Abstract
Automating the process of neural circuit reconstruction on a large-scale is one of the foremost challenges in the field of neuroscience. In this study we examine the methodology for circuit reconstruction from three-dimensional light microscopy (LM) stacks of images. We show how the minimal error-rate of an ideal reconstruction procedure depends on the density of labeled neurites, giving rise to the fundamental limitation of an LM based approach for neural circuit research. Circuit reconstruction procedures typically involve steps related to neuron labeling and imaging, and subsequent image pre-processing and tracing of neurites. In this study, we focus on the last step – detection of traces of neurites from already pre-processed stacks of images. Our automated tracing algorithm, implemented as part of the Neural Circuit Tracer software package, consists of the following main steps. First, image stack is filtered to enhance labeled neurites. Second, centerline of the neurites is detected and optimized. Finally, individual branches of the optimal trace are merged into trees based on a cost minimization approach. The cost function accounts for branch orientations, distances between their end-points, curvature of the merged structure, and its intensity. The algorithm is capable of connecting branches which appear broken due to imperfect labeling and can resolve situations where branches appear to be fused due the limited resolution of light microscopy. The Neural Circuit Tracer software is designed to automatically incorporate ImageJ plug-ins and functions written in MatLab and provides roughly a 10-fold increases in speed in comparison to manual tracing.
Keywords: tracing, segmentation, axon, dendrite, confocal, stack
INTRODUCTION
It is evident, that the complete understanding of the brain function can only be derived from a substantially detailed account of synaptic connectivity in the underlying neural circuit. Can such an account be given in the form of a complete connectome of the circuit (Lichtman and Sanes 2008, Sporns et al. 2005)? While the connectomes of small invertebrate circuits can be fully determined electron-microscopically (EM) (Chen et al. 2006, White et al. 1986), this technique, in spite of a number of impressive developments in recent years [see e.g. (Briggman and Denk 2006, Denk and Horstmann 2004, Hayworth et al. 2006, Mishchenko et al. 2010)], still lacks the capacity to reconstruct connectivity on a larger scale. Consider a hypothetical serial-section EM reconstruction of a 1 mm3 brain tissue. Volume of this size roughly corresponds to a blowfly brain or a mammalian neocortical column. At 5 nm x 5 nm x 50 nm spatial resolution, the amount of data required to describe the volume completely is on the order of 800 TB, which is too large to be easily processed by modern day computers. More importantly, even at very low probabilities of errors in following small axon profiles from one serial section to the next, say 0.01%, cumulative probability of error in connectivity over a distance of about 1 mm (or 20,000 serial sections) is virtually 100%, making the resulting connectivity diagram unusable. This fundamental limitation of the technique makes the complete connectome description of large circuits unrealistic, at least for the time being.
Other issues further compound the problem (Stepanyants and Chklovskii 2005). First, synaptic connectivity in large circuits can change over time due to the formation and elimination of synapses [see e.g. (Grutzendler et al. 2002, Trachtenberg et al. 2002, Yuste and Bonhoeffer 2001)]. Second, synaptic connectivity in large circuits is bound to be variable from one brain to another (Bohland et al. 2009). Hence, the connectome of a large neural circuit is not likely to be time and brain invariant. To understand the extent of the variability it would be necessary to compare connectomes of different brains. However, this is not a trivial task since neurons in large brain circuits may not be identifiable from one animal to another. In the end, one will be forced to abandon the idea of the complete connectome description, and instead resort to a partial or statistical/probabilistic account of connectivity.
As an alternative to a statistical connectome description one can base the account of connectivity on features of the circuit that are more stable over time and may be more invariant among different brains. Since a synaptic contact requires physical proximity between axonal and dendritic branches of pre- and postsynaptic neurons, such an account can be made in terms of the relative layout of branches, or potential synapses (Stepanyants and Chklovskii 2005, Stepanyants et al. 2002, Stepanyants et al. 2008). Description of connectivity in terms of potential synapses, though far from being complete, is particularly useful for characterizing large circuits. This is because while actual synaptic connectivity may change over time, potential connectivity is typically much more stable (Holtmaat et al. 2005, Trachtenberg et al. 2002). What is more, potential connectivity in different animals (same species, age, brain area, neuron classes, etc.) appears to be less variable than actual connectivity (Jefferis et al. 2007, Stepanyants et al. 2008). This is because while the number of potential synapses between neurons depends mainly on their class and positions in the brain, the number of actual synapses, in addition, depends on neurons’ functional properties (e.g. orientation preference in primary visual cortex). Finally, because potential connectivity is defined by the appositions between axonal and dendritic branches on a micrometer scale, reconstructions of potential connectivity can be done with light confocal or two-photon microscopy (LM), taking advantage of the multitude of imaging and cell labeling techniques (Wilt et al. 2009).
With this study, we are aiming to develop a tool which will automate the process of large-scale 3D reconstruction of neurites. A typical circuit reconstruction procedure contains several general steps (Meijering 2010, Russ 2007): neuron labeling and imaging, image pre-processing, tracing of the wires, and post-processing. The pre-processing stage may include alignment of individual images within the stack, deconvolution of the stack, and the application of various noise reduction and feature enhancement filters. The post-processing stage could contain detection of features such as spines, boutons and synapses, 3D rendering of neuronal branches, and tiling of the reconstructions of individual stacks. In this study, we will mainly focus on the wire tracing part, as it is arguably the main obstacle on the way to automating the circuit reconstruction process. As neurites of many cell types can span the entire brain of an animal (e.g. cortical pyramidal cell axons) or the entire animal itself (e.g. C. elegans), our ultimate goal is to be able to perform reconstructions on a large-scale, and to recover the traces of axonal and dendritic arbors of sparsely labeled populations of neurons in their entirety. But, how sparse should the labeling be?
ANALYSIS
How does the minimal LM reconstruction error-rate depend on the sparseness of labeled neurites?
It is clear that densely labeled neurites are more difficult to reconstruct, potentially leading to larger numbers of errors in the reconstructed image. Assuming that labeling and imaging are performed ideally, errors in the reconstruction process would most commonly occur when the length density of labeled neurites, ρ, is high. Length density is defined as the combined length of all processes per unit volume (Escobar et al. 2008). At high ρ neurites may appear fused in 3D (Figure 1A) and it is often difficult to resolve their individual branches or the branching pattern. Assuming that branches are distributed isotropically and uniformly in the stack of volume V, and that their typical diameter in the image is d (apparent, not real diameter), we can estimate the expected number of places in the stack where two branches appear to be in contact. This number is πdρ2V/2 (Escobar et al. 2008). In these locations one would have to rely on additional characteristics of labeled neurites in order to tell them apart. Such characteristics may include relative branch orientations, branch thicknesses, types (e.g. axons or dendrites), brightness of the label, and color. Below we examine the possibility of discriminating fused branches based on their relative orientations.
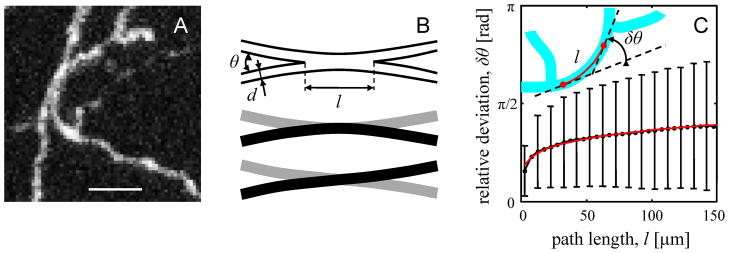
Figure 1.
Estimating the minimal error-rate of LM based circuit reconstruction procedures. A. Maximum intensity projection of a small part of an image stack containing GFP labeled mouse neocortical axons of layer 6 neurons (De Paola et al. 2006). The axonal braches appear fused in 3D and it is not possible to resolve with certainty their individual traces or the branching pattern. Such neurite clusters are the main contributors to the error-rate of an LM based circuit reconstruction. Scale bar is 5 μm. B. Top: model of the scenario shown in A. If two branches appear fused and run side-by-side over a relatively long distance l, their individual traces cannot be resolved (middle or bottom). C. Scaling of the deviation in neurite orientation with path length. Black line shows the dependence of the deviation on path length derived from the reconstructions of neocortical layer 6 axons. Error-bars indicate standard deviations of δθ. Red line is the best fit with Eq. 1. Inset illustrates how the deviation in neurite orientation is calculated for a tree segment which does not traverse through branch points (red segment of cyan tree).
