined mechanisms by which CD151 might affect MDA-MB-231 cell invasion (Yang et al., 2008) and morphology (Fig. 1A) in Matrigel, which is largely composed of laminin-1. MDA-MB-231 cells treated with control or CD151-specific siRNAs showed little adhesion to a surface coated with BSA, and similar adhesion to laminin-1 (higher dose). However, cells lacking CD151 showed significantly less static adhesion to surfaces coated with a lower dose of laminin-1 (mediated by α6β1 integrin) or coated with two different doses of laminin-5 (Fig. 1C). Two different CD151 siRNAs yielded similar decreases in MDA-MB-231 adhesion (not shown). By contrast, knockdown of CD151 (by >90%) did not affect adhesion of MCF-10A cells to laminin-5 (mediated by α3β1 and α6β4 integrins) or fibronectin (Fig. 1D). MCF-10A adhesion to laminin-1 was insufficiently elevated above background to allow for evaluation of CD151-knockdown effects (not shown).
Fig. 1.
CD151 effects on mammary cell morphology and adhesion. (A) MDA-MB-231 cells treated with siRNAs were seeded at 5×104 cells/well on Matrigel-coated 48-well plates and photographed after 18 hours. Scale bar: 5 mm. Results are representative of many experiments. Numbers indicate nodes with three or more branches. (B) MDA-MB-231 cells were treated with siRNAs for 5 days, and then the surface expression of α6 integrin and CD151 was determined by flow cytometry, counting at least 5000 cells/experiment. MFI, mean fluorescence intensity. (C) After treatment with siRNAs for 5 days, MDA-MB-231 cells were seeded in triplicate into 96-well plates precoated with ECM proteins. After 45 minutes, static cell adhesion was assessed. Values are means ± s.e.m.; n=3; *P<0.02. (D) After siRNA treatment the MCF-10A cells were seeded in triplicate on the indicated ECM proteins for 30 minutes, and then static cell adhesion was assessed.
To gain insight into the mechanism of the different CD151 effects on MDA-MB-231 and MCF-10A cell adhesion, we analyzed integrin–CD151 complexes in those cells, using metabolic labeling with [3H]palmitate. Immunoprecipitation of α3 and α6 (but not α2) integrins yielded [3H]palmitate-labeled CD151 (supplementary material Fig. S1, lanes 1–4), and immunoprecipitation of CD151 yielded a mixture of α3 and α6 integrins (supplementary material Fig. S1, lanes 5, 6*). Hence, CD151 is associated with α3 and α6 integrins, as expected. Relevant to the cell adhesion results (Fig. 1C,D), fewer α6 integrins were present in MDA-MB-231 (supplementary material Fig. S1, lane 4) than in MCF-10A cells (lane 3). Cells containing fewer α6 integrins may be more dependent on CD151 for optimizing adhesion avidity on laminin-1 and laminin-5, especially at lower coating doses (Fig. 1C). By contrast, high levels of α6β4 integrin, together with α3β1 integrin, appear to support adhesion of MCF-10A cells to laminin-5, even when CD151 was ablated (Fig. 1D). The abundance of α3β1 integrin in MDA-MB-231 cells (supplementary material Fig. S1, lane 2) should not affect adhesion to laminin-1, because α3β1 integrin interacts minimally with laminin-1 (Delwel et al., 1994). The relative abundance of α3 and α6 subunits was confirmed by immunoblotting (supplementary material Fig. S1, lower panels). In a control experiment, no [3H]palmitate-labeled proteins associated with α2 integrin (supplementary material Fig. S1, lane 7), even though α2 is as abundant as α3 and α6 integrins in MCF-10A cells (not shown).
CD151 does not affect α6 integrin expression or activation, but may affect diffusion
Despite nearly complete removal of CD151 from MDA-MB-231 cells, surface levels of integrin α6, α3 and β1 subunits and tetraspanin CD82 did not decrease (supplementary material Fig. S2). Depletion of tetraspanin CD82 also did not diminish surface expression of CD151 or integrin subunits. Furthermore, the percentage of β1 integrin expressing the 9EG7 neoepitope [sometimes associated with integrin activation (Nilsson et al., 2006)] was also not decreased (9.9% increased to 11.5%; supplementary material Fig. S2).
Because CD151 can affect the molecular organization of integrin complexes (Takeda et al., 2007; Yang et al., 2008) and affect laminin adhesion selectively in a cell expressing low levels of α6 integrin (Fig. 1C,D), and we found that it especially affected adhesion to lower doses of laminin (Fig. 1C) and did not affect integrin expression or activation, we considered that CD151 might affect integrin lateral diffusion. To test this hypothesis, we first analyzed antibody-induced capping, which depends on integrin diffusion (Yauch et al., 1997). For capping experiments, we used MCF-10A cells, which express α6β4 integrin at sufficiently high levels (supplementary material Fig. S1) to be readily detected upon antibody-induced capping. When CD151 (green) was present in MCF-10A cells (Fig. 2A,C,E), >40% of cells showed large clusters of α6β4 integrin (red), colocalized with CD151 (yellow). However, upon CD151 silencing, <10% of cells showed integrin clusters (Fig. 2B,D,F). Cells with little remaining CD151 showed the most diffuse (and therefore less obvious) α6β4 staining (white arrowheads, Fig. 2B). In a control experiment, CD151 absence did not affect CD9 staining (Fig. 2G,H).
Fig. 2.
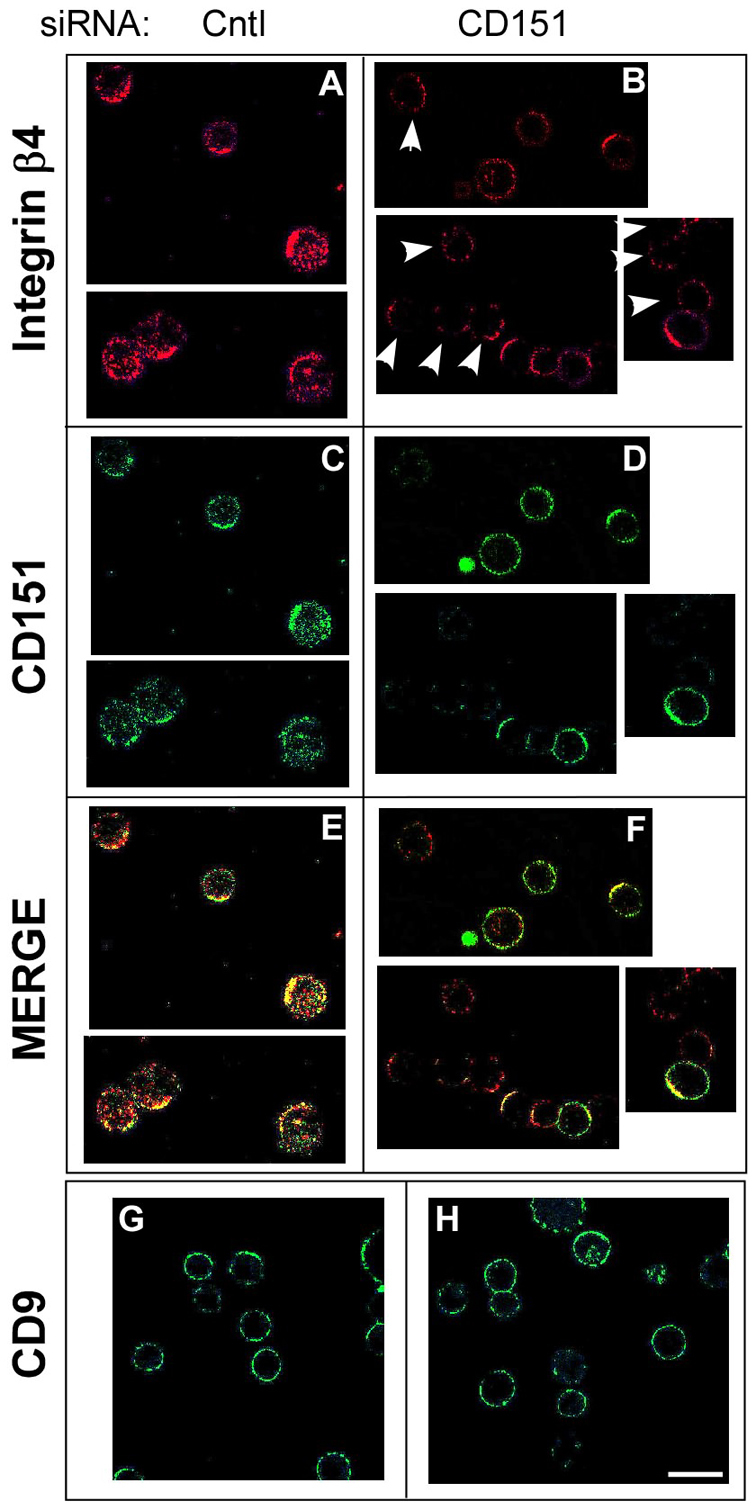
CD151 affects integrin clustering. (A-H) To induce α6β4 integrin clustering, siRNA-treated MCF-10A cells were detached and incubated with anti-β4 antibody (ASC-8) for 45 minutes (at 4°C), followed by staining with Alexa-Fluor-594-conjugated red secondary antibody at 37°C for 10 minutes (A,B). After washing, these cells were further stained with FITC-conjugated anti-CD151 (11G5A; C,D) or CD9 (MM2-57; G,H), allowed to adhere to a poly-lysine-coated coverslip, and then mounted with Prolong anti-fade reagent for examination by confocal microscopy. (E) Merged image of A and C; (F) merged image of B and D. Scale bar: 20 μm.
