Tumor-suppressor genes are necessary for maintaining cell growth control. When these genes fail to function properly, deregulated growth—a defining feature of cancer cells—ensues. The p53 tumor suppressor, first described in 1979 as an interacting protein with the transforming large T-antigen of simian virus 40, has emerged as the most commonly altered gene and growth suppressing pathway in human cancer. p53 protein is stabilized in response to a number of stresses, including exposure of cells to DNA damaging agents, hypoxia, or inappropriate expression or activation of a number of cellular or viral oncogenes. The upstream regulatory interactions and downstream events that follow from p53 stabilization provide a complex network of signals ultimately leading to cell cycle arrest, apoptosis, and tumor suppression (1, 2). We know relatively little, however, about the cell cycle dependence of the p53 response, and the role of p53 in the response to replication blockade in S-phase of the cell cycle. Studies reported by Gottifredi et al. (3) in this issue of PNAS shed light on the p53 response in S-phase, differentiate between passage through S-phase and blockade in S-phase in terms of the events downstream of p53 activation, and provide evidence that the transcriptional program downstream of “activated” p53 is drastically attenuated if DNA synthesis is blocked. Interesting questions arise regarding how and why cells stabilize and inhibit p53 during S-phase blockade and parallels can be drawn to the transforming adenovirus E1A protein or the hypoxia inducible factor activating drug desferoxamine, both of which also stabilize p53 while inhibiting its transactivation potential.
It has become clear that, although the p53 protein can interact with a number of cellular proteins or repress gene expression, the ability of p53 to activate transcription is of critical importance to its function in tumor suppression (4). Following exposure to DNA damaging agents, one of the most important effects of p53 stabilization, in nearly all mammalian cell types, is a block in the cell-division cycle (Fig. 1). The p53 protein binds directly to genomic p53 response elements and stimulates the expression of p21WAF1/CIP1, an inhibitor of cyclin-dependent kinases (CDKs). CDKs are key regulators of the cell cycle, working together with their partners, cyclin proteins, to make sure that, for example, DNA replication (S-phase) follows smoothly from the cellular growth phase known as G1. Through its negative effects on various CDKs, p21WAF1/CIP1 inhibits both the G1–S and the G2–mitosis transitions. Recent studies show clearly that p53 is required to maintain a durable arrest in G1 following DNA damage, whereas other events such as cyclin D1 or cdc25A degradation may be more important for rapid (p53-independent) G1 arrest initiation (5, 6). Other effectors of p53, including the glycosylated protein Reprimo, can also arrest cells in G2 phase through effects on Cdc2 kinase activity and cyclin B1 nuclear translocation (7). In epithelial cells—those that line organs such as the intestine and bladder—p53 also stimulates the expression of protein 14–3-3σ, which sequesters cyclin B1–CDK1 complexes outside the nucleus and thereby helps to maintain a G2 block (8, 9). Interestingly, the inhibition of 14–3-3σ can, in a single step, make primary human epithelial cells grow indefinitely in culture (10). This immortality may be a key feature distinguishing tumor cells from normal cells. It is clear that in G2, p53 and its targets p21 and 14–3-3σ are required to maintain cell cycle arrest (8).
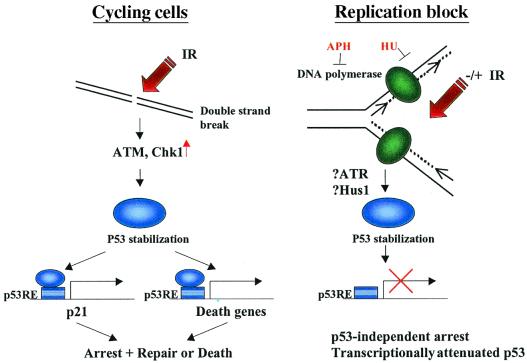
Figure 1.
