Abstract
GPR120 is a long-chain fatty acid receptor that stimulates incretin hormone release from colonic endocrine cells and is implicated in macrophage and adipocyte function. The functional consequences of long (L) and short (S) human GPR120 splice variants, which differ by insertion of 16 amino acids in the third intracellular loop, are currently unknown. Here we compare signaling and intracellular trafficking of GPR120S and GPR120L receptors, using calcium mobilization and dynamic mass redistribution (DMR) assays, together with quantitative imaging measurements of β-arrestin2 association and receptor internalization. FLAG- or SNAP-tagged GPR120S receptors elicited both intracellular calcium mobilization and DMR responses in human embryonic kidney 293 cells, when stimulated with oleic acid, myristic acid, or the agonist 4-[[(3-phenoxyphenyl)methyl]amino]benzenepropanoic acid (GW9508). Responses were insensitive to pertussis toxin, but increases in intracellular calcium were attenuated by 2-aminoethoxydiphenyl borate, an inhibitor of store inositol trisphosphate receptors. Despite equivalent cell surface expression of SNAP-tagged GPR120L receptors, no specific calcium or DMR responses were observed in cells transfected with this isoform. However, agonist-stimulated GPR120S and GPR120L receptors both recruited β-arrestin2 and underwent robust internalization, with similar agonist potencies in each case. After oleic acid-induced internalization, neither GPR120 isoform recycled rapidly to the cell surface. In both cases, confocal microscopy indicated receptor targeting to lysosomal compartments. Thus, the third intracellular loop insertion in GPR120L prevents G protein-dependent intracellular calcium and DMR responses, but this receptor isoform remains functionally coupled to the β-arrestin pathway, providing one of the first examples of a native β-arrestin-biased receptor.
Introduction
It is now recognized that long-chain free fatty acid (FFA) nutrients exert important effects as signaling molecules, beyond the consequences of mitochondrial β-oxidation (Yaney and Corkey, 2003). As ligands for the transcription factor family of peroxisome proliferator-activated receptors (PPARs), they regulate expression of genes that influence metabolism (Varga et al., 2011). Two cell surface G protein-coupled receptors (GPCRs) are also capable of mediating rapid responses to long-chain (C12–C22) saturated and unsaturated FFAs (Stoddart et al., 2008; Talukdar et al., 2011). FFA1, also known as GPR40, is predominantly expressed in pancreatic β-cells (Briscoe et al., 2003; Itoh et al., 2003). It mediates short-term FFA effects in enhancing glucose-stimulated insulin secretion (Briscoe et al., 2003, 2006; Itoh et al., 2003; Steneberg et al., 2005) and may also have longer term actions that contribute to β-cell dysfunction (Steneberg et al., 2005; Talukdar et al., 2011).
The second GPCR, GPR120, also binds C14 to C18 saturated or C16 to 22 mono- and polyunsaturated long-chain FFAs (Hirasawa et al., 2005) but is not closely related in amino acid sequence to FFA1 (Fredriksson et al., 2003; Stoddart et al., 2008). This receptor is a nutrient sensor on colonic L-type enteroendocrine cells, where its stimulation by luminal FFAs releases incretins such as glucagon-like peptide-1 and cholecystokinin into the circulation (Hirasawa et al., 2005; Tanaka et al., 2008), indirectly leading to increased insulin secretion. FFA1, which is coexpressed in similar cells, may contribute to these effects (Edfalk et al., 2008), and both FFA1 and GPR120 also act as FFA detectors in taste buds (Cartoni et al., 2010). However, independent effects of GPR120 have been identified in adipocytes and macrophages (Gotoh et al., 2007; Oh et al., 2010). In adipocytes, GPR120 activation increased glucose uptake and adipogenesis and GPR120 signaling in macrophages exerted anti-inflammatory actions (Gotoh et al., 2007; Oh et al., 2010). Similar in vivo dual effects of plasma ω-3 FFAs, acting via GPR120, were confirmed through comparison of wild-type and GPR120 knockout mice (Oh et al., 2010). Thus, as novel diabetes treatments, GPR120 agonists might be unique in improving insulin sensitivity while also reducing “metabolic” inflammation implicated in the disease pathogenesis (Talukdar et al., 2011).