CD151 minimally affects diffusion rates
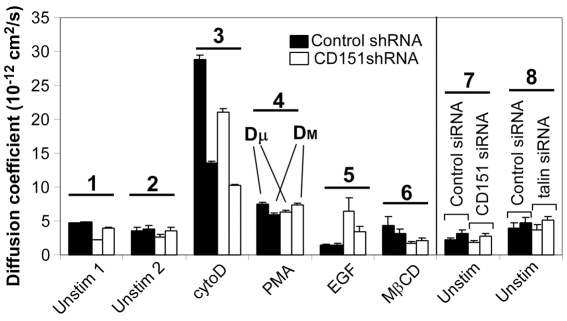
Next, we used SPT (Cairo et al., 2006) to assess CD151 effects on α6 integrin diffusion in the plasma membrane. We used MDA-MB-231 cells because the lower density of α6 integrin on MDA-MB-231, compared with MCF-10A cells (supplementary material Fig. S1), facilitates bead tracking. Both short-range (Dμ) and long-range (DM) diffusion coefficients showed minimal change when CD151 was ablated stably (Fig. 3, groups 1, 2) or transiently (group 7). Dμ is determined over a short time interval and DM over a longer time interval, and therefore measure the local and long-range mobility of α6 integrin, respectively (Mirchev and Golan, 2001). Diffusion coefficients measured at 37°C (Dμ =3.3±0.48× 10−12 cm2/second, DM =4.0±1.2×10−12 cm2/second; n=96) were similar to those measured at room temperature (Fig. 3, groups 1, 2, 7). Treatment of cells with cytoD increased diffusion rates (Fig. 3, group 3; supplementary material Fig. S3), consistent with integrin uncoupling from the actin cytoskeleton (Kucik et al., 1996; Yauch et al., 1997). Diffusion rates for α6 integrin were less affected by other agents [phorbol 12-myristate 13-acetate (PMA), epidermal growth factor (EGF), methyl-β-cyclodextrin (MβCD)]. Diffusion of α6 integrin in CD151-knockdown cells was either slightly lower (groups 3, 6), not different (group 4), or elevated (group 5), compared with controls. Mean Dμ values from knockdown cells (Fig. 3, first white bar, groups 1–7) were ~20% lower than in control cells (first black bar, groups 1–7). Similarly, the mean DM in knockdown cells (second white bar, groups 1–7) was only ~7% lower than in controls (second black bar, groups 1–7). Hence, CD151 is not a major regulator of the α6 integrin diffusion rate.
Fig. 3.
CD151 has little effect on diffusion coefficients. Diffusion coefficients (Dμ and DM), determined as described in the Materials and Methods, are indicated for each condition. Data supporting these bar graphs are shown in supplementary material Table S1, Table S2 and Fig. S3. Left panel: effect of CD151 shRNA; right panel: effect of CD151 siRNA (plus talin siRNA). Prior to adding beads, cells were either unstimulated, or treated with cytochalasin D (cytoD, 10 μM, 1 hour), phorbol 12-myristate 13-acetate (PMA, 0.2 μg/ml, 30 minutes), epidermal growth factor (EGF, 0.02 μg/ml, 1 hour) or methyl-β-cyclodextrin (MβCD, 2 mM, 1 hour). Values are means ± s.e.m. The numbers 1–8 indicate the sample groups.
CD151 affects diffusion mode
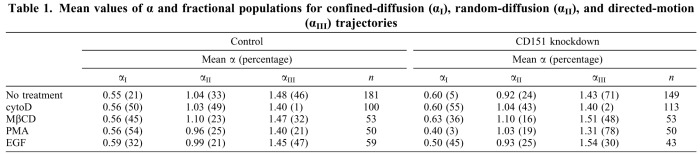
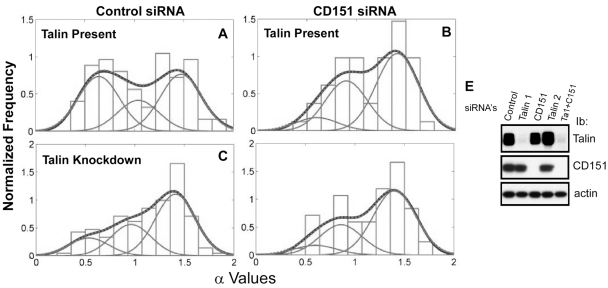
CD151-knockdown effects were much more pronounced when the mode of integrin diffusion was analyzed. SPT analysis discerned three distinct modes of α6 integrin diffusion (representative tracks shown in supplementary material Fig. S4). Correspondingly, we define three populations (I, II, III) with mean α-values of 0.4–0.6, 0.9–1.1 and 1.3–1.5. Population I trajectories represent weakly confined diffusion; population II includes random (Brownian) diffusion trajectories; and population III contains trajectories that show diffusion combined with directional (assisted) motion. Fractional populations from one experiment (Table 1) were statistically identical to mean fractional populations from three experiments (supplementary material Table S3), indicating reproducibility of results.
Table 1.
Mean values of α and fractional populations for confined-diffusion (αI), random-diffusion (αII), and directed-motion (αIII) trajectories
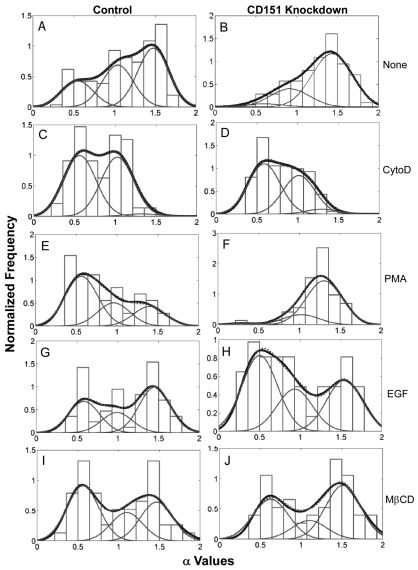
Control cells showed 21% confined diffusion, 33% random diffusion and 46% directed motion of α6 integrins on the cell surface (Fig. 4A; Table 1). In MDA-MD-231 cells lacking CD151, the confined population dropped to 5% and the directed-motion population rose to 71% (Fig. 4B; Table 1), indicating that CD151 enhances confined diffusion while diminishing DMO. Upon siRNA knockdown (targeting a different CD151 RNA sequence), confined diffusion of α6 integrin again decreased (from 38 to 9%), whereas random (from 22 to 36%) and directed (from 40 to 55%) motion increased (Fig. 5A; Table 2). CD151 did not affect αv integrin diffusion in MDA-MB-231 cells. Both with and without CD151 ablation, αv diffusion was mostly confined (84–88%), with a small random fraction (12–16%) and no directed motion (supplementary material Fig. S5A,B). CD151 ablation also did not change αv diffusion coefficients (Dμ and DM; supplementary material Fig. S5C–F).