Impaired p53-mediated transcriptional response during S-phase blockade in the absence or presence of additional concomitant DNA damaging exposures. In the case of cycling cells, double-strand breaks lead to p53 stabilization through the activation of ATM and Chk1 kinases. Stabilized and activated p53 binds to the regulatory regions of target genes, which mediate its effects (Left). However, when cells are blocked in S-phase, p53 fails to induce some of its target genes even though it is stabilized (Right). Moreover, even x-ray irradiation cannot restore the transcriptional activation of p53 despite phosphorylation, acetylation, and stabilization of p53. ATR and Hus1 are conserved PI-3 kinase-like and PCNA-like, respectively, proteins that have been implicated in the cellular response to UV exposure and replication blockade (11). p53RE, consensus p53 DNA binding sites; HU and APH, hydroxyurea and aphidicolin, respectively.
Although it is evident that p53 plays an important role in G1/G2 checkpoints to conserve genomic integrity, the contribution of p53 for an S-phase checkpoint is less well understood (11). It is well known that the upstream regulator of p53, ATM, is required for an S-phase checkpoint, and that ATM-deficient cells display both radiosensitivity and radioresistant DNA synthesis (12). Two other pathways that ultimately lead to p53 stabilization are known to exist: (i) Following inappropriate oncogenic signals, the ARF protein sequesters MDM2 away from p53 so that it becomes stabilized, and (ii) In response to UV light, other kinases related to ATM (such as ATR), but not ATM, may phosphorylate p53, leading to p53 stabilization through release from MDM2 binding. All three pathways inhibit the degradation of p53 protein, thus stabilizing p53 at a high expression level. Under normal circumstances, the MDM2 protein interacts with p53 and functions as an E3 ubiquitin ligase, which targets p53 for ubiquitination, nuclear export, and degradation (13). The checkpoint kinases phosphorylate the amino terminus of p53, which affects its affinity for MDM2, leading to p53 stabilization. Increased expression of p53 allows it to carry out its major function: to bind to particular DNA sequences and activate the expression of adjacent genes (Fig. 1). These genes, directly or indirectly, mediate the biological effects of p53 in checkpoint control and tumor suppression (2).
Gottifredi et al. show that when cells are treated with the ribonucleotide reductase inhibitor hydroxyurea (HU), or the DNA polymerase inhibitor aphidicolin (APH), the transactivation of p53 target molecules is impaired even though phosphorylation, acetylation, and stabilization of p53 are observed (Fig. 1). Moreover, this phenomenon (i.e., transcriptionally attenuated p53 during DNA replication block) is not rescued by γ-irradiation. The finding that DNA replication blockade results in stabilization of a transactivation-impaired p53 protein reveals a cell cycle-specific regulatory circuit that adds complexity to our understanding of the p53 response (Fig. 1). Moreover, it then becomes important to understand how p53 is stabilized during a replication blockade and why it is transcriptionally impaired.
The pathways initiated when DNA replication is stalled are not as well understood at present as those initiated after γ-irradiation (11). However, with respect to the observations made, the source of the block itself is likely to be less crucial because HU and APH inhibit DNA synthesis through different mechanisms. Strand breaks, considered the main cause for activation of p53 after γ-irradiation, are also common events when DNA replication forks are stalled. It is not, however, understood whether a stalled replication fork without any breaks can initiate the signaling cascade, or whether the nature of the breaks when DNA synthesis stops is different from those induced by γ-irradiation. It is of interest that the DNA replication checkpoint prevents mitosis if DNA replication is either in progress or blocked, and it appears that the signal may be mediated through RNA synthesis by the Primase activity of DNA polymerases (14). Whatever the initiating signals, Gottifredi et al. show that phosphorylation of residues S15, S20, and S46, and acetylation of K382 occur both after γ-irradiation and when DNA synthesis is inhibited. These modifications in p53 predicted that p53 might be transcriptionally active and so it was surprising that activation of certain p53 target genes such as p21 or MDM2 was not observed in the case of DNA replication blockade. Moreover, the pathways leading to such modifications could be very different: The authors and other investigators (15) have found that accumulation of p53 after DNA synthesis blockade is ATM-independent, whereas it is well documented that following γ-irradiation, stabilization of p53 requires functional ATM kinase. A number of alternate candidates, such as ATR, might be involved in phosphorylation of S15 when DNA replication is stalled (16). Furthermore, studies in fission yeast and mammalian systems have reported that activation of Cds1/Chk2 occurs when DNA synthesis is blocked by HU (17, 18), and that hChk2 can phosphorylate p53 at multiple sites including S15 and S20 (19, 20). Another potential mediator of the HU arrest signal is Hus1, the mammalian homologue of yeast hus1 (hydroxyurea sensitive), which is required for DNA replication checkpoints (21). It is interesting that in Hus1-null embryonic fibroblasts, the expression of several p53 target genes was found to be up-regulated (21). Whatever the pathways involved, because γ-irradiation fails to rescue the effect of HU or APH, it was suggested that modifications of p53 may not be the primary cause of its transcriptional defect.