When two branches appear to be in contact, they can still be resolved some distance away from the place of contact if their relative orientations are significantly different (Figure 1B). Branch orientations that cannot be resolved will lead to ambiguities or errors in the trace. To estimate the fraction of such branch orientations we note that over short distances (< 150 μm) tortuous paths of axons and dendrites can be described with an empirical scaling relation (Figure 1C):
| (1) |
This expression shows how the average deviation of neurite’s orientation from a straight path, δθ, depends on the path length along the neurite, l. Parameter lp is referred to as the effective persistence length. Over this length branch orientation changes on average by 1 radian (or 57°), assuming that there is no branching along the way.
Figure 1C shows the dependence of δθ on l for mouse axons of neocortical layer 6 neurons (sample reconstructions are shown in Figure 8). In calculating δθ we used the reconstructed traces of neurites and uniformly sampled from them neurite segments which do not traverse through branch points. Deviation in orientation was calculated for every such segment (see inset in Figure 1C) and the results were combined within groups of similar length segments (black error-bars in Figure 1C). The effective persistence length, lp, and the scaling exponent, γ, were determined from the nonlinear list squares regression of this data with Eq. 1: lp = 2050 ± 190 μm (mean ± S.D.) and γ = 0.18 ± 0.01. The adjusted R2 = 0.985 illustrates the goodness of the fit (red line).
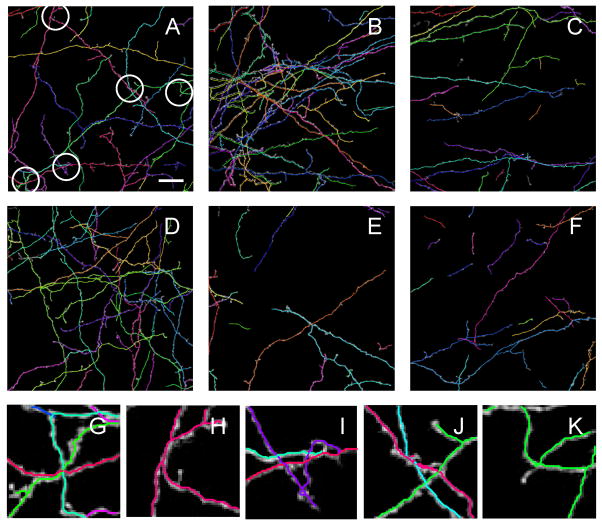
Figure 8.
Examples of automated tracing of mouse neocortical axons of sparsely labeled layer 6 neurons (De Paola et al. 2006). A – F. Automated traces are superimposed on the maximum projections of the image stacks. Based on Eq. 2 and length densities of reconstructed neurites in individual stacks the expected numbers of errors or ambiguities are: 8 in A, 21 in B, 4 in C, 12 in D, 2 in E, and 2 in F. Circles in A highlight five such regions. In these regions axonal braches appear fused in 3D and it is difficult to resolve their individual traces or the branching pattern with certainty. Close-ups of these regions are shown in panels G – K. Scale bar in A is 20 μm.
Two branches which appear to be in contact cannot be resolved if the deviations in their individual orientations, δθ, over the length of the contact, l, are comparable to the angle of branch incidence, θ ≈ 2d/l (Figure 1B). As a result, contacting branches, incident at θ < θc = (2d/lp)γ/(1+γ) are unlikely be resolved. Assuming isotropy, the fraction of such branch orientations in 3D is equal to (1−cos(θc))/2. As θc < 1, this expression is well approximated with the leading term of its expansion over θc, resulting in the fraction of not resolvable relative branch orientations of (2d/lp)2γ/(1+γ)/4.
Multiplying the number of fused branches with the fraction of not resolvable relative branch orientations, we estimate the expected minimal volume density of errors (or ambiguities) in the stack:
| (2) |
This minimal expected error-rate depends strongly (quadratically) on the density of labeled neurites. The expected number of errors per unit length of labeled neurites can be calculated from Eq. 2, by dividing both sides of this expression with ρ:
| (3) |
This calculation makes it possible to estimate the lower bound of the error-rate for different levels of label sparseness. As the combined length of an axonal arbor, La, is much larger than that of a dendritic arbor for many central neuron classes, in labeling neurites for circuit level analyses it is essential to ensure that the minimal error-rate does not exceed about one error per axon, or eLLa < 1. Otherwise, results of automated, or even manual reconstruction procedures, may be unreliable.
Consider an attempt to reconstruct potential connectivity on the scale of the entire mouse cerebral cortex. The total length density of axons in the mouse neocortex is estimated at about 4 μm−2 (Stepanyants et al. 2009). Assuming that the apparent axon diameter d ≈ 1 μm, the typical values of the effective persistence length of cortical axons (most of which are the axons of pyramidal neurons) and the scaling exponents, γ, are similar to those obtained from Figure 1C, we estimate that reconstruction of 0.01% of excitatory neurons uniformly labeled throughout the cortex (only 1 out of 10,000 neurons labeled, ρ = 0.0001 x 4 μm−2) can be done reliably. This reconstruction would result in about 8 errors per mm3 of cortical gray matter according to Eq. 2, or 1 error per 50 mm of axon length according to Eq. 3. As the average axon length per neuron in mouse neocortex is about 40 mm (Braitenberg and Schüz 1998), the above error-rate is sufficiently low, making reconstructions suitable for circuit level analyses of connectivity. It is worth noting that by using multiple color labels (Lichtman et al. 2008, Livet et al. 2007) one can increase the fraction of labeled neurons many-fold without affecting the error-rate significantly.
METHODS
Outline of the automated tracing algorithm
There are numerous approaches to automated tracing of linear structures of neurites from 3D LM images [see (Meijering 2010) for review]. In general, these approaches fall into two main categories. The first category of algorithms is based on local neurite tracking (or tracing) (Al-Kofahi et al. 2002, Bas and Erdogmus 2010a, b, Can et al. 1999, Srinivasan et al. 2007, Wang et al. 2007). Tracking is typically initiated from a seed point provided by the user and the algorithm steps along the structures of neurites by analyzing intensity distribution in a small neighborhood of the current location. Tracking based algorithms are computationally inexpensive and can successfully interpolate through small intensity gaps in the image. On the downside, tracking based algorithms usually run into problems at branch points or branch crossover regions. This is because, at a given step of a tracking algorithm, information about neurite structures contained in the image is limited only to the structures previously visited by the algorithm.
The second group of algorithms is based on image segmentation. Here, some measure of tubularity is calculated for every voxel in the image in order to delineate voxels belonging to the neurites from the ones belonging to the background. Next, centerline of the segmented regions is determined by means of skeletonization (or thinning) (Lee et al. 1994, Palagyi and Kuba 1998, Weaver et al. 2004), voxel coding (Vasilkoski and Stepanyants 2009, Zhou and Toga 1999, Zhou et al. 1998), or by optimally connecting a set of maximum tubularity seed points (Deschamps and Cohen 2001, Dijkstra 1959, González Germán et al. 2008, González G. et al. 2010, Sethian 1999, Wink et al. 2002, Xie et al. 2010). Segmentation based algorithms are computationally expensive because they operate on the entire image. The resulting centerline is often broken into multiple pieces, which need to be interconnected, and contains numerous spurious branches, which must be pruned. Additional algorithms are invoked to accomplish these tasks.
Our algorithm for automated tracing of neurites is rooted in the notion that in order to reconstruct completely one or several structures from a sparsely labeled stack of images, it is necessary to reconstruct all of the neurites contained in the stack first and, only after, extract the structures of interest. Consequently, our algorithm is based on image segmentation rather than tracking. It consists of several steps and relies on a number of parameters (Table 1), some of which are supplied by the user, while others are learned during interactive training. The main steps of the algorithm are outlined below and are illustrated in Figure 2:
Table 1.
