Characterization of a novel monocyte subset that suppresses CD8+ proliferation during HIV and SIV infection.
Keywords: macrophages, AIDS
Abstract
Monocytes have been categorized in three main subpopulations based on CD14 and CD16 surface expression. Classical monocytes express the CD14++CD16−CCR2+ phenotype and migrate to inflammatory sites by quickly responding to CCL2 signaling. Here, we identified and characterized the expansion of a novel monocyte subset during HIV and SIV infection, which were undistinguishable from classical monocytes, based on CD14 and CD16 expression, but expressed significantly lower surface CCR2. Transcriptome analysis of sorted cells demonstrated that the CCR2low/neg cells are a distinct subpopulation and express lower levels of inflammatory cytokines and activation markers than their CCR2high counterparts. They exhibited impaired phagocytosis and greatly diminished chemotaxis in response to CCL2 and CCL7. In addition, these monocytes are refractory to SIV infection and suppress CD8+ T cell proliferation in vitro. These cells express higher levels of STAT3 and NOS2, suggesting a phenotype similar to monocytic myeloid-derived cells, which suppress expansion of CD8+ T cells in vivo. They may reflect an antiproliferative response against the extreme immune activation observed during HIV and SIV infections. In addition, they may suppress antiviral responses and thus, have a role in AIDS pathogenesis. Antiretroviral therapy in infected macaque and human subjects caused this population to decline, suggesting that this atypical phenotype is linked to viral replication.
Introduction
Monocytes have often been described as transitional cells of myeloid origin, whose main function is to replenish the phagocyte population in tissues through differentiation into macrophages and other specialized cells, such as microglia, osteoclasts, and DCs [1]. However, during their short life in the blood, monocytes are far from bystanders; they are constantly patrolling the vasculature, detecting pathogens and unwanted cells, and clearing them from the circulation [2]. They also quickly respond to chemokine signals by trafficking to tissues upon stimulation, as well as mediating important functions in immune responses by producing copious amounts of cytokines [3, 4].
Circulating monocytes derive from hematopoietic stem cells in the bone marrow and can be easily distinguished from other leukocytes when analyzed by flow cytometry, presenting a CD3negCD20negCD11bhighCD14high/low phenotype [5–7]. They can also be separated into different subpopulations with distinct surface markers and biological functions [1]. Based on expression levels of CD14 (LPS receptor) and CD16 (FcγRIII), three well-defined monocyte subpopulations have been identified in humans and nonhuman primates: CD14+CD16−, referred to as classical or inflammatory; CD14lowCD16high (or CD14dimCD16+), referred to as nonclassical, resident, or proinflammatory; and a transitional subpopulation of CD14highCD16+, referred to as intermediate. Similar functional subpopulations are found in mice and rats, although other surface markers (Ly6C, 7/4) are used for such classification [5, 8, 9].
Classical monocytes express CCR2 and migrate into the circulation and toward inflammatory sites, mainly through CCL2 (or MCP-1)-dependent signaling. Upon reaching the target tissues, they differentiate into macrophages or in some sites, DCs [10–12]. Conversely, nonclassical monocytes appear to represent a more advanced stage of differentiation, closer to resident macrophages, and they are the main subpopulation supplying resident macrophages and DCs to healthy tissues [13]. They migrate into tissues in response to MIP-1α (or CCL3), CCL5 (or RANTES), and CX3CL1 (or fractalkine in humans) but not CCL2, as they lack CCR2 on their surface [1, 5]. In humans, this CD14dimCD16+ subset patrols the endothelium of blood vessels and responds to viruses via TLR7 and -8, in contrast to classical and intermediate monocytes, which respond to bacteria-associated signals [2].
Several novel blood monocyte phenotypes have been described recently, indicating that their heterogeneity is not restricted to the three classical subtypes. Monocytes expressing the angiopoietin receptor Tie-2 exhibit marked proangiogenic activity and account for 35–75% of the nonclassical subset in the blood of human cancer patients [14]. CD16highCCR2− monocytes, producing high levels of HO-1, may play an anti-inflammatory role during Kawazaki disease and influenza virus infection [15, 16], and a subset of CD14lowCD16+ cells expressing the FcγRI CD64, in conjunction with high levels of HLA-DR, CD11c, and CD86, possibly represents an intermediate phenotype between monocytes and DCs [17]. CD14+HLA-DRlow/neg immunosuppressive monocytes have been identified in many cancer patients [18, 19], although cells presenting a similar phenotype have been reported in other neoplasias and were classified as monocytic MDSCs [20, 21].
In HIV and SIV infection, the expansion of specific monocyte subsets has been associated with the development of HAND, and a high rate of monocyte turnover correlates with AIDS progression in SIV-infected macaques [7, 22, 23]. In this study, the analysis of monocyte subpopulations during acute SIV infection using our consistent, accelerated animal model for HAND and HAART led to the discovery of a CD14+CD16− monocyte subset that lacks surface CCR2, rendering them unresponsive to CCL2. These cells express a unique transcriptome, impaired phagocytosis and chemotaxis, and are refractory to viral infection. They also express higher levels of STAT3 and NOS2 and are able to interfere with the proliferation of CD8+ T cells in vitro, which leads us to suggest that this novel subset could be classified as MDSC, with an antiproliferative role during SIV infection. In parallel, we demonstrate that HIV-1-infected human subjects also have this monocyte subset and that efficacious antiretroviral treatment, in macaques as in humans, is able to revert the frequency of this population back to preinfection levels. We demonstrate that HIV and SIV infection lead to changes in monocyte phenotype and that these changes impact early immune responses, which may affect virus pathogenesis.
MATERIALS AND METHODS
Animals
Forty-five juvenile pigtailed macaques (Macaca nemestrina) were inoculated i.v. with SIV/DeltaB670 and SIV/17E-Fr, as described previously [24]. Beginning on Day 12 p.i., six animals were treated with a combination of four antiretroviral drugs (PMPA, saquinavir, atazanavir, and the integrase inhibitor L-870812) and were euthanized after 160–186 days p.i., as described elsewhere [25]. Groups of SIV-infected, untreated animal (six each) were euthanized after 7, 14, and 21 days p.i. All other animals were euthanized between 50 and 90 days p.i. Two macaques were mock-inoculated and used as procedure controls. Before euthanasia, animals were perfused with sterile PBS, and tissues were harvested for fixing and freezing. All procedures were done in accordance with federal and institutional policies.
Patients
Samples were selected from a cohort designed to recruit recently HIV-1-infected people in 2002 in São Paulo, Brazil [26, 27]. All procedures adopted in this study were performed according to the terms agreed by the Institutional Review Board from the Hospital das Clínicas, University of São Paulo (CAPPesq, Research Projects Ethics Committee, São Paulo, Brazil). This study was approved by CAPPesq under Protocol 0148/11, and written, informed consents were obtained from all volunteers. Recent HIV infection was determined by the serological test algorithm for recent HIV seroconversion. Subjects were included in the study when they had a negative, desensitized ELISA HIV-1 test. Antiretroviral treatment was initiated when CD4+ T cell count was <300 cells/μl or upon identification of any AIDS-defining condition. We randomly selected 12 subjects to evaluate the early infection monocyte subpopulations at baseline, 3 months, and post-6 months. Another group of six subjects was evaluated before and after initiation of antiretroviral therapy.
Samples
Macaque blood samples were collected at multiple time-points before and after inoculation. Whole blood was used for cytometry analysis. For some experiments, PBMCs, previously collected at different time-points, were rapidly thawed in 25% FBS RPMI and kept on ice to maintain monocyte viability. After a cold wash with 10% FBS RPMI, viable cells were enriched using the Annexin V MicroBead kit (Miltenyi Biotec, Auburn, CA, USA) and aliquoted for FACS analysis and functional assays, as described below. For human samples, PBMCs were thawed, checked for viability, and used for flow cytometry assays, also described below. For in vitro evaluation of CCR2 expression, PBMCs were transferred to low cell-binding HydroCell plates (Nunc, Rochester, NY, USA), incubated overnight in 10% FBS RPMI containing 100 ng/ml CCL2 (ProSpec, Charlotte, NC, USA) or 1 ng/ml LPS (Sigma-Aldrich, St. Louis, MO, USA), and analyzed by flow cytometry.
Flow cytometry
Macaque blood samples were stained with FITC-conjugated anti-CD14, PE-conjugated anti-CCR2, and PE-Cy5-conjugated anti-CD16 antibodies (see Supplemental Table 1) for 20 min at room temperature. Cells were fixed for 10 min with Lyse/Fix buffer (Becton Dickinson, Franklin Lakes, NJ, USA), and for intracellular staining, cells were permeabilized using Cytofix/Cytoperm (Becton Dickinson). Analyses were performed on a FACSCalibur flow cytometer using CellQuest software. PBMCs from frozen samples were stained with different antibody combinations for 30 min on ice. Panels with information for all antibodies can be found in Supplemental Table 1. After fixation, samples were analyzed on a BD LSRFortessa cell analyzer using DIVA software. Human PBMCs and cells used in other experiments were processed similarly and evaluated on a BD FACSCanto or LSRFortessa flow cytometer using DIVA software. All data acquired were analyzed using FlowJo software. Positivity for each antibody was determined by FMO.
Quantification of SIV virions
Viral RNA was isolated from plasma using the QIAamp Viral RNA Mini Kit (Qiagen, Valencia, CA, USA) and analyzed by qPCR, as described previously [28].
Quantification of CCL2
Levels of plasma CCL2 were measured using the Quantikine Human for MCP-1 ELISA Kit (R&D Systems, Minneapolis, MN, USA), following the manufacturer's protocol.
Sorting
Macaque PBMCs were thawed as described above and stained with anti-CD3 and anti-CD20 (FITC), anti-CD14 (PerCP-Cy5.5), anti-CD16 (PE), and anti-CCR2 (allophycocyanin) antibodies (Supplemental Table 1). After 30 min on ice, cells were fixed with 4% formaldehyde and sorted on a BDAria Special Order cytometer. Purity of sorted populations varied from 95% to 98%.