The finding that DNA replication blockade results in stabilization of a transactivation-impaired p53 protein reveals a cell cycle-specific regulatory circuit that adds complexity to our understanding of the p53 response.
It is of interest to determine how p53 is transcriptionally impaired when DNA synthesis is blocked. It is clear that the p53 targets examined by Gottifredi et al. are very poorly activated even in the presence of high levels of p53 after treatment of cells with either HU or APH. The inability of γ-irradiation to rescue this impairment suggests that stalled DNA synthesis actively represses stabilized p53. This repression is not likely due to an overall reduction in cellular mRNA synthesis, because c-fos and cyclin E mRNA are up-regulated in HU-treated cells. It is also apparent that p53-dependent transactivation may not be completely inhibited during replication blockade. The authors observe that PIG3, a known target of p53 involved in the generation of reactive oxygen species, is induced by HU and APH exposure and mention that this induction is not observed in p53-deficient cells (3). Similarly, a late p21 up-regulation in replication-blocked cells is also apparently p53-dependent. Finding that p53 is localized in the nuclei of cells with replication blockade [i.e., having excluded improper subcellular localization (22) as the underlying cause for altered transcriptional potential] a number of possibilities are put forth to attempt to explain the inhibition of p53-dependent transcription: (i) Upstream events activated by inhibition of blocked DNA replication could lead to a repressing modification of p53. (ii) Inhibition of DNA synthesis could prevent one or more critical kinases from phosphorylating p53. In this regard, the authors showed that HU treatment does not affect ATM kinase activity suggesting that this upstream pathway is intact. Studies in yeast, however, have led to the proposal that HU-induced activation of Cds1 leads to inactivation of Chk1 (23). If this could be applied in mammalian cells, and if p53 function requires both hChk kinases for its full activity, then this could explain their results. (iii) Inhibition of DNA synthesis may result in an event that selectively inactivates p53, which is downstream of the signaling/modification pathways. This could be the result of an interaction with a corepressor, such as mSin3A (24) or HDAC1 (25), or due to the action of an as yet unidentified p53-specific repressor.
There are other possibilities for inactivation of p53-dependent transcription. It is well known, for example, that for p53 to activate downstream genes it must first bind to their specific regulatory DNA response elements. Thus, one clue to the underlying defect may emerge when p53 derived from cells with DNA replication blockade is tested for its ability to bind specific response elements. If the binding is lost this may suggest a model wherein p53-interacting proteins or conformational changes due to unknown posttranslational modifications may inhibit p53 binding to DNA. If the binding to DNA response elements remains intact, then the authors could be correct in their prediction that other interactions or modifications may impact directly on transactivation of DNA-bound p53. In this regard, in addition to the possibilities mentioned, another scenario for transcriptionally inactive p53 may involve the reduced local availability of coactivators, such as p300 or the basal transcriptional machinery at genomic p53 target sites. Some support for this idea comes from observations that adenovirus E1A, which stabilizes p53, inhibits its transactivation potential through its p300-interacting domain (26). Other support comes from recent observations that desferoxamine, which induces HIF1 and stabilizes p53, also leads to transcriptionally defective p53 (27). HIF1 is known to recruit p300 during the transcriptional response to hypoxia (28). Direct testing of this hypothesis as well as more open-ended screens for p53-interacting proteins in cells with DNA replication blockade may offer new possibilities. Of note, to our knowledge, the role of p300 as a coactivator for p53 has not been dissociated from its role in p53 acetylation. Thus, it is formally possible that even though p53 becomes acetylated, its ability to activate downstream genes may be weakened by decreased availability of coactivators. It is also conceivable that Hus1 may signal repression of p53 targets during S-phase (21). Whatever the mechanism of regulation of p53, the observations by Gottifredi et al. provide an intriguing example of a stimulus that can stabilize p53 and yet actively block the p53 transcriptional activating program that is induced by γirradiation.