Numerical values of parameters and processing times of automated tracing procedures leading to the results shown in Figures 7 – 9. The average processing time was measured on Intel Core 2 Duo, 2.20 GHz, 4 GB, 64-bit computer. Image stacks, containing neuromuscular projection fibers, were reduced 3-fold with a median filter along x, y, and z dimensions before tracing. Subscripts of parameters α, β, γ correspond to 2, 3, and 4 branch end-points mergers. NA for 4 branch end-points mergers indicates that this merger type was suppressed.
| Dataset | Stack size (voxels) | Average processing time (seconds) | CSF size (voxels) | Intensity threshold (% of max) | Region size threshold (voxels) | Branch length threshold (voxels) | Trace optimization | Branch merger | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| α2 | α3 | α4 | ε | |||||||||
| a1 | b1 | β2 | β3 | β4 | ||||||||
| a2 | b2 | γ2 | γ3 | γ4 | ||||||||
| δ2 | δ3 | δ4 | ||||||||||
| 0.1 | 0.1 | 0.1 | ||||||||||
| Olfactory | 512 | 0.5 | 0.05 | 2.0 | 0.5 | 0 | ||||||
| axons | 512 | 190 | [1,1.5,2] | 7 | 200 | 6 | 1.0 | |||||
| (Figure 7) | 60 | 0.2 | 0.05 | 1.0 | 1.0 | 3.0 | ||||||
| 10 | 0 | 0 | ||||||||||
| 0.1 | 0.2 | NA | ||||||||||
| Neocortical | 512 | 0.5 | 0.05 | 1.0 | 1.0 | NA | ||||||
| axons | 512 | 125 | 1 | 10 | 200 | 10 | 1.5 | |||||
| (Figure 8) | 46 | 0.2 | 0.05 | 1.0 | 1.0 | NA | ||||||
| 1.5 | 1.0 | NA | ||||||||||
| 0.1 | 0.2 | NA | ||||||||||
| Neuromuscular | 341 | 0.5 | 0.05 | 2.0 | 2.0 | NA | ||||||
| axons | 341 | 195 | 2.5 | 5 | 200 | 15 | 2.0 | |||||
| (Figure 9) | 54 | 0.2 | 0.05 | 1.0 | 1.0 | NA | ||||||
| 10 | 10 | NA | ||||||||||
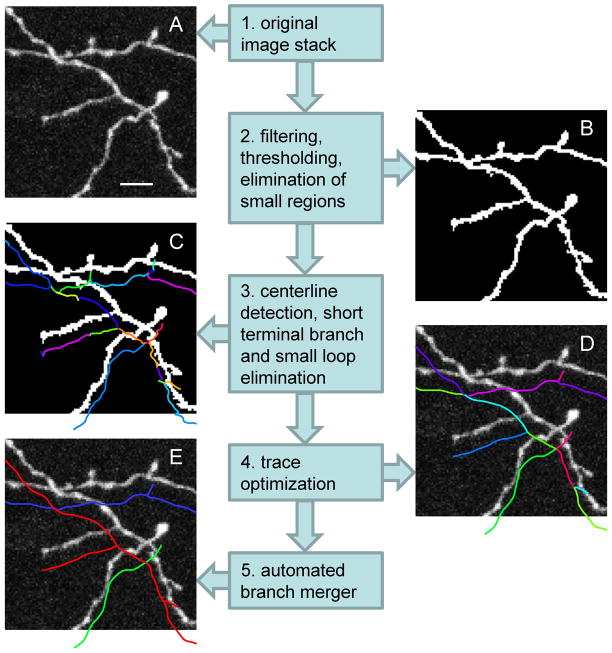
Figure 2.
Schematic illustration of the main steps of the automated tracing algorithm. A. Maximum intensity projection of a small part of an image stack containing GFP labeled mouse neocortical axons of layer 6 neurons (De Paola et al. 2006). Scale bar is 5 μm. B. Same image after the application of the CSF (Eq. 4, σ = 1 voxel size), intensity thresholding (at 25 out of 255), and elimination of small regions (less than 200 voxels in volume). C. Voxel coded binary image and the initial trace showing individual branches in different colors. Short terminal branches and small nested loops were removed. The trace is shifted down to improve visibility. D. The initial trace is optimized with two consecutive optimization algorithms. E. Result of the automated branch merger algorithm.
Image stack (see Figure 2A) is pre-processed if necessary. This step may include alignment of images within the stack, stack deconvolution, conversion of color formats to gray scale, elimination of cell bodies and other non neurite structures. Pre-processing steps strongly dependent on the details of neuron labeling and imaging and are not addressed in this study.
The pre-processed image is filtered to enhance the tracing process and binarized (thresholded). Separate regions containing small numbers of voxels are eliminated. The outcome of this step is illustrated in Figure 2B.
Centerline of the remaining structures in the binary image is detected. This centerline is represented with a graph consisting of short connected straight line segments. Due to noise and imperfect labeling centerline usually contains short erroneous terminal branches and small nested loops. Erroneous terminal branches are eliminated by setting a branch length threshold. Similarly, every small nested loop cluster is replaced with a single vertex located at the cluster’s center of intensity. The result is referred to as the initial trace. The outcome of this step is illustrated in Figure 2C.
The initial trace is optimized with two sequential modified active contour methods (Kass et al. 1988, Vasilkoski and Stepanyants 2009), Figures 2D. End-points of the trace remain fixed during the first optimization, but the branch- and intermediate-points are allowed to move to their optimal positions. The second optimization algorithm improves the placement of the branch- and end-points of the trace.
Individual branches contained in the optimal trace are merged into branching tree structures. Branch here is defined as a neurite connecting the root or a branch-point to a successive branch- or an end-point. This step is based on a novel cost function optimization method. Special care is taken to prevent the formation of loops at every step of the branch merging process. The result is illustrated in Figure 2E.
In the following subsections we provide more details for the steps 2, 3, and 4 of the algorithm.
Filtering tubular structures of neurites
Multi-scale filters can be used successfully to enhance tubular structures. Some such filters are based on the analyses of eigenvalues of the Hessian matrix (Frangi et al. 1998, Lorenz et al. 1997, Sato et al. 1998, Streekstra and van Pelt 2002). These eigenvalues are calculated at different spatial scales for every voxel in the image, and the filter is constructed in such a way that voxels with high intensity variations along one direction relative to the orthogonal directions produce high responses. Filter responses at different spatial scales are combined by taking the maximum over the range of scales. An alternative approach is to use a bank of steerable filters designed to enhance different orientations and take the maximum over the range of orientations (Freeman et al. 1991, Gonzalez et al. 2009, Jacob and Unser 2004). This approach is computationally very expensive as it is usually necessary to use hundreds of filters to resolve orientations of neurites in 3D.
In this study, we use multi-scale center surround filters (CSF) as means for enhancing linear structures. The main advantage of the CSFs over Hessian or steerable filters is in the processing speed. Because of their isotropy, CSFs perform equally well on neurites of different orientations. When the size of a filter, σ, is matched with the caliber of neurites, CSF can smooth out the intensity within the boundaries of the neurites, sharpen the boundaries, and reduce the background noise. A popular choice of a CSF used in detecting linear structures is the Laplacian of Gaussian (LoG) (Marr and Hildreth 1980). In 3D LoG has the form:
| (4) |
Numerical coefficients in this equation are chosen in such a way that both positive and negative components of the filter are normalized to unity and the filter as a whole is normalized to zero.
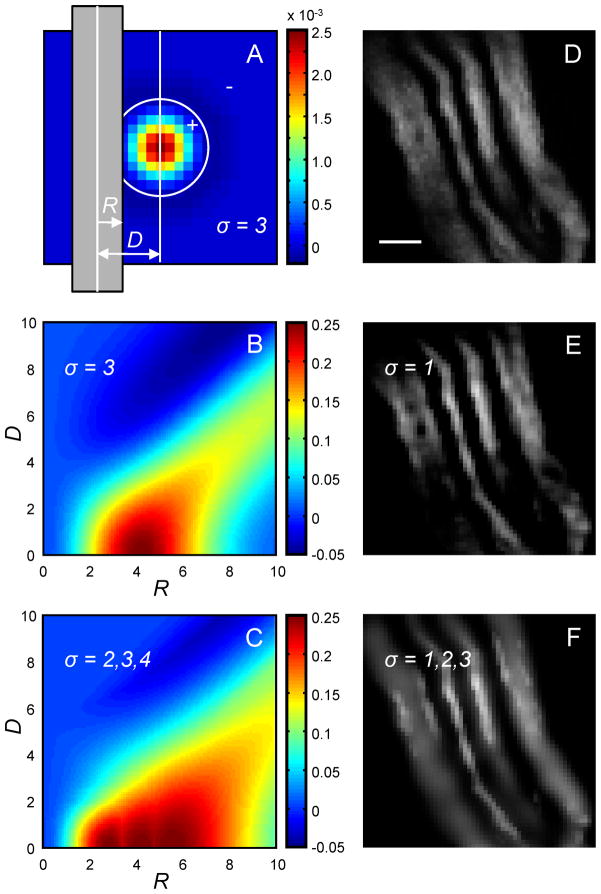
The LoG filter, for σ = 3 voxel sizes, is shown in Figure 3A. If applied to a cylindrical neurite of a uniform intensity (intensity of 1 in Figure 3A), the LoG filter produces the response shown in Figure 3B. As it becomes evident from examining the figure, a problem will arises if the image stack contains neurites of different calibers. LoG will reduce the intensity inside thick branches, effectively curving out their interiors (e.g. R = 8 voxel sizes), as well as reduce the intensity of very thin branches (e.g. R = 1 voxel size), which may lead to branch breaking. To circumvent these problems one may combine the outputs from different size filters applied to an image, I, by taking the maximum output at every voxel:
| (5) |
Figure 3.