Fig. 4.
Effects of stable CD151 knockdown on α6 integrin diffusion mode. Distribution of the diffusion-mode-parameter α in control MDA-MB-231 cells (left panels) and in CD151-knockdown cells (right panels) that were untreated (A,B) or treated with cytoD (C,D), PMA (E,F), EGF (G,H) or MβCD (I,J). The thick line in each panel represents the envelope (smoothing) curve of each distribution, and was calculated using all measured α-values as described in the text. The thick curve was fit with a sum of three Gaussian curves (thin lines), which produced the dotted line. Visibility of dotted lines is partially limited because of the excellent agreement between experimental data and curve-fit data. For comparison, a histogram of the experimental data is plotted in the background. The numbers of α values included in each analysis were: A, n=181; B, n=149; C, n=100; D, n=113; E, n=50; F, n=50; G, n=59; H, n=43; I, n=53; J, n=53.
Fig. 5.
Effects of CD151 and talin siRNAs on α6 integrin diffusion mode. (A–D) Distribution of the diffusion-mode-parameter α in control-siRNA-treated MDA-MB-231 cells (left panels) and in CD151-siRNA-treated cells (right panels), which were further untreated (A,B) or treated with talin knockdown siRNA (C,D). The curve designation is as in Fig. 4. The numbers of α-values included in each analysis were: A, n=87; B, n=57; C, n=148; D, n=59. (E) Cells were treated with siRNAs to knockdown talin 1, CD151, talin 2 and talin 1+CD151. Talin 1, CD151 and actin were then western blotted from whole cell lysates.
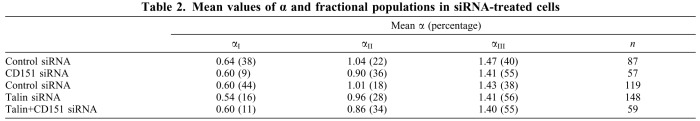
Table 2.
Mean values of α and fractional populations in siRNA-treated cells
The effect of the cytoskeleton on diffusion mode
Directed motion of α6 integrin in the cell membrane implies a role for the cytoskeleton in regulating integrin diffusion. Disruption of the actin cytoskeleton organization by cytochalasin D (cytoD) removed almost completely the directed-motion population (leaving 1–2%), while increasing confined (50–55%) and random (43–49%) populations (Fig. 4C,D; Table 1). Hence, directed motion may involve α6 integrin binding to the cytoskeleton and translocation along actin fibers. Removal of the cytoskeletal protein talin promoted DMO and diminished confined diffusion, thus mimicking removal of CD151 (Fig. 5A,C; Table 2). However, cells lacking CD151 were altered such that talin knockdown showed no additional DMO-inducing effects (Table 2; compare Fig. 5B and 5D). Thus, DMO in CD151-ablated cells has the unusual property of being actin-dependent but having no apparent contribution from talin.
Loss of response to phorbol ester stimulation
Although CD151 knockdown and control cells stimulated with PMA did not differ much in the rate of integrin diffusion (Fig. 3), they differed markedly in diffusion mode. In control MDA-MB-231 cells, PMA decreased α6 integrin DMO (from 46 to 21%) and increased confined diffusion (from 21 to 54%; Fig. 4A,E; Table 1). However, CD151-knockdown cells became unresponsive to PMA. As indicated, DMO was essentially unchanged (from 71 to 78%) and confined diffusion did not increase (from 5 to 3%; Fig. 4B,F; Table 1). In sharp contrast to PMA, other agents (i.e. EGF, MβCD) reversed DMO in CD151-knockdown cells. EGF decreased DMO (from 71 to 30%), with a corresponding major increase in confined diffusion (from 5 to 45%; Fig. 4B,H; Table 1). Treatment with MβCD (a cholesterol-depleting agent) decreased DMO for both control cells (from 46 to 32%) and CD151-ablated cells (from 71 to 48%), decreased random diffusion, and caused corresponding increases in confined populations (Fig. 4I,J; Table 1).
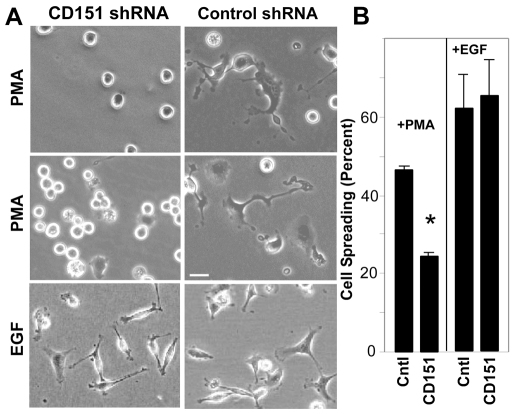
Integrin engagement with extracellular matrix ligands leads to PKC-dependent cell spreading, which exerts major control over cell proliferation and signaling (Assoian and Klein, 2008; Defilippi et al., 1999). Integrin PKC-dependent signaling can also overlap with EGF–EGFR signaling events to affect cell morphology (Cabodi et al., 2004; Rabinovitz et al., 2004; Wilhelmsen et al., 2007). We found that differential CD151 effects on PMA- and EGF-induced DMO translate to differential effects on cell spreading. Ablation of CD151 caused a pronounced decrease in PMA-induced [i.e. cAMP-dependent protein kinase (PKC)-dependent] MDA-MB-231 cell spreading on laminin-1, but minimally affected EGF-induced cell spreading (Fig. 6A,B). Thus, the inability of PMA (but not EGF) to reverse α6 integrin DMO may provide a mechanism for the loss of cell spreading function in PMA-treated (but not EGF-treated) CD151-knockdown cells.
Fig. 6.
CD151-knockdown cells show an aberrant spreading response to PMA. (A) MDA-MB-231 cells, stably expressing control vector or CD151 shRNA, were allowed to spread on plastic surfaces coated with laminin-1, for 1 hour, and then for 1 additional hour in the presence of 20 ng/ml PMA or 10 ng/ml EGF. Cells were then photographed. Results from two independent experiments are shown for the PMA treatment. Scale bar: 20 μm. (B) The percentage of cell spreading for each condition was determined. Values are means ± s.d. n=3; *P<0.002, t-test. The difference between the EGF-treated samples is not significant.
Discussion
It is well established that CD151 associates closely with laminin-binding integrins to fundamentally affect their functions (see Introduction). However, at a basic molecular level it has been unclear how this occurs. Because CD151 does not affect integrin expression or activation, we considered that it might affect integrin diffusion, which is known to affect integrin functions (Kucik et al., 1996; Yauch et al., 1997). Unexpectedly, CD151 had minimal effect on α6 integrin diffusion magnitude, but instead markedly affected diffusion mode. Our results suggest that CD151 restricts α6 integrin to random and confined (RCD) modes of diffusion, thereby making the integrin more available to participate in adhesion, spreading and other activities. That is, RCD confers on α6 integrin more freedom to reach newly formed adhesion sites and enforce and/or enlarge them, as it is released from other sites of disrupted adhesion; in contrast, DMO indicates inability of α6 integrin to be ‘recycled’ through diffusion in the membrane because it can serve in only a single or limited number of binding sites. In CD151-knockdown cells, α6 integrin showed dysregulated actin-dependent ‘directed’ motion, which could not be reversed by phorbol ester treatment. Hence, the absence of CD151 not only causes dysregulated α6-directed motion, but also appears to dysregulate conventional PKC isoforms proximal to α6 integrins.
