Abstract
The β2-adrenergic receptor (β2AR) plays important physiological roles in the heart and lung and is the primary target of β-agonists, the mainstay asthma drugs. Activation of β2AR by β-agonists is attenuated by receptor down-regulation, which ensures transient stimulation of the receptor but reduces the efficacy of β-agonists. Here we report the identification, through a functional genome-wide RNA interference (RNAi) screen, of new genes critically involved in β2AR down-regulation. We developed a lentivirus-based RNAi library consisting of 26-nt short-hairpin RNAs (shRNAs). The library was generated enzymatically from a large collection of expressed sequence tag (EST) DNAs corresponding to ∼20,000 human genes and contains on average ∼6 highly potent shRNAs (>75% knockdown efficiency) for each gene. Using this novel shRNA library, together with a robust cell model for β2AR expression, we performed fluorescence-activated cell sorting and isolated cells that, as a consequence of shRNA-mediated gene inactivation, exhibited defective agonist-induced down-regulation. The screen discovered several previously unrecognized β2AR regulators, including farnesyl diphosphate synthase (FDPS). We showed that inactivation of FDPS by shRNA, small interfering RNA, or the highly specific pharmaceutical inhibitor alendronate inhibited β2AR down-regulation. Notably, in human airway smooth muscle cells, the physiological target of β-agonists, alendronate treatment functionally reversed agonist-induced endogenous β2AR loss as indicated by an increase in cAMP production. FDPS inactivation interfered with β2AR internalization into endosomes through disrupting the membrane localization of the Rab5 small GTPase. Furthermore, Rab5 overexpression reversed the deficient receptor down-regulation induced by alendronate, suggesting that FDPS regulates receptor down-regulation in a Rab5-dependent manner. Together, our findings reveal a FDPS-dependent mechanism in the internalization and down-regulation of β2AR, identify FDPS as a potential target for improving the therapeutic efficacy of β-agonists, and demonstrate the utility of the unique EST-derived shRNA library for functional genetics studies.—Jiang, X., Pan, H., Nabhan, J. F., Krishnan, R., Koziol-White, C., Panettieri, R. A., Lu, Q. A novel EST-derived RNAi screen reveals a critical role for farnesyl diphosphate synthase in β2-adrenergic receptor internalization and down-regulation.
Keywords: alendronate, β-agonist, genetic screen, short-hairpin RNA
The β2 adrenergic receptor (β2AR) is a prototypic member of the G-protein coupled receptor (GPCR) family and plays important physiological functions in the heart and lung (1, 2). β2ARs expressed in the lung epithelial and airway smooth muscle cells are the targets of catecholamine hormones. Binding of hormones or other agonists to the receptor activates the associated stimulatory G protein and, consequently, a signaling cascade that includes adenylyl cyclase activation, cyclic adenosine monophosphate (cAMP) formation, and protein kinase A (PKA) activation. Activated PKA phosphorylates multiple downstream target proteins and leads to a variety of physiological responses, including airway smooth muscle relaxation and bronchodilation (1). Because of their effects in the lung, β2AR agonists are widely used to alleviate the symptoms of asthma (3), a major chronic lung disease characterized by airway constriction and narrowing (4).
Activation of β2AR signaling by β-agonists is usually attenuated by desensitization and down-regulation (5, 6). Such attenuation prevents persistent activation of the receptor and ensures cell and tissue homeostasis. Immediately (usually within seconds) after agonist activation, β2ARs are rapidly phosphorylated by G-protein-coupled receptor kinases (GRKs; refs. 7, 8), leading to the recruitment of cytosolic β-arrestins to the membrane receptors (9, 10). The interaction of β-arrestins with β2AR uncouples the activated G-protein complex from the receptors, thus effectively terminating the G-protein-dependent signaling cascade. Unlike the acute short-term agonist exposure, prolonged exposure to agonists leads to down-regulation or loss of functional β2AR on the cell surface. β2AR down-regulation involves endocytosis-mediated internalization of the receptor (11). Clathrin-mediated endocytosis promotes agonist-induced internalization of the receptor and is essential for β2AR down-regulation (11, 12). After being internalized into early endosomes, the receptor can either recycle back to the cell surface or be sorted into late endosomes or lysosomes for degradation (11, 13). A prerequisite for efficient sorting into lysosomes is β2AR ubiquitination (14–17), which is mediated by a ubiquitin E3 ligase and an adaptor protein (18, 19).
Down-regulation through lysosomal degradation leads to permanent removal of β2AR from the cell surface and has important clinical consequences. Reduced levels of cell surface β2AR slow the responsiveness of target tissue cells to agonists, thus severely limiting the efficacy of agonists in asthma therapy. Many adverse effects, such as loss of asthma control and longer durations of asthma exacerbation, may also be attributed to the down-regulation of receptors caused by prolonged agonist use (20). The importance of such receptor down-regulation is also supported by studies of β2AR polymorphisms (21, 22). Products of β2AR alleles that display accelerated degradation in vitro are associated with altered sensitivity to β-agonists in patients with asthma (23–25). Because of the important role of β2AR down-regulation in determining the clinical responses of β-agonists, it is critical to fully understand the molecular mechanisms underlying agonist-induced β2AR down-regulation.
Genome-wide functional genetic screen offers a powerful approach for unraveling the molecular basis of human diseases and biological processes (26, 27). RNA interference (RNAi)-based gene silencing relies on the potent and specific gene suppression activity exhibited by small interfering RNA (siRNA) of 19–21-nt (28, 29) or by short-hairpin RNAs (shRNAs; refs. 30–32). The use of RNAi libraries, together with phenotype-based assays, has led to identification of genes critical for a variety of biological processes, including cell growth, senescence, and tumor development (33–36). Application of the powerful RNAi tools to study the βAR signaling has the potential to reveal novel mechanisms but encounters 2 challenges. First, current RNAi methods, while powerful, are costly, have limited gene coverage, and exhibit unpredictable and often inefficient knockdown activity. Second, robust cellular β2AR down-regulation models, which are genetically tractable for high-throughput genome-wide screens, are lacking. In this report we have addressed both of these challenges, performed a genome-wide functional screen, and identified new regulators of β2AR down-regulation. We focus on the functional characterization of one of the RNAi hits that targets farnesyl diphosphate synthase (FDPS).
MATERIALS AND METHODS
Chemical reagents and enzymes
Unless otherwise indicated, all chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA); restriction enzymes, DNA polymerases, and T4 ligases were from New England Biolabs (Ipswich, MA. USA).
Enzymatic conversion of cDNA into shRNA-expressing structures
Step 1: cDNA fragmentation
cDNA fragments (2.5 μg) were partially digested with 0.2 U DNase I at 25°C for 10–15 min. The optimal digestion time was empirically determined so that the average size of final digests was ∼100–300-nt. The ends of the digests were repaired with 5 U T4 DNA polymerase and 200 μM dNTPs. The addition of an adenine to 3′ ends (3′-A overhang) was achieved using 1 U Taq polymerase and 200 μM dATP and incubating at 60°C for 3 h.
Step 2: U adaptor ligation and EcoP15I cleavage
DNA fragments with 3′-A overhang were ligated to 50 pmol of a hairpin-shaped oligonucleotide, U adaptor (5′-pCTGCTGCAGGATCCAAATCCTGCAGCAGT-3′) that was PAGE-purified, 5′-phosphorylated, and self-annealed before use. The adaptor has overlapping recognition sequences for PstI (CTGCAG) and EcoP15I (CAGCAG). The ligation product was digested with EcoP15I, and the resulting U adaptor-linked short DNA fragments were purified as ∼40-bp species by PAGE.
Step 3: priming adaptor ligation and primer extension
The U adaptor-linked short DNA fragments were ligated with a priming adaptor made up of 3 oligonucleotides (10 pmol of each): a priming oligo (red line in Fig. 1A, 5′-ACATTTTGCTGCCGGTC-3′); a ligation sense oligo (purple line, 5′-GGATCGATAAGTCAAAAA-3′); and a ligation antisense oligo (green line, 5′-pNNTTTTTGACTCATCGATGGGACCGGCAGCAAAATGTTCG-3′, 5′-phosphorylated PAGE-purified). Both the sense and antisense ligation oligos contain the recognition site for ClaI (ATCGAT), but only the antisense ligation oligo has the recognition site for MlyI (GAGTC, antisense mutated to GACTC). The ligation products were then converted into palindromic dsDNAs by primer extension and stand displacement with 1 U Bst DNA polymerase large fragment and 200-ng single-stranded DNA binding protein. The extension products were identified as ∼140-bp species by PAGE.
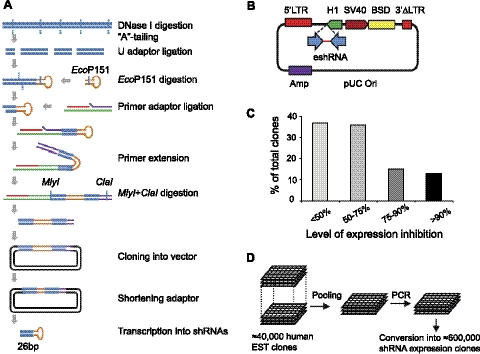
Figure 1.