Nucleic acid extraction
Genomic DNA and total RNA were isolated from sorted cells using the Allprep DNA RNA formalin-fixed, paraffin-embedded kit (Qiagen) with modifications. Briefly, cells were pelleted and resuspended in PKD buffer with 10 μl proteinase K. Samples were incubated for 15 min at 55°C and then for 5 min on ice. Lysates were centrifuged at 10,000 g for 15 min. Pellets were resuspended in ATL buffer, and DNA was extracted, according to the manufacturer's instructions. Supernatants containing the RNA were incubated for 15 min at 80°C and then mixed with two volumes of 100% ethanol and 1 μg carrier RNA (Qiagen). After 30 min at −20°C, RNA was pelleted by centrifugation and resuspended in 6 μl RNase-free water.
mRNA quantification
CodeSets for 90 macaque genes were designed according to NanoString specifications, based on rhesus macaque (Macaca mulata)-published sequences. RNA samples were hybridized for 16 h with the CodeSet, and genes were quantitated using the nCounter Digital Analyzer [29]. Data were normalized initially by the average of the values from spiked, positive standards and then by the geometric mean of four housekeeping genes. Raw data with values lower than the background controls were considered negative. Results were deposited at the Gene Expression Omnibus database, under Accession Number GSE27559 (http://www.ncbi.nlm.nih.gov).
Viral DNA quantification
DNA samples were analyzed for SIV gag by qPCR, as described previously [30]. SIV circular 2-LTR was quantitated in the same reaction tubes using the following set of primers and probe: 2LTR forward-GAAGACCCTGGTCTGTTAGGACC; 2LTR reverse-CTTGCACTGTAATAAATCCCTTCCA; 2LTR-HEX-TTTGGGAAACCGAAGCAGGAAAATCC. Copy numbers were determined by extrapolation from standard curves generated by serial dilution of plasmids containing the specific DNA sequences for each amplicon. Results were normalized to copy numbers of a single-copy cellular gene (IFN-β).
Phagocytosis assay
Macaque PBMCs were thawed as described above. Cells (2×105) were resuspended in 10% FBS RPMI and placed in HydroCell low-adherence plates (Nunc), mixed with 20 μl Escherichia coli particles, covered with pHrodo (Life Technologies, Carlsbad, CA, USA), a nonfluorescent dye that gains fluorescence in acidic milieu, such as lysosomes. After 30 min at 37°C, cells were placed on ice and stained with conjugated antibodies for FACS analysis. Besides plating cells without pHrodo for viability control, cells were also plated with pHrodo but kept on ice to prevent phagocytosis and were used as negative control.
Chemotaxis assay
BD BioCoat Matrigel Invasion Chambers were used for chemotaxis experiments, following the manufacturer's instructions. This transwell system contains a top insert containing a 10-μM pore membrane covered with a gelatinous matrix that prevents transmigration of noninvasive cells. Top inserts holding 500 μl, containing 105 thawed PBMCs, were transferred to a HydroCell low-adherence, 24-well plate with 100 ng/ml CCL2, CCL3, or CCL7 (ProSpec) in 500 μl 10% FBS RPMI. Wells containing only media were used as background control for each sample. After 24 h of incubation at 37°C, cells that migrated to the bottom wells were split in two groups: 30% were counted in a hemocytometer, and 70% were stained for FACS analysis.
Lymphoproliferation assay
Lymphocytes and other nonmonocytic cells were isolated from fresh PBMCs from two uninfected macaques using anti-CD14 magnetic beads and labeled with a tracer dye (Vybrant DiI, Life Technologies) for 10 min. After two washes, cells were cultivated in RPMI with 10% macaque serum, 25 μg/ml PHA (Sigma-Aldrich), 2 U/ml IL-2 (ProSpec), and 5 μM ZDV. Concomitantly, CD14highCD16−CCR2low/neg monocytes were isolated from thawed PBMCs, collected from three SIV-infected animals, euthanized at 14 days p.i., using the Monocyte Isolation Kit II (Miltenyi Biotec) in combination with biotinylated anti-CCR2 (R&D Systems). Once purified, CD14highCD16−CCR2low/neg monocytes were added to labeled cells (10,000 monocytes for 100,000 nonmonocytic cells) in proliferation media. Cell sorting by magnetic beads resulted in a 95% pure population of CCR2low/neg classical monocytes but only a 30–40% pure population of CCR2high cells. Therefore, as control, we used monocytes CD14+CD16−CCR2+ similarly isolated from thawed PBMCs from three uninfected macaques using the same protocol minus the anti-CCR2 antibody. After 72 h, the supernatant was stored for IFN-γ quantification, and cells were collected and stained for FACS. Proliferation levels were analyzed by FlowJo.
IFN-γ quantification
Supernatants collected from the lymphoproliferation assay were incubated with anti-IFN-γ cytometric beads (Becton Dickinson), according to the company's protocol. Data were acquired on a BD LSRFortessa and analyzed by FCAP array software (Becton Dickinson).
Statistical analysis
Differences in absolute numbers of cells and in CCR2hi/CCR2low/neg classical monocyte ratios at different time-points were analyzed by Wilcoxon paired rank test. Bivariate analyses were done using Spearman's rank correlation test. All other results were analyzed using Mann-Whitney t test. Outliers were defined by applying the Grubbs test [31]. Calculations were performed using Prism software (GraphPad Software, La Jolla, CA, USA), and statistical significance was set as P < 0.05.
RESULTS
Circulating monocyte subpopulations change during acute SIV infection
In the consistent, accelerated SIV model for HIV/AIDS and CNS disease, pigtailed macaques are inoculated with the immunosuppressive swarm SIV/DeltaB670 and the neurotropic molecular clone SIV/17E-Fr, leading to the onset of AIDS and encephalitis within 3 months [30]. Viral mRNA in the brain can be detected as early as 4 days p.i., and macrophages, derived from monocytes that recently migrated from the peripheral blood, represent the main source of viral replication [32]. To investigate the contribution of circulating monocytes in the early events observed in the brain, blood samples from 18 SIV-infected macaques were collected before virus inoculation and at 7, 10, 14, 21, and 28 days p.i. and analyzed by flow cytometry. Monocytes were initially distinguished from other leukocytes by FSC and SSC and then assessed according to their levels of CD14 and CD16 expression (Fig. 1A). As proposed by the Nomenclature Committee of the International Union of Immunological Societies, classical monocytes were defined as CD14+CD16−, intermediate monocytes as CD14highCD16+, and nonclassical monocytes as CD14dimCD16+ [33]. Cells expressing low levels of CD14 and no CD16 have been identified previously as a heterogeneous population of nonmonocytic leukocytes and were not considered in our analyses [8].
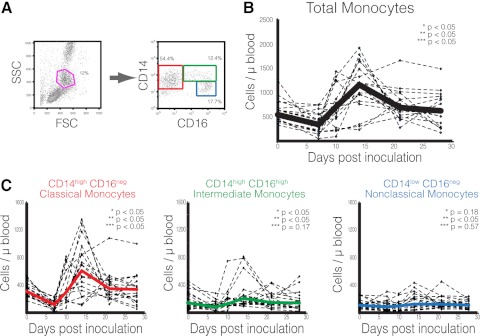
Figure 1. Changes in monocyte subpopulations during SIV acute infection.
(A) Gating strategy used to discriminate monocyte subsets [classical monocytes (red), intermediate monocytes (green), and nonclassical monocytes (blue)]. (B) Absolute number of total circulating monocytes. Solid line represents the median of 18 SIV-infected macaques. (C) Absolute number of each monocyte subset. Solid lines represent the median of all animals. (B and C) Asterisks denote Wilcoxon paired rank test; P values between absolute number of cells at the following time-points: *uninfected versus 7 days p.i.; **7 days versus 14 days p.i.; and ***14 days versus 21 days p.i.
A significant change in the total number of circulating monocytes was observed in the first 28 days p.i. and was characterized by three distinct phases: an initial period of decline, followed by expansion, and a return to preinoculation levels (Fig. 1B). Although all individual subsets presented similar trends (Figs. 1C), their contribution to the overall population in each of the three phases were noticeably different. At 7 days p.i., the absolute number of classical and intermediate monocytes was reduced significantly (P<0.05 for both), indicating that they were the main subsets driving the initial depletion observed in the total population. Both subsets were CD14high, whereas no significant change was observed in the CD14dimCD16+ nonclassical subgroup (P=0.31). After this transient decrease, all subpopulations expanded significantly (Fig. 1B), leading to a pronounced, albeit temporary, monocytosis at 14 days p.i. In the third phase, the total monocyte decline appeared to be the result of classical monocyte fluctuations (P=0.03), as no significant variation was detected in subpopulations expressing CD16 (P=0.12 for intermediate, and P=0.65 for nonclassical monocytes; Fig. 1C). These results suggest that CD14+CD16− cells were the main driver of monocyte changes during acute infection.
CCR2 is down-regulated in classical monocytes during SIV acute infection
It is well established that CD14+CD16− monocytes express high levels of CCR2 on their surface [3]. As we were characterizing changes in the monocyte population during acute infection, we observed a dramatic down-regulation of surface CCR2 expression, specifically in the CD14+CD16− subset as early as 7 days p.i. (Fig. 2A and B). In contrast, intermediate CD14highCD16+ monocytes, which constitute a heterogeneous population regarding the expression of several surface proteins, manifested no significant change in CCR2 expression when compared with steady-state levels.