Initial clues pointing to altered diffusion
We focused on MDA-MB-231 and MCF-10A basal-type mammary epithelial cells because CD151 markedly affects several α6 integrin functions in those cells (Sadej et al., 2009; Yang et al., 2008). Five observations led to the investigation of CD151 effects on integrin diffusion. First, we observed decreased cable formation by MDA-MB-231 cells, which is caused by decreased adhesion and/or adhesion strengthening on the laminin-1 Matrigel matrix. Consistent with this, CD151 knockdown diminished MDA-MB-231 cell adhesion to laminin-1.
Second, the contribution of CD151 to cell adhesion was more evident upon adhesion to lower doses of laminin-1 and laminin-5, in agreement with laminin adhesion results seen elsewhere (Winterwood et al., 2006). Under suboptimal integrin and/or ligand density conditions, integrin diffusion would become more crucial. Conversely, CD151 knockdown did not affect MCF-10A cell adhesion to laminin-5, probably because of the abundance of α6 integrins on those cells. This would elevate adhesion to a level above the threshold at which CD151 effects on diffusion may be detected in this type of assay.
Third, CD151 removal did not affect α6 integrin expression or activation. As seen here and elsewhere (Baleato et al., 2008; Sachs et al., 2006; Takeda et al., 2007; Wright et al., 2004), α6 expression remained unchanged. Furthermore, in contrast to another report (Iwase et al., 2003), the presence of CD151 did not promote the appearance of integrin ‘activation’ neoepitopes, suggesting little effect on the conformation and activation of laminin-binding integrins. In this regard, laminin-binding integrins are more stable than other integrins, and therefore less likely to undergo conformational changes during activation (Bazzoni et al., 1998). It was also previously noted that CD151 can affect α6 integrin adhesion strengthening without affecting ligand binding (Lammerding et al., 2003), again pointing to a mode of action independent of the effects on integrin ligand binding affinity and/or activation.
Fourth, ablation of CD151 caused a reduction in antibody-induced α6β4 clustering in MCF-10A cells. As shown previously, diminished clustering may arise as a result of altered integrin lateral diffusion (Yauch et al., 1997).
Fifth, removal of CD151 altered the spectrum of cell-surface partner proteins for α6 integrins, indicative of disconnection from tetraspanin-enriched microdomains (TEMs). As seen elsewhere, residence within TEMs can affect cell surface diffusion (Espenel et al., 2008).
Diffusion rate is minimally affected
Most integrin diffusion studies have focused on factors affecting diffusion magnitude (i.e. diffusion coefficients). However, knockdown of CD151 only marginally affected α6 integrin diffusion coefficients (Dμ and DM), as seen from results compiled using seven different experimental conditions. Stimulation with cytoD and PMA increased diffusion rates for both control and CD151-knockdown cells. Stimulation of α6 integrin diffusion rate by cytoD and PMA is consistent with results seen for other integrins (Kucik et al., 1996; Yauch et al., 1997; Zhou and Li, 2000). EGF stimulation differentially affected diffusion rates for control and CD151-knockdown cells, which may help to explain altered responses to EGF by CD151-knockdown cells (Yang et al., 2008). Nonetheless, despite a few small differences, the overall effect of CD151 knockdown on diffusion rates was minimal.
Diffusion measured for α6 integrin in MDA-MB-231 cells (2–10×10−12 cm2/second) is relatively slow compared with diffusion rates (50–1000×10−12 cm2/second) for other integrins in various cells (Barreiro et al., 2008; Grabham et al., 2000; Kucik et al., 1996; Yauch et al., 1997). However, in at least one other situation (Chen et al., 2007), rates of integrin diffusion approach the low levels seen here. Similar diffusion coefficients were measured at room temperature and 37°C, indicating that protein–lipid interactions are unlikely to be responsible for the low magnitude of α6 diffusion. More likely, α6 diffusion is constrained by the actin-based cytoskeleton (as mentioned above) and/or by β4 interactions with the intermediate filament cytoskeleton (Rabinovitz et al., 2004; Wilhelmsen et al., 2007).
CD151 ablation results in dysregulated DMO
Removal of CD151, using both shRNA and siRNA, consistently shifted α6 integrin from RCD to DMO. Conversely, disruption of the cytoskeleton with cytoD almost completely removed moderate DMO (from control cells) and excess DMO (from CD151-ablated cells), while enhancing RCD in both cases. Hence, α6 integrin DMO occurs along actin fibers, as typically seen for DMO of other integrins (Grabham et al., 2000; Bauer et al., 1993).
However, control and CD151-ablated cells showed notable differences in the regulation of α6 integrin DMO. Talin appears to play an active role in maintaining α6 in RCD mode (when CD151 is present) because talin ablation caused α6 integrin to shift away from RCD mode. Because talin links integrins with the actin cytoskeleton (Critchley, 2009), we expected that talin ablation would especially affect α6 integrin DMO in both control and CD151-ablated cells, because DMO is so dependent on the actin cytoskeleton. However, only control cells were sensitive to talin ablation, as seen by an increase in DMO trajectories. When CD151 is absent, talin no longer appears to play a role, because knockdown of talin had no effect on the percentage of trajectories in DMO mode. The insensitivity of DMO trajectories in CD151-ablated cells to talin ablation identifies an atypical case of an integrin–actin connection independent of talin, and supports the idea that α6 is undergoing dysregulated DMO in CD151-ablated cells. Loss of talin sensitivity and dysregulated DMO are both consistent with α6 being functionally less available to contribute to cell adhesion and spreading (see next section).
CD151-knockdown cells were also impervious to PMA, showing essentially no shift of DMO towards RCD. By contrast, α6 integrin DMO in control cells was shifted mostly to RCD upon PMA treatment. PMA activates and translocates novel and conventional PKC isozymes (e.g. PKCα and β). CD151 can recruit conventional PKCs to proximity with α6 integrins (Zhang et al., 2001). Consistent with this, CD151 removal decreases the association of PKCα with α6 integrins (Q. Li, X. H. Yang, F. Xu, C. Sharma, H.-X. Wang, K. Knoblich, I. Rabinovitz, S. R. Granter and M. E. Hemler, unpublished). This helps to explain why α6 integrin DMO becomes insensitive to PMA in CD151-ablated cells. By contrast, control cell CD151 can recruit PKC, which may lead to localized disruption of the actin cytoskeleton (Cuchillo-Ibanez et al., 2004), thus resulting in diminished α6 integrin DMO.
Integrin α6 DMO in CD151-knockdown cells remained somewhat sensitive to cholesterol depletion, perhaps because cholesterol depletion can disrupt cellular actin (Kwik et al., 2003) in a PKC-independent manner. Control cells, with constitutively more RCD and less DMO, were less affected by cholesterol depletion. In this regard, RCD of tetraspanin CD9 was also mostly unaffected by cholesterol depletion (Espenel et al., 2008). CD151-knockdown cells also retained sensitivity to EGF, with respect to shifting DMO towards RCD. This makes sense because EGF stimulates cell migration partly by signaling the transient disassembly of the actin cytoskeleton (Chang et al., 1995), in a PKC-independent manner.
Functional consequences of dysregulated α6 DMO
Three sets of results support the functional relevance of dysregulated α6 integrin DMO. First, the increased appearance of α6 integrin DMO in CD151-ablated MDA-MB-231 cells correlates with diminished adhesion to laminin-1 and diminished cable formation. Second, PMA stimulated CD151-knockdown cells failed to shift α6 away from DMO and failed to spread on laminin-1. By contrast, EGF stimulation of CD151-ablated cells triggered a substantial DMO to RCD shift, in parallel with abundant spreading on laminin-1. Third, ablation of CD151, which is not a partner for integrin αv and does not affect αv function (Winterwood et al., 2006), did not change either the diffusion mode or diffusion magnitude of αv integrin. These results help to rule out non-specific membrane perturbation effects, and reinforce the functional connection between CD151 and α6 integrin diffusion mode.