Development of an EST-derived 26-nt shRNA library. A) Schematic outline of the enzymatic procedure to convert double-stranded DNA fragments into structures expressing 26-nt shRNAs. Details are given in Materials and Methods. B) Lentiviral shRNA-expressing vector pLentiSuper2. 5′LTR, 5′ long terminal repeat; 3′ΔLTR, promoter-deleted 3′ long terminal repeat; H1, RNA polymerase III H1 promoter; SV40, SV40 viral prompter; BSD, blasticidin selection marker; eshRNA, enzymatic prepared shRNA; Amp, ampicillin selection marker. C) Knockdown efficiency of shRNAs from a single gene library. HEK293T cells were cotransfected with 800 ng DNA of 60 individual shRNA-expressing clones or empty vector pLentiSuper2, together with 200 ng DNA of target gene (ARRDC1-GFP) and 50 ng DNA of nontarget control. Expression of both target and control genes were detected by immunoblotting using an anti-GFP antibody. Expression level of ARRDC1-GFP was normalized to that of GFP. Knockdown efficiency by shRNA was calculated as percentage relative to empty vector controls. D) Pooling and conversion of a large collection of ESTs into the shRNA library.
Step 4: cloning into the lentiviral vector
The extension products were digested with MlyI and ClaI. The digested products were purified by PAGE and ligated with SmaI and ClaI-digested vector pLS2. A SmaI site was placed just downstream of the H1 promoter to allow transcription of the 26-bp palindrome (30). The ligation products were transformed into the ultracompetent bacteria XL-10 Gold (Stratagene, La Jolla, CA, USA).
Step 5: truncation of excessive hairpin linker
Plasmid DNA was digested with PstI to remove most of the U adaptor sequence. The digested plasmid DNA was purified by agarose gel and recircularized by self-ligation. Ligation products were transformed into ultracompetent bacteria XL-10 Gold, and plasmid DNA was made by midprep as library stocks.
ShRNA library construction
For expressed sequence tag (EST) library-derived shRNA-expressing library, the EST fragments were prepared as described previously (37). The single-gene cDNA or the collection of the EST fragments was used as starting DNA material in the above 5-step enzymatic procedure to generate shRNA-expressing libraries.
Plasmid vectors and expression constructs
The backbone vector of our lentiviral shRNA expressing library is pLS2, which was made by cloning the shRNA expression cassette from pRetroSuper vector (30) into a lentivirus-based vector pLEST (37). pLS2 contains a blasticidin-resistant marker and expresses shRNA from the H1 promoter. Other DNA vectors include pEGFP-C1 and pEGFP-N1 (Clontech, Madison, WI, USA). pEGFP-N1-ARRDC1 expresses human ARRDC1 with C-terminal fusion of EGFP. pEGFP-Rab5 constructs (wild type and S34N mutant) and pCDNARab5 construct were kindly provided by Dr. Stephen G. Ferguson (University of Western Ontario, London, ON, Canada).
Mammalian cell culture and transfection
Human embryonic kidney HEK293T and 293β2AR cells were cultured in DMEM (Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS; Invitrogen), 4 mM glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. The HEK293 cells stably expressing full-length N-terminal Flag-tagged β2AR (293β2ARWT) and stably expressing N-terminal Flag-tagged recycle-deficient β2AR (293β2AR) were kindly provided by Dr. Mark von Zastrow (University of California, San Francisco, CA, USA; ref. 38). Primary human airway smooth muscle (HASM) cells were maintained in Ham's F12 medium with 10% fetal calf serum (FCS) and antibiotics, and grown in DMEM supplemented with 1% FBS for isoproterenol (ISO) treatment. Passages 4 to 7 HASM cells were used in all experiments. Transfection of plasmid DNA into HEK293 cells was done using FuGene 6 reagent (Roche, Indianapolis, IN, USA). Transfection of plasmid DNA into 293β2AR cells was performed using TurboFect reagent (Fermentas, Glen Burnie, MD, USA). Transfection of siRNA was done using DharmaFECT 1 reagent (Thermo Scientific, Rockford, IL, USA). Nontargeting control siRNA and FDPS-specific siRNA (sense 5′-ccauguacauggcaggaau(dT)(dT)-3′, antisense 5′-auuccugccauguacaugg(dT)(dT)-3′) were from Sigma-Aldrich (Mission siRNA).
Immunoblot analysis
At 48 h after DNA transfection, cells were washed with PBS and lysed with Nonidet P-40 lysing buffer (50 mM Tris-HCl, pH7.5; 150 mM NaCl; and 0.5% Nonidet P-40) containing protease inhibitor cocktail (Roche). Protein samples were separated on NuPAGE 4–12% Bis-Tris gels (Invitrogen) and transferred to PVDF membranes (Bio-Rad, Hercules, CA, USA). Immunoblot signals were developed using SuperSignal West Pico/Femto Chemiluminescent Substrates (Pierce, Rockford, IL, USA) and quantified by a luminescence imager or exposed to X-ray films. Antibodies used in this study included Flag M2 antibody (Sigma-Aldrich), FDPS antibody (Abcam, Cambridge, MA, USA), Rab5 antibody (Cell Signaling, Danvers, MA, USA), and green fluorescent protein (GFP) antibody (Clontech). Antibodies for ubiquitin, β2AR(H20), p β2AR(Ser-355/356), β-actin, and EGFR were from Santa Cruz Biotechnology (Santa Cruz, CA, USA). HRP-conjugated goat anti-rabbit and anti-mouse IgG antibodies were from Zymed Laboratories (South San Francisco, CA, USA).
Lentivirus packaging and transduction
Lentiviral packaging was done as described previously (37). Briefly, HEK293T cells were cotransfected with the pLentiSuper2 backbone lentiviral vector together with the vector encoding packaging proteins Gag-Pol and Rev and the vector encoding G protein of vesicular stomatitis virus. The virus supernatants were collected 24, 36, 48, and 60 h after transfection, pooled, and filtered through 0.45-μm filters. Viral titers were determined by the number of blasticidin-resistant cell colonies transduced with serial dilutions of lentivirus supernatants. For transduction of the shRNA library, 293β2AR cells were seeded in 10-cm tissue culture dishes (2.5×106 cells/dish). After 24 h, each dish of cells was infected with 1 × 107 PFU lentivirus of the shRNA library in medium containing 5 μg/ml polybrene (Sigma-Aldrich). At 2 d post-transduction, cells were selected in medium containing 5 μg/ml blasticidin for 8 d.
Flow cytometry analysis and fluorescence-activated cell sorting (FACS)
ShRNA library transduced 293β2AR cells (5×106) were cultured for 1 d in Opti-MEM (Invitrogen) containing 1% FBS and 10 μM ISO for 16 h to induce the down-regulation of β2AR. Cells were washed with PBS and incubated with diluted TrypLE Express Enzyme (1:10 dilution in PBS; Invitrogen) for exactly 1 min at room temperature before inactivation with complete medium. Cells were washed once with PBS containing 1% FBS to remove residual traces of TrypLE. Cells (1×107) were suspended in 1 ml PBS containing 1% FBS and stained with 10 μg/ml FITC-conjugated anti-Flag M2 antibody (Sigma-Aldrich) on a rotary mixer for 30 min. Cells were washed again and passed through 50-μm cell strainers to remove cell clumps before being sorted on a FACS Aria multicolor high speed sorter (BD Biosciences, San Jose, CA, USA). During FACS analysis, gating was adjusted to measure the FITC (green) signal. Background of autofluorescence was identified by plotting FITC with PE (red) or SSC (side scatter) signal. FITC-positive cells were collected in calcium and magnesium-free PBS with 20% FBS. Flow cytometry analysis was performed as above with counterstaining using 2 μg/ml propidium iodide (PI) before analysis on a FACS Canto II system (BD Biosciences).
Genomic PCR and shRNA sequencing
Genomic DNA was isolated using the Gentra Puregene Cell Kit (Qiagen, Valencia, CA, USA). PCR was performed using 200 ng genomic DNA, 0.5 μM of primers H1–2F (5′-CAGGAAGATGGCTGTGAGGGAC-3′) and H1–4R (5′-CGGATCTCGACGGTATCGGT-3′), 5 U Taq polymerase (NEB, Ipswich, MA, USA) and 0.25 U Pfu polymerase (Stratagene), and 200 μM each of dNTP and 1X Pfu buffer in a 50-μl reaction. The 83-bp shRNA expressing sequences were released from the 362-bp PCR products by digestion with BamHI and ClaI, gel purified, and inserted into the BamHI and ClaI sites on pLentiSuper2. BDX chemistry (Sequetech, Mountain View, CA) was used for DNA sequencing with primers H1–1F (5′-CTTTGGATTTGGGAATCTTA-3′) and/or H1–1R (5′-GCAACAGACATACAAACTAAAG-3′).
cAMP assay
Cellular cAMP concentrations were measured using a method adapted from published procedures (39–41). HASM cells were plated at 2 × 105 cells/well, in 6-well plates. Cells were treated with either control vehicle or 30 μM ISO for 3 d. After washing 3 times with PBS, cells were cultured in ISO-free, low serum (1% FBS) medium for 4 h. The medium was then replaced with 0.5 ml of medium containing 0.1 mM IBMX (to prevent degradation of cAMP by phophodiesterases). After 30 min, a single concentration of 10 μM ISO and a single concentration of forskolin (10−6 to 10−4 M) or diluent were added to the cells. After 10 min, HASM cells were extracted with 0.1 M HCl. Cell extracts were neutralized, acetylated, and used for cAMP measurement using the Biovision cAMP Assay kit (Biovision, Mountain View, CA, USA). cAMP concentration was calculated based on a cAMP standard curve.