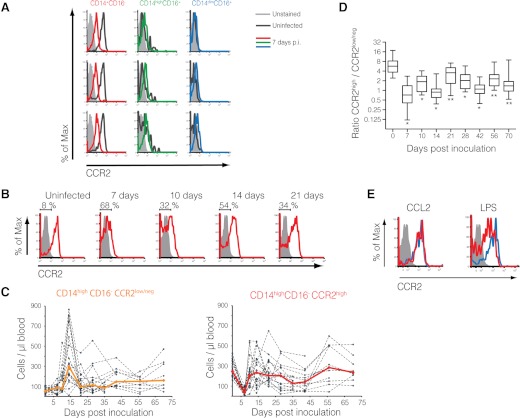
Figure 2. CCR2 is down-regulated in classical monocytes during acute infection.
(A) Histograms demonstrate the specific down-regulation of CCR2 expression on CD14+CD16− monocytes at 7 days p.i. Unstained control (defined by FMO) is represented in light gray. Uninfected samples are represented by dark gray lines and 7 days p.i., by colored lines. Each row represents one macaque. Representative of 18 animals. (B) CCR2 surface expression on classical monocytes during acute SIV infection. Brackets represent the percentage of CCR2low/neg. Unstained control is represented in light gray. Representative of 18 animals. (C) Absolute number of CCR2low/neg and CCR2high classical monocytes in 18 SIV-infected macaques until 70 days p.i. Solid lines represent median. (D) Ratio between CCR2high and CCR2low/neg classical monocytes for each time-point. Median differences between uninfected and each time-point were analyzed by Wilcoxon paired rank test. *P < 0.01; **P < 0.05. (E) Surface CCR2 expression on uninfected classical monocytes exposed overnight to CCL2 (100 ng/ml) and LPS (1 ng/ml). Unstained control is represented in light gray. Untreated controls are represented by blue lines and treated cells by red lines. Representative of three experiments.
CD14highCD16−CCR2low/neg monocytes expanded considerably at 14 days p.i. (Fig. 2C) but were reduced at Day 21 p.i., remaining higher than preinfected levels throughout infection. CCR2high classical monocytes also expanded from 7 to 14 days p.i. but slowly declined to preinfected levels after 21 days p.i., with a distinct longitudinal profile when compared with the CCR2low/neg subset (Fig. 2B and C).
During acute SIV infection, the ration between CCR2high and CCR2low/neg subsets changed dramatically, and by Day 7 p.i. >50% of circulating classical monocytes were CCR2low/neg. Even with the increase of CD14highCD16−CCR2high cells after Day 10 p.i., the ratios between CCR2high and CCR2low/neg classical monocytes were not restored to preinfected levels and remained significantly lower throughout infection (Fig. 2D).
To evaluate the role of CCL2 in the down-regulation of surface CCR2 expression, PBMCs from uninfected macaques were cultivated overnight in RPMI media with CCL2 (100 ng/ml). As positive control, cells were exposed to bacterial endotoxin (LPS), a strong CCR2 expression modulator [34]. Although CCL2 has been reported to quickly down-regulate CCR2 surface expression in RBL-2H3-transfected cells [35], no significant change was observed in primary monocytes under CCL2 treatment overnight (Fig. 2E). Similar results were observed in human embryonic kidney-293T cells transfected with a rhesus CCR2 construct [36] treated under the same conditions (data not shown), indicating that long-term treatment with CCL2 does not alter CCR2 surface expression significantly and that the lower detection of CCR2 expression that we observed in vivo was not caused by interactions between agonist and receptor, which could block the access of cytometry antibodies to their specific epitopes. In vivo analysis corroborated these finding, showing a small correlation (r=0.18; P<0.05) between levels of plasma CCL2 measured by ELISA and the absolute number of CD14highCD16−CCR2low/neg cells (Supplemental Fig. 1A). Other biological parameters frequently measured during HIV/SIV infection, such as viral load and levels of CD4+ and CD8+ T cells, did not correlate with the frequency of this new CCR2low/neg subset (Supplemental Fig. 1B–D).
The loss of surface CCR2 could impact cell trafficking to tissues and efficient responses during infection and inflammation. Therefore, we postulated that in addition to the dramatic down-regulation of CCR2 expression, other changes might be occurring in this subset and that CD14highCD16−CCR2low/neg monocytes represent a distinct, functional phenotype when compared with their CCR2high counterparts.
CD14highCD16−CCR2low/neg monocytes display a transcriptome distinct from classical monocytes
To investigate the mRNA expression profile of CCR2high and CCR2low/neg classical monocytes, cells from six SIV-infected macaques were FACS-sorted according to their surface marker phenotype (Supplemental Fig. 2). Longitudinally acquired blood samples were limited in volume; therefore, viably frozen PBMCs collected during necropsy from macaques euthanized at 14 days p.i. were used. Each subset's transcriptome was analyzed by NanoString, using a panel of 90 probes specifically designed for the detection and quantitation of macaque genes associated with inflammatory and immuneregulatory processes. The specificity of the probe set was confirmed using spleen and brain tissue from uninfected and SIV-infected pigtailed macaques (data not shown).
CD14highCD16−CCR2low/neg presented a unique phenotype, based on 78 genes that were expressed in at least one subset. Levels of expression of 41 genes were distinct between CCR2high and CCR2low/neg classical monocytes with P values < 0.05 (Fig. 3A). Among the 41 transcripts with differential expression between the two subsets, 31 (34.4% of all genes in the array) were expressed at lower levels in CD14highCD16−CCR2low/neg monocytes, including the typical inflammatory cytokines TNF-α, IL-6, IL-7, IL-1β, IL-27, CCL3, CCL4, CCL19, CXCL3, CXCL9, CXCL10, XCL1, IFN-γ, and IFN-α2. Accordingly, they also expressed lower levels of IRF1, IRF7, STAT5a, and IFN-stimulated genes, including OAS2 and ZBP1.
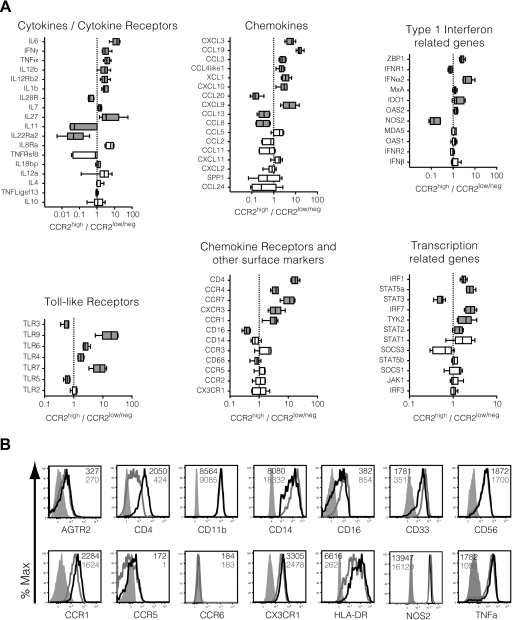
Figure 3. Relative mRNA expression between sorted CD14+CD16−CCR2high and CD14+CD16−CCR2low/neg monocytes from six SIV-infected macaques at 14 days p.i.
(A) Classical monocytes from PBMCs collected at 14 day p.i. were FACS-sorted according to their CCR2 surface expression. Total RNA from both subsets was isolated and analyzed by Nano-String technology. Graphics illustrate relative fold change in gene expression, comparing CCR2high with CCR2low/neg classical monocytes. Genes are separated in families and listed according to their P values. Gray boxes denote statistical significance between the two subsets (Wilcoxon paired rank test P>0.05). TNFRsf8, TNFR superfamily 8; TNFLigsf13, TNF ligand superfamily 13; SPP1, secreted phosphoprotein 1; MDA5, melanoma differentiation-associated gene 5; TYK2, tyrosine kinase 2; SOCS3, suppressor of cytokine signaling 3. (B) Protein expression of various genes in CCR2high (black lines) and CCR2low/neg (dark gray lines) classical monocytes evaluated by flow cytometry. Unstained controls are illustrated in light gray-filled histograms. Numbers inside the graphics represent the mean fluorescence intensity of each subset, shaded as described above. Representative of three animals.
CD14highCD16−CCR2low/neg monocytes presented higher levels of TLR3 and TLR5 and lower levels of TLR6, TLR4, and TLR7 when compared with their CCR2high counterpart, as well as undetectable levels of TLR9 (Supplemental Fig. 3A). In addition to TLR3 and TLR5, CD14highCD16−CCR2low/neg monocytes presented higher levels of transcripts for two IFN-related receptors (IL-28R, IFNR1) and less-characterized cytokines (CCL20, CCL13, CCL8). They also presented significantly higher levels of STAT3 and NOS2, suggesting that these cells have an immunosuppressive phenotype, similar to MDSCs [21].
Thirty-six of the analyzed mRNAs demonstrated no significant change between the two populations, although in some cases, P values were >0.05 as a result of a single outlier, defined by Grubbs test [31]. Removal of the outlier made CCR5 and IL-12A significantly higher in classical monocytes (from P=0.11 and P=0.26, respectively, to P<0.05). We observed no relative change in the expression of IL-4, IL-10, IL-18bp, CCL2, CCL5, CCL24, CXCL2, or CXCL11. Some cytokines were undetectable for both subsets in all samples (IL-9, IL-13, IL-17, IL-25, IL-28A/B, IL-29, CCL1, and CCL7).
To corroborate our transcriptome findings, we evaluated the expression of several proteins in both subpopulations by flow cytometry, including some genes that were not part of our NanoString array, such as AGTR2, CD11b, CD33, CD56, CCR6, and HLA-DR. The combination of data from RNA and protein analyses confirmed the monocytic phenotype of the CCR2low/neg cells. They expressed the myeloid markers CD33 and CD11b and slightly higher levels of CD14 and CD16. Both populations expressed similar levels of CD68 and also CD56, a surface marker typically associated with NK cells in humans but that identifies monocytes in macaques [37]. HLA-DR was also down-regulated in these monocytes, similarly to what has been reported in monocytic MDSCs in cancer [18, 19]. Lower levels of CCR2 mRNA in CD14highCD16−CCR2low/neg monocytes were observed in three of the six animals (Supplemental Fig. 3B), indicating that other factors, such as protein turnover, might be involved in the reduction of membrane expression. Although lacking CCR2 on their membranes, these cells still express receptors known to respond to chemokines in monocytes, such as AGTR2, CCR1, and CX3CR1 [38–40]. We also observed a slight but not significant increase of NOS2 protein expression in the CCR2low/neg cells.