For technical reasons, α6 integrin diffusion was measured on the cell apical surface. Results obtained are indicative of global cellular responses to ‘outside-in’ signaling (e.g. as a result of treatment with EGF or MCD) or ‘inside-out’ signaling (e.g. as a result of perturbation by cytoD or talin siRNA). Furthermore, in dynamic situations, such as cell spreading and migration, the mode of apical α6 integrin diffusion should markedly affect function. In particular, RCD should confer more freedom for α6 integrin to reach newly formed adhesion sites and reinforce and/or enlarge them; by contrast, DMO could indicate the inability of α6 integrin to diffuse to new adhesion sites because the directionality of the diffusion could indicate strong integrin attachment to cytoskeletal binding sites. Thus, both RCD and DMO are likely to be necessary for normal cell function, and CD151 appears to have a crucial role in this regulation.
Summary and further functional implications
CD151 promotion of α6 integrin diffusion in RCD modes may be at least partly due to recruitment of α6 integrins into tetraspanin-enriched microdomains (TEMs) (Hemler, 2005; Nydegger et al., 2006; Yanez-Mo et al., 2009). Within TEMs, CD151 can associate with many other tetraspanins, including CD9 and CD82, which themselves display RCD, but not DMO (Danglot et al., 2010; Espenel et al., 2008). Localization of CD9 within TEMs correlates with confined diffusion (Espenel et al., 2008), whereas tetraspanin CD82 appears to help activated EGFR switch from confined to random diffusion (Danglot et al., 2010). These results reinforce the idea that TEMs may promote RCD while preventing DMO.
Molecules with type I (confined) and II (random) trajectories should be capable of ‘exploring’ larger areas in the cell membrane. Hence, by supporting RCD, CD151 may make α6 integrins more available to participate in cell adhesion, spreading and other functions. By contrast, α6 integrin that is diverted to type III (directed) trajectories, because of CD151 ablation, is in a dysregulated state. This state is characterized by aberrant association with the actin cytoskeleton, as evidenced by loss of sensitivity to both talin ablation and PMA stimulation. Normal integrin adhesion and spreading functions are typically dependent on an integrin–talin–actin connection, and stimulated by PMA. In the absence of CD151, α6 integrin appears to have lost both talin and PMA sensitivity, as well as the ability to ‘explore’ the cell membrane, thus explaining diminished adhesion and spreading functions. We predict that results seen here with MDA-MD-231 cells will extrapolate to most, if not all, of the many other cell types where α6 integrin is associated with CD151. Control of α6 integrin diffusion mode is probably also a principal factor in the subsequent effects of CD151 in kidney and skin development, skin wound healing, tumor growth, metastasis, angiogenesis and other functions. Our new mechanistic understanding also should help to explain why cells and mice lacking CD151 show substantial alterations in phorbol-ester-stimulated cell signaling and oncogenesis (Q. Li, X. H. Yang, F. Xu, C. Sharma, H.-X. Wang, K. Knoblich, I. Rabinovitz, S. R. Granter and M. E. Hemler, unpublished).
Materials and Methods
Cells and reagents
Immortalized MCF-10A and malignant MDA-MB-231 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) or Roswell Park Memorial Institute (RPMI) 1640 with 10% FCS (Life Sciences Technologies, Inc.), 10 mmol/l HEPES and antibiotics (penicillin and streptomycin). Anti-CD151 antibodies included 5C11 (Yauch et al., 1998), 1A5 (Testa et al., 1999) and FITC-conjugated 11G5A (GeneTex, Inc.). Other tetraspanin antibodies recognized CD9 (MM2/57 unconjugated and FITC-conjugated; from Biosource) and CD82 (M104, from Osamu Yoshi, Shionogi Institute, Osaka, Japan). Anti-integrin antibodies (Bazzoni et al., 1995; Bergelson et al., 1994; Lee et al., 1995; Weitzman et al., 1993) recognized α3 (A3-X8), α6 (GoH3, A6-ELE), α2 (IIE10), β1 (TS2/16), activated β1 (9EG7), and β4 (ASC-8) subunits. The anti-αv antibody was RMV-7 (eBiosciences). Targeting of CD151 (using siRNA#4 and siRNA#2) and CD82 (Yang et al., 2008) was achieved using oligonucleotides purchased from Dharmacon, and shRNA knockdown of CD151 was described previously (Yang et al., 2008).
Cell cable formation and adhesion assays
For cell cable formation, cells were plated on the surface of a layer of 3D Matrigel as described previously (Zhang et al., 2002). Cell images were acquired with a monochrome CCD camera (RT SPOT, Diagnostic Instruments, Sterling Heights, MI) on an Axiovert 135 inverted microscope (Zeiss). To measure static cell adhesion, 96-well plates were coated (at 5–20 μg/ml, for 12 hours) with laminin-1, laminin-5, BSA or fibronectin; and then cells were added, and attached cells were quantified using a Cytofluor 2300 measurement system (Millipore) as described previously (Bazzoni et al., 1995).
Immunoprecipitation and [3H]palmitate labeling
Because tetraspanins (e.g. CD151) and many of their partner proteins (e.g. α3, α6 and β4 integrin subunits) undergo post-translational palmitoylation, labeling with [3H]palmitate has been useful for evaluation of tetraspanin complexes (Takeda et al., 2007; Yang et al., 2004). Cells (siRNA-treated, 80–90% confluent) were washed in PBS, serum starved for 3–4 hours, pulsed for 1–2 hours in medium containing 0.2–0.3 mCi/ml [3H]palmitate plus 5% dialyzed fetal bovine serum, and then lysed in 1% Brij-96 for 5 hours at 4°C. Immunoprecipitation and protein detection were then carried out as described previously (Takeda et al., 2007; Yang et al., 2002; Yang et al., 2004).
Single particle tracking (SPT)
Beads (1 μm carboxylated polystyrene microspheres) were coated with anti-α6 antibody (A6-ELE) as described previously (Karnchanaphanurach et al., 2009). Briefly, beads in MES buffer, pH 6.1, were sonicated for 5 minutes and the carboxyl groups were sensitized by incubation in the presence of 100 mM N-hydroxysuccinimide and 0.4% 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride for 20 minutes at room temperature. After washing in MES, a mixture of anti-α6 antibody (A6-ELE) and isotype control IgG was incubated with the beads under constant shaking for 2 hours at room temperature. We used the minimum amount of specific antibody required to achieve selective binding of beads to cells, mixed with a sufficient amount of isotype IgG to ensure complete coverage of the bead surface. The conjugation reaction was terminated by adding ethanolamine to 100 mM final concentration for 15 minutes, followed by addition of BSA to 1% for 15 minutes. The beads were washed twice in PBS with 0.1% BSA and resuspended at 0.05% (m/v). Then, the beads were incubated for 1 hour with MDA-MB-231 cells, yielding 15–20% of cells with one or two attached beads. Under identical conditions, control beads coated with isotype IgG bound to less than 2% of cells. In some experiments, cells were also treated with cytochalasin D (cytoD; 10 μM, 1 hour), methyl-β-cyclodextrin (MβCD; 2 mM, 1 hour), phorbol 12-myristate 13-acetate (PMA; 0.2 μg/ml, 30 minutes) or epidermal growth factor (EGF; 0.02 μg/ml, 1 hour) prior to adding the beads. For single particle tracking (Cairo et al., 2006), video recordings at 30 frames/second were obtained for ~70 seconds, and 30 beads were tracked in each experiment. Video data were processed with MetaMorph (Molecular Devices) and converted to trajectories, then analyzed with mean square displacement (MSD) analysis implemented in a custom MATLAB (MathWorks) program. The microdiffusion coefficient (Dμ) was calculated by performing a linear fit to MSD=4Dμt, using the first four increments of the MSD versus time interval curve. The macrodiffusion coefficient (DM) was calculated by fitting the initial third of the MSD versus time interval curve to the equation MSD=4DMtα. The parameter α classifies the mode of diffusion (Mirchev and Golan, 2001). Diffusion trajectories were grouped based on population analysis as described previously (Cairo et al., 2006). Briefly, a kernel-smoothing probability density calculation was used to smooth the normalized distribution of α-values for each experimental condition. This envelope (smoothing) curve was then fitted to the sum of three Gaussian distributions, which represented three populations of diffusion trajectories. For all of the α-value distributions obtained in control cells under the various experimental conditions, the 3-Gaussian fit gave a better fit than a 2-Gaussian fit, as determined by applying the F-statistic at 95% to test the significance of the goodness of fit. For consistency, we applied a 3-Gaussian fit to all experimental conditions (Tables 1,2; Figs 4,5; supplementary material Tables S1–S3, Figs. S3,S5). The three Gaussians had intersection points in the ranges of 0.7–0.9 (leftmost and middle Gaussian curves) and 1.1–1.2 (middle and rightmost Gaussian curves), giving experimental thresholds to classify trajectories based on their α-values. Thus, α<0.8 (leftmost Gaussian curve) represented confined or corralled motion, 0.8<α<1.2 (middle Gaussian curve) was consistent with Brownian diffusion, and α>1.2 (rightmost Gaussian curve) represented directed diffusion. Two normal distributions were considered statistically different if: [(σ12 + σ22)/(σ12)(σ22)](μ1–μ2)2>8, where μ and σ denote the mean and standard deviation of each distribution (Johnson et al., 2004). The fractional percentage of each population was calculated from the normalized weighting factor by which each Gaussian is multiplied in the best-fitted sum. If, instead of using empirically determined thresholds derived directly from experimental data, we used fixed thresholds (α<0.8; 0.8<α<1.2; α>1.2) throughout, the fractional percentages assigned to each population (confined, Brownian, directed; Table 1, Table 2; supplementary material Fig. S5) would change only slightly (typically 3–4%), and none of the conclusions of the study would be affected.