Multi-scale center-surround filter. A. Central cross-section of the LoG (Eq. 4, σ = 3 voxel sizes) is shown as a colormap. Gray bar represents the cross-section of a cylindrical neurite of radius R located distance D away from the center of the filter. B. D – R colormap of intensity in the filtered image. C. D – R colormap of intensity in the multi-scale filtered image (Eq. 5, σ = 2, 3, and 4 voxel sizes) covers a broader range of neurite calibers, R. D. Small part of an image containing mouse neuromuscular projection fibers (Lu et al. 2009). Scale bar is 1 μm. E. Single-scale CSF may create holes in thick neurites or break the thin ones. F. Multi-scale CSF sharpens the boundaries of neurites and smoothes out their intensity.
Asterisk in this expression denotes convolution. The response of such multi-scale CSF, Figure 3C (σ = 2, 3, and 4 voxel sizes), encompasses a larger range of neurite calibers, R, which is advantageous. For real neurites, Figure 3D, comparison of the results of single- and multi-scale LoG filters is shown in Figures 3E and 3F.
Extracting initial traces of neurites
There are multiple methods that can be used for detecting the centerline of neurites. Some methods are based on skeletonization or thinning of binary images [see e.g. (Lee et al. 1994, Palagyi and Kuba 1998, Weaver et al. 2004)]. The basic idea behind these methods is an iterative removal of voxels from the surface of the segmented image in a way that preserves the topology of the contained structure and does not erode terminal branches. This can be accomplished by fixing the tips of the terminal branches and monitoring the number of disconnected structures at every step of the algorithm. The algorithm proceeds until no more voxels can be removed from the image without increasing the number of regions. Alternatively, the centerline of neurites can be obtained by using the Minimum Cost Path methods [see e.g. (Deschamps and Cohen 2001, Dijkstra 1959, González Germán et al. 2008, González G. et al. 2010, Sethian 1999, Wink et al. 2002, Xie et al. 2010)].
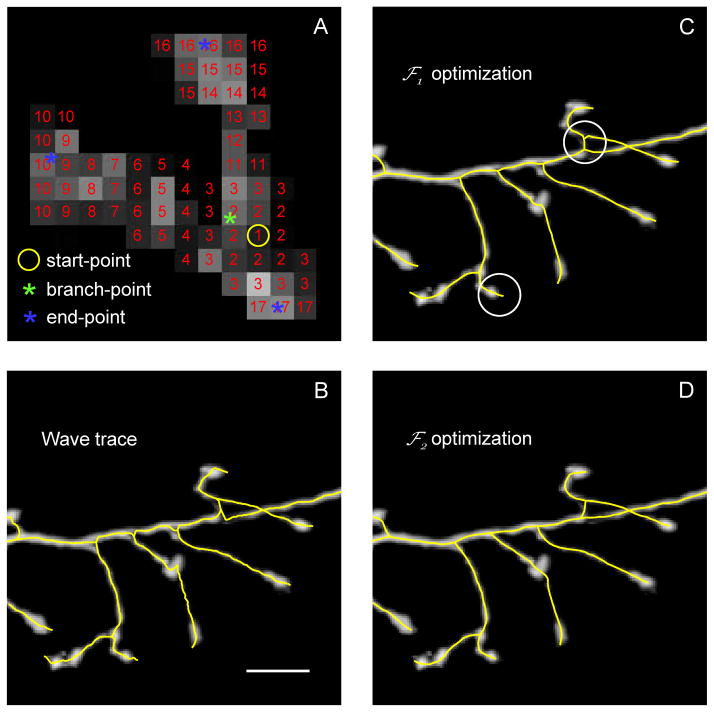
In this study we use voxel coding algorithm (Vasilkoski and Stepanyants 2009, Zhou and Toga 1999, Zhou et al. 1998) as the means for centerline detection (see Figure 4A). Here, a 3D wave of consecutive numerical labels is initiated at an arbitrary voxel in the image. At every step of the algorithm a new wave front is determined as a set of yet unlabeled image voxels in the 3×3×3 neighborhood of the current front’s voxels. After the wave of labels propagates through the first connected region in the image, another wave is initiated at a different location, and the algorithm proceeds until all the regions are labeled. Voxel coding may be initiated at seed points provided by the user, however, this is not essential and random seeds were used throughout this study.
Figure 4.
Voxel coding and optimization of the trace. A. Maximum intensity projection of a small part of a dendritic arbor. A 3D wave of consecutive numerical labels is initiated at an arbitrary voxel (yellow circle) in the image. At every subsequent step of the algorithm a new wave front is determined as a set of yet unlabeled image voxels in the neighborhood (3×3×3 in the figure) of the current front’s voxels. Modified with permission from (Vasilkoski and Stepanyants 2009) (their Figures 2A). B. The centers of intensity of consecutive wave fronts are connected with straight segments to form the wave trace (yellow line). The branch-points of the wave trace are defined as the centers of intensity of the dividing fronts (green asterisk in A). The end-points are the centers of intensity of the terminal wave fronts (blue asterisks in A). C. The initial trace is optimized with Eq. 7 for a more accurate representation of the neurites. Typical flaws of this optimization include imprecise placements of the branch- and end-points (circled regions). D. These problems are fixed with a subsequent optimization of the trace with Eq. 9. Scale bar in B is 10 μm.
The centers of intensity of consecutive wave fronts (graph nodes) are connected with straight line segments (graph edges) to form a graph. The branch-points of the graph correspond to the centers of intensity of the dividing wave fronts, and end-points are the centers of intensity of the terminal wave fronts (see Figure 4A). Due to noise and imperfect labeling, voxel coding algorithm may result in a graph containing short erroneous terminal branches and small nested loops. Erroneous terminal branches are eliminated by setting a branch length threshold. Similarly, every small nested loop cluster is replaced with a single vertex located at the cluster’s center of intensity. This reduced graph is referred to as the initial trace of neurites. It usually consists of multiple unconnected sub-graphs which, in turn, may contain multiple branches and branch segments (see Figure 4B).
Trace optimization
The initial trace, obtained with the voxel coding algorithm, usually represents the structure of neurites only approximately (Figure 4B). A smother and more accurate representation can be achieved with fitness function optimization algorithms. Because the initial trace typically lies sufficiently close to the optimal solution, such optimization can be performed with a fast iterative gradient ascent procedure (Boyd and Vandenberghe 2004).
In the previous version of the optimization algorithm (Vasilkoski and Stepanyants 2009) the fitness function, Φ1, was constructed from intensity integrated along the trace (average intensity along the trace times the trace length) and the elastic energy of the trace:
| (6) |
Vectors r⃗k in this expression specify the positions of vertices of the trace and R⃗k denote the positions of voxel centers in the image stack. Parameter s denotes voxel size which, for simplicity, is assumed to be the same in all 3 dimensions. Index kl in the second term enumerates all the vertices of the trace that are connected to the vertex k. For example, if vertex k is an intermediate node, kl enumerates the 2 of its neighboring vertices, and if vertex k is a bifurcating node, kl describes 3 vertices. Parameter λ denotes the average density of segments along the trace (number of segments per unit length of the trace), and a1 > 0 controls the tension in the trace. Parameter σ should be of the order of the typical radius of neurites contained in the image, and, due to the fast decay of the Gaussian pre-factor, summation over i in Eq. 6 can be restricted to a small number of voxels in the vicinity of the trace.
Maximization of the functional Φ1 can be achieved with the following gradient ascent procedure:
| (7) |
Here, superscript n enumerates the steps of the algorithm and parameter b1 > 0 controls the step size. Terminal vertices of the trace must remain fixed during this procedure to prevent the subtrees within the trace from collapsing to single points. Details related to the stability of the algorithm and the appropriate range of values of parameters a1 and b1 are addressed in (Vasilkoski and Stepanyants 2009).
Trace, resulting from optimization of Eq. 6, captures very precisely the layout of neuronal branches (Figure 4C), but the placement of the branch- and end-points in the trace can be further improved with a secondary optimization procedure. In this optimization, a new fitness function Φ2, reflecting the intensity and straightness of the trace is maximized:
| (8) |
Index k in this expression runs over the intermediate vertices only, and dot in the second term denotes the scalar product. At every intermediate vertex, k, the notion of straightness is captured by the cosine of the angle between segments connecting k with its two first order neighbors (denoted with k−1 and k+1 in Eq. 8). The gradient ascent procedure for this functional is:
| (9) |
Positions of all vertices of the trace, including branch- and end-points, are updated during this procedure. For brevity, Eq. 9 is written in a way where it is only applicable to vertices, k, with strictly two first and second order neighbors, i.e. k−2, k−1 and k+1, k+2. To describe the steps of other types of vertices, Eq. 9 must be modified in the following way: (i) if some of the neighboring vertices of k do not exist (e.g. if k is an end-point vertex), or (ii) if the number of first or second order neighbors of k is greater than two due to branching (e.g. if k is a bifurcation point it has 3 first order vertices) then the terms corresponding to such neighboring vertices must be dropped from Eq. 9.