CD14highCD16−CCR2low/neg monocytes also presented a dramatic reduction in the mRNA and surface protein levels of CD4, which is known to be expressed in monocytes in certain circumstances [41]. These results, together with the down-regulation of surface CCR5, suggest that these cells might be resistant to SIV and HIV infection.
These results strongly support that CD14highCD16−CCR2low/neg cells are a distinct subset within classical monocytes. As several of these variations could interfere with functions normally ascribed to monocytes, we investigated whether CD14highCD16−CCR2low/neg monocytes were able to properly phagocytize and respond to chemoattractants.
CD14highCD16−CCR2low/neg monocytes present impaired phagocytosis and chemotaxis
To investigate whether CD14highCD16−CCR2low/neg cells retained phagocytic abilities when compared with classical monocytes from healthy subjects, viably frozen PBMCs from uninfected and SIV-infected macaques euthanized at 7, 14, and 21 days p.i. were exposed to E. coli covered in pHrodo, a dye that is not fluorescent at neutral pH but fluoresces when located in acidic milieu, such as phagosomes and lysosomes.
More than 80% of monocytes from uninfected macaques were positive for pHrodo fluorescence, in contrast to 58% and 51% of monocytes from 7- and 14-day p.i. samples, respectively (Fig. 4A). Control monocytes presented higher mean fluorescence, which indicates a larger intake of particles/cell (Fig. 4B). There was no significant difference between uninfected and 21-day-p.i. samples, suggesting that monocytes regained their ability to efficiently phagocytize after acute infection.
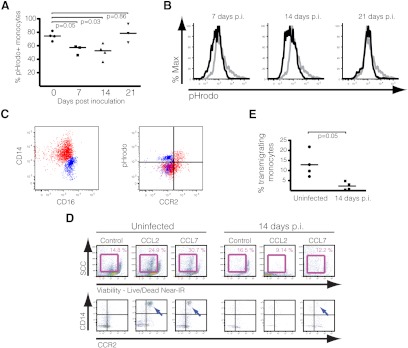
Figure 4. CD14highCD16−CCR2low/neg monocytes present impaired phagocytosis and chemotaxis.
(A) Percentage of monocytes positive for pHrodo fluorescence at different time-points. Bars represent medians, and P values were calculated by Mann-Whitney t test. (B) Histograms demonstrate the intensity of pHrodo red fluorescence in total monocytes from uninfected (gray line) and infected macaques (black line) at different time-points. Graphics are representative of four experiments for each of the time-points. (C) Total monocytes from a 14-day-p.i. macaque, depicting the expression of CD14 versus CD16 and pHrodo versus CCR2. Classical and intermediate monocytes are displayed in red and nonclassical in blue, demonstrating that the vast majority of CCR2low/neg cells expressing the phagocytosis red fluorescent marker includes nonclassical monocytes. Similar results were observed at 7 and 21 days p.i. Graphics are representative of four experiments. (D) Graphs depict cells collected from the bottom of a transwell system containing control media or media with CCL2 or CCL7 and stained for FACS analysis. After gating on viable cells (Live/Dead Near-IR dye) with the approximate size for monocytes (upper panels), populations were analyzed for CD14 and CCR2 expression (bottom wells; lower panels). Positivity was determined by FMO. Bottom wells from uninfected samples presented CD14+CCR2low monocytes (blue arrows; lower panels) for both experimental conditions, whereas few cells from 14 days p.i. transmigrated. The figure is representative of four experiments. (E) Graphic shows the percentage of monocytes that transmigrated after being exposed to CCL2-containing media. Total PBMCs were counted using a hemocytometer, and monocytes were identified by flow cytometry. After 18 h, all media were collected from the bottom wells, and cells were stained for lineage markers (CD3, CD8, CD20), CD14, CD16, and CCR2. Monocyte numbers were estimated by flow cytometry, after all events were counted for each sample. Bars represent medians, and P values were calculated by Mann-Whitney t test.
The maintenance of PBMCs in culture at 37°C for the duration of this experiment was enough to increase the surface expression of CD16 in classical monocytes, even in the absence of pHrodo, likely as part of their programming to differentiate into macrophages [42]. Distinctions between classical and intermediate monocytes were harder to establish, but it was noticeable that at all time-points, only CCR2high cells among the two CD14++ subtypes were also positive for pHrodo (Fig. 4C). Expression of CCR2, as expected, was not essential for efficient phagocytosis, as 50% of nonclassical monocytes CD14dimCD16+, which do not express CCR2, were also pHrodo-positive. These data indicate that monocytes, in general, present a transitory phase of impaired phagocytosis after SIV infection and that the CD14highCD16−CCR2low/neg monocytes are affected specifically by this dysfunction in all time-points examined.
We also analyzed the ability of monocytes, at 14 days p.i., to respond to CCL2 signaling using a transwell migration assay. PBMCs from uninfected or infected macaques at 14 days p.i. were placed on top inserts and transferred to wells containing medium with 100 ng/ml CCL2, CCL3, or CCL7 [43]. After 24 h, cells were collected from the bottom wells and stained for FACS analyses.
In samples from uninfected monkeys, PBMCs started transmigrating to the lower chambers as early as 4 h after plating. Although we observed transmigrated cells even in media containing no chemoattractant, only wells with CCL2 and CCL7 contained CD14+ monocytes (Fig. 4D). Levels of CD14 and CCR2 appeared lower than what we normally observed in fresh PBMCs, suggesting that monocytes down-regulated surface expression during or after transmigration. In the wells holding 14-day-p.i. PBMCs, as shown in Fig. 4D and E, we observed a significant reduction in transmigrated cells, independent of the chemoattractant. We observed almost no CD14+CCR2high leukocytes, paralleling the previous observation that monocytes, in general, have impaired function during acute infection and that CD14highCD16−CCR2low/neg cells, specifically, do not respond to chemokine signaling in this assay. Samples from infected animals at 14 days p.i. still contained a large number of CCR2high cells before transmigrating (20–50% of all classical monocytes), suggesting that besides lower surface-receptor expression, other factors might play a role in their dysfunctional chemotactic response. Similar results were observed with monocytes at 7 and 21 days p.i. (data not shown).
CD14highCD16−CCR2low/neg monocytes impair in vitro proliferation of CD8+, but not CD4+, T cells, and harbor fewer copies of SIV DNA genome
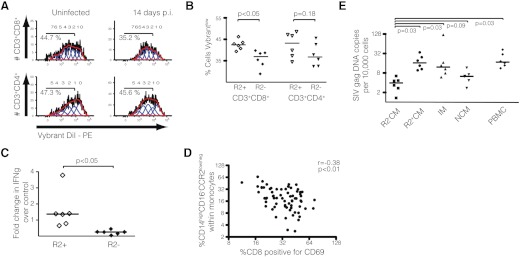
A population of CD14+HLA-DRlow/neg cells with a suppressive phenotype has been identified in patients with B cell non-Hodgkin's lymphoma [18]. To assess a functional role of CD14highCD16−CCR2low/neg monocytes in lymphoproliferation, we exposed proliferating lymphocytes from uninfected macaques to classical monocytes isolated from other uninfected animals or from CCR2low/neg classical monocytes from SIV-infected macaques at 14 days p.i. (Fig. 5A). To prevent virus spread from infected monocytes, cells were cultivated in 5 μM ZDV. After 72 h, we observed a significant decrease in the proliferation of CD8+ T cells exposed to CD14highCD16−CCR2low/neg monocytes (P<0.05), although no significant change was observed in CD4+ T cells (Fig. 5B). Levels of IFN-γ in the coculture supernatants were also significantly lower in the wells containing CD14highCD16−CCR2low/neg monocytes (Fig. 5C), even with the removal of the outlier. Bivariate analysis of in vivo data showed a significant negative correlation between the percentage of monocytes expressing the CD14highCD16−CCR2low/neg phenotype and the percentage of CD69+ CD8+ T cells, but not CD4+ T cells (data not shown), in infected macaques from 7 to 28 days p.i. (r=−0.38; P<0.01), suggesting that the expansion of this unique monocyte subpopulation might interfere with the development of adaptive immune responses early during infection (Fig. 5D).
Figure 5. CD14+CD16−CCR2− monocytes impair the proliferation of CD8+ T cells and harbor fewer copies of SIV genomic DNA.
(A) Histograms showing the percentage of proliferated lymphocytes exposed to classical monocytes from uninfected macaques or CD14highCD16−CCR2low/neg monocytes from SIV-infected macaques at Day 14 p.i. for 72 h under proliferative conditions. Representative of three lymphocyte donors cocultivated with monocytes from three uninfected or three infected animals. (B) Lymphocytes CD8+, but not CD4+, cocultivated with CD14highCD16−CCR2low/neg monocytes (R2−) presented significantly fewer cells with low expression of the tracer dye than when cocultivated with classical monocytes from uninfected macaques (R2+). (C) Levels of IFN-γ in the supernatant of lymphocytes cocultivated in the presence of CD14highCD16−CCR2low/neg monocytes (R2−) are reduced when compared with classical monocytes (R2+). Values represent fold change over levels of IFN-γ collected from PBMCs cocultivated in proliferation media but without monocytes. (B and C) Bars represent medians, and P values were calculated by Mann-Whitney t test. (D) Percentage of monocytes expressing the phenotype CD14highCD16−CCR2low/neg was analyzed against the percentage of CD8 T cells positive for CD69. Each point represents one time-point (7, 10, 14, 21 days p.i.) for each of 14 animals. Correlations were calculated using Spearman's rank correlation test. (E) DNA was isolated from sorted monocytes, and SIV gag was quantitated by qPCR and normalized by 10,000 cells (see Materials and Methods). Bars represent medians, and P values were calculated by Wilcoxon paired rank test. Each point represents an average of two isolations from each animal [CCR2− classical monocytes (R2−CM); CCR2+ classical monocytes (R2+CM); intermediate monocytes (IM); nonclassical monocytes (NCM); PBMC].