Immunofluorescence microscopy
Immunostaining was performed according as described previously (15). In brief, cells were grown on coverslips, fixed in 4% paraformaldehyde, permeabilized in 0.2% Triton X-100, and incubated with primary and secondary antibodies. Primary antibodies were anti-Flag (rabbit polyclonal; Sigma-Aldrich) at 1:100, anti-LAMP2 (mouse monoclonal; Cell Signaling) and anti-EEA1 (mouse monoclonal; BD Biosciences) at 1:200. Secondary antibodies were TRITC-conjugated goat anti-rabbit IgG and FITC-conjugated goat anti-mouse IgG (Sigma-Aldrich). The coverslips were mounted onto slides with Vectashield mounting medium (Vector, Burlingame, CA, USA), and cells were examined under a Leica TCSNT laser confocal microscope (Leica Microsystems, Wetzlar, Germany).
Membrane protein extraction and analysis
Preparation of membrane protein extracts was done using the Mem-PER Mammalian Membrane Protein Extraction Kit (Pierce Protein Research; Thermo Scientific). Briefly, 293β2AR cells mock-treated or treated with alendronate (ALN) were lysed with an initial detergent. A second detergent was then added to solubilize the membrane proteins. After a quick centrifugation, the mixture was incubated at 37°C to separate the hydrophobic proteins from the hydrophilic proteins through phase partitioning. Fractionated membrane extracts as well as whole-cell extract were used for Western blotting to detect Rab5.
RESULTS
Development of a novel RNAi library consisting of 26-nt shRNAs
ShRNAs are siRNA-like structures that are often expressed from a vector and can be integrated into the genome, allowing for stable, long-lasting gene inactivation (30–32). Because several studies have shown that relatively longer shRNAs (26 nt) exhibit much more potent gene silencing activity than regular 19- to 21-nt shRNAs (42, 43), we sought to develop a new gene inactivation libraries comprising 26-nt shRNAs. We first designed a procedure (Fig. 1A) that converts double-stranded cDNA molecules into structures expressing 26-nt shRNAs. Our approach took advantage of a type IIS restriction enzyme, EcoP15I, that cuts ∼24 and 26 nt away from its recognition site (44). Randomly fragmented cDNA sequences were ligated to a hairpin-shaped adaptor containing an embedded EcoP15I recognition site. Digestion of the ligated DNAs with EcoP15I generated fragments of 26-bp cDNA sequences linked to the hairpin-shaped adaptor. Primer extension and stand displacement were used to convert the hairpin constructs into palindromic double-stranded DNAs (dsDNAs). Subsequent expression of the palindromic dsDNAs from the polymerase III promoter H1 leads to production of cDNA-specific 26-nt shRNAs (Fig. 1A, B).
To test the knockdown efficiency of 26-nt shRNAs generated by the enzymatic method, we constructed a shRNA library for a single human gene, ARRDC1. The ARRDC1-specific shRNA library contains several thousand of individual shRNAs, covering most of the possible targeting sequences. Individual shRNAs were randomly picked and used to transduce HEK293T cells expressing either ARRDC1-GFP or GFP. We analyzed a total of 60 individual infected cell clones and determined the ARRDC1-GFP level relative to GFP. About 60% of selected shRNAs reduced ARRDC1-GFP expression by >50%, and >20% of shRNAs reduced the expression by ≥75% (Fig. 1C). These data indicate that the enzymatic procedure efficiently generated shRNAs with potent knockdown activity.
We next applied the method to construct a genome-wide 26-nt shRNA library. We used a large collection of >40,000 mostly unique EST sequences representing >20,000 human genes (45). These EST DNA clones were pooled into 10 fractions, and each fraction was individually amplified by PCR and converted into shRNA-expressing structures (Fig. 1D). The final library contains ∼600,000 individual shRNA-expressing constructs (∼30 different shRNAs per gene on average). Based on the knockdown efficiency of the single-gene shRNA library (Fig. 1C), we estimate that the library contains ∼6 highly efficient (>75% suppression) shRNAs for each gene.
A cell-based functional RNAi screen identified new regulators of β2AR down-regulation
We next applied our newly developed shRNA library in a functional genetic screen to identify novel regulators of β2AR down-regulation. To establish a cellular β2AR down-regulation assay amenable for a high-throughput screen, we used an HEK293 cell line (designated as 293β2AR) stably expressing a modified β2AR that is deficient in recycling and thus undergoes efficient down-regulation on the β-agonist ISO stimulation (ref. 38 and Fig. 2A). The β2AR is N-terminally Flag-tagged and thus can be easily detected in flow cytometry using anti-Flag antibodies. As shown in Fig. 2B, consistent with the Western blotting result (Fig. 2A), a significant decrease (∼5-fold) in surface fluorescence was detected by flow cytometry in cells treated with ISO (Fig. 2B). Our flow cytometry-based assay provides a highly quantitative and efficient way to isolate cells with enriched amounts of cell surface β2AR.
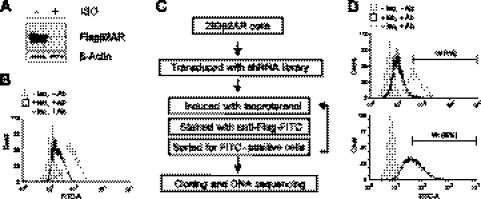
Figure 2.
A FACS-based shRNA screen identified new genes required for β2AR down-regulation. A) Efficient down-regulation of β2AR in 293β2AR cells. Cells were treated with 10 μM ISO for 16 h, and the receptor expression was determined by immunoblotting. B) Flow cytometry analysis of β2AR level at the cell surface. 293β2AR cells were mock-treated or treated with 10 μM ISO for 16 h and stained with FITC-conjugated anti-Flag antibody. Cells not stained with the antibody were used as a negative control. C) Schematic of a FACS-based shRNA screen for genes required for β2AR down-regulation. D) FACS enrichment of 293β2AR cells with a high level of surface β2AR. Cells were infected with the EST-derived shRNA lentiviral library, treated with 10 μM ISO for 16 h, and stained with FITC-conjugated anti-Flag antibody. Four rounds of sorting were performed to enrich FITC-bright cells. Graphs show the flow cytometry analyses of cell populations prior to the first or the fourth round of sorting with gating conditions indicated. See Table 1 for list of validated gene hits from the RNAi screen.
We next constructed a high-coverage shRNA library in the 293β2AR cells. Since our shRNA library was lentivirus based, we transduced 293β2AR cells with high-titer lentiviruses generated from the shRNA library to produce a cellular library with an estimated >5×106 individual viral integration events. This represents >8-fold coverage of the ∼600,000 shRNAs in the library. The established cell shRNA library cells were treated with ISO and underwent FACS to isolate cells that maintain a high level of surface β2AR even after ISO stimulation (Fig. 2C). The gating conditions for the first round of FACS were set so that 1% of the cell population was isolated (Fig. 2D, left panel). After sorting, the cells were cultured, expanded, and subjected to 3 additional rounds of FACS to enrich for cells with a higher amount of cell surface β2AR. In the final (fourth) round of FACS sorting, cells with a higher amount of undegraded receptor represented ∼29% of the population (Fig. 2D, right panel), indicating a substantial enrichment of cells with higher amounts of β2AR by the FACS procedure.
The FACS-enriched cells potentially contain shRNAs that inactivate genes critically involved in agonist-induced β2AR down-regulation. To identify the shRNAs, we extracted genomic DNA from the FACS-sorted cells and performed genomic PCR. We sequenced ∼100 shRNAs, many of which correspond to the same target genes. In total, we identified 16 human genes from the screen (Table 1). Although most of the RNAi hits have not been previously linked to β2AR regulation, one of the hits, ARRDC3, encodes a novel protein that is homologous to β-arrestins, which are well known for their role in terminating β2AR signaling (9, 17). We have recently shown that ARRDC3 recruits a specific ubiquitin ligase to mediate the ubiquitination and subsequent degradation of β2AR (18). To initially validate our screen result, we transduced the shRNAs for 9 of the RNAi gene hits into 293β2AR cells and assessed the effect on agonist-induced β2AR down-regulation. ShRNAs targeting ARRDC3 and ≥4 other hits (FDPS, CaMKK2, KIAA0786, and AI075761) exhibited inhibitory effects on ISO-induced β2AR down-regulation (data not shown), suggesting these genes as new putative regulators of β2AR down-regulation.
Table 1.