At every iteration step of the gradient ascent algorithms of Eqs. 7 and 9 positions of all vertices are synchronously updated. Long segments are subdivided and short segments are combined to ensure stability of the trace and adequate representation of intensity in the image (Vasilkoski and Stepanyants 2009). The sequential optimization scheme was designed to improve trace precision while maintaining stability. The initial trace is stretched in the course of the first optimization, straightening the zigzags resulting from voxel coding (Figure 4C). This step is essential, as the second optimization, which improves the placement of branch- and end-points (Figure 4D), results in instability if applied directly to the initial (wave) trace. Numerical values of parameters used in both optimization procedures are shown in Table 1. For the 3 datasets examined in this study, both optimizations converged to their optimal solutions (as judged by monitoring the fitness functions) within 50 iteration steps. Optimization improves the placement of branch- and end-points by about 2 – 3 μm over the initial trace (Vasilkoski and Stepanyants 2009) (compare Figures 4B and 4D). What is however more important, optimization smoothes out zigzags present in the initial trace. This is essential for the next step of the algorithm, during which individual branches are merged into tree structures based on their orientations, intensities, and curvatures. Calculation of these quantities relies on smoothness of the trace.
Automated branch merger
Branch merging is the most important stage of the described automated tracing procedure. At this stage we are invariably risking to create erroneously connected neurites. Such false positive mergers are difficult to find and to correct. Hence, the best reconstruction strategy in our opinion is to merge branches only when the confidence level is relatively high. Regions with complex merger patterns are better handled by trained users.
In merging branches of the optimal trace we first identify the end-points of all the branches and group them into spatially segregated clusters. For this, we consider a graph in which the nodes represent the branch end-points and the edges are placed only between spatially neighboring nodes (distance less than a threshold set by the user). Branch end-point clusters are sorted based on the number of contained end-points and the algorithm proceeds from merging 2 end-point clusters (in the order of increasing cost, see Eq. 10), to 3 end-point clusters, to the higher order clusters.
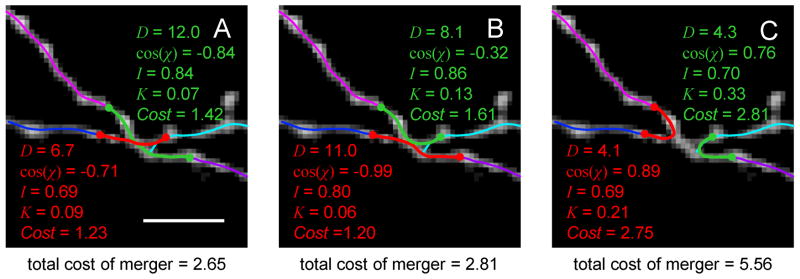
Within each cluster of branch end-points, all possible merger scenarios leading to 2 branch mergers, 3 branch mergers (bifurcations), and 4 branch mergers (trifurcations) are considered. Figure 5 shows an example of 4 branch end-point merger. In this case, there are 15 merger scenarios in total: 1 scenario resulting in 4 separate branches (no merger), 4 scenarios leading to 1 bifurcation and 1 separate branch, 3 scenarios resulting in 2 separate branches (Figures 5A–C), 6 scenarios leading to 3 separate branch, and 1 scenario corresponding to 1 trifurcation. For each scenario the optimal merging trace is determined and the cost of the merger is evaluated. The algorithm performs the lowest cost merger and moves to the next cluster, unless the merger results in the formation of a loop. In this case the algorithm will attempt the subsequent lowest cost merger.
Figure 5.
Local optimization based branch merging strategy. Maximum intensity projection of an image stack showing a cluster of 4 end-points of the optimal trace (thin lines). All possible branch merger scenarios are considered by the algorithm. Three of these scenarios are shown in A, B, and C. The associated distances between branch end-points [D (in pixels)], cosines of angles made by the merging branch pairs [cos(χ)], average normalized intensities (I), curvatures (K) of the optimally connecting paths (thick red and green lines), and the corresponding costs are shown. Merger scenario shown in A seems to be the best choice as judged by the total cost of merger. Scale bar in A is 5 μm.
During manual tracing, trained human operators discriminate between correct and erroneous tracing scenarios by combining information about multiple features of neurites. Such features may include distance between branches, branch orientations, average intensities, variations of intensities, branch thicknesses, curvatures, tortuosities (Stepanyants et al. 2004), colors, presence of spines or boutons, etc. Likewise, to evaluate different merger scenarios within a given cluster of branch end-points, we designed the cost function which contains 4 of the above mentioned features:
| (10) |
Index i in this expressions enumerates separate branch end-point mergers within the considered merger scenario. Di denotes the total distance between all merging branch end-point pairs within the i-th merger. χij represents the angle made by the branch pair j within the i-th merger. It is calculated as the angle between tangents to the two branches in the vicinity of the considered end-points. By default, χi0 equals 180° for 2 branch end-points mergers, 120° for bifurcations, and 90° for trifurcations. Ii is the average intensity along the optimally connecting traces of the i-th merger, and I0 is the average intensity along the optimal trace of the entire image. These quantities are calculated by dividing the first terms in Eqs. 6 or 7 with the trace length. Ki is the average curvature of the optimally connecting traces of merger i. As before, the curvature is calculated only for the intermediate vertices. N represents the number of remaining (not merged) branch end-points. Figure 5 shows individual components of the cost for 3 out of 15 possible merger scenarios involving 4 branch end-points.
The cost function in Eq. 10 depends on 13 non-negative parameters (Table 1): three sets of parameters α, β, γ, δ – one set for each allowed merger type (2, 3, and 4 branch end-point mergers), as well as the parameter ε. These parameters are determined with the perceptron learning algorithm (Engel and Broeck 2001) during user assisted branch merging procedure. Here, for every cluster of branch end-points in the training stack of images, the user is sequentially presented with different merger scenarios and has to identify the correct one. Once the correct merger is identified, numerical values of D, χ, I, K, and N for this merger are associated with 1 (or correct merger) and those for all other merger scenarios within the cluster are associated with −1 (or incorrect merger). The algorithm proceeds to other clusters of endpoints until training is complete. Perceptron is trained to identify the correct merger within each cluster by solving the system of inequalities which ensures that costs of the correct mergers are the lowest within each group.
Numerical values of the cost function parameters α, β, γ, and δ are obtained from the perceptron learning, during which more than 95% of mergers are typically recognized correctly. Reconstructions are not very sensitive to the exact values of α, β, γ, and δ, and the parameter sets generalize well on test images obtained under similar experimental conditions. For example, in reconstructing datasets of drosophila olfactory neuron axons (Brown et al. 2011) used in the DIADEM challenge (Brown et al. 2011), we trained the algorithm on 3 training image stacks and tested its performance on 3 qualifier image stacks by using the DIADEM metric. The metric scores were not statistically different between the training (0.89, 0.89, and 0.67) and test groups (0.66, 0.79, and 0.90).
IMPLEMENTATION
Parameters and data formats
Our algorithm depends on a number of parameters, some of which are set by the user while others are learned from the training data and are generalized on images of the same type (see Table 1). Parameters set by the user include: the multi-scale CSF sizes, thresholds for intensity, region size, and branch length, as well as trace optimization parameters. There are no strict guidelines on how to choose the values of these parameters, and the best choice of parameters is dependent on the image type.
In general, the multi-scale CSF sizes must span the range of neurites’ radii. Because voxel coding algorithm works on binary images, intensity thresholding is a necessary step. The threshold must be high enough to eliminate the background and separate neurites as much as possible without breaking them into many small regions. Region size threshold is used to eliminate small noise remaining in the image after filtering and thresholding. Typically, few hundred voxels are sufficient to remove such noise without affecting regions of neurites. Branch length threshold is used to eliminate short terminal branches. This is usually necessary for simplifying the topology of branch merging patterns. It is recommended to set this threshold to at least few voxel sizes in order to reduce artifacts of the voxel coding algorithm. For neurites containing short terminal branches (2 – 5 μm), spines, or filopodia, branch length threshold should be high enough to eliminate the traces of such structures. These structures must be detected with separate algorithms, after the main trace of neurites is determined. The choice of optimization parameters was previously described (Vasilkoski and Stepanyants 2009). Best values of parameters a1,2, which control the stiffness and straightness of the trace, depend on the class of neurites. For example, in the neocortex axons of many inhibitory neuron classes are much more tortuous than axons of excitatory neurons (Stepanyants et al. 2004), and therefore they must be reconstructed by using lower values of a1,2. More studies are needed to determine the best values of the trace optimization parameters for different neuron classes.