Monocytes are targets of HIV and SIV infection and may support active replication in vivo [44]. Some groups have identified the CD14++CD16+ subset as more susceptible to HIV and SIV infection [8, 45]. To investigate which monocyte subsets harbor viral DNA in our model at 14 days p.i., genomic DNA samples isolated from classical (CD14highCD16−CCR2high), nonclassical (CD14lowCD16+), intermediate (CD14highCD16+), and CD14highCD16−CCR2low/neg monocytes, sorted by cytometry, were analyzed by qPCR using sets of primers and probes specific for SIV gag and circular 2-LTR sequences. Genomic DNA samples from fixed PBMCs were used as positive control.
There was no significant difference between the number of SIV gag DNA copies detected in intermediate and classical monocytes (P=0.58), whereas both subsets had higher levels than nonclassical monocytes (P=0.03 for both; Fig. 5E). Levels of SIV gag DNA in CD14highCD16−CCR2low/neg monocytes were a median of 9.3× lower than CD14highCD16−CCR2high monocytes (P=0.03), 2.9× lower than CD14highCD16+ monocytes (P=0.03), and 1.8× lower than CD14lowCD16+ monocytes (P=0.09). We observed no difference in levels of gag DNA between PBMCs and classical or intermediate monocytes (P=0.43 and P=0.68, respectively). However, there were twice as many copies of SIV gag DNA in PBMCs than in total monocytes (17.3 vs. 9.4, respectively), when considering the median of all values together, corrected by the proportion of each subset (data not shown). No SIV 2-LTR DNA was detected in any monocyte subpopulation, although they could be quantitated in PBMCs (median of 2.45 copies/10,000 cells), suggesting that monocytes were infected but not sustaining detectable viral replication. These results, together with data indicating that CD14highCD16−CCR2low/neg express less CD4 and CCR5, suggest that this novel subset may be less susceptible to SIV infection.
Monocyte subpopulations revert to uninfected levels in HAART-treated, SIV-infected macaques
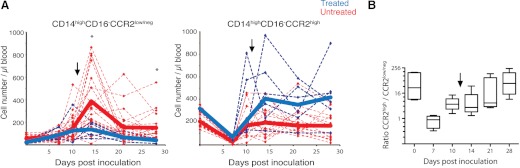
Given that the CD14highCD16−CCR2low/neg subpopulation rapidly expanded in response to SIV infection, we postulated that a reverse process would follow as animals initiated treatment with HAART. Six SIV-infected macaques were treated with a potent four antiretroviral regimen starting at 12 days p.i., which falls within the expansion phase that we observed for all monocyte subsets. Phenotypic analysis of classical monocytes demonstrated that at 14 days p.i., the CCR2low/neg subset was reduced significantly when compared with untreated animals (Fig. 6A). Conversely, the CCR2high subgroup continued to expand after treatment, and by 21 days p.i., the ratio between classical and CD14highCD16−CCR2low/neg monocytes returned to steady-state levels found in uninfected macaques (Fig. 6B).
Figure 6. Antiretroviral therapy controls the expansion of CCR2low/neg classical monocytes in SIV-infected macaques.
(A) Absolute number of CCR2low/neg and CCR2high classical monocytes in 18 untreated (red) and six HAART-treated (blue), SIV-infected macaques. Solid lines represent median. Asterisks denote significant difference (Wilcoxon paired rank test; P<0.05) between treated and untreated animals at 14 and 28 days p.i. (B) Ratio between CCR2high and CCR2low/neg classical monocytes in treated macaques for each time-point. Arrow marks the beginning of treatment at 12 days p.i.
These changes correlate more closely to the suppression of viral replication, as a result of HAART, than to the decaying levels of circulating virus, as the median (interquartiles) of SIV RNA copy equivalent/mL plasma, from 3 to 9 days post-treatment (14 and 21 days p.i.), was 0.75 (0.12−1.8) × 106. These data suggest that the driving force behind the surge of CD14highCD16−CCR2low/neg monocytes is active replication not the presence of viral particles.
CD14highCD16−CCR2low/neg monocytes are also detected in HIV-infected patients and decrease in numbers after HAART therapy
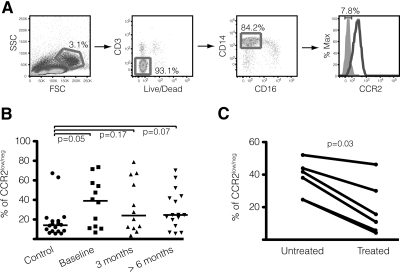
To investigate whether humans infected by HIV also presented an increase of classical monocytes lacking CCR2 surface expression, we analyzed samples from a cohort of recently HIV-1-infected patients [27] and compared them with PBMCs collected from negative control individuals (Supplemental Table 2). Uninfected subjects presented a median of 12.9% of CD14highCD16−CCR2low/neg monocytes, with the exception of two samples, where levels were as high as in some infected patients (Fig. 7A and B). We attributed these results to a combination of genetic variability and the lack of information about the general health status of these individuals. It is possible that this novel subset is not unique to HIV/SIV infection and may also increase during other pathological or infectious conditions.
Figure 7. CD14highCD16−CCR2low/neg monocytes are observed in humans and expand after HIV infection.
(A) Strategy for cytometry analysis of human PBMCs. Monocytes were gated initially according to size and granularity. Cells expressing the viability dye (Live/Dead) and CD3 were excluded. The last panel shows the percentage of CCR2low/neg classical monocytes (black line) compared with unstained sample (gray filled histogram) determined by FMO. (B) Comparison between the percentage of CCR2low/neg cells among classical monocytes in uninfected and HIV-infected patients, where Baseline designates the time when a first HIV+ diagnostic was made, and the other two groups represent the period elapsed after Baseline. Bars represent medians, and P values were calculated by Mann-Whitney t test. (C) Frequency of CD14highCD16−CCR2low/neg monocytes declines after HAART treatment. Graphic depicts the percentage of CCR2low/neg cells among classical monocytes in six patients, 1–2 months before and 1–2 months after starting treatment, and P value was calculated by Wilcoxon paired rank test.
The difference between uninfected and HIV-1-infected individuals, however, was evident, increasing from 12.9% to 39.1% in samples acquired when a positive diagnosis was first reported (P<0.05). Samples collected from the same patients on two subsequent occasions demonstrated no significant change in the frequency of this subset (P=0.67 for 3 months, and P=0.27 for >6 months after baseline). However, as observed in our macaque model, patients treated with HAART (Supplemental Table 3) also presented a decrease in the percentage of circulating CCR2low/neg cells among classical monocytes (Fig. 7C), from a median of 39.9% to levels similar to uninfected individuals (13.25%; P=0.03), which suggests that similar mechanisms occur in the modulation of this phenotype during HIV infection. We were unable to establish an association between CD14highCD16−CCR2low/neg monocytes and changes in the absolute number of other circulating leukocyte populations, including CD4+ and CD8+ T cells (data not shown), which is another important similarity between the results from our SIV model and the human cohort.
DISCUSSION
While characterizing monocyte subpopulations in our accelerated SIV model for AIDS and HIV-associated neurocognitive disorder, we observed a profound change in the expression of CCR2 in classical CD14+CD16− monocytes, starting early after inoculation and lasting throughout infection. A similar monocyte subset has been reported in patients with ulcerative colitis after their blood was depleted of excess granulocytes and monocytes by extracorporeal adsorption, a treatment that also leads to the reduction of proinflammatory cytokines and the increase of anti-inflammatory factors [46]. Therefore, the existence of circulating CCR2low/neg classical monocytes is not, in itself, a novel finding, but the sizable expansion and persistence of this subset during HIV and SIV infections had not been identified previously.
Phenotypic changes in blood monocytes during SIV infection have been described by several groups in different macaque models, but no study has reported longitudinal alterations in CCR2 surface expression [7, 8, 47–49]. Supporting some of the findings from other groups, we observed in our model a reproducible variation in the absolute number of circulating monocytes during acute infection. These were likely associated with events related to viral replication, including change in monocyte turnover, death of infected progenitor cells in the bone marrow, and cell migration toward organs harboring CCL2-expressing, activated cells [50]. These population changes were driven predominantly by CD14+CD16− cells, which are the most abundant monocyte subset in the blood, and normally express high levels of surface CCR2. As early as 7 days p.i., however, we observed an increase in CCR2low/neg classical monocytes. Initially, it appeared that the change was only proportional to CCR2high cells, but further analysis showed that this novel subgroup was also increasing in absolute numbers and was maintained in circulation throughout infection. Besides CD14, these cells also expressed CX3CR1 and other myeloid markers, such as CD11b and CD33, confirming their monocytic phenotype.
Although it was impossible to differentiate between these two subsets based solely on their levels of CD14 and CD16 expression by flow cytometry, comparative transcriptome analysis of samples collected at 14 days p.i. showed that they represented two distinct populations once they were categorized by their levels of surface CCR2. Based on the mRNA expression levels of cytokines, however, it was unclear whether they constituted an anti-inflammatory population, as they expressed lower levels of the anti-inflammatory cytokine IL-10 and higher levels of CCL20, CCL13, and CCL8, which are involved in the recruitment of leukocytes [51–53].