Gene hits from the RNAi screen
| Gene | Description |
|---|---|
| FDPS | Farnesyl diphosphate synthase |
| ARRDC3 | Arrestin domain containing 3 |
| CaMKK2 | Calcium/calmodulin-dependent kinase kinase 2 (β) |
| TPPP3 | Tubulin polymerization-promoting protein family member 3 |
| KIAA0786 | Latrophilin 2 isoform |
| LRP5 | Low-density lipoprotein receptor-related protein 5 |
| AI075761 | Transcribed locus Hs 52264 |
| PELP1 | Proline, glutamate, and leucinerich protein 1 |
| SNAPC5 | Small nuclear RNA activating complex, polypeptide 5 |
| FBXO44 | F-box protein 44 |
| BZW1L1 | Basic leucine zipper and W2 domain 1-like 1 |
| GGN | Gametogenetin |
| GGT7 | γ-Glutamyltransferease 7 |
| NPIP | Nuclear pore complex interacting protein |
| AI015265 | cDNA IMAGE clone 1641211 |
| GNL3L | Guanine nucleotide binding protein 3-like |
FDPS is required for agonist-induced β2AR down-regulation
We focused on FDPS in part because highly potent and specific pharmaceutical inhibitors for the enzyme already exist (46, 47) and therefore can be used to inhibit β2AR down-regulation to improve β-agonist therapy. FDPS is an enzyme of the cellular mevalonate synthetic pathway that catalyzes the formation of farnesyl diphosphate, which serves as a precursor for cholesterol synthesis and a substrate for protein prenylation (48). To further confirm the role of FDPS on β2AR down-regulation, we tested the effect of FDPS siRNA. We transiently transfected 293β2AR cells with a FDPS siRNA targeting a sequence different from the shRNA identified in the initial RNAi screen. As shown in Fig. 3A, the cell surface β2AR level after ISO (10 μM for 3 h) treatment in FDPS siRNA-transfected cells was significantly higher than that in control cells transfected with nontargeting siRNA. The siRNA reduced the level of FDPS protein efficiently by ∼75% (Fig. 3B). Consistent with the increase of cell surface level of β2AR, a significant amount of β2AR, as determined by Western blotting, remained undegraded in FDPS siRNA-transfected cells after ISO stimulation (Fig. 3B). These results indicate that FDPS is required for the efficient agonist-induced β2AR down-regulation. In addition, FDPS siRNA treatment also, to a lesser extent, increased the basal (non-ISO treatment) β2AR level (Fig. 3A, B), suggesting that FDPS may also affect steady-state receptor expression.
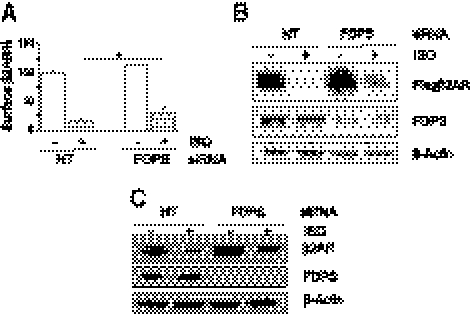
Figure 3.
FDPS is required for β2AR down-regulation. A) Suppression of FDPS expression by siRNA increased cell surface β2AR level on ISO stimulation. 293β2AR cells were transfected with FDPS-specific siRNA or nontargeting siRNA (NT), and 72 h later cells were treated with 10 μM ISO for 3 h. Amount of surface β2AR was determined by flow cytometry. Three independent analyses were done. *P < 0.05. B) Inhibition of β2AR degradation by FDPS siRNA. 293β2AR cells were treated as in A, and the corresponding lysates were subjected to immunoblotting. C) SiRNA-mediated knockdown of FDPS inhibited endogenous β2AR down-regulation. Primary HASM cells transfected with control or FDPS siRNAs were cultured in the absence or the presence of 30 μM ISO for 3 d. Immunoblotting was done using the indicated antibodies.
Since our screen and validation assays used a modified β2AR with a fast down-regulation phenotype, we next tested whether FDPS inhibition exerts similar effects on wild-type β2AR. We used the primary HASM cells, which express a high level of endogenous β2AR and is a physiological tissue target of β-agonists (49, 50). Using an antibody that can detect endogenous β2AR (Supplemental Fig. S1), we determined the effect of FDPS knockdown on the receptor down-regulation in HASM cells. Because of the efficient recycling of wild-type β2AR, we treated the HASM cell with ISO at a high concentration (30 μM) and for an extended period (3 d). Such ISO treatment induced moderately efficient β2AR down-regulation in control HASM cells (Fig. 3C). However, in FDPS siRNA-treated HASM cells, β2AR after ISO treatment was at a much higher level compared to that in the control siRNA-treated cells, suggesting impaired receptor down-regulation (Fig. 3C). The basal β2AR level in FDPS knockdown cells also moderately increased compared to control cells. These results indicate that, similar to its effects on the fast-degrading β2AR, FDPS is also required for the down-regulation of endogenous wild-type β2AR.
Chemical inhibition of FDPS by ALN blocks β2AR down-regulation and improves receptor function
FDPS is the direct target of a group of inhibitory drugs known as nitrogen-containing bisphosphonates (46, 47), among which ALN is a highly specific and potent inhibitor and an FDA-approved drug (51, 52). We investigated whether FDPS inhibition by ALN affects β2AR down-regulation. As shown in Fig. 4A, 48 h of ALN treatment in 293β2AR cells markedly increased the amount of undegraded β2AR after ISO stimulation, indicating an impairment in β2AR down-regulation. ALN treatment also exhibited an inhibitory effect on the down-regulation of the full-length β2AR (Fig. 4B), leading to a higher level of cell surface β2A (Supplemental Fig. S2). We next examined the effect of ALN on down-regulation of endogenous β2AR in HASM cells. As shown in Fig. 4C, D, ALN exhibited a dose and time-dependent inhibitory effect on β2AR down-regulation. Together, these results demonstrated that chemical inhibition of FDPS by ALN blocks β2AR degradation, consistent with the notion that FDPS is required for receptor down-regulation.
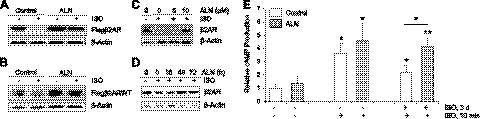
Figure 4.
Effects of FDPS inhibitor ALN on β2AR down-regulation and β-agonist-induced cAMP formation. A) Effect of ALN on down-regulation of the fast-degrading β2AR. 293β2AR cells were treated with 50 μM ALN for 48 h and then exposed to 10 μM ISO for 3 h, and cell lysates were subjected to immunoblotting. B) Effects of ALN on full-length β2AR down-regulation. HEK293 cells stably expressing Flag-tagged full-length β2AR were treated with 50 μM ALN and 10 μM ISO for 48 h. β2AR expression was determined by immunoblotting. C) Concentration-dependent effects of ALN on β2AR down-regulation. HASM cells were treated with indicated concentrations of ALN in the absence and presence of 30 μM ISO for 3 d. Cell lystaes were analyzed by immunoblotting using indicated antibodies. D) Time-dependent effects of ALN on β2AR down-regulation. HASM cells were treated with 10 μM ALN for the indicated time in the absence and presence of 30 μM ISO for 3 d. Cell lysates were analyzed by immunoblotting. E) Effects of ALN on agonist-stimulated cAMP production. HASM cells were treated with 3 d ISO in the absence or presence of ALN (10 μM, 3 d), washed with PBS, and cultured in ISO-free medium for 4 h before subjected to acute ISO stimulation for 10 min. cAMP level was measured as described in Materials and Methods. *P < 0.05; **P < 0.01.
The important functional outcome of β2AR activation by β-agonists is an increase of intracellular cyclic AMP (cAMP), which serves as a second messenger to promote subsequent cell signaling and physiological responses (50, 53). To assess the effect of altered FDPS activity on β2AR signaling, we examined the effect of ALN on ISO-stimulated cAMP production (Fig. 4E). In control HASM cells, short-term ISO stimulation led to ∼3.5-fold elevation of cAMP level. Pretreatment of HASM cells with ISO for 3 d reduced the cAMP elevation to only ∼2-fold, consistent with desensitization of β2AR signaling by prolonged ISO stimulation. However, in HASM cells treated with ALN, short-term ISO stimulation induced cAMP production by ∼4-fold. This cAMP production level is similar to that in control cells not predesensitized with ISO, indicating that ALN reversed the down-regulating effect of pre-ISO treatment. The effect is specific for β2AR-dependent cAMP formation, as forskolin-induced cAMP formation was not altered by ALN treatment (data not shown). Together, our results demonstrated that chemical inhibition of FDPS by ALN blocked β2AR down-regulation and consequently enhanced β2AR signaling.
FDPS inactivation interferes with β2AR internalization
β2AR down-regulation is a multistep process that includes receptor ubiquitination (14–17), internalization through endocytosis (11, 12), and eventual trafficking to lysosomes for degradation (11, 54). While ubiquitination plays a major role in the down-regulation and lysosomal degradation of β2AR (15, 17, 55, 56), FDPS knockdown did not interfere with agonist-induced β2AR ubiquitination (Supplemental Fig. S3A). This result indicates that FDPS does not play a significant role in modulating receptor ubiquitination or phosphorylation.
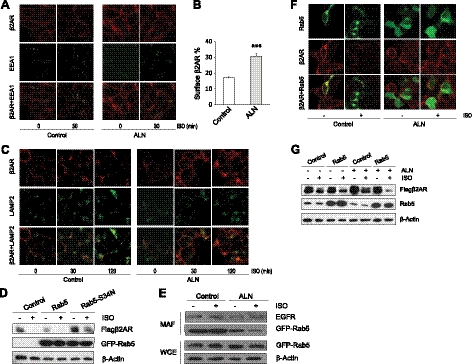
We next examined the role of FDPS in β2AR internalization and trafficking into endosomes. Short-term ISO treatment induces internalization but little degradation of β2AR, thus allowing specific assessment of the level of internalized β2AR. We first used immunofluorescence imaging to visualize β2AR internalization. On 30 min exposure to ISO, a majority of β2AR was internalized from cell membrane into cytosol and colocalized with early endosome marker EEA1 in control cells (Fig. 5A). In ALN-treated cells, however, β2AR internalization was much less efficient, with a significant amount of receptor remaining on the plasma membrane (Fig. 5A). Consistent with this result, flow cytometry analysis showed more cell surface β2AR in ALN-treated cells (Fig. 5B). We next examined the colocalization of β2AR with late endosome marker LAMP2. As shown in Fig. 5C, ALN-treated cells contain significantly more β2AR at the cell surface after 30 min ISO stimulation, and the overall β2AR signal (both plasma membrane and cytoplasmic fractions) was much higher in ALN-treated cells than in control cells. However, most of β2AR was able to internalize after long-time (120 min) ISO stimulation, and the majority of the internalized receptor colocalized well with LAMP2 in ALN-treated cells, indicating that ALN slowed but did not prevent the trafficking of β2AR into late endosomes. Together, our data support a model in which FDPS inhibition by ALN slows B2AR internalization into early endosomes, which in turn delays the subsequent trafficking of B2AR into late endosomes, leading to the overall inhibitory effect on B2AR down-regulation.