Oh et al. (2010) also revealed that GPR120 responses in adipocytes and macrophages relied on distinct cell signaling mechanisms. In adipocytes, enhancement of glucose uptake was dependent on Gq/11 protein activation. However, the macrophage anti-inflammatory effect instead required the recruitment of β-arrestin adaptor proteins, which have classic roles in receptor desensitization and internalization and are also signaling scaffolds (Gurevich and Gurevich, 2006). Internalized GPR120-β-arrestin2 complexes prevented proinflammatory cascades by sequestering a component of the inflammatory pathway in an inactive form (Oh et al., 2010). This functional requirement for different GPR120 signaling cascades raises questions about whether they may be activated selectively by agonist-GPR120 complexes in different cell types.
It is therefore intriguing that human GPR120 exists in two splice variants (Moore et al., 2009), which, for example, are coexpressed in colonic endocrine cells (Galindo et al., 2012). The short isoform (GPR120S) contains 361 residues, whereas the long isoform (GPR120L) contains 16 additional residues between positions 231 and 247 in intracellular loop 3 (ICL3) (Fig. 1). For rhodopsin-like GPCRs, ICL3 is a critical cytoplasmic element of the receptor involved in both G protein and β-arrestin recognition (Gurevich and Gurevich, 2006; Rosenbaum et al., 2009), and ICL3 insertion in GPR120L could result in significant changes in receptor signaling properties. Investigations of GPR120L pharmacology have in general required overexpression of promiscuous or chimeric Gα subunits (Hirasawa et al., 2005; Galindo et al., 2012). In the absence of this method of facilitating G protein coupling, most recent studies have focused on human and rodent equivalents of GPR120S (Tanaka et al., 2008; Moore et al., 2009; Oh et al., 2010; Galindo et al., 2012).
Fig. 1.
The amino acid sequence of human GPR120S, also indicating the position of the additional 16 residues found only in ICL3 of the long isoform GPR120L (*). Putative N-linked glycosylation (white on black N) and palmitoylation sites (white on black C) are shown, together with the two basic Arg residues (black on gray R) mutated in this study.
Here we compare GPR120S and GPR120L receptors in the same defined recombinant system. We demonstrate that in human embryonic kidney (HEK) 293 cells, GPR120L is impaired in G protein coupling compared with GPR120S, in response to FFAs and the synthetic agonist 4-[[(3-phenoxyphenyl)methyl]amino]benzenepropanoic acid (GW9508) (Briscoe et al., 2006), as assessed in Ca2+ signaling and label-free dynamic mass redistribution (DMR) assays (Schröder et al., 2010). However, by using quantitative imaging analysis of β-arrestin recruitment and receptor internalization (Kilpatrick et al., 2010), we show that both GPR120 isoforms efficiently engage the β-arrestin pathway and exhibit intracellular trafficking similar to that of lysosomes. Thus, alternative splicing generates a GPR120 isoform with signaling specificity tailored toward arrestin-dependent pathways.
Materials and Methods
Materials.
FFAs were purchased from Sigma-Aldrich (Poole, UK) and were stored as 100 mM stock aliquots in DMSO. From these aliquots, fresh 1.8 mM experimental stocks were prepared daily in the appropriate media. GW9508 was from Tocris Bioscience (Bristol, UK), and pertussis toxin (PTX) was from Calbiochem (La Jolla, CA). Cell culture media and selection antibiotics were from Sigma-Aldrich, Lonza Wokingham Ltd. (Wokingham, Berkshire, UK), or Invitrogen (Paisley, UK). Molecular biology reagents were from Sigma-Aldrich, Promega (Southampton, UK), New England Biolabs (Hitchin, UK), or Fermentas (St. Leon-Rot, Germany).
Molecular Biology.