Traces of neurites in this study are built out of short connected straight line segments. Their topological structure is conveniently represented with an adjacency matrix. This format makes it possible to describe structures containing loops, which are often present at the initial stages of the algorithm. This data format also allows for fast traversing through the trace and identifying its branch- and end-points. The final trace of all the neurites contained in the image stack can be converted from the adjacency matrix format to a more traditional SWC format of neuron morphology (Cannon et al. 1998).
The Neural Circuit Tracer software
The Neural Circuit Tracer is open source software built using Java (Sun Microsystems) and Matlab (MathWorks, Inc., Natick MA). It is based on the core of ImageJ (http://rsbweb.nih.gov/ij) and the graphic user interface has been developed by using Java Swings. The software combines a number of functionalities of ImageJ with several newly developed functions for automated and manual tracing of neurites. The Neural Circuit Tracer is designed in a way that will allow the users to add any plug-ins developed for ImageJ. More importantly, functions written in MatLab and converted into Java with Matlab JA toolbox can also be added to the Neural Circuit Tracer.
Current version of the software includes basic ImageJ components and plug-ins, such as image stack view, ImageJ 3D viewer plug-in (http://rsb.info.nih.gov/ij/plugins/3d-viewer), and a number of 3D filters. Additional features included in the current version are:
Loading a stack of images with the option to reduce the stack in xy or xyz dimensions. The reduction can be accompanied with 3D median, mean, minimum, maximum, or standard deviation filtering of the stack. Image stack can be loaded in Tagged Image File (.tif) and MatLab (.mat) formats.
Multi-scale CSF and a region size filters (described above).
Automated tracing module. This function is based on the algorithm described above.
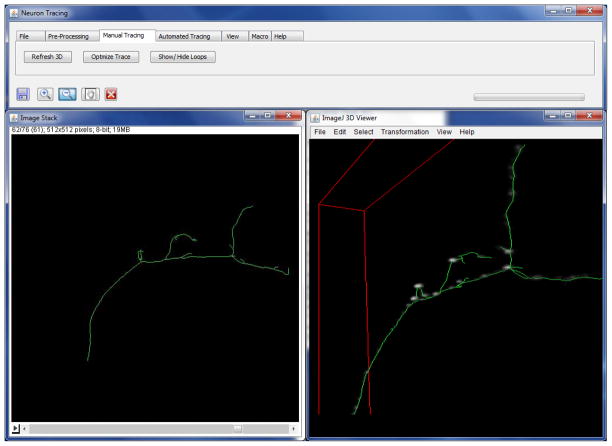
Manual editing module. The trace is conveniently displayed in 2D on top of the ImageJ stack view and in 3D inside the ImageJ 3D viewer plug-in (see Figure 6). Trace can be zoomed in/out and rotated in 3D. Individual tree segments can be added and deleted. Tracing can be continued from any vertex of the trace. Trees can be connected, disconnected and deleted.
Two trace optimization functions (described above).
Saving and loading of the stack and the trace in MatLab (.mat) format.
Exporting the trace in SWC format (Cannon et al. 1998).
Adding Java and MatLab based plug-ins.
Building, saving, and running macros. User can save a group of functions as macros and run them consecutively.
Figure 6.
Graphic user interface of the Neural Circuit Tracer. The image stack viewer (left) is based on ImageJ core. It displays the z projection of the trace superimposed on a given z-slice of the image stack. The image stack viewer makes it possible to manually trace neurites or edit automated traces in 3D. The trace can be displayed inside the ImageJ 3D viewer plug-in to facilitate manual tracing (right).
RESULTS AND DISCUSSION
Performance of the automated tracing algorithm presented in this study was evaluated on 3 DIADEM challenge datasets (Brown et al. 2011). Neurites in the selected datasets were fluorescently labeled and imaged with confocal or two-photon microscopy. Other DIADEM datasets, obtained with brightfield microscopy, were not examined due to the beaded appearance of neurites. Voxel coding algorithm cannot be used for extracting the initial traces of neurites from such images and must be replaced with the Minimum Cost Path or tracking based methods. After the initial trace is detected, optimization and branch merging steps of the algorithm can be applied without changes.
Automated and manual (gold standard) traces can be compared quantitatively by using the DIADEM metric (Gillette et al. 2011) to evaluate the performance of the algorithm. However, this comparison is hindered by several considerations. First, the DIADEM metric relies heavily on the topologies of the reconstructions, yet it only compares connected tree structures. Broken trees score lower because only the tree sections connected to the roots contribute to the score. In general, topological errors which occur closer to the root of the reconstruction receive larger penalties. This seems reasonable if the root is selected at or close to the neuron’s cell body, but is somewhat arbitrary otherwise (see e.g. Figure 8). Second, gold standard reconstructions obtained manually are usually not perfect. For example, comparisons of reconstructions performed manually by different users do not result in perfect scores (Gillette et al. 2011). Hence, automated reconstructions, even if more accurate than the gold standard, will not receive perfect scores. Finally, scoring automated reconstructions with the DIADEM metric becomes meaningless in situations where the numbers of theoretically expected ambiguities or errors are high.
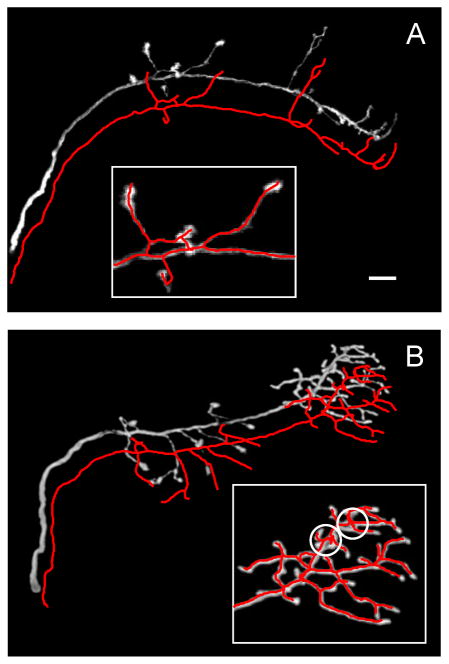
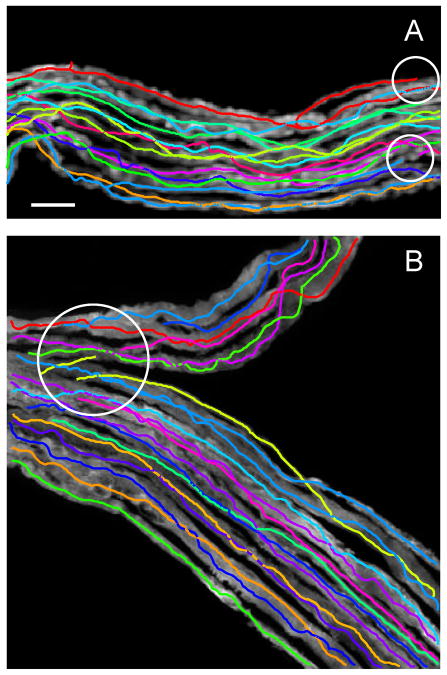
Figure 7 shows two automated tracing examples of drosophila olfactory neuron axons (Jefferis et al. 2007). Six such image stacks were traced automatically and the results were evaluated by using the DIADEM metric and the gold standards. Metric scores were generally high, 0.80 ± 0.11 (mean ± SD), indicating good overall quality of automated reconstructions. Typical errors for this dataset included broken branches and other topological mistakes which can be classified as branch stealing (circled regions in the inset of Figure 7B). Such errors were minor and could be easily corrected in the editing module of the Neural Circuit Tracer, requiring 1 – 2 minute of user involvement per image stack. This editing time is significantly shorter than the average time required for manual reconstruction of these images, which is estimated at about 20 minutes per stack (Brown et al. 2011).
Figure 7.
Examples of automated tracing of drosophila olfactory neuron axons (Jefferis et al. 2007). The traces are shifted with respect to the projected image stacks to improve visibility. Insets in A and B zoom in on regions of high complexity. This region in B contains two possible errors (circled areas) which can be classified as branch stealing. Scale bar in A is 10 μm.