Figure 5.
FDPS modulates β2AR internalization and down-regulation in a Rab5-dependent manner. A) Effect of ALN on β2AR internalization into early endosomes. 293β2AR cells were treated with 50 μM ALN for 48 h and then stimulated with 10 μM ISO for 30 min. Cells were then fixed, permeablized, and stained with primary antibodies (polyclonal anti-Flag antibody for Flag-β2AR and a monoclonal antibody for EEA1) and respective TRIC- or FITC-conjugated secondary antibodies. Images were visualized under confocal microscopy. B) Effect of ALN on β2AR internalization. 293β2AR were mock-treated or treated with ALN and stimulated with 10 μM ISO for 30 min to induce internalization. Flow cytometry was used to measure amount of β2AR at the cell surface. Percentages of β2AR remained at the cell surface on ISO stimulation were calculated relative to surface receptor level without ISO treatment. ***P < 0.001. C) Effect of ALN on β2AR trafficking into late endosomes. 293β2AR cells were treated as in A and then stimulated with 10 μM ISO for indicated time. Immunostaining was performed to detect Flag-β2AR and LAMP2. D) Effect of wild-type or mutant Rab5 expression on β2AR down-regulation. 293β2AR cells were transfected with wild-type or mutant (S34N) GFP-Rab5 expression plasmids and after 48 h, cells were treated with 10 μM ISO for 3 h. Cell extracts were subjected to immunoblotting to detect receptor levels. Anti-GFP antibody was used to detect the expression of GFP-Rab5 fusion protein. E) Effect of ALN on Rab5 membrane association. 293β2AR cells were treated with 50 μM ALN for 48 h followed by 10 μM ISO stimulation for 30 min. Cells were lysed, and membrane-associated fractions were isolated using the Mem-PER Mammalian Membrane Protein Extraction Kit (Pierce). Immunoblotting was done on the membrane-associated fraction (MAF) and the whole cell extract (WCE) using indicated antibodies. F) Effect of ALN on Rab5 localization. 293β2AR cells transfected with GFP-Rab5 were mock-treated or treated with ALN for 48 h and then subjected to 10 μM ISO stimulation (or control vehicle treatment) for 30 min. Cells were fixed, permeabilized, and stained with anti-Flag antibody. Images were visualized with a confocal fluorescence microscope. G) Effect of Rab5 overexpression on the inhibition by ALN on β2AR down-regulation. 293β2AR cells were transfected with Rab5 expression plasmid or control plasmid pCDNA3.1, and after 48 h cells were treated with 25 μM ALN or control vehicle for 24 h. Cell were then subjected to 10 μM ISO stimulation (or control vehicle treatment) for 2 h, and extracts were analyzed by immunoblotting.
FDPS regulates β2AR down-regulation in a Rab5-dependent manner
FDPS catalyzes the formation of farnesyl diphosphate, which is a precursor for both cholesterol synthesis (48) and protein prenylation (farnesylation and geranylgeranylation; ref. 46). We examined which branch of the pathway may be responsible for the effects of FDPS deficiency on β2AR down-regulation. β2AR resides in cholesterol-rich lipid rafts, and cellular cholesterol contents could regulate β2AR signaling (57–59). However, the cholesterol-lowering drug pravastatin had little effect on β2AR down-regulation (Supplemental Fig. S3B). Moreover, ALN treatment did not significantly decrease cellular cholesterol level (data not shown). These results suggest that the inhibition of cholesterol synthesis does not account for the effect of FDPS inactivation on β2AR down-regulation.
FDPS inactivation affects protein prenylation, a modification essential for membrane localization of many small GTPases (60), including Rab family GTPases (61). Rab GTPases are known to regulate β2AR endocytosis, recycling, and endosomal sorting (62–65). In particular, studies have demonstrated that Rab5 is required for β2AR internalization (64, 65). Rab5 localizes to the plasma membrane and/or early endosomes (66, 67), and the extent of membrane association is determined by modification of the prenylation motif in Rab5 (68). Consistent with these studies, expression of a catalytically inactive Rab5 significantly inhibited β2AR down-regulation (Fig. 5D). We reasoned that FDPS inactivation may disrupt Rab5 membrane localization to regulate β2AR internalization and down-regulation. To test this hypothesis, we examined the Rab5 localization in control and ALN-treated cells. We transiently transfected 293β2AR cells with a GFP-Rab5 fusion construct and assessed Rab5 localization by both membrane fractionation and fluorescence imaging. As shown in Fig. 5E, the amount of Rab5 associated with membrane fractions of cell extracts was significantly lower in ALN-treated cells than that in the control cells, while ALN did not alter the membrane association of a control plasma membrane protein EGFR.
Consistent with the membrane fractionation result, confocal imaging (Fig. 5F) showed that GFP-Rab5 was mainly localized on the plasma membrane and endosomes (as indicated by cytosolic aggregates) and colocalized with cell surface β2AR. In response to ISO (30 min) treatment, most surface β2AR was internalized and colocalized with endosomal Rab5. Strikingly, Rab5 exhibited a diffused cytosolic distribution in ALN-treated cells both before and after ISO treatment (Fig. 5F). Our data suggest that FDPS affects β2AR internalization and down-regulation by modulating the membrane localization of Rab5. We further tested whether overexpression of wild-type Rab5 compensates for the functional loss of Rab5 and thus reverses the inhibitory effect of ALN on β2AR down-regulation. As shown in Fig. 5G, in Rab5 transfected cells, β2AR receptor was as efficiently down-regulated on ISO stimulation in the presence of ALN treatment, indicating that Rab5 mediates the effect of FDPS on β2AR down-regulation. Together, our data support the notion that FDPS regulates β2AR down-regulation in a Rab5-dependent manner.
DISCUSSION
Using a novel RNAi platform consisting of enzymatically generated 26-nt shRNAs, together with a robust flow cytometry-based β2AR assay, we performed a genome-wide genetic screen to identify genes whose inactivation leads to defective β2AR down-regulation. Our approach discovered several genes that are previously unrecognized for their role in β2AR down-regulation, suggesting novel mechanisms of β2AR regulation. One of the novel regulators is FDPS, an enzyme in the cellular mevalonate pathway. We demonstrated that FDPS is required for β2AR down-regulation and acts on receptor internalization through a Rab5 GTPase-dependent mechanism. Moreover, we showed that ALN, a potent FDPS inhibitor, inhibits β2AR down-regulation and functionally reverses agonist-induced down-regulation of β2AR signaling in human airway smooth muscle cells, suggesting that FDPS inhibition by the drug may be used to improve the efficacy of β2AR agonists in asthma therapy. Our successful β2AR screen also provides a proof-of-principle demonstration for the application of the EST-derived shRNA library in facilitating the loss-of-function genome-wide screens.
The unique EST-derived 26-nt shRNA library established in this report has several advantages over existing RNAi platforms. Several laboratories have previously reported enzyme-mediated methods for generating 19–21 bp shRNA constructs from DNA coding gene of interest or pool of genes (69–71). Our library expresses longer (26-bp) shRNAs that are more efficient in knocking down target gene expression compared to regular 19- to 21-bp shRNAs (42, 43). The library was generated from a large EST collection that contains not only protein-coding but also noncoding RNAs (37), thus expanding the coverage of current RNAi technologies. Indeed, one of our RNAi hits may encode a noncoding RNA, suggesting a completely novel mechanism of action in β2AR down-regulation. In addition, our library contains an average of ∼30 shRNAs for each gene. The increased number of shRNA per target gene improves the probability of efficient knockdown and may help exclude the possibility of “off-target” effects. However, we note that due to the randomness and unbiased nature of the enzymatic process of generating 26-nt shRNAs, our library also contains inefficient shRNA constructs, which may increase background noise in a screen. Such a problem may be alleviated in the future development of improved RNAi libraries by selecting the rare and yet extremely potent shRNA sequences using a very recently developed methodology (72).