GPR120S (GenBank accession number BC101175) and GPR120L (GenBank accession number NM_181745) receptor cDNAs were inserted between restriction sites BamHI (5′) and XhoI (3′) in the tetracycline repressor (TR)-regulated expression vector pcDNA4/TO (Invitrogen). cDNA sequences lacked the start Met codon and included the stop codon when appropriate. Insertion of a Kozak element and appropriate coding sequences between KpnI and BamHI into GPR120 vectors generated receptors tagged at the N terminus with 1) the FLAG epitope tag (DYKDDDDK) or 2) a 5HT3 receptor-derived signal sequence followed by the SNAP tag (New England Biolabs). For some receptor constructs, the 3′ STOP codon was removed to also allow C-terminal receptor fusion (between XhoI/XbaI, linker LE) to green fluorescent protein (GFP) containing superfolder mutations S30R and Y39N (Pédelacq et al., 2006). For bimolecular fluorescence complementation (BiFC) experiments, FLAG-tagged GPR120 cDNAs lacking STOP codons were transferred to pCMV FLAG (Stratagene, Edinburgh, UK), and a venus yellow fluorescent protein fragment 155 to 238 (Yc) was inserted in frame between 3′ XhoI and ApaI sites (linker LE), generating GPR120S-Yc and GPR120L-Yc fusion proteins. R99A (2.64 in Ballesteros-Weinstein numbering) and R178A (4.65) mutations in GPR120S were generated using QuikChange II site-directed mutagenesis (Stratagene). The identities of all constructs were confirmed by sequencing.
Cell Culture.
Cells were maintained in a humidified environment at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS) and passaged when confluent by trypsinization (0.25% w/v in Versene). Transfections into HEK293 cells were performed using Lipofectamine (Invitrogen) according to the manufacturer's instructions. For inducible expression, GPR120 receptor cDNAs in pcDNA4/TO were introduced into 293TR cells (Invitrogen, which are transfected with the TR protein to allow inducible expression), and mixed population stable lines were selected by resistance to blasticidin (TR vector, 5 μg/ml; Invitrogen) and phleomycin (Zeocin; receptor plasmid, 20 μg/ml; Invitrogen). BiFC cell lines were generated by selection of GPR120-Yc-transfected cells (G418; 0.8 mg/ml) on a clonal phleomycin-resistant HEK293 cell line expressing β-arrestin2-venus Yn (2–173) that we have described previously (Kilpatrick et al., 2010). Routine culturing of stable transfected cell lines included lower media concentrations of blasticidin (2.5 μg/ml), phleomycin (5 μg/ml), or G418 (0.2 mg/ml) to maintain selection pressure. For receptor-inducible expression, cells were seeded onto experimental plates or coverslips 48 h before experiments, and, as appropriate, DMEM containing 1 μg/ml tetracycline was added 24 h later. Otherwise, cells were seeded on the day before experiments.
Fluo4 Calcium Mobilization Assay.
Confluent cell layers on poly-d-lysine-coated 96-well clear-bottomed black plates (Costar 3904; Thermo Fisher Scientific, Loughborough, UK) were loaded with the intracellular calcium indicator Fluo4. In brief, loading was performed in DMEM/10% FBS containing 2.5 mM probenecid (Sigma-Aldrich), 1.5 μM Fluo4 AM (Invitrogen), and 0.023% Pluronic acid F127 (45 min at 37°C; Invitrogen). Cells were washed and then incubated in HEPES-buffered saline solution (HBSS; 25 mM HEPES, 10 mM glucose, 146 mM NaCl, 5 mM KCl, 1 mM MgSO4, 2 mM sodium pyruvate, and 1.3 mM CaCl2, pH 7.45) containing 2.5 mM probenecid and 0.02% FFA-free bovine serum albumin (BSA) (20 min at 37°C; Sigma). Where appropriate, inhibitors were added during this equilibration step. Fluo4 fluorescence was measured on a FlexStation II (Molecular Devices, Sunnyvale, CA) for 165 s, with agonist additions (in HBSS/0.02% BSA, maximum 0.3% DMSO) made from a compound plate at 15 s.
Automated Imaging of Receptor Internalization and β-Arrestin2 Recruitment.
Cells were used at 80 to 95% confluence on poly-d-lysine-coated 96-well thin-bottomed plates (Greiner 655090; Greiner Bio-One, Stonehouse, UK). For receptor imaging, SNAP-GPR120 cell lines were labeled in DMEM-10% FBS containing 0.1 μM SNAP-Surface BG-AF488 (20 min at 37°C, unless otherwise stated; New England Biolabs). Cells were washed and then incubated in HBSS-0.2% BSA containing FFAs and synthetic agonists at the indicated concentrations (30 min at 37°C, unless otherwise stated). For measurement of receptor-arrestin BiFC, cells were directly treated with agonists in HBSS-0.2% BSA at 37°C at the times and concentrations indicated (for more details, see Kilpatrick et al., 2010).