Results of automated tracing of mouse neocortical axons of layer 6 neurons (De Paola et al. 2006) are shown in Figure 8. Here, six stacks of images were traced individually. These reconstructions may be stitched together manually to form a larger-scale mosaic. Reconstruction errors include branch breaking due to uneven labeling and branch/tree stealing. The stacks also contain branch crossover regions which could not be resolved with high confidence (Figure 8G–J). Because neurites in these images are distributed roughly uniformly and isotropically, we can invoke Eqs. 2 and 3 to estimate the theoretically expected numbers of such regions. This calculation resulted in 8 expected ambiguities/error in A, 21 in B, 4 in C, 12 in D, 2 in E, and 2 in F, which is in good agreement (up to a factor of 2) with counts of such sites in individual stacks. Due to the high numbers of expected ambiguities/errors in this dataset, the DIADEM metric could not be used in a meaningful way to evaluate the quality of the results. The estimated average editing time necessary to fix reconstruction errors (~10 minutes per stack) is much shorter than the manual tracing time of several hours per stack (Brown et al. 2011).
The mouse neuromuscular projection fiber dataset (Lu et al. 2009) is a mosaic of 152 stacks of images. Image stacks were reduced 3-fold in all dimensions and traced individually. Two examples of such tracings are shown in Figure 9. Automated reconstruction errors included branch breaking due to uneven labeling or due to branch proximity to stack surfaces (circled regions in Figure 9), as well as fusing of nearby axons. The average editing and trace stitching time (~10 minutes per stack) is much shorter than the manual tracing time, which is estimated based on (Brown et al. 2011) to be in the range of 80 – 100 minutes per stack.
Figure 9.
Examples of automated tracing of mouse neuromuscular projection fibers (Lu et al. 2009). The automated traces in A and B are superimposed on the maximum projections of the image stacks. Circled regions highlight typical errors in automated tracing. These errors include branch breaking due to non-uniform labeling of neurites and due to branch proximity to the stack surfaces, and erroneous branch mergers due to fusing of neurites (see Figure 1). Scale bar in A is 10 μm.
CONCLUSIONS
In this study we describe a methodology for automated tracing of neurites from 3D LM stacks of images. Our algorithm and software can be used for tracing neurites labeled with fluorescent and non-fluorescent markers and imaged with confocal, two-photon, or brightfield microscopy. We estimate how the minimal error-rate of an LM based reconstruction approach depends on the sparseness of labeled neurites. Reconstructions can only be useful for circuit level analyses if the labeling is sufficiently sparse. If the threshold of label sparseness is exceeded, neurites cannot be reconstructed with confidence, even manually by trained operators.
Automated reconstruction tools, including the one described here can substantially aid more time consuming manual reconstruction techniques, but are unlikely to replace them completely, at least in the near future. Major improvements in the reconstruction methodology may be easier done at the experimental rather than computational end of the problem. Because there are no automated tools that perform well on diverse experimental datasets, the optimal reconstruction strategy may be to extract the initial traces of neurites, but merge branches automatically into tree like structures only when the confidence for such mergers is relatively high. The remaining mergers can be done by the user without significant time investment. It is usually much faster and easier to connect broken branches with editing software than to find and repair erroneous connections.
Acknowledgments
We would like to acknowledge Dr. Zlatko Vasilkoski’s contribution to the implementation of the voxel coding algorithm and discussions related to the subject of this study. This work was supported by the NIH grant NS063494.
Footnotes
INFORMATION AND SHARING AGREEMENT
More information on the Neural Circuit Tracer software can be found at http://www.neurogeometry.net.
References
- Al-Kofahi KA, Lasek S, Szarowski DH, Pace CJ, Nagy G, Turner JN, Roysam B. Rapid automated three-dimensional tracing of neurons from confocal image stacks. IEEE Trans Inf Technol Biomed. 2002;6:171–187. doi: 10.1109/titb.2002.1006304. [DOI] [PubMed] [Google Scholar]
- Bas E, Erdogmus D. Piecewise linear cylinder models for 3-dimensional axon segmentation in Brainbow imagery. Biomedical Imaging: From Nano to Macro, 2010 IEEE International Symposium on.2010a. pp. 1297–1300. [Google Scholar]
- Bas E, Erdogmus D. Principal curve tracing. ESANN 2010 proceedings, European Symposium on Artificial Neural Networks - Computational Intelligence and Machine Learning; Bruges, Belgium. 2010b. pp. 405–410. [Google Scholar]
- Bohland JW, et al. A proposal for a coordinated effort for the determination of brainwide neuroanatomical connectivity in model organisms at a mesoscopic scale. PLoS Comput Biol. 2009;5:e1000334. doi: 10.1371/journal.pcbi.1000334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd SP, Vandenberghe L. Convex optimization. Cambridge, UK ; New York: Cambridge University Press; 2004. [Google Scholar]
- Braitenberg V, Schüz A. Cortex: statistics and geometry of neuronal connectivity. Berlin; New York: Springer; 1998. [Google Scholar]
- Briggman KL, Denk W. Towards neural circuit reconstruction with volume electron microscopy techniques. Curr Opin Neurobiol. 2006;16:562–570. doi: 10.1016/j.conb.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Brown KM, Barrionuevo G, Canty AJ, De Paola V, Hirsch JA, Jefferis GS, Lu J, Snippe M, Sugihara I, Ascoli GA. The DIADEM Data Sets: Representative Light Microscopy Images of Neuronal Morphology to Advance Automation of Digital Reconstructions. Neuroinformatics. 2011 doi: 10.1007/s12021-010-9095-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Can A, Shen H, Turner JN, Tanenbaum HL, Roysam B. Rapid automated tracing and feature extraction from retinal fundus images using direct exploratory algorithms. IEEE Trans Inf Technol Biomed. 1999;3:125–138. doi: 10.1109/4233.767088. [DOI] [PubMed] [Google Scholar]
- Cannon RC, Turner DA, Pyapali GK, Wheal HV. An on-line archive of reconstructed hippocampal neurons. J Neurosci Methods. 1998;84:49–54. doi: 10.1016/s0165-0270(98)00091-0. [DOI] [PubMed] [Google Scholar]
- Chen BL, Hall DH, Chklovskii DB. Wiring optimization can relate neuronal structure and function. Proc Natl Acad Sci U S A. 2006;103:4723–4728. doi: 10.1073/pnas.0506806103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paola V, Holtmaat A, Knott G, Song S, Wilbrecht L, Caroni P, Svoboda K. Cell type-specific structural plasticity of axonal branches and boutons in the adult neocortex. Neuron. 2006;49:861–875. doi: 10.1016/j.neuron.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Denk W, Horstmann H. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol. 2004;2:e329. doi: 10.1371/journal.pbio.0020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps T, Cohen LD. Fast extraction of minimal paths in 3D images and applications to virtual endoscopy. Medical Image Analysis. 2001;5:281–299. doi: 10.1016/s1361-8415(01)00046-9. [DOI] [PubMed] [Google Scholar]
- Dijkstra EW. A note on two problems in connexion with graphs. Numerische Mathematik. 1959;1:269–271. [Google Scholar]
- Engel A, Broeck Cvd. Statistical mechanics of learning. Cambridge, UK ; New York, NY: Cambridge University Press; 2001. [Google Scholar]
- Escobar G, Fares T, Stepanyants A. Structural plasticity of circuits in cortical neuropil. J Neurosci. 2008;28:8477–8488. doi: 10.1523/JNEUROSCI.2046-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangi AF, Niessen WJ, Vincken KL, Viergever MA. Multiscale vessel enhancement filtering. Medical Image Computing and Computer-Assisted Intervention - Miccai’98. 1998;1496:130–137. [Google Scholar]
- Freeman WT, Adelson EH Massachusetts Institute of Technology. Media Laboratory. Vision and Modeling Group. The design and use of steerable filters. Cambridge, Mass: Vision and Modeling Group, Media Laboratory, Massachusetts Institute of Technology; 1991. [Google Scholar]