Our RNAi screen identified FDPS as a new regulator of β2AR internalization and down-regulation. FDPS is an essential enzyme of the cellular mevalonate synthetic pathway, which is required for protein prenylation and cholesterol synthesis (48). Given that prenylation is required for membrane localization of small GTPase such as Rab5 (66, 68), and the well-established role of Rab5 GTPase in β2AR internalization (64, 65), we hypothesized that FDPS affects β2AR internalization and subsequent down-regulation by modulating the membrane localization of Rab5 GTPase. Indeed, consistent with our hypothesis, FDPS inhibition by ALN disrupts the membrane localization of Rab5. More importantly, the overexpression of Rab5 reversed receptor down-regulation defect caused by FDPS inactivation. Although FDPS inhibition likely affects protein prenylation in general and may induce pleiotropic effects, our results support a major role of Rab5 in the process. In addition to protein prenylation, FDPS and the mevalonate pathway also control the biosynthesis of cholesterol. Cholesterol is known to play a role in stabilizing β2AR structure in plasma membrane and in regulating β2AR signaling (57–59). However, we found that the FDPS inhibitor ALN did not significantly reduce cholesterol level in cells. Moreover, we failed to detect a significant effect of pravastatin, a cholesterol-lowering drug, on β2AR down-regulation. These results suggest that the effect of FDPS inactivation on β2AR is unlikely an outcome of decreased cholesterol synthesis. Together these studies revealed that FDPS regulates β2AR internalization and down-regulation through a Rab5-dependent and largely cholesterol-independent mechanism.
β2AR down-regulation is a multistep process that includes receptor ubiquitination (14–17), internalization through endocytosis (11, 12), and eventual trafficking to lysosomes for degradation (11, 54). Despite a major role of ubiquitination in the down-regulation and lysosomal degradation of β2AR (15, 17, 55, 56), FDPS knockdown did not affect β2AR ubiquitination. Rather, our data support a model in which FDPS inhibition slows β2AR internalization into early endosomes, which in turn delays the subsequent trafficking of β2AR into late endosomes, leading to the overall inhibitory effect on β2AR down-regulation. A previous study showed the β2AR internalization process was biphasic with respect to agonist concentration, with only the high-affinity component possibly linked to internalization (and GRKs and arrestin; ref. 73). While the precise role of internalization remains incompletely resolved, it is clear that internalization is involved in receptor down-regulation. For example, one study (12) found that expression of dynamin mutant (which blocks endocytic internalization) inhibited β2AR down-regulation. Our study thus further supports a critical role of internalization in the β2AR down-regulation process. However, given that β2AR internalization occurs much faster (5–10 min) than down-regulation and thus may not be the rate-limiting step, the impairment of internalization by ALN may not fully explain its effect on β2AR down-regulation. We investigated further the effect of ALN on endocytic trafficking of β2AR, which is an important and likely rate-limiting step in the down-regulation process (Fig. 5A, C). Although it remains to be determined whether FDPS inhibition directly affects the endocytic trafficking of β2AR, the indirect interference by ALN on β2AR trafficking may partly account for its inhibitory effect on β2AR down-regulation.
Agonists of β2AR have become a mainstay of asthma therapy (3, 74) but have important limitations. First, chronic use of β-agonists down-regulates β2AR, reducing the efficacy of the therapy (75, 76). Second, there is significant variability in response to this therapy among patients (77–79). Both limitations may be in part attributable to altered functional β2AR at the cell surface. A reduction in functional β2AR level slows the responsiveness of target lung tissue cells to β-agonists and causes loss of bronchodilation and protection against bronchoconstriction (80–82). Many other adverse effects such as longer durations of asthma exacerbation may also be attributed to a decrease in receptor number (20). Targeted inhibition of genes required for β2AR down-regulation provides an approach to improving the efficacy and safety of β-agonists in asthma therapy. ALN is a highly specific inhibitor of FDPS and is widely used in the treatment of bone-degenerative diseases (47). We found that FDPS inhibition by ALN blocked agonist-induced β2AR down-regulation. Notably, ALN functionally reversed β2AR loss induced by prolonged agonist stimulation in primary airway smooth muscle cells. Our results provide the rationale for further evaluation of the effect of ALN and other FDPS inhibitors on the β-agonist response using in vitro lung tissues such as precision-cut lung slices (83), as well as in animal models. Such studies will further establish the augmenting effect of FDPS inhibition on the physiological function of β-agonists and could eventually lead to the therapeutic use of ALN and other FDPS inhibitors in asthma therapy.
In addition to FDPS, we have identified several other new regulators of β2AR down-regulation. The RNAi hits we report here likely represent only a partial list of new genes critically required for β2AR down-regulation, as our screen is by no means complete. Indeed, we did not identify some of the well-known regulators of β2AR down-regulation, such as the NEDD4 E3 ligase (19) and β-arrestins (84). This may be partly due to the fact that we sequenced only a limited number of shRNAs from the final sorted cell population of the RNAi screen. Application of the now commonly available and increasingly affordable deep-sequencing technology should help us uncover more genes that encode known or unknown potential regulators of β2AR. Functional characterization of these genes and the already identified and validated RNAi hits will reveal novel insights into mechanisms underlying β2AR down-regulation and could provide more therapeutic targets to improve β-agonist-based therapy.
Supplementary Material
Acknowledgments
The authors thank Drs. Lester Kobzik and Marianne Wessling-Resnick for critical reading of the manuscript. The authors are also grateful for the advice and help from other members of the Q.L. laboratory.
Q.L. was supported by an American Lung Association Biomedical Research grant, a Harvard-National Institute of Environmental Health Sciences (NIEHS) Center grant (P30ES000002), and a NIEHS Superfund Research Program grant (P42ES016454). R.A.P. was supported by U.S. National Institutes of Health (NIH) grants HL097796 and P30ES013508. X.J. was supported by a NIH training grant (5T32HL7118-35).
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
- ALN
- alendronate
- β2AR
- β2-adrenergic receptor
- cAMP
- cyclic adenosine monophosphate
- dsDNA
- double-stranded DNA
- EST
- expressed sequence tag
- FACS
- fluorescence-activated cell sorting
- FBS
- fetal bovine serum
- FCS
- fetal calf serum
- FDPS
- farnesyl diphosphate synthase
- GFP
- green fluorescence protein
- GPCR
- G-protein coupled receptor
- GRK
- G-protein-coupled receptor kinase
- HASM
- human airway smooth muscle
- ISO
- isoproterenol
- PKA
- protein kinase A
- RNAi
- RNA interference
- shRNA
- short-hairpin RNA
- siRNA
- small interfering RNA.
REFERENCES
- 1. Johnson M. (1998) The beta-adrenoceptor. Am. J. Respir. Crit. Care Med. 158, S146–153 [DOI] [PubMed] [Google Scholar]
- 2. Pierce K. L., Premont R. T., Lefkowitz R. J. (2002) Seven-transmembrane receptors. Nat. Rev. Mol. Cell Biol. 3, 639–650 [DOI] [PubMed] [Google Scholar]
- 3. Barnes P. J. (2004) New drugs for asthma. Nat. Rev. Drug Discov. 3, 831–844 [DOI] [PubMed] [Google Scholar]
- 4. Tattersfield A. E., Knox A. J., Britton J. R., Hall I. P. (2002) Asthma. Lancet 360, 1313–1322 [DOI] [PubMed] [Google Scholar]
- 5. Dohlman H. G. (2002) Diminishing returns. Nature 418, 591. [DOI] [PubMed] [Google Scholar]
- 6. Premont R. T., Gainetdinov R. R. (2007) Physiological roles of G protein-coupled receptor kinases and arrestins. Annu. Rev. Physiol. 69, 511–534 [DOI] [PubMed] [Google Scholar]
- 7. Benovic J. L., DeBlasi A., Stone W. C., Caron M. G., Lefkowitz R. J. (1989) Beta-adrenergic receptor kinase: primary structure delineates a multigene family. Science 246, 235–240 [DOI] [PubMed] [Google Scholar]
- 8. Liggett S. B. (1991) Desensitization of the beta-adrenergic receptor: distinct molecular determinants of phosphorylation by specific kinases. Pharmacol. Res. 24(Suppl. 1), 29–41 [DOI] [PubMed] [Google Scholar]
- 9. DeWire S. M., Ahn S., Lefkowitz R. J., Shenoy S. K. (2007) Beta-arrestins and cell signaling. Annu. Rev. Physiol. 69, 483–510 [DOI] [PubMed] [Google Scholar]