Internalization and arrestin BiFC assays were terminated by fixation in 3% paraformaldehyde in phosphate-buffered saline (10 min at room temperature), followed after phosphate-buffered saline washes by H33342 (5 μM; 20 min) treatment to stain cell nuclei. Images of BiFC fluorescence or SNAP-labeled receptors were acquired from four central sites per well on the IX Ultra confocal plate reader (Molecular Devices), using a Plan Fluor 40× NA0.6 extra long working distance objective, with 405 nm (H33342) and 488 nm (BG-AF488 and BiFC) laser excitation (Kilpatrick et al., 2010). Supplemental Figure 1 also shows similar images acquired from the IX Microplate Reader (Molecular Devices) using the same protocol.
DMR.
Cells were seeded on fibronectin-coated 384-well plates (Corning Life Sciences, Corning, NY). Two hours before the experiment, plates underwent a 5-wash buffer exchange (HBSS-0.02% BSA-0.5% DMSO) and were left to equilibrate (2 h at 26°C) in the Corning Epic Biosensor. If inhibitors were used, these were added 1 h after buffer exchange, and plates were left to equilibrate for 1 h. Agonists were added in quadruplicate and readings were taken every 40 s for 50 min.
Receptor Imaging Using Confocal Microscopy.
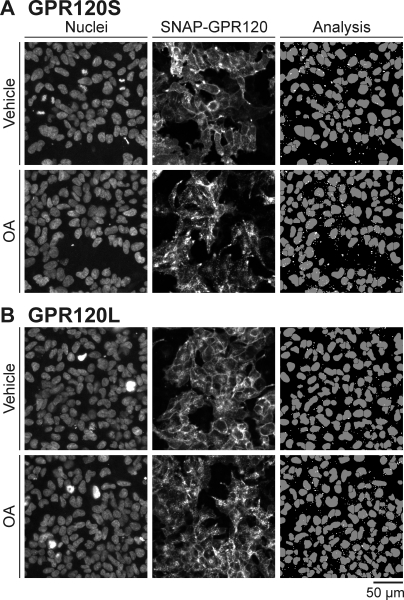
Cells at 50 to 70% confluence on poly-d-lysine-coated eight-well plates (Nunc Lab-Tek; Thermo Fisher Scientific) were labeled with 0.5 μM SNAP-Surface BG-AF488 as described previously, followed by treatment with vehicle or 300 μM oleic acid in HBSS-0.1% FFA-free BSA (60 min at 37°C). In addition, incubation media contained either 10 nM LysoTracker Red (60 min at 37°C; Invitrogen) throughout, or 5 μg/ml transferrin-AF633 (Invitrogen) was added 15 min before imaging. Live cell images were taken using a Zeiss LSM 510 laser scanning microscope (Carl Zeiss, Welwyn, UK) using a 63× Plan-Apochromat oil objective, numerical aperture 1.4, with argon 488 nm (BG-AF488) and helium-neon 543 nm (LysoTracker Red) and 633 nm (transferrin-AF647) laser lines for excitation. Images were taken sequentially with band passes of 505 to 530 nm (BG-AF488) and 560 to 615 nm (LysoTracker Red) and a long bandpass of 650 nm (transferrin-AF647). Pinhole diameter was set to 1 Airy unit for the longest wavelength, with the detector gain and amplifier offset adjusted to ensure that images were not saturated; equivalent settings were maintained for control and agonist-treated cell images.
Data Analysis.