- Gillette TA, Brown KM, Ascoli GA. The DIADEM metric: comparing multiple reconstructions of the same neuron. Neuroinformatics. 2011 doi: 10.1007/s12021-011-9117-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez G, Aguet F, Fleuret F, Unser M, Fua P. Steerable features for statistical 3D dendrite detection. Med Image Comput Comput Assist Interv. 2009;12:625–632. doi: 10.1007/978-3-642-04271-3_76. [DOI] [PubMed] [Google Scholar]
- González G, Fleuret F, Fua P. Automated Delineation of Dendritic Networks in Noisy Image Stacks. In: Forsyth D, Torr P, Zisserman A, editors. Computer Vision – ECCV 2008. Vol. 5305. Springer; Berlin/Heidelberg: 2008. pp. 214–227. [Google Scholar]
- González G, Turetken E, Fleuret F, Fua P. Delineating trees in noisy 2D images and 3D image-stacks. Pages 2799–2806. Computer Vision and Pattern Recognition (CVPR), 2010 IEEE Conference on.2010. [Google Scholar]
- Grutzendler J, Kasthuri N, Gan WB. Long-term dendritic spine stability in the adult cortex. Nature. 2002;420:812–816. doi: 10.1038/nature01276. [DOI] [PubMed] [Google Scholar]
- Hayworth KJ, Kasthuri N, Schalek R, Lichtman JW. Automating the collection of ultrathin serial sections for large volume TEM reconstructions. Microscop Microanal. 2006;12:86–87. [Google Scholar]
- Holtmaat AJ, Trachtenberg JT, Wilbrecht L, Shepherd GM, Zhang X, Knott GW, Svoboda K. Transient and persistent dendritic spines in the neocortex in vivo. Neuron. 2005;45:279–291. doi: 10.1016/j.neuron.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Jacob M, Unser M. Design of steerable filters for feature detection using canny-like criteria. IEEE Trans Pattern Anal Mach Intell. 2004;26:1007–1019. doi: 10.1109/TPAMI.2004.44. [DOI] [PubMed] [Google Scholar]
- Jefferis GS, Potter CJ, Chan AM, Marin EC, Rohlfing T, Maurer CR, Jr, Luo L. Comprehensive maps of Drosophila higher olfactory centers: spatially segregated fruit and pheromone representation. Cell. 2007;128:1187–1203. doi: 10.1016/j.cell.2007.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass M, Witkin A, Terzopoulos D. Snakes: Active contour models. International Journal of Computer Vision. 1988;1:321–331. [Google Scholar]
- Lee TC, Kashyap RL, Chu CN. Building skeleton models via (3-D) medial surface/axis thinning algorithms. . CVGIP: Graph. Models Image Process. 1994;56:462–478. [Google Scholar]
- Lichtman JW, Sanes JR. Ome sweet ome: what can the genome tell us about the connectome? Curr Opin Neurobiol. 2008;18:346–353. doi: 10.1016/j.conb.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtman JW, Livet J, Sanes JR. A technicolour approach to the connectome. Nat Rev Neurosci. 2008;9:417–422. doi: 10.1038/nrn2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, Lichtman JW. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- Lorenz C, Carlsen IC, Buzug TM, Fassnacht C, Weese J. Multi-scale line segmentation with automatic estimation of width, contrast and tangential direction in 2D and 3D medical images. Cvrmed-Mrcas’97. 1997;1205:233–242. [Google Scholar]
- Lu J, Tapia JC, White OL, Lichtman JW. The interscutularis muscle connectome. PLoS Biol. 2009;7:e32. doi: 10.1371/journal.pbio.1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr D, Hildreth E. Theory of edge detection. Proc R Soc Lond B Biol Sci. 1980;207:187–217. doi: 10.1098/rspb.1980.0020. [DOI] [PubMed] [Google Scholar]
- Meijering E. Neuron tracing in perspective. Cytometry A. 2010;77:693–704. doi: 10.1002/cyto.a.20895. [DOI] [PubMed] [Google Scholar]
- Mishchenko Y, Hu T, Spacek J, Mendenhall J, Harris KM, Chklovskii DB. Ultrastructural analysis of hippocampal neuropil from the connectomics perspective. Neuron. 2010;67:1009–1020. doi: 10.1016/j.neuron.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palagyi K, Kuba A. A 3D 6-subiteration thinning algorithm for extracting medial lines. Pattern Recognition Letters. 1998;19:613–627. [Google Scholar]
- Russ JC. The image processing handbook. Boca Raton: CRC/Taylor and Francis; 2007. [Google Scholar]
- Sato Y, Nakajima S, Shiraga N, Atsumi H, Yoshida S, Koller T, Gerig G, Kikinis R. Three-dimensional multi-scale line filter for segmentation and visualization of curvilinear structures in medical images. Med Image Anal. 1998;2:143–168. doi: 10.1016/s1361-8415(98)80009-1. [DOI] [PubMed] [Google Scholar]
- Sethian JA. Level set methods and fast marching methods : evolving interfaces in computational geometry, fluid mechanics, computer vision, and materials science. Cambridge, U.K. ; New York: Cambridge University Press; 1999. [Google Scholar]
- Sporns O, Tononi G, Kotter R. The human connectome: A structural description of the human brain. PLoS Comput Biol. 2005;1:e42. doi: 10.1371/journal.pcbi.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan R, Zhou X, Miller E, Lu J, Litchman J, Wong ST. Automated axon tracking of 3D confocal laser scanning microscopy images using guided probabilistic region merging. Neuroinformatics. 2007;5:189–203. doi: 10.1007/s12021-007-0013-4. [DOI] [PubMed] [Google Scholar]
- Stepanyants A, Chklovskii DB. Neurogeometry and potential synaptic connectivity. Trends Neurosci. 2005;28:387–394. doi: 10.1016/j.tins.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Stepanyants A, Hof PR, Chklovskii DB. Geometry and structural plasticity of synaptic connectivity. Neuron. 2002;34:275–288. doi: 10.1016/s0896-6273(02)00652-9. [DOI] [PubMed] [Google Scholar]
- Stepanyants A, Tamas G, Chklovskii DB. Class-specific features of neuronal wiring. Neuron. 2004;43:251–259. doi: 10.1016/j.neuron.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Stepanyants A, Martinez LM, Ferecsko AS, Kisvarday ZF. The fractions of short- and long-range connections in the visual cortex. Proc Natl Acad Sci U S A. 2009;106:3555–3560. doi: 10.1073/pnas.0810390106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanyants A, Hirsch JA, Martinez LM, Kisvarday ZF, Ferecsko AS, Chklovskii DB. Local potential connectivity in cat primary visual cortex. Cereb Cortex. 2008;18:13–28. doi: 10.1093/cercor/bhm027. [DOI] [PubMed] [Google Scholar]
- Streekstra GJ, van Pelt J. Analysis of tubular structures in three-dimensional confocal images. Network. 2002;13:381–395. [PubMed] [Google Scholar]
- Trachtenberg JT, Chen BE, Knott GW, Feng G, Sanes JR, Welker E, Svoboda K. Long-term in vivo imaging of experience-dependent synaptic plasticity in adult cortex. Nature. 2002;420:788–794. doi: 10.1038/nature01273. [DOI] [PubMed] [Google Scholar]
- Vasilkoski Z, Stepanyants A. Detection of the optimal neuron traces in confocal microscopy images. J Neurosci Methods. 2009;178:197–204. doi: 10.1016/j.jneumeth.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhou X, Lu J, Lichtman J, Chang S-F, Wong STC. Dynamic local tracing for 3D axon curvilinear structure detection from microscopic image stack. IEEE, International Symposium of Biomedical Imaging (ISBI); Washington D. C. 2007. [Google Scholar]
- Weaver CM, Hof PR, Wearne SL, Lindquist WB. Automated algorithms for multiscale morphometry of neuronal dendrites. Neural Comput. 2004;16:1353–1383. doi: 10.1162/089976604323057425. [DOI] [PubMed] [Google Scholar]
- White J, Southgate E, Thomson J, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Wilt BA, Burns LD, Wei Ho ET, Ghosh KK, Mukamel EA, Schnitzer MJ. Advances in light microscopy for neuroscience. Annu Rev Neurosci. 2009;32:435–506. doi: 10.1146/annurev.neuro.051508.135540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wink O, Frangi AF, Verdonck B, Viergever MA, Niessen WJ. 3D MRA coronary axis determination using a minimum cost path approach. Magnetic Resonance in Medicine. 2002;47:1169–1175. doi: 10.1002/mrm.10164. [DOI] [PubMed] [Google Scholar]
- Xie J, Zhao T, Lee T, Myers E, Peng H. Automatic neuron tracing in volumetric microscopy images with anisotropic path searching. Med Image Comput Comput Assist Interv. 2010;13:472–479. doi: 10.1007/978-3-642-15745-5_58. [DOI] [PubMed] [Google Scholar]
- Yuste R, Bonhoeffer T. Morphological changes in dendritic spines associated with long- term synaptic plasticity. Annu Rev Neurosci. 2001;24:1071–1089. doi: 10.1146/annurev.neuro.24.1.1071. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Toga AW. Efficient Skeletonization of Volumetric Objects. IEEE Transactions on Visualization and Computer Graphics. 1999;05:196–209. doi: 10.1109/2945.795212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Kaufman A, Toga AW. Three-dimensional skeleton and centerline generation based on an approximate minimum distance field. Visual Computer. 1998;14:303–314. [Google Scholar]