Cell spreading
Cell spreading was recorded using a Nikon Eclipse Ti Series inverted microscope, equipped with a humidified 37°C CO2 chamber, automated mobile stage and focusing system, and capability for simultaneously capturing cell movements in real-time in 24 bright fields (in a 24-well plate) under a 20× objective. Cells, in the presence of 0.1% fetal bovine serum, were plated for 1 hour on laminin-1-coated surfaces, in the presence of either 10 ng/ml EGF or 20 ng/ml PMA, as indicated.
Footnotes
Funding
This work was supported by National Institutes of Health [grant numbers CA42368 to M.E.H. and HL32854 to D.E.G.]; and by a S.G. Komen Career Catalyst Award [grant number KG081481 to X.H.Y.] and a DOD Concept Award [grant number W81XWH-07-1-0568 to X.H.Y.]. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.093963/-/DC1
References
- Ang J., Lijovic M., Ashman L. K., Kan K., Frauman A. G. (2004). CD151 protein expression predicts the clinical outcome of low-grade primary prostate cancer better than histologic grading: a new prognostic indicator? Cancer Epidemiol. Biomarkers Prev. 13, 1717-1721 [PubMed] [Google Scholar]
- Ashman L. K. (2002). CD151. J. Biol. Regul. Homeost. Agents 16, 223-226 [PubMed] [Google Scholar]
- Assoian R. K., Klein E. A. (2008). Growth control by intracellular tension and extracellular stiffness. Trends Cell Biol. 18, 347-352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin G., Novitskaya V., Sadej R., Pochec E., Litynska A., Hartmann C., Williams J., Ashman L., Eble J. A., Berditchevski F. (2008). Tetraspanin cd151 regulates glycosylation of alpha3beta1 integrin. J. Biol. Chem. 283, 35445-35454 [DOI] [PubMed] [Google Scholar]
- Baleato R. M., Guthrie P. L., Gubler M. C., Ashman L. K., Roselli S. (2008). Deletion of CD151 results in a strain-dependent glomerular disease due to severe alterations of the glomerular basement membrane. Am. J. Pathol. 173, 927-937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro O., Zamai M., Yanez-Mo M., Tejera E., Lopez-Romero P., Monk P. N., Gratton E., Caiolfa V. R., Sanchez-Madrid F. (2008). Endothelial adhesion receptors are recruited to adherent leukocytes by inclusion in preformed tetraspanin nanoplatforms. J. Cell Biol. 183, 527-542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer J. S., Varner J., Schreiner C., Kornberg L., Nicholas R., Juliano R. L. (1993). Functional role of the cytoplasmic domain of the integrin α5 subunit. J. Cell Biol. 122, 209-221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzoni G., Shih D.-T., Buck C. A., Hemler M. E. (1995). MAb 9EG7 defines a novel β1 integrin epitope induced by soluble ligand and manganese, but inhibited by calcium. J. Biol. Chem. 270, 25570-25577 [DOI] [PubMed] [Google Scholar]
- Bazzoni G., Ma L., Blue M.-L., Hemler M. E. (1998). Divalent cations and ligands induce conformational changes that are highly divergent among β1 integrins. J. Biol. Chem. 273, 6670-6678 [DOI] [PubMed] [Google Scholar]
- Belkin A. M., Stepp M. A. (2000). Integrins as receptors for laminins. Microsc. Res. Tech. 51, 280-301 [DOI] [PubMed] [Google Scholar]
- Berditchevski F. (2001). Complexes of tetraspanins with integrins: more than meets the eye. J. Cell Sci. 114, 4143-4151 [DOI] [PubMed] [Google Scholar]
- Berditchevski F., Gilbert E., Griffiths M. R., Fitter S., Ashman L., Jenner S. J. (2001). Analysis of the CD151-alpha3beta1 integrin and CD151-tetraspanin interactions by mutagenesis. J. Biol. Chem. 276, 41165-41174 [DOI] [PubMed] [Google Scholar]
- Berditchevski F., Odintsova E., Sawada S., Gilbert E. (2002). Expression of the palmitoylation-deficient CD151 weakens the association of alpha 3beta 1 integrin with the tetraspanin-enriched microdomains and affects integrin-dependent signalling. J. Biol. Chem. 277, 36991-37000 [DOI] [PubMed] [Google Scholar]
- Bergelson J. M., St. John N. F., Kawaguchi S., Pasqualini R., Berdichevsky F., Hemler M. E., Finberg R. W. (1994). The I domain is essential for echovirus 1 interaction with VLA-2. Cell Adhes. Commun. 2, 455-464 [DOI] [PubMed] [Google Scholar]
- Cabodi S., Moro L., Bergatto E., Boeri E. E., Di Stefano P., Turco E., Tarone G., Defilippi P. (2004). Integrin regulation of epidermal growth factor (EGF) receptor and of EGF-dependent responses. Biochem. Soc. Trans. 32, 438-442 [DOI] [PubMed] [Google Scholar]
- Cairo C. W., Mirchev R., Golan D. E. (2006). Cytoskeletal regulation couples LFA-1 conformational changes to receptor lateral mobility and clustering. Immunity 25, 297-308 [DOI] [PubMed] [Google Scholar]
- Chang J. H., Gill S., Settleman J., Parsons S. J. (1995). c-Src regulates the simultaneous rearrangement of actin cytoskeleton, p190RhoGAP, and p120RasGAP following epidermal growth factor stimulation. J. Cell Biol. 130, 355-368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Titushkin I., Stroscio M., Cho M. (2007). Altered membrane dynamics of quantum dot-conjugated integrins during osteogenic differentiation of human bone marrow derived progenitor cells. Biophys. J. 92, 1399-1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Critchley D. R. (2009). Biochemical and structural properties of the integrin-associated cytoskeletal protein talin. Annu. Rev. Biophys. 38, 235-254 [DOI] [PubMed] [Google Scholar]
- Cuchillo-Ibanez I., Lejen T., Albillos A., Rose S. D., Olivares R., Villarroya M., Garcia A. G., Trifaro J. M. (2004). Mitochondrial calcium sequestration and protein kinase C cooperate in the regulation of cortical F-actin disassembly and secretion in bovine chromaffin cells. J. Physiol. 560, 63-76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danglot L., Chaineau M., Dahan M., Gendron M. C., Boggetto N., Perez F., Galli T. (2010). Role of TI-VAMP and CD82 in EGFR cell-surface dynamics and signaling. J. Cell Sci. 123, 723-735 [DOI] [PubMed] [Google Scholar]
- Defilippi P., Olivo C., Venturino M., Dolce L., Silengo L., Tarone G. (1999). Actin cytoskeleton organization in response to integrin-mediated adhesion. Microsc. Res. Tech. 47, 67-78 [DOI] [PubMed] [Google Scholar]
- Delwel G. O., de Melker A. A., Hogervorst F., Jaspars L. H., Fles D. L., Kuikman I., Lindblom A., Paulsson M., Timpl R., Sonnenberg A. (1994). Distinct and overlapping ligand specificities of the alpha 3A beta 1 and alpha 6A beta 1 integrins: recognition of laminin isoforms. Mol. Biol. Cell 5, 203-215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espenel C., Margeat E., Dosset P., Arduise C., Le Grimellec C., Royer C. A., Boucheix C., Rubinstein E., Milhiet P. E. (2008). Single-molecule analysis of CD9 dynamics and partitioning reveals multiple modes of interaction in the tetraspanin web. J. Cell Biol. 182, 765-776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaborski T. R., Clark A., Jr, Waugh R. E., McGrath J. L. (2008). Membrane mobility of beta2 integrins and rolling associated adhesion molecules in resting neutrophils. Biophys. J. 95, 4934-4947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabham P. W., Foley M., Umeojiako A., Goldberg D. J. (2000). Nerve growth factor stimulates coupling of beta1 integrin to distinct transport mechanisms in the filopodia of growth cones. J. Cell Sci. 113, 3003-3012 [DOI] [PubMed] [Google Scholar]