Calcium and DMR measurement data were analyzed in terms of the peak agonist response (maximum − minimum), with data points from individual experiments performed in triplicate or quadruplicate. Data were normalized (as a percentage of the 300 μM oleic acid response) when comparisons were made between data sets from the same cell line and experiment set. For quantitative analysis of internalization and receptor-arrestin BiFC, a granularity algorithm (MetaXpress 2.0; Molecular Devices) detected vesicles of a specified diameter range in images from the IX Ultra confocal plate reader; the threshold intensity for classification was set with reference to the plate positive (300 μM oleic acid) and negative controls. Several parameters normalized to cell count (number of nuclei stained by H33342) were obtained (vesicle count, area, and intensity per cell) for triplicate wells (12 sites per data group). These all provided similar results, and, in the figures, the normalized data from vesicle average intensity per cell are presented throughout. Comparisons of SNAP-GPR120S and SNAP-GPR120L expression were estimated by analysis of images acquired under identical settings and from the same plate. The SNAPreceptor images (vehicle-treated, from time course internalization experiments) were processed by the multiwavelength cell scoring algorithm (MetaXpress 2.0). Segmentation defined regions of the image covered by individual cells, on the basis of both the location of H33342-stained nuclei and boundaries of SNAPreceptor fluorescence staining. The mean pixel intensity within these regions (covering both plasma membrane and cytoplasm) was then averaged for all cells in each image. This measure of cellular fluorescence intensity was pooled between experiments (mean ± S.E.M.) and used as an indicator of expression levels.
Concentration-response curves were fitted to the pooled data points (mean ± S.E.M.) by nonlinear least-squares regression (Prism 5.02; GraphPad Software, Inc., San Diego, CA), and EC50 values are quoted if a maximum agonist response could be defined. Time course data are described by single-phase association kinetics, after a manually adjusted latency period where appropriate. This latency accounted for the observation that FFA internalization and arrestin recruitment measurements showed a short delay (typically 2–5 min) after agonist addition before any response was observed. Two-way analysis of variance and Bonferroni post-tests were used to define statistical significance between concentration-response relationships (GraphPad Prism).
Results
Only the Short GPR120 Splice Variant Is Coupled to Calcium Mobilization in HEK293 Cells.
We initially investigated signaling by FLAG-tagged short and long GPR120 isoforms in 293TR cells, in which receptor expression was controlled by a tetracycline-inducible promoter. After tetracycline treatment, GPR120-mediated responses were measured as peak increases in the intracellular Ca2+ concentration (Fig. 2). The unsaturated C18:1 FFA oleic acid (OA), the saturated C14:0 FFA myristic acid (Myr), and the synthetic agonist GW9508 (Briscoe et al., 2006) were all full agonists in FLAG-GPR120S-expressing cells (Fig. 2A), with respective pEC50 values of 4.7 ± 0.3, 4.2 ± 0.3, and 5.5 ± 0.3 (n = 4–5). In the absence of tetracycline induction, no effects of OA, at up to 300 μM, were observed (Fig. 2A), indicating that FFA responses were mediated by the transfected GPR120S receptor. Surprisingly, however, no Ca2+ signals were obtained from FLAG-GPR120L receptors, expressed in the same system, to OA, Myr, or GW9508 (Fig. 2B).
Fig. 2.
Intracellular calcium mobilization in response to GPR120S or GPR120L receptor activation. Peak intracellular calcium responses to increasing concentrations of OA, Myr, and GW9508 were measured from fluorescence changes in Fluo4 indicator-loaded 293TR cells expressing FLAG-GPR120S (A), FLAG-GPR120L (B), SNAP-GPR120S (C), and SNAP-GPR120L receptors (D). OA responses were also assessed in cells without prior tetracycline induction of receptor expression (No induction). Vehicle was medium containing 0.3% DMSO. The curve fits for FLAG- or SNAP-GPR120S GW9508 responses (from A or C) are also reproduced as dotted lines for comparison in the GPR120L graphs (B and D). Pooled data represent the mean ± S.E.M. of 3 to 13 experiments performed in triplicate, which yielded pEC50 values quoted in the text. E, representative images of the cell surface expression of SNAP-GPR120S and SNAP-GPR120L receptors in induced 293TR cells (acquired on the IX Ultra plate reader after labeling with membrane-impermeant SNAP-AF488 dye, as Fig. 6). The average cellular fluorescence intensity was calculated from these images (see Data Analysis) as a guide to the relative expression levels of the two isoforms, and the resulting pooled data (n = 6) is shown in histogram F. *, P < 0.05 (unpaired Student's t test). RFU, relative fluorescence units.