Notably, the CCR2low/neg subset expressed higher levels of STAT3, a transcription factor whose phosphorylation has been related to cell maturation and anti-inflammatory responses in macrophages [54]. The latter, together with the observation that these cells also expressed higher levels of IL-22Rα2 (a receptor that heterodimerizes with IL-10R), prompted us to analyze its ability to perform efficient phagocytosis. It has been reported that monocyte-derived macrophages from HIV-1-infected patients exhibited defective induction of antiparasite activity and that monocytes from similar patients were able to block autophagy in bystander cells through an IL-10/STAT3 signaling [55]. Classical monocytes express high levels of CD14, TLR4, and TLR2 and are therefore very efficient at responding to bacterial infection [2]. We observed that although in lower levels than classical monocytes, these CD14highCD16−CCR2low/neg cells also expressed LPS-sensing receptors, which should render them responsive to bacterial elements. However, our results demonstrated that these cells specifically presented impaired phagocytosis and that all monocytes at 7 and 14 days p.i. responded weakly to E. coli products. They were also defective in chemotaxis, confirming the above observations that infection by HIV/SIV can interfere with monocyte functionality and thwart their ability to react to, capture, and destroy microbes. As various surface receptors were still detectable, including CCR1, CX3CR1, and AGTR2, these cells might potentially respond to other chemokines instead of CCR2 agonists.
Most importantly, this novel subset was also able to suppress CD8+ T cell proliferation and IFN-γ release in vitro, and its expansion in the blood correlated negatively with the percentage of in vivo-activated CD8+ T cells during acute SIV infection. Monocytes presenting a similar immunosuppressive CD14+HLA-DRlow/neg phenotype have been previously reported to suppress lymphocyte proliferation in B cell non-Hodgkin's lymphoma and prostate cancer [18, 19] and have been classified by some groups as monocytic MDSCs [56]. Besides showing lower levels of surface HLA-DR, CD14highCD16−CCR2low/neg monocytes share two other distinctive features with MDSCs. First, they express higher levels of STAT3, the main transcription factor that regulates MDSC expansion [57]. Second, they also present higher expression of cytokine iNOS (or NOS2), which has been historically associated with the suppressive activity of MDSCs, specifically the monocytic subset [58]. Despite these similarities, future studies are necessary for a confirmatory classification of these CD14highCD16−CCR2low/neg cells, as no definite parameters have been determined to distinguish conclusively monocytic MDSCs from some monocyte subsets [59].
Besides, the transcriptome results and the gradual loss of surface CCR2 and HLA-DR within the classical monocyte population lead us to postulate that contrary to what is proposed for MDSCs, this novel subset described here may not originate directly from monoblasts but from CD14+CCR2high monocytes already egressed from the bone marrow, a process highly dependent on the CCL2/CCR2 pathway [60]. It is conceivable that the differentiation itself happens while classical monocytes shuttle back to the bone marrow, as described previously for nonclassical monocytes in mouse [61], or while they traffic through secondary lymphoid organs. Independent of their origin and nomenclature, the expansion of these suppressor cells appears to be important for the regulation of the immune system during a period of damaging hyperactivation. However, they may also interfere with antiviral processes by inhibiting CD8+ T cell activation in vivo and retarding the development of an efficient adaptive immune response. The fact that we were able to detect this unusual subpopulation in HIV-infected patients, and a return to uninfected levels upon HAART therapy, indicates that these cells arise in both lentivirus infections and that suppression of viral replication can return their levels to normal.
The role of monocytes in HIV infection is not fully understood. Monocytes and macrophages present a variety of restriction factors that can hinder viral replication, including down-regulation of surface receptors and expression of intrinsic immune proteins and microRNAs [44]. Less than 0.1% of monocytes from patients contain HIV DNA [62], which is in accordance with the results presented in this study. On the other hand, replication-competent virus can be recovered from blood monocytes, even from patients undergoing HAART [63], which makes these cells pivotal components for the establishment of virus latency in tissues and the development of eradication therapies. A specific monocyte subset, CD14+CD16++, has been reported to expand during AIDS in humans and macaques [4, 8, 23, 47]. This intermediate subpopulation, phenotypically located between classical and nonclassical subsets, expresses higher levels of CCR5 and CD4 and may be preferentially more susceptible to HIV and SIV infection than the other subgroups. We observed no significant difference in the levels of SIV DNA between intermediate and classical monocytes, although there was a dramatic difference between both subsets and CD14highCD16−CCR2low/neg cells. As most studies consider CD14+CD16− a uniform population, it is possible that the large number of the CCR2low/neg cells during infection is masking the actual prevalence of SIV DNA in authentic classical monocytes. Furthermore, CD14highCD16−CCR2low/neg monocytes also present higher levels of CCL20, which binds to CCR6 and is able to inhibit HIV replication by inducing apolipoprotein B mRNA-editing, enzyme-catalytic, polypeptide-like 3G in lymphocytes [64]. An autocrine response could explain why these cells are refractory to SIV infection, but CCR6 surface was not detectable in macaque monocytes (Fig. 3B). The dramatic difference in the number of SIV DNA copies between classical CCR2high and CCR2low/neg classical monocytes could corroborate the theory that these latter cells originate from monoblasts and are originally resistant to infection because of their low levels of CD4 and CCR5 surface expression. We hypothesize that the discrepancy between these subpopulations may simply reflect the variable ratios by which classical monocytes differentiate into other phenotypes [12], such as inflammatory tissue macrophages, DCs, intermediate monocytes, and the CCR2low/neg subset, under inflammatory conditions. In addition, SIV- and HIV-infected monocytes are reported to undergo phenotypic changes, including apoptosis, modulation of MHC proteins, and even up-regulation of surface CCR2 expression [43, 65, 66], which could also interfere with differentiation and trafficking.
We have previously reported a rapid increase in several proinflammatory cytokines in the brain of SIV-infected macaques during acute infection, followed by a coordinated down-regulation by 14 days p.i [32]. Accordingly, levels of cytokines in the blood are also subjected to a similar regulation pattern in HIV patients, where an intense but transient expression of high levels of inflammatory molecules, called a “cytokine storm”, can be observed in the first 10 days of infection [67]. The surge of these suppressor monocytes in our model appears to substantiate these observations, leading us to postulate that phenotypic changes in classical monocytes may be part of an anti-inflammatory reaction in response to the vigorous inflammatory process caused during HIV and SIV acute infection.
As AIDS is fundamentally an imbalance between immune activation and immune suppression and as monocytes exhibit plasticity, it is likely that CD14highCD16−CCR2low/neg cells do not represent a homogeneous subgroup but a heterogeneous and mutable population that varies throughout the infectious process or even presents different expression profiles depending on the viral strain involved. Nonetheless, their ability to suppress lymphocyte proliferation, even before strong adaptive responses are developed, emphasizes the importance of these cells in understanding the early events of HIV and SIV infection.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by National Institutes of Health grants MH070306, NS055648, and MH085554. This study was partially supported with funding from the Brazilian Program for STD and AIDS, Ministry of Health (914/BRA/3014-UNESCO/Kallas), the São Paulo City Health Department (2004–0.168.922–7/Kallas), and the Fundação de Amparo a Pesquisa do Estado de São Paulo (04/15856-9/Kallas). K.I.C.'s scholarship is supported by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Brazilian Ministry of Education, through the Programa Nacional de Pós Doutorado (PNPD) program. We thank Brandon B. Bullock for technical assistance as well as the rest of the Retrovirus Laboratory for helpful discussion. Special thanks to Jasmeet Sethi and the Microarray Core at the Johns Hopkins School of Medicine for help with NanoString data acquisition and Ada Tam for assistance with cytometry sorting. Medical Editor Michael E. Linde made substantial contributions to the editing of the manuscript. We also thank Daniela S. Rosa, Bianca N. Santos, Helena Tomiyama, Priscilla Costa, and Claudia Tomiyama at Laboratório de Investigação Médica 60, School of Medicine, University of São Paulo, for valuable support in the sample repository constitution and the monocyte experiments.
The online version of this paper, found at www.jleukbio.org, includes supplemental information.
- AGTR
- angiotensin receptor
- CAPPesq
- Comissão para Análise de Projetos de Pesquisa
- FMO
- fluorescence minus one
- FSC
- forward-scatter
- HAART
- highly active antiretroviral therapy
- HAND
- HIV-associated neurocognitive disorders
- IL-18bp
- IL-18 binding protein
- IRF
- IFN regulatory factor
- MDSC
- myeloid-derived suppressor cell
- OAS2
- 2′-5′-oligoadenylate synthetase 2
- p.i.
- postinoculation
- PMPA
- (1R)-9-(2-phosphonylmethoxypropyl)-adenine
- qPCR
- quantitative PCR
- RPMI
- Roswell Park Memorial Institute medium
- SSC
- side-scatter
- ZBP1
- Z-DNA-binding protein 1
- ZDV
- zidovudine
AUTHORSHIP
L.G., J.E.C., and E.G.K. conceived of and designed the experiments. L.G., E.N.S., J.N.R., M.L., K.I.C., and S.E.Q. performed the experiments. L.G., E.N.S., K.I.C., J.K., M.C.Z., J.E.C., and E.G.K. analyzed the data. L.G., J.E.C., and E.G.K. wrote the paper.
DISCLOSURES
The authors have no conflicts of interest to declare. This work has not been presented or currently submitted to other journal.