- Haeuw J. F., Goetsch L., Bailly C., Corvaia N. (2011). Tetraspanin CD151 as a target for antibody-based cancer immunotherapy. Biochem. Soc. Trans. 39, 553-558 [DOI] [PubMed] [Google Scholar]
- Hemler M. E. (2005). Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 6, 801-811 [DOI] [PubMed] [Google Scholar]
- Hirata H., Ohki K., Miyata H. (2005). Mobility of integrin alpha5beta1 measured on the isolated ventral membranes of human skin fibroblasts. Biochim. Biophys. Acta 1723, 100-105 [DOI] [PubMed] [Google Scholar]
- Iwase H., Omoto Y., Toyama T., Yamashita H., Hara Y., Sugiura H., Zhang Z. (2003). Clinical significance of AIB1 expression in human breast cancer. Breast Cancer Res. Treat. 80, 339-345 [DOI] [PubMed] [Google Scholar]
- Johnson J. L., Winterwood N., DeMali K. A., Stipp C. S. (2009). Tetraspanin CD151 regulates RhoA activation and the dynamic stability of carcinoma cell-cell contacts. J. Cell Sci. 122, 2263-2273 [DOI] [PubMed] [Google Scholar]
- Johnson N. L., Kotz S., Balakrishnan N. (2004). Continuous Univariate Distributions. New York: Wiley; [Google Scholar]
- Kagan A., Feld S., Chemke J., Bar-Khayim Y. (1988). Occurrence of hereditary nephritis, pretibial epidermolysis bullosa and beta-thalassemia minor in two siblings with end-stage renal disease. Nephron 49, 331-332 [DOI] [PubMed] [Google Scholar]
- Karamatic Crew V., Burton N., Kagan A., Green C. A., Levene C., Flinter F., Brady L. R., Daniels G., Anstee D. J. (2004). CD151, the first member of the tetraspanin (TM4) superfamily detected on erythrocytes, is essential for the correct assembly of human basement membranes in kidney and skin. Blood 104, 2217-2223 [DOI] [PubMed] [Google Scholar]
- Karnchanaphanurach P., Mirchev R., Ghiran I., Asara J. M., Papahadjopoulos-Sternberg B., Nicholson-Weller A., Golan D. E. (2009). C3b deposition on human erythrocytes induces the formation of a membrane skeleton-linked protein complex. J. Clin. Invest. 119, 788-801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazarov A. R., Yang X., Stipp C. S., Sehgal B., Hemler M. E. (2002). An extracellular site on tetraspanin CD151 determines α3 and α6 integrin-dependent cellular morphology. J. Cell Biol. 158, 1299-1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno M., Hasegawa H., Miyake M., Yamamoto T., Fujita S. (2002). CD151 enhances cell motility and metastasis of cancer cells in the presence of focal adhesion kinase. Int. J. Cancer 97, 336-343 [DOI] [PubMed] [Google Scholar]
- Kucik D. F., Dustin M. L., Miller J. M., Brown E. J. (1996). Adhesion-activating phorbol ester increases the mobility of leukocyte integrin LFA-1 in cultured lymphocytes. J. Clin. Invest. 97, 2139-2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwik J., Boyle S., Fooksman D., Margolis L., Sheetz M. P., Edidin M. (2003). Membrane cholesterol, lateral mobility, and the phosphatidylinositol 4,5-bisphosphate-dependent organization of cell actin. Proc. Natl. Acad. Sci. USA 100, 13964-13969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammerding J., Kazarov A. R., Huang H., Lee R. T., Hemler M. E. (2003). Tetraspanin CD151 regulates alpha6beta1 integrin adhesion strengthening. Proc. Natl. Acad. Sci. USA 100, 7616-7621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R. T., Berditchevski F., Cheng G. C., Hemler M. E. (1995). Integrin-mediated collagen matrix reorganization by cultured human vascular smooth muscle cells. Circ. Res. 76, 209-214 [DOI] [PubMed] [Google Scholar]
- Liu L., He B., Liu W. M., Zhou D., Cox J. V., Zhang X. A. (2007). Tetraspanin CD151 promotes cell migration by regulating integrin trafficking. J. Biol. Chem. 282, 31631-31642 [DOI] [PubMed] [Google Scholar]
- Michaelson J. S., Cho S., Browning B., Zheng T. S., Lincecum J. M., Wang M. Z., Hsu Y. M., Burkly L. C. (2005). Tweak induces mammary epithelial branching morphogenesis. Oncogene 24, 2613-2624 [DOI] [PubMed] [Google Scholar]
- Mirchev R., Golan D. E. (2001). Single-particle tracking and laser optical tweezers studies of the dynamics of individual protein molecules in membranes of intact human and mouse red cells. Blood Cells Mol. Dis. 27, 143-147 [DOI] [PubMed] [Google Scholar]
- Neve R. M., Chin K., Fridlyand J., Yeh J., Baehner F. L., Fevr T., Clark L., Bayani N., Coppe J. P., Tong F., et al. (2006). A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10, 515-527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson S., Kaniowska D., Brakebusch C., Fassler R., Johansson S. (2006). Threonine 788 in integrin subunit beta1 regulates integrin activation. Exp. Cell Res. 312, 844-853 [DOI] [PubMed] [Google Scholar]
- Nishiuchi R., Sanzen N., Nada S., Sumida Y., Wada Y., Okada M., Takagi J., Hasegawa H., Sekiguchi K. (2005). Potentiation of the ligand-binding activity of integrin alpha3beta1 via association with tetraspanin CD151. Proc. Natl. Acad. Sci. USA 102, 1939-1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitskaya V., Romanska H., Dawoud M., Jones J. L., Berditchevski F. (2010). Tetraspanin CD151 regulates growth of mammary epithelial cells in three-dimensional extracellular matrix: implication for mammary ductal carcinoma in situ. Cancer Res. 70, 4698-4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nydegger S., Khurana S., Krementsov D. N., Foti M., Thali M. (2006). Mapping of tetraspanin-enriched microdomains that can function as gateways for HIV-1. J. Cell Biol. 173, 795-807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovitz I., Tsomo L., Mercurio A. M. (2004). Protein kinase C-alpha phosphorylation of specific serines in the connecting segment of the beta 4 integrin regulates the dynamics of type II hemidesmosomes. Mol. Cell. Biol. 24, 4351-4360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasekhar V. K., Studer L., Gerald W., Socci N. D., Scher H. I. (2011). Tumour-initiating stem-like cells in human prostate cancer exhibit increased NF-kappaB signalling. Nat. Commun. 2, 162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanska H. M., Berditchevski F. (2011). Tetraspanins in human epithelial malignancies. J. Pathol. 223, 4-14 [DOI] [PubMed] [Google Scholar]
- Sachs N., Kreft M., van den Bergh Weerman M. A., Beynon A. J., Peters T. A., Weening J. J., Sonnenberg A. (2006). Kidney failure in mice lacking the tetraspanin CD151. J. Cell Biol. 175, 33-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadej R., Romanska H., Baldwin G., Gkirtzimanaki K., Novitskaya V., Filer A. D., Krcova Z., Kusinska R., Ehrmann J., Buckley C. D., et al. (2009). CD151 regulates tumorigenesis by modulating the communication between tumor cells and endothelium. Mol. Cancer Res. 7, 787-798 [DOI] [PubMed] [Google Scholar]