Another important question is: Why is p53 selectively held in check when DNA replication is blocked? The authors speculate that during a normal S-phase there are likely to be strand breaks or stalled forks that can initiate the signaling events that stabilize p53, but in S-phase the E2F-1 protein is active and when combined with a fully active p53 is likely to induce cell death (29). E2F1 is itself a known upstream regulator of p53 and its family member p73 (30). In the case of p53 stabilization, this occurs through E2F1-dependent transactivation of the ARF protein and ARF-dependent sequestration of MDM2. Although it is possible that E2F1 may ultimately impact on the observed replication blockade phenotype of p53, there is no data to suggest that the observed posttranslational modifications of p53 occur in the ARF pathway. Nonetheless, to avoid a catastrophic response, such as apoptosis, to what is likely to be a commonly occurring stall or break that would normally be repaired or resolved, the cell must disable p53. It is not yet clear, however, that p53 is completely disabled during replication blockade. There are some hints that some p53 targets are still up-regulated (3), and it is possible that other effects of p53 [e.g., in DNA repair, repression of S-phase active genes such as BRCA1 (31), or the maintenance of genomic stability] may not be disrupted in cells with replication blockade.
Gottifredi et al. caution that in cancer treatment some combination therapies may not always synergize and in some cases could result in noncooperative or even counteracting outcomes. It could also be suggested that novel therapeutic strategies may emerge from the knowledge that cells blocked in S-phase stabilize a transcriptionally impaired p53, and that coexposure to DNA damaging agents is “remembered” by the cells [i.e., they subsequently arrest if the replication blockade is reversed (3)]. It is possible that such cells may be more likely to accumulate irreversible DNA damage and might be more likely to die if they attempt to divide following exposure to DNA damaging agents and subsequent removal of a DNA replication blockade. Such effects may have some relevance for drug development strategies in cancer, for example in chronic myelogenous leukemia where HU is routinely used as a cytoreductive agent.
Footnotes
See companion article on page 1036.
References
- 1.Vousden K H. Cell. 2000;103:691–694. doi: 10.1016/s0092-8674(00)00171-9. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Lane D, Levine A J. Nature (London) 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 3.Gottifredi V, Shieh S-Y, Taya Y, Prives C. Proc Natl Acad Sci USA. 2001;98:1036–1041. doi: 10.1073/pnas.021282898. . (First Published January 16, 2001; 10.1073/pnas.021282898) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jimenez G S, Nister M, Stommel J M, Beeche M, Barcarse E A, Zhang X Q, O'Gorman S, Wahl G M. Nat Genet. 2000;26:37–43. doi: 10.1038/79152. [DOI] [PubMed] [Google Scholar]
- 5.Agami R, Bernards R. Cell. 2000;102:55–66. doi: 10.1016/s0092-8674(00)00010-6. [DOI] [PubMed] [Google Scholar]
- 6.Mailand N, Falck J, Lukas C, Syljuasen R G, Welcker M, Bartek J, Lukas J. Science. 2000;288:1425–1429. doi: 10.1126/science.288.5470.1425. [DOI] [PubMed] [Google Scholar]
- 7.Ohki R, Nemoto J, Murasawa H, Oda E, Inazawa J, Tanaka N, Taniguchi T. J Biol Chem. 2000;275:22627–22630. doi: 10.1074/jbc.C000235200. [DOI] [PubMed] [Google Scholar]