REFERENCES
- 1. Gordon S., Taylor P. R. (2005) Monocyte and macrophage heterogeneity. Nat. Rev. Immunol. 5, 953–964 [DOI] [PubMed] [Google Scholar]
- 2. Cros J., Cagnard N., Woollard K., Patey N., Zhang S. Y., Senechal B., Puel A., Biswas S. K., Moshous D., Picard C., Jais J. P., D'Cruz D., Casanova J. L., Trouillet C., Geissmann F. (2010) Human CD14dim monocytes patrol and sense nucleic acids and viruses via TLR7 and TLR8 receptors. Immunity 33, 375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Weber C., Belge K. U., von Hundelshausen P., Draude G., Steppich B., Mack M., Frankenberger M., Weber K. S., Ziegler-Heitbrock H. W. (2000) Differential chemokine receptor expression and function in human monocyte subpopulations. J. Leukoc. Biol. 67, 699–704 [DOI] [PubMed] [Google Scholar]
- 4. Strauss-Ayali D., Conrad S. M., Mosser D. M. (2007) Monocyte subpopulations and their differentiation patterns during infection. J. Leukoc. Biol. 82, 244–252 [DOI] [PubMed] [Google Scholar]
- 5. Sunderkotter C., Nikolic T., Dillon M. J., Van Rooijen N., Stehling M., Drevets D. A., Leenen P. J. (2004) Subpopulations of mouse blood monocytes differ in maturation stage and inflammatory response. J. Immunol. 172, 4410–4417 [DOI] [PubMed] [Google Scholar]
- 6. Bigley V., Haniffa M., Doulatov S., Wang X. N., Dickinson R., McGovern N., Jardine L., Pagan S., Dimmick I., Chua I., Wallis J., Lordan J., Morgan C., Kumararatne D. S., Doffinger R., van der Burg M., van Dongen J., Cant A., Dick J. E., Hambleton S., Collin M. (2011) The human syndrome of dendritic cell, monocyte, B and NK lymphoid deficiency. J. Exp. Med. 208, 227–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Burdo T. H., Soulas C., Orzechowski K., Button J., Krishnan A., Sugimoto C., Alvarez X., Kuroda M. J., Williams K. C. (2010) Increased monocyte turnover from bone marrow correlates with severity of SIV encephalitis and CD163 levels in plasma. PLoS Pathog. 6, e1000842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim W. K., Sun Y., Do H., Autissier P., Halpern E. F., Piatak M., Jr., Lifson J. D., Burdo T. H., McGrath M. S., Williams K. (2010) Monocyte heterogeneity underlying phenotypic changes in monocytes according to SIV disease stage. J. Leukoc. Biol. 87, 557–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Geissmann F., Jung S., Littman D. R. (2003) Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity 19, 71–82 [DOI] [PubMed] [Google Scholar]
- 10. Ancuta P., Liu K. Y., Misra V., Wacleche V. S., Gosselin A., Zhou X., Gabuzda D. (2009) Transcriptional profiling reveals developmental relationship and distinct biological functions of CD16+ and CD16− monocyte subsets. BMC Genomics 10, 403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cheong C., Matos I., Choi J. H., Dandamudi D. B., Shrestha E., Longhi M. P., Jeffrey K. L., Anthony R. M., Kluger C., Nchinda G., Koh H., Rodriguez A., Idoyaga J., Pack M., Velinzon K., Park C. G., Steinman R. M. (2010) Microbial stimulation fully differentiates monocytes to DC-SIGN/CD209(+) dendritic cells for immune T cell areas. Cell 143, 416–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yona S., Jung S. (2010) Monocytes: subsets, origins, fates and functions. Curr. Opin. Hematol. 17, 53–59 [DOI] [PubMed] [Google Scholar]
- 13. Geissmann F., Auffray C., Palframan R., Wirrig C., Ciocca A., Campisi L., Narni-Mancinelli E., Lauvau G. (2008) Blood monocytes: distinct subsets, how they relate to dendritic cells, and their possible roles in the regulation of T-cell responses. Immunol. Cell Biol. 86, 398–408 [DOI] [PubMed] [Google Scholar]
- 14. Venneri M. A., De Palma M., Ponzoni M., Pucci F., Scielzo C., Zonari E., Mazzieri R., Doglioni C., Naldini L. (2007) Identification of proangiogenic TIE2-expressing monocytes (TEMs) in human peripheral blood and cancer. Blood 109, 5276–5285 [DOI] [PubMed] [Google Scholar]
- 15. Yachie A., Toma T., Mizuno K., Okamoto H., Shimura S., Ohta K., Kasahara Y., Koizumi S. (2003) Heme oxygenase-1 production by peripheral blood monocytes during acute inflammatory illnesses of children. Exp. Biol. Med. (Maywood) 228, 550–556 [DOI] [PubMed] [Google Scholar]
- 16. Mizuno K., Toma T., Tsukiji H., Okamoto H., Yamazaki H., Ohta K., Kasahara Y., Koizumi S., Yachie A. (2005) Selective expansion of CD16high CCR2− subpopulation of circulating monocytes with preferential production of haem oxygenase (HO)-1 in response to acute inflammation. Clin. Exp. Immunol. 142, 461–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grage-Griebenow E., Zawatzky R., Kahlert H., Brade L., Flad H., Ernst M. (2001) Identification of a novel dendritic cell-like subset of CD64(+)/CD16(+) blood monocytes. Eur. J. Immunol. 31, 48–56 [DOI] [PubMed] [Google Scholar]
- 18. Lin Y., Gustafson M. P., Bulur P. A., Gastineau D. A., Witzig T. E., Dietz A. B. (2011) Immunosuppressive CD14+HLA-DRlow/− monocytes in B-cell non-Hodgkin lymphoma. Blood 117, 872–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vuk-Pavlovíc S., Bulur P. A., Lin Y., Qin R., Szumlanski C. L., Zhao X., Dietz A. B. (2010) Immunosuppressive CD14+HLA-DRlow/− monocytes in prostate cancer. Prostate 70, 443–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Filipazzi P., Huber V., Rivoltini L. (2010) Phenotype, function and clinical implications of myeloid-derived suppressor cells in cancer patients. Cancer Immunol. Immunother. 61, 255–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nagaraj S., Collazo M., Corzo C. A., Youn J. I., Ortiz M., Quiceno D., Gabrilovich D. I. (2009) Regulatory myeloid suppressor cells in health and disease. Cancer Res. 69, 7503–7506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vanham G., Edmonds K., Qing L., Hom D., Toossi Z., Jones B., Daley C. L., Huebner B., Kestens L., Gigase P., Ellner J. J. (1996) Generalized immune activation in pulmonary tuberculosis: co-activation with HIV infection. Clin. Exp. Immunol. 103, 30–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pulliam L., Gascon R., Stubblebine M., McGuire D., McGrath M. S. (1997) Unique monocyte subset in patients with AIDS dementia. Lancet 349, 692–695 [DOI] [PubMed] [Google Scholar]
- 24. Zink M. C., Suryanarayana K., Mankowski J. L., Shen A., Piatak M., Jr., Spelman J. P., Carter D. L., Adams R. J., Lifson J. D., Clements J. E. (1999) High viral load in the cerebrospinal fluid and brain correlates with severity of simian immunodeficiency virus encephalitis. J. Virol. 73, 10480–10488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dinoso J. B., Rabi S. A., Blankson J. N., Gama L., Mankowski J. L., Siliciano R. F., Zink M. C., Clements J. E. (2009) A simian immunodeficiency virus-infected macaque model to study viral reservoirs that persist during highly active antiretroviral therapy. J. Virol. 83, 9247–9257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bassichetto K. C., Bergamaschi D. P., Oliveira S. M., Deienno M. C., Bortolato R., de Rezende H. V., Arthur T., Tomiyama H., Watkins C., Mesquita F., Abbate M. C., Kallas E. G. (2008) Elevated risk for HIV-1 infection in adolescents and young adults in São Paulo, Brazil. PLoS One 3, e1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kallas E. G., Bassichetto K. C., Oliveira S. M., Goldenberg I., Bortoloto R., Moreno D. M., Kanashiro C., Chaves M. M., Sucupira M. C., Diniz A., Mesquita F. C. (2004) Establishment of the serologic testing algorithm for recent human immunodeficiency virus (HIV) seroconversion (STARHS) strategy in the city of São Paulo, Brazil. Braz. J. Infect. Dis. 8, 399–406 [DOI] [PubMed] [Google Scholar]
- 28. Barber S. A., Herbst D. S., Bullock B. T., Gama L., Clements J. E. (2004) Innate immune responses and control of acute simian immunodeficiency virus replication in the central nervous system. J. Neurovirol. 10 (Suppl. 1), 15–20 [DOI] [PubMed] [Google Scholar]
- 29. Geiss G. K., Bumgarner R. E., Birditt B., Dahl T., Dowidar N., Dunaway D. L., Fell H. P., Ferree S., George R. D., Grogan T., James J. J., Maysuria M., Mitton J. D., Oliveri P., Osborn J. L., Peng T., Ratcliffe A. L., Webster P. J., Davidson E. H., Hood L., Dimitrov K. (2008) Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 26, 317–325 [DOI] [PubMed] [Google Scholar]
- 30. Clements J. E., Babas T., Mankowski J. L., Suryanarayana K., Piatak M., Jr., Tarwater P. M., Lifson J. D., Zink M. C. (2002) The central nervous system as a reservoir for simian immunodeficiency virus (SIV): steady-state levels of SIV DNA in brain from acute through asymptomatic infection. J. Infect. Dis. 186, 905–913 [DOI] [PubMed] [Google Scholar]
- 31. Velasco F., Verma S., Guevara M. (2000) Comparison of the performance of fourteen statistical tests for detection of outlying values in Geochemical Reference Material Databases. Math. Geol. 32, 439–464 [Google Scholar]
- 32. Witwer K. W., Gama L., Li M., Bartizal C. M., Queen S. E., Varrone J. J., Brice A. K., Graham D. R., Tarwater P. M., Mankowski J. L., Zink M. C., Clements J. E. (2009) Coordinated regulation of SIV replication and immune responses in the CNS. PLoS One 4, e8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ziegler-Heitbrock L., Ancuta P., Crowe S., Dalod M., Grau V., Hart D. N., Leenen P. J., Liu Y. J., MacPherson G., Randolph G. J., Scherberich J., Schmitz J., Shortman K., Sozzani S., Strobl H., Zembala M., Austyn J. M., Lutz M. B. (2010) Nomenclature of monocytes and dendritic cells in blood. Blood 116, e74–e80 [DOI] [PubMed] [Google Scholar]