- 10. Ferguson S. S., Downey W. E., 3rd, Colapietro A. M., Barak L. S., Menard L., Caron M. G. (1996) Role of beta-arrestin in mediating agonist-promoted G protein-coupled receptor internalization. Science 271, 363–366 [DOI] [PubMed] [Google Scholar]
- 11. Hanyaloglu A. C., von Zastrow M. (2008) Regulation of GPCRs by endocytic membrane trafficking and its potential implications. Annu. Rev. Pharmacol. Toxicol. 48, 537–568 [DOI] [PubMed] [Google Scholar]
- 12. Gagnon A. W., Kallal L., Benovic J. L. (1998) Role of clathrin-mediated endocytosis in agonist-induced down-regulation of the beta2-adrenergic receptor. J. Biol. Chem. 273, 6976–6981 [DOI] [PubMed] [Google Scholar]
- 13. von Zastrow M., Sorkin A. (2007) Signaling on the endocytic pathway. Curr. Opin. Cell Biol. 19, 436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liang W., Austin S., Hoang Q., Fishman P. H. (2003) Resistance of the human beta 1-adrenergic receptor to agonist-mediated down-regulation. Role of the C terminus in determining beta-subtype degradation. J. Biol. Chem. 278, 39773–39781 [DOI] [PubMed] [Google Scholar]
- 15. Liang W., Fishman P. H. (2004) Resistance of the human beta1-adrenergic receptor to agonist-induced ubiquitination: a mechanism for impaired receptor degradation. J. Biol. Chem. 279, 46882–46889 [DOI] [PubMed] [Google Scholar]
- 16. Saksena S., Sun J., Chu T., Emr S. D. (2007) ESCRTing proteins in the endocytic pathway. Trends Biochem. Sci. 32, 561–573 [DOI] [PubMed] [Google Scholar]
- 17. Shenoy S. K., McDonald P. H., Kohout T. A., Lefkowitz R. J. (2001) Regulation of receptor fate by ubiquitination of activated beta 2-adrenergic receptor and beta-arrestin. Science 294, 1307–1313 [DOI] [PubMed] [Google Scholar]
- 18. Nabhan J. F., Pan H., Lu Q. (2010) Arrestin domain-containing protein 3 recruits the NEDD4 E3 ligase to mediate ubiquitination of the beta2-adrenergic receptor. EMBO Rep. 11, 605–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shenoy S. K., Xiao K., Venkataramanan V., Snyder P. M., Freedman N. J., Weissman A. M. (2008) Nedd4 mediates agonist-dependent ubiquitination, lysosomal targeting, and degradation of the beta2-adrenergic receptor. J. Biol. Chem. 283, 22166–22176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shore S. A., Drazen J. M. (2003) Beta-agonists and asthma: too much of a good thing? J. Clin. Invest. 112, 495–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Drysdale C. M., McGraw D. W., Stack C. B., Stephens J. C., Judson R. S., Nandabalan K., Arnold K., Ruano G., Liggett S. B. (2000) Complex promoter and coding region beta 2-adrenergic receptor haplotypes alter receptor expression and predict in vivo responsiveness. Proc. Natl. Acad. Sci. U. S. A. 97, 10483–10488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Turki J., Pak J., Green S. A., Martin R. J., Liggett S. B. (1995) Genetic polymorphisms of the beta 2-adrenergic receptor in nocturnal and nonnocturnal asthma. Evidence that Gly16 correlates with the nocturnal phenotype. J. Clin. Invest. 95, 1635–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hancox R. J., Sears M. R., Taylor D. R. (1998) Polymorphism of the beta2-adrenoceptor and the response to long-term beta2-agonist therapy in asthma. Eur. Respir. J. 11, 589–593 [PubMed] [Google Scholar]
- 24. Israel E., Drazen J. M., Liggett S. B., Boushey H. A., Cherniack R. M., Chinchilli V. M., Cooper D. M., Fahy J. V., Fish J. E., Ford J. G., Kraft M., Kunselman S., Lazarus S. C., Lemanske R. F., Martin R. J., McLean D. E., Peters S. P., Silverman E. K., Sorkness C. A., Szefler S. J., Weiss S. T., Yandava C. N. (2000) The effect of polymorphisms of the beta(2)-adrenergic receptor on the response to regular use of albuterol in asthma. Am. J. Respir. Crit. Care Med. 162, 75–80 [DOI] [PubMed] [Google Scholar]
- 25. Taylor D. R., Drazen J. M., Herbison G. P., Yandava C. N., Hancox R. J., Town G. I. (2000) Asthma exacerbations during long term beta agonist use: influence of beta(2) adrenoceptor polymorphism. Thorax 55, 762–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Paddison P. J., Silva J. M., Conklin D. S., Schlabach M., Li M., Aruleba S., Balija V., O'Shaughnessy A., Gnoj L., Scobie K., Chang K., Westbrook T., Cleary M., Sachidanandam R., McCombie W. R., Elledge S. J., Hannon G. J. (2004) A resource for large-scale RNA-interference-based screens in mammals. Nature 428, 427–431 [DOI] [PubMed] [Google Scholar]
- 27. Schlabach M. R., Luo J., Solimini N. L., Hu G., Xu Q., Li M. Z., Zhao Z., Smogorzewska A., Sowa M. E., Ang X. L., Westbrook T. F., Liang A. C., Chang K., Hackett J. A., Harper J. W., Hannon G. J., Elledge S. J. (2008) Cancer proliferation gene discovery through functional genomics. Science 319, 620–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Elbashir S. M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498 [DOI] [PubMed] [Google Scholar]
- 29. Elbashir S. M., Lendeckel W., Tuschl T. (2001) RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15, 188–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brummelkamp T. R., Bernards R., Agami R. (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science 296, 550–553 [DOI] [PubMed] [Google Scholar]
- 31. Chang A. C., Zsak L., Feng Y., Mosseri R., Lu Q., Kowalski P., Zsak A., Burrage T. G., Neilan J. G., Kutish G. F., Lu Z., Laegreid W., Rock D. L., Cohen S. N. (2006) Phenotype-based identification of host genes required for replication of African swine fever virus. J. Virol. 80, 8705–8717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stegmeier F., Hu G., Rickles R. J., Hannon G. J., Elledge S. J. (2005) A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 102, 13212–13217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kolfschoten I. G., van Leeuwen B., Berns K., Mullenders J., Beijersbergen R. L., Bernards R., Voorhoeve P. M., Agami R. (2005) A genetic screen identifies PITX1 as a suppressor of RAS activity and tumorigenicity. Cell 121, 849–858 [DOI] [PubMed] [Google Scholar]
- 34. Wajapeyee N., Serra R. W., Zhu X., Mahalingam M., Green M. R. (2008) Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell 132, 363–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Westbrook T. F., Martin E. S., Schlabach M. R., Leng Y., Liang A. C., Feng B., Zhao J. J., Roberts T. M., Mandel G., Hannon G. J., Depinho R. A., Chin L., Elledge S. J. (2005) A genetic screen for candidate tumor suppressors identifies REST. Cell 121, 837–848 [DOI] [PubMed] [Google Scholar]
- 36. Zender L., Xue W., Zuber J., Semighini C. P., Krasnitz A., Ma B., Zender P., Kubicka S., Luk J. M., Schirmacher P., McCombie W. R., Wigler M., Hicks J., Hannon G. J., Powers S., Lowe S. W. (2008) An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell 135, 852–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lu Q., Wei W., Kowalski P., Chang A., Cohen S. (2004) EST-based genome-wide gene inactivation identifies ARAP3 as a host protein affecting cellular susceptibility to anthrax toxin. Proc. Natl. Acad. Sci. . U. S. A. 101, 17246–17251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cao T. T., Deacon H. W., Reczek D., Bretscher A., von Zastrow M. (1999) A kinase-regulated PDZ-domain interaction controls endocytic sorting of the beta2-adrenergic receptor. Nature 401, 286–290 [DOI] [PubMed] [Google Scholar]
- 39. Laporte J. D., Moore P. E., Panettieri R. A., Moeller W., Heyder J., Shore S. A. (1998) Prostanoids mediate IL-1beta-induced beta-adrenergic hyporesponsiveness in human airway smooth muscle cells. Am. J. Physiol. 275, L491–501 [DOI] [PubMed] [Google Scholar]
- 40. Moore P. E., Laporte J. D., Abraham J. H., Schwartzman I. N., Yandava C. N., Silverman E. S., Drazen J. M., Wand M. P., Panettieri R. A., Jr., Shore S. A. (2000) Polymorphism of the beta(2)-adrenergic receptor gene and desensitization in human airway smooth muscle. Am. J. Respir. Crit. Care Med. 162, 2117–2124 [DOI] [PubMed] [Google Scholar]
- 41. Moore R. H., Hall H. S., Rosenfeld J. L., Dai W., Knoll B. J. (1999) Specific changes in beta2-adrenoceptor trafficking kinetics and intracellular sorting during downregulation. Eur. J. Pharmacol. 369, 113–123 [DOI] [PubMed] [Google Scholar]
- 42. Siolas D., Lerner C., Burchard J., Ge W., Linsley P. S., Paddison P. J., Hannon G. J., Cleary M. A. (2005) Synthetic shRNAs as potent RNAi triggers. Nat. Biotechnol. 23, 227–231 [DOI] [PubMed] [Google Scholar]
- 43. Kim D. H., Behlke M. A., Rose S. D., Chang M. S., Choi S., Rossi J. J. (2005) Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat. Biotechnol. 23, 222–226 [DOI] [PubMed] [Google Scholar]
- 44. Moncke-Buchner E., Mackeldanz P., Kruger D. H., Reuter M. (2004) Overexpression and affinity chromatography purification of the type III restriction endonuclease EcoP15I for use in transcriptome analysis. J. Biotechnol. 114, 99–106 [DOI] [PubMed] [Google Scholar]