- 8.Chan T A, Hermeking H, Lengauer C, Kinzler K W, Vogelstein B. Nature (London) 1999;401:616–620. doi: 10.1038/44188. [DOI] [PubMed] [Google Scholar]
- 9.Laronga C, Yang H Y, Neal C, Lee M H. J Biol Chem. 2000;275:23106–23112. doi: 10.1074/jbc.M905616199. [DOI] [PubMed] [Google Scholar]
- 10.Dellambra E, Golisano O, Bondanza S, Siviero E, Lacal P, Molinari M, D'Atri S, De Luca M. J Cell Biol. 2000;149:1117–1130. doi: 10.1083/jcb.149.5.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou B B, Elledge S J. Nature (London) 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 12.Xu Y, Ashley T, Brainerd E E, Bronson R T, Meyn M S, Baltimore D. Genes Dev. 1996;10:2411–2422. doi: 10.1101/gad.10.19.2411. [DOI] [PubMed] [Google Scholar]
- 13.Geyer R K, Yu Z K, Maki C G. Nat Cell Biol. 2000;2:569–573. doi: 10.1038/35023507. [DOI] [PubMed] [Google Scholar]
- 14.Michael W M, Ott R, Fanning E, Newport J. Science. 2000;289:2133–2137. doi: 10.1126/science.289.5487.2133. [DOI] [PubMed] [Google Scholar]
- 15.Khanna K K, Lozano G. Oncogene. 1993;8:3307–3312. [PubMed] [Google Scholar]
- 16.Sarkaria J N, Tibbetts R S, Busby E C, Kennedy A P, Hill D E, Abraham R T. Cancer Res. 1998;58:4375–4378. [PubMed] [Google Scholar]
- 17.Matsuoka S, Huang M, Elledge S J. Science. 1998;282:1893–1897. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- 18.Chaturvedi P, Eng W K, Zhu Y, Mattern M R, Mishra R, Hurle M R, Zhang X, Annan R S, Lu Q, Faucette L F, et al. Oncogene. 1999;18:4047–4054. doi: 10.1038/sj.onc.1202925. [DOI] [PubMed] [Google Scholar]
- 19.Chehab N H, Malikzay A, Appel M, Halazonetis T D. Genes Dev. 2000;14:278–288. [PMC free article] [PubMed] [Google Scholar]
- 20.Shieh S Y, Ahn J, Tamai K, Taya Y, Prives C. Genes Dev. 2000;14:289–300. [PMC free article] [PubMed] [Google Scholar]
- 21.Weiss R S, Enoch T, Leder P. Genes Dev. 2000;14:1886–1898. [PMC free article] [PubMed] [Google Scholar]
- 22.Giannakakou P, Sackett D L, Ward Y, Web-ster K R, Blagosklonny M V, Fojo T. Nat Cell Biol. 2000;2:709–717. doi: 10.1038/35036335. [DOI] [PubMed] [Google Scholar]
- 23.Brondello J M, Boddy M N, Furnari B, Russell P. Mol Cell Biol. 1999;19:4262–4269. doi: 10.1128/mcb.19.6.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy M, Ahn J, Walker K K, Hoffman W H, Evans R M, Levine A J, George D L. Genes Dev. 1999;13:2490–2501. doi: 10.1101/gad.13.19.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo J, Su F, Chen D, Shiloh A, Gu W. Nature (London) 2000;16:377–381. doi: 10.1038/35042612. [DOI] [PubMed] [Google Scholar]
- 26.Somasundaram K, El-Deiry W S. Oncogene. 1997;14:1047–1057. doi: 10.1038/sj.onc.1201002. [DOI] [PubMed] [Google Scholar]
- 27.Ashcroft M, Taya Y, Vousden K H. Mol Cell Biol. 2000;20:3224–3233. doi: 10.1128/mcb.20.9.3224-3233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arany Z, Huang L E, Eckner R, Bhattacharya S, Jiang C, Goldberg M A, Bunn H F, Livingston D M. Proc Natl Acad Sci USA. 1996;93:12969–12973. doi: 10.1073/pnas.93.23.12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu X, Levine A J. Proc Natl Acad Sci USA. 1994;91:3602–3606. doi: 10.1073/pnas.91.9.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Irwin M, Marin M C, Phillips A C, Seelan R S, Smith D I, Liu W, Flores E R, Tsai K Y, Jacks T, Vousden K H, et al. Nature (London) 2000;407:645–648. doi: 10.1038/35036614. [DOI] [PubMed] [Google Scholar]
- 31.MacLachlan T K, Dash B C, Dicker D T, El-Deiry W S. J Biol Chem. 2000;275:31869–31875. doi: 10.1074/jbc.M003338200. [DOI] [PubMed] [Google Scholar]