- 34. Sica A., Saccani A., Borsatti A., Power C. A., Wells T. N., Luini W., Polentarutti N., Sozzani S., Mantovani A. (1997) Bacterial lipopolysaccharide rapidly inhibits expression of C-C chemokine receptors in human monocytes. J. Exp. Med. 185, 969–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xu L., Khandaker M. H., Barlic J., Ran L., Borja M. L., Madrenas J., Rahimpour R., Chen K., Mitchell G., Tan C. M., DeVries M., Feldman R. D., Kelvin D. J. (2000) Identification of a novel mechanism for endotoxin-mediated down-modulation of CC chemokine receptor expression. Eur. J. Immunol. 30, 227–235 [DOI] [PubMed] [Google Scholar]
- 36. Margulies B. J., Hauer D. A., Clements J. E. (2001) Identification and comparison of eleven rhesus macaque chemokine receptors. AIDS Res. Hum. Retroviruses 17, 981–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Carter D. L., Shieh T. M., Blosser R. L., Chadwick K. R., Margolick J. B., Hildreth J. E., Clements J. E., Zink M. C. (1999) CD56 identifies monocytes and not natural killer cells in rhesus macaques. Cytometry 37, 41–50 [PubMed] [Google Scholar]
- 38. Swirski F. K., Nahrendorf M., Etzrodt M., Wildgruber M., Cortez-Retamozo V., Panizzi P., Figueiredo J. L., Kohler R. H., Chudnovskiy A., Waterman P., Aikawa E., Mempel T. R., Libby P., Weissleder R., Pittet M. J. (2009) Identification of splenic reservoir monocytes and their deployment to inflammatory sites. Science 325, 612–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li L., Huang L., Sung S. S., Vergis A. L., Rosin D. L., Rose C. E., Jr., Lobo P. I., Okusa M. D. (2008) The chemokine receptors CCR2 and CX3CR1 mediate monocyte/macrophage trafficking in kidney ischemia-reperfusion injury. Kidney Int. 74, 1526–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gong X., Gong W., Kuhns D. B., Ben-Baruch A., Howard O. M., Wang J. M. (1997) Monocyte chemotactic protein-2 (MCP-2) uses CCR1 and CCR2B as its functional receptors. J. Biol. Chem. 272, 11682–11685 [DOI] [PubMed] [Google Scholar]
- 41. Kazazi F., Mathijs J. M., Foley P., Cunningham A. L. (1989) Variations in CD4 expression by human monocytes and macrophages and their relationships to infection with the human immunodeficiency virus. J. Gen. Virol. 70, 2661–2672 [DOI] [PubMed] [Google Scholar]
- 42. Merino A., Buendia P., Martin-Malo A., Aljama P., Ramirez R., Carracedo J. (2011) Senescent CD14+CD16+ monocytes exhibit proinflammatory and proatherosclerotic activity. J. Immunol. 186, 1809–1815 [DOI] [PubMed] [Google Scholar]
- 43. Eugenin E. A., Osiecki K., Lopez L., Goldstein H., Calderon T. M., Berman J. W. (2006) CCL2/monocyte chemoattractant protein-1 mediates enhanced transmigration of human immunodeficiency virus (HIV)-infected leukocytes across the blood-brain barrier: a potential mechanism of HIV-CNS invasion and NeuroAIDS. J. Neurosci. 26, 1098–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bergamaschi A., Pancino G. (2010) Host hindrance to HIV-1 replication in monocytes and macrophages. Retrovirology 7, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Crowe S., Zhu T., Muller W. A. (2003) The contribution of monocyte infection and trafficking to viral persistence, and maintenance of the viral reservoir in HIV infection. J. Leukoc. Biol. 74, 635–641 [DOI] [PubMed] [Google Scholar]
- 46. Takeda S., Sato T., Katsuno T., Nakagawa T., Noguchi Y., Yokosuka O., Saito Y. (2010) Adsorptive depletion of α4 integrin(hi)- and CX3CR1hi-expressing proinflammatory monocytes in patients with ulcerative colitis. Dig. Dis. Sci. 55, 1886–1895 [DOI] [PubMed] [Google Scholar]
- 47. Bissel S. J., Wang G., Trichel A. M., Murphey-Corb M., Wiley C. A. (2006) Longitudinal analysis of activation markers on monocyte subsets during the development of simian immunodeficiency virus encephalitis. J. Neuroimmunol. 177, 85–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Clay C. C., Rodrigues D. S., Ho Y. S., Fallert B. A., Janatpour K., Reinhart T. A., Esser U. (2007) Neuroinvasion of fluorescein-positive monocytes in acute simian immunodeficiency virus infection. J. Virol. 81, 12040–12048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Otani I., Akari H., Nam K. H., Mori K., Suzuki E., Shibata H., Doi K., Terao K., Yosikawa Y. (1998) Phenotypic changes in peripheral blood monocytes of cynomolgus monkeys acutely infected with simian immunodeficiency virus. AIDS Res. Hum. Retroviruses 14, 1181–1186 [DOI] [PubMed] [Google Scholar]
- 50. Zink M. C., Coleman G. D., Mankowski J. L., Adams R. J., Tarwater P. M., Fox K., Clements J. E. (2001) Increased macrophage chemoattractant protein-1 in cerebrospinal fluid precedes and predicts simian immunodeficiency virus encephalitis. J. Infect. Dis. 184, 1015–1021 [DOI] [PubMed] [Google Scholar]
- 51. Garcia-Zepeda E. A., Combadiere C., Rothenberg M. E., Sarafi M. N., Lavigne F., Hamid Q., Murphy P. M., Luster A. D. (1996) Human monocyte chemoattractant protein (MCP)-4 is a novel CC chemokine with activities on monocytes, eosinophils, and basophils induced in allergic and nonallergic inflammation that signals through the CC chemokine receptors (CCR)-2 and -3. J. Immunol. 157, 5613–5626 [PubMed] [Google Scholar]
- 52. Hieshima K., Imai T., Opdenakker G., Van Damme J., Kusuda J., Tei H., Sakaki Y., Takatsuki K., Miura R., Yoshie O., Nomiyama H. (1997) Molecular cloning of a novel human CC chemokine liver and activation-regulated chemokine (LARC) expressed in liver. Chemotactic activity for lymphocytes and gene localization on chromosome 2. J. Biol. Chem. 272, 5846–5853 [DOI] [PubMed] [Google Scholar]
- 53. Islam S. A., Chang D. S., Colvin R. A., Byrne M. H., McCully M. L., Moser B., Lira S. A., Charo I. F., Luster A. D. (2011) Mouse CCL8, a CCR8 agonist, promotes atopic dermatitis by recruiting IL-5+ T(H)2 cells. Nat. Immunol. 12, 167–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Heinrich P. C., Behrmann I., Muller-Newen G., Schaper F., Graeve L. (1998) Interleukin-6-type cytokine signalling through the gp130/Jak/STAT pathway. Biochem. J. 334, 297–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Van Grol J., Subauste C., Andrade R. M., Fujinaga K., Nelson J., Subauste C. S. (2010) HIV-1 inhibits autophagy in bystander macrophage/monocytic cells through Src-Akt and STAT3. PLoS One 5, e11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gabrilovich D. I., Nagaraj S. (2009) Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 9, 162–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nefedova Y., Nagaraj S., Rosenbauer A., Muro-Cacho C., Sebti S. M., Gabrilovich D. I. (2005) Regulation of dendritic cell differentiation and antitumor immune response in cancer by pharmacologic-selective inhibition of the Janus-activated kinase 2/signal transducers and activators of transcription 3 pathway. Cancer Res. 65, 9525–9535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rodriguez P. C., Ochoa A. C. (2008) Arginine regulation by myeloid derived suppressor cells and tolerance in cancer: mechanisms and therapeutic perspectives. Immunol. Rev. 222, 180–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Greten T. F., Manns M. P., Korangy F. (2011) Myeloid derived suppressor cells in human diseases. Int. Immunopharmacol. 11, 802–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tsou C. L., Peters W., Si Y., Slaymaker S., Aslanian A. M., Weisberg S. P., Mack M., Charo I. F. (2007) Critical roles for CCR2 and MCP-3 in monocyte mobilization from bone marrow and recruitment to inflammatory sites. J. Clin. Invest. 117, 902–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Varol C., Landsman L., Fogg D. K., Greenshtein L., Gildor B., Margalit R., Kalchenko V., Geissmann F., Jung S. (2007) Monocytes give rise to mucosal, but not splenic, conventional dendritic cells. J. Exp. Med. 204, 171–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lewin S. R., Kirihara J., Sonza S., Irving L., Mills J., Crowe S. M. (1998) HIV-1 DNA and mRNA concentrations are similar in peripheral blood monocytes and alveolar macrophages in HIV-1-infected individuals. AIDS 12, 719–727 [DOI] [PubMed] [Google Scholar]
- 63. Mikovits J. A., Lohrey N. C., Schulof R., Courtless J., Ruscetti F. W. (1992) Activation of infectious virus from latent human immunodeficiency virus infection of monocytes in vivo. J. Clin. Invest. 90, 1486–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lafferty M. K., Sun L., DeMasi L., Lu W., Garzino-Demo A. (2010) CCR6 ligands inhibit HIV by inducing APOBEC3G. Blood 115, 1564–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kamp W., Breij E. C., Nottet H. S., Berk M. B. (2001) Interactions between major histocompatibility complex class II surface expression and HIV: implications for pathogenesis. Eur. J. Clin. Invest. 31, 984–991 [DOI] [PubMed] [Google Scholar]
- 66. Laforge M., Campillo-Gimenez L., Monceaux V., Cumont M.C., Hurtrel B., Corbeil J., Zaunders J., Elbim C., Estaquier J. (2011) HIV/SIV infection primes monocytes and dendritic cells for apoptosis. PLoS Pathog. 7, e1002087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Stacey A. R., Norris P. J., Qin L., Haygreen E. A., Taylor E., Heitman J., Lebedeva M., DeCamp A., Li D., Grove D., Self S. G., Borrow P. (2009) Induction of a striking systemic cytokine cascade prior to peak viremia in acute human immunodeficiency virus type 1 infection, in contrast to more modest and delayed responses in acute hepatitis B and C virus infections. J. Virol. 83, 3719–3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.