- Stahl S., Weitzman S., Jones J. C. (1997). The role of laminin-5 and its receptors in mammary epithelial cell branching morphogenesis. J. Cell Sci. 110, 55-63 [DOI] [PubMed] [Google Scholar]
- Sterk L. M., Geuijen C. A., van Den Berg J. G., Claessen N., Weening J. J., Sonnenberg A. (2002). Association of the tetraspanin CD151 with the laminin-binding integrins alpha3beta1, alpha6beta1, alpha6beta4 and alpha7beta1 in cells in culture and in vivo. J. Cell Sci. 115, 1161-1173 [DOI] [PubMed] [Google Scholar]
- Stipp C. S., Kolesnikova T. V., Hemler M. E. (2003a). EWI-2 regulates alpha3beta1 integrin-dependent cell functions on laminin-5. J. Cell Biol. 163, 1167-1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stipp C. S., Kolesnikova T. V., Hemler M. E. (2003b). Functional domains in tetraspanin proteins. Trends Biochem. Sci. 28, 106-112 [DOI] [PubMed] [Google Scholar]
- Takeda Y., Kazarov A. R., Butterfield C. E., Hopkins B. D., Benjamin L. E., Kaipainen A., Hemler M. E. (2007). Deletion of tetraspanin Cd151 results in decreased pathologic angiogenesis in vivo and in vitro. Blood 109, 1524-1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa J. E., Brooks P. C., Lin J. M., Quigley J. P. (1999). Eukaryotic expression cloning with an antimetastatic monoclonal antibody identifies a tetraspanin (PETA-3/CD151) as an effector of human tumor cell migration and metastasis. Cancer Res. 59, 3812-3820 [PubMed] [Google Scholar]
- Tokuhara T., Hasegawa H., Hattori N., Ishida H., Taki T., Tachibana S., Sasaki S., Miyake M. (2001). Clinical significance of CD151 gene expression in non-small cell lung cancer. Clin. Cancer Res. 7, 4109-4114 [PubMed] [Google Scholar]
- Weitzman J. B., Pasqualini R., Takada Y., Hemler M. E. (1993). The function and distinctive regulation of the integrin VLA-3 in cell adhesion, spreading and homotypic cell aggregation. J. Biol. Chem. 268, 8651-8657 [PubMed] [Google Scholar]
- Wilhelmsen K., Litjens S. H., Kuikman I., Margadant C., van Rheenen J., Sonnenberg A. (2007). Serine phosphorylation of the integrin beta4 subunit is necessary for epidermal growth factor receptor induced hemidesmosome disruption. Mol. Biol. Cell 18, 3512-3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterwood N. E., Varzavand A., Meland M. N., Ashman L. K., Stipp C. S. (2006). A critical role for tetraspanin CD151 in alpha3beta1 and alpha6beta4 integrin-dependent tumor cell functions on laminin-5. Mol. Biol. Cell 17, 2707-2721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright M. D., Geary S. M., Fitter S., Moseley G. W., Lau L. M., Sheng K. C., Apostolopoulos V., Stanley E. G., Jackson D. E., Ashman L. K. (2004). Characterization of mice lacking the tetraspanin superfamily member CD151. Mol. Cell. Biol. 24, 5978-5988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanez-Mo M., Barreiro O., Gordon-Alonso M., Sala-Valdes M., Sanchez-Madrid F. (2009). Tetraspanin-enriched microdomains: a functional unit in cell plasma membranes. Trends Cell Biol. 19, 434-446 [DOI] [PubMed] [Google Scholar]
- Yang X., Claas C., Kraeft S. K., Chen L. B., Wang Z., Kreidberg J. A., Hemler M. E. (2002). Palmitoylation of tetraspanin proteins: modulation of CD151 lateral interactions, subcellular distribution, and integrin-dependent cell morphology. Mol. Biol. Cell 13, 767-781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Kovalenko O. V., Tang W., Claas C., Stipp C. S., Hemler M. E. (2004). Palmitoylation supports assembly and function of integrin-tetraspanin complexes. J. Cell Biol. 167, 1231-1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. H., Richardson A. L., Torres-Arzayus M. I., Zhou P., Sharma C., Kazarov A. R., Andzelm M. M., Strominger J. L., Brown M., Hemler M. E. (2008). CD151 accelerates breast cancer by regulating α6 integrin functions, signaling, and molecular organization. Cancer Res. 68, 3204-3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. H., Flores L. M., Li Q., Zhou P., Xu F., Krop I. E., Hemler M. E. (2010). Disruption of laminin-integrin-CD151-focal adhesion kinase axis sensitizes breast cancer cells to ErbB2 antagonists. Cancer Res. 70, 2256-2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauch R. L., Felsenfeld D., Kraeft S.-K., Chen L. B., Sheetz M., Hemler M. E. (1997). Mutational evidence for control of cell adhesion through integrin recruitment, independent of ligand binding. J. Exp. Med. 186, 1347-1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauch R. L., Berditchevski F., Harler M. B., Reichner J., Hemler M. E. (1998). Highly stoichiometric, stable and specific association of integrin α3β1 with CD151 provides a major link to phosphatidylinositol 4-kinase and may regulate cell migration. Mol. Biol. Cell 9, 2751-2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauch R. L., Kazarov A. R., Desai B., Lee R. T., Hemler M. E. (2000). Direct extracellular contact between integrin α3β1 and TM4SF protein CD151. J. Biol. Chem. 275, 9230-9238 [DOI] [PubMed] [Google Scholar]
- Zevian S., Winterwood N. E., Stipp C. S. (2011). Structure-function analysis of tetraspanin CD151 reveals distinct requirements for tumor cell behaviors mediated by alpha3beta1 versus alpha6beta4 integrin. J. Biol. Chem. 286, 7496-7506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. A., Bontrager A. L., Hemler M. E. (2001). TM4SF proteins associate with activated PKC and Link PKC to specific beta1 integrins. J. Biol. Chem. 276, 25005-25013 [DOI] [PubMed] [Google Scholar]
- Zhang X. A., Kazarov A. R., Yang X., Bontrager A. L., Stipp C. S., Hemler M. E. (2002). Function of the tetraspanin CD151-a6b1 integrin complex during cellular morphogenesis. Mol. Biol. Cell 13, 1-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Li J. (2000). Macrophage-enriched myristoylated alanine-rich C kinase substrate and its phosphorylation is required for the phorbol ester-stimulated diffusion of beta 2 integrin molecules. J. Biol. Chem. 275, 20217-20222 [DOI] [PubMed] [Google Scholar]
- Zijlstra A., Lewis J., Degryse B., Stuhlmann H., Quigley J. P. (2008). The inhibition of tumor cell intravasation and subsequent metastasis via regulation of in vivo tumor cell motility by the tetraspanin CD151. Cancer Cell 13, 221-234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo H. J., Lin J. Y., Liu Z. Y., Liu W. F., Liu T., Yang J., Liu Y., Wang D. W., Liu Z. X. (2010). Activation of the ERK signaling pathway is involved in CD151-induced angiogenic effects on the formation of CD151-integrin complexes. Acta Pharmacol. Sin. 31, 805-812 [DOI] [PMC free article] [PubMed] [Google Scholar]