- 45. Lu Q., Wei W., Kowalski P. E., Chang A. C., Cohen S. N. (2004) EST-based genome-wide gene inactivation identifies ARAP3 as a host protein affecting cellular susceptibility to anthrax toxin. Proc. Natl. Acad. Sci. U. S. A. 101, 17246–17251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Roelofs A. J., Thompson K., Gordon S., Rogers M. J. (2006) Molecular mechanisms of action of bisphosphonates: current status. Clin. Cancer Res. 12, 6222s–6230s [DOI] [PubMed] [Google Scholar]
- 47. Russell R. G. (2007) Bisphosphonates: mode of action and pharmacology. Pediatrics 119(Suppl. 2), S150–162 [DOI] [PubMed] [Google Scholar]
- 48. Buhaescu I., Izzedine H. (2007) Mevalonate pathway: a review of clinical and therapeutical implications. Clin. Biochem. 40, 575–584 [DOI] [PubMed] [Google Scholar]
- 49. Penn R. B., Benovic J. L. (2008) Regulation of heterotrimeric G protein signaling in airway smooth muscle. Proc. Am. Thorac. Soc. 5, 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shore S. A., Moore P. E. (2003) Regulation of beta-adrenergic responses in airway smooth muscle. Respir. Physiol. Neurobiol. 137, 179–195 [DOI] [PubMed] [Google Scholar]
- 51. Bergstrom J. D., Bostedor R. G., Masarachia P. J., Reszka A. A., Rodan G. (2000) Alendronate is a specific, nanomolar inhibitor of farnesyl diphosphate synthase. Arch. Biochem. Biophys. 373, 231–241 [DOI] [PubMed] [Google Scholar]
- 52. Fisher J. E., Rogers M. J., Halasy J. M., Luckman S. P., Hughes D. E., Masarachia P. J., Wesolowski G., Russell R. G., Rodan G. A., Reszka A. A. (1999) Alendronate mechanism of action: geranylgeraniol, an intermediate in the mevalonate pathway, prevents inhibition of osteoclast formation, bone resorption, and kinase activation in vitro. Proc. Natl. Acad. Sci. U. S. A. 96, 133–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Billington C. K., Penn R. B. (2003) Signaling and regulation of G protein-coupled receptors in airway smooth muscle. Respir. Res. 4, 2. [PMC free article] [PubMed] [Google Scholar]
- 54. Ferguson S. S. (2001) Evolving concepts in G protein-coupled receptor endocytosis: the role in receptor desensitization and signaling. Pharmacol. Rev. 53, 1–24 [PubMed] [Google Scholar]
- 55. Shenoy S. K. (2007) Seven-transmembrane receptors and ubiquitination. Circ. Res. 100, 1142–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Xiao K., Shenoy S. K. Beta2-adrenergic receptor lysosomal trafficking is regulated by ubiquitination of lysyl residues in two distinct receptor domains. J. Biol. Chem. 286, 12785–12795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Insel P. A., Head B. P., Ostrom R. S., Patel H. H., Swaney J. S., Tang C. M., Roth D. M. (2005) Caveolae and lipid rafts: G protein-coupled receptor signaling microdomains in cardiac myocytes. Ann. N. Y. Acad. Sci. 1047, 166–172 [DOI] [PubMed] [Google Scholar]
- 58. Pontier S. M., Percherancier Y., Galandrin S., Breit A., Gales C., Bouvier M. (2008) Cholesterol-dependent separation of the beta2-adrenergic receptor from its partners determines signaling efficacy: insight into nanoscale organization of signal transduction. J. Biol. Chem. 283, 24659–24672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Xiang Y., Rybin V. O., Steinberg S. F., Kobilka B. (2002) Caveolar localization dictates physiologic signaling of beta 2-adrenoceptors in neonatal cardiac myocytes. J. Biol. Chem. 277, 34280–34286 [DOI] [PubMed] [Google Scholar]
- 60. Zhang F. L., Casey P. J. (1996) Protein prenylation: molecular mechanisms and functional consequences. Annu. Rev. Biochem. 65, 241–269 [DOI] [PubMed] [Google Scholar]
- 61. Pereira-Leal J. B., Hume A. N., Seabra M. C. (2001) Prenylation of Rab GTPases: molecular mechanisms and involvement in genetic disease. FEBS Lett. 498, 197–200 [DOI] [PubMed] [Google Scholar]
- 62. Moore R. H., Millman E. E., Alpizar-Foster E., Dai W., Knoll B. J. (2004) Rab11 regulates the recycling and lysosome targeting of beta2-adrenergic receptors. J. Cell Sci. 117, 3107–3117 [DOI] [PubMed] [Google Scholar]
- 63. Parent A., Hamelin E., Germain P., Parent J. L. (2009) Rab11 regulates the recycling of the beta2-adrenergic receptor through a direct interaction. Biochem. J. 418, 163–172 [DOI] [PubMed] [Google Scholar]
- 64. Seachrist J. L., Anborgh P. H., Ferguson S. S. (2000) beta 2-adrenergic receptor internalization, endosomal sorting, and plasma membrane recycling are regulated by rab GTPases. J. Biol. Chem. 275, 27221–27228 [DOI] [PubMed] [Google Scholar]
- 65. Seachrist J. L., Ferguson S. S. (2003) Regulation of G protein-coupled receptor endocytosis and trafficking by Rab GTPases. Life Sci. 74, 225–235 [DOI] [PubMed] [Google Scholar]
- 66. Chavrier P., Parton R. G., Hauri H. P., Simons K., Zerial M. (1990) Localization of low molecular weight GTP binding proteins to exocytic and endocytic compartments. Cell 62, 317–329 [DOI] [PubMed] [Google Scholar]
- 67. Konstantinopoulos P. A., Karamouzis M. V., Papavassiliou A. G. (2007) Post-translational modifications and regulation of the RAS superfamily of GTPases as anticancer targets. Nat. Rev. Drug Discov. 6, 541–555 [DOI] [PubMed] [Google Scholar]
- 68. Gomes A. Q., Ali B. R., Ramalho J. S., Godfrey R. F., Barral D. C., Hume A. N., Seabra M. C. (2003) Membrane targeting of Rab GTPases is influenced by the prenylation motif. Mol. Biol. Cell 14, 1882–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sen G., Wehrman T. S., Myers J. W., Blau H. M. (2004) Restriction enzyme-generated siRNA (REGS) vectors and libraries. Nat. Genet. 36, 183–189 [DOI] [PubMed] [Google Scholar]
- 70. Shirane D., Sugao K., Namiki S., Tanabe M., Iino M., Hirose K. (2004) Enzymatic production of RNAi libraries from cDNAs. Nat. Genet. 36, 190–196 [DOI] [PubMed] [Google Scholar]
- 71. Shtutman M., Maliyekkel A., Shao Y., Carmack C. S., Baig M., Warholic N., Cole K., Broude E. V., Harkins T. T., Ding Y., Roninson I. B. (2010) Function-based gene identification using enzymatically generated normalized shRNA library and massive parallel sequencing. Proc. Natl. Acad. Sci. U. S. A. 107, 7377–7382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Fellmann C., Zuber J., McJunkin K., Chang K., Malone C. D., Dickins R. A., Xu Q., Hengartner M. O., Elledge S. J., Hannon G. J., Lowe S. W. (2011) Functional identification of optimized RNAi triggers using a massively parallel sensor assay. Mol. Cell. 41, 733–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Williams B. R., Barber R., Clark R. B. (2000) Kinetic analysis of agonist-induced down-regulation of the beta(2)-adrenergic receptor in BEAS-2B cells reveals high- and low-affinity components. Mol. Pharmacol. 58, 421–430 [DOI] [PubMed] [Google Scholar]
- 74. Walker J. K., Penn R. B., Hanania N. A., Dickey B. F., Bond R. A. New perspectives regarding beta(2)-adrenoceptor ligands in the treatment of asthma. Br. J. Pharmacol. 163, 18–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Haney S., Hancox R. J. (2006) Recovery from bronchoconstriction and bronchodilator tolerance. Clin. Rev. Allergy Immunol. 31, 181–196 [DOI] [PubMed] [Google Scholar]
- 76. Haney S., Hancox R. J. (2007) Overcoming beta-agonist tolerance: high dose salbutamol and ipratropium bromide. Two randomised controlled trials. Respir. Res. 8, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Abramson M. J., Walters J., Walters E. H. (2003) Adverse effects of beta-agonists: are they clinically relevant? Am. J. Respir. Med. 2, 287–297 [DOI] [PubMed] [Google Scholar]
- 78. Sharma S., Litonjua A. A., Tantisira K. G., Fuhlbrigge A. L., Szefler S. J., Strunk R. C., Zeiger R. S., Murphy A. J., Weiss S. T. (2008) Clinical predictors and outcomes of consistent bronchodilator response in the childhood asthma management program. J. Allergy Clin. Immunol. 122, 921–928, e924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Tantisira K. G., Weiss S. T. (2005) The pharmacogenetics of asthma: an update. Curr. Opin. Mol. Ther. 7, 209–217 [PubMed] [Google Scholar]
- 80. Lipworth B. J. (1997) Airway subsensitivity with long-acting beta 2-agonists. Is there cause for concern? Drug Safety 16, 295–308 [DOI] [PubMed] [Google Scholar]
- 81. Nelson H. S., Dorinsky P. M. (2006) Safety of long-acting beta-agonists. Ann. Intern. Med. 145, 706; author reply 708–710 [DOI] [PubMed] [Google Scholar]
- 82. Nelson H. S., Weiss S. T., Bleecker E. R., Yancey S. W., Dorinsky P. M. (2006) The Salmeterol Multicenter Asthma Research Trial: a comparison of usual pharmacotherapy for asthma or usual pharmacotherapy plus salmeterol. Chest 129, 15–26 [DOI] [PubMed] [Google Scholar]
- 83. Cooper P. R., Lamb R., Day N. D., Branigan P. J., Kajekar R., San Mateo L., Hornby P. J., Panettieri R. A., Jr. (2009) TLR3 activation stimulates cytokine secretion without altering agonist-induced human small airway contraction or relaxation. Am. J. Physiol. Lung Cell. Mol. Physiol. 297, L530–L537 [DOI] [PubMed] [Google Scholar]
- 84. Shenoy S. K., Lefkowitz R. J. (2005) Receptor regulation: beta-arrestin moves up a notch. Nat. Cell Biol. 7, 1159–1161 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.