To confirm cell surface expression of the GPR120 isoforms and to also study their intracellular trafficking, the N terminal FLAG epitope on the receptors was replaced with a signal sequence (from the 5HT3 receptor) followed by the SNAP-tag protein. The SNAP-tag is an 18-kDa protein based on the enzyme O6-alkylguanine DNA alkyltransferase, which can be covalently labeled by a range of fluorophores conjugated to its BG substrate (Keppler et al., 2003). We first examined the expression of SNAP-GPR120S receptors that were also fused at the C terminus to GFP. Monitoring GFP fluorescence in induced 293TR SNAP-GPR120S-GFP cells demonstrated that the receptors were predominantly targeted to the plasma membrane. Thus, addition of the N-terminal SNAP-tag did not adversely affect receptor processing and maturation (Supplemental Fig. 1). Plasma membrane and some intracellular GFP immunofluorescence also colocalized with SNAP-tag labeling by membrane impermeant BG conjugated fluorophores, indicating cell surface expression with a low level of constitutive internalization (Supplemental Fig. 1). The identification of the SNAP-tagged receptors by BG fluorophores (0.1–1 μM) was specific, with little labeling in cells that had not been exposed to tetracycline. In these cells, SNAP-GPR120S activation initiated transient Ca2+ responses with pEC50 values for OA (4.6 ± 0.2), Myr (4.2 ± 0.3), and GW9508 (5.5 ± 0.2) equivalent to those observed for the FLAG-GPR120S receptor (Fig. 2C; for examples of time courses, see Supplemental Fig. 2). However, as before, no calcium responses were observed in HEK293TR SNAP-GPR120L cells (Fig. 2D). Similar plasma membrane localization and overall expression levels for SNAP-GPR120S and SNAP-GPR120L receptors were observed using plate reader imaging, after the transfected 293TR cells were labeled with SNAP fluorophore (Fig. 2, E and F; see also Fig. 6).
Fig. 6.
Quantitative analysis of agonist-induced SNAP-GPR120S (A) and SNAP-GPR120L (B) internalization. An example of an experiment (n = 4) is illustrated in which 293TR cells induced to express each receptor isoform were compared on the same analysis plate. Cells were labeled with 0.1 μM membrane-impermeant SNAP-Surface BG-AF488 (30 min, 37°C), before stimulation with vehicle or 300 μM OA (60 min, 37°C). After fixation, cell nuclei were labeled with H33342. Representative images of H33342 and receptor labeling (from at least four experiments) are illustrated, representing a magnified portion (25% area) of original images acquired and analyzed by the IX Ultra plate reader. A granularity algorithm (right panels) identified punctate receptor vesicles 2 to 5 μm in diameter (white dots) and the cell nuclei (gray). This analysis quantified the extent of receptor internalization as described under Materials and Methods.
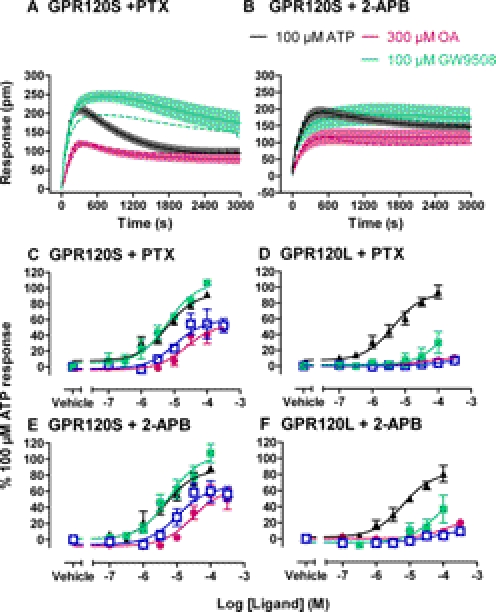
Coupling of SNAP-GPR120S to intracellular Ca2+ elevations seemed to be independent of Gi/o proteins, because agonist responses were unaffected by an 18-h pretreatment with PTX (100 nM) (Fig. 3A; Supplemental Fig. 2). The effect of 2-aminoethoxydiphenyl borate (2-APB) (50 μM) as an inhibitor of intracellular calcium release at a concentration greater than its IC50 value (42 μM) for store IP3 receptors but below that reported to elicit nonselective rises in cytoplasmic Ca2+ (>90 μM) was also examined (Maruyama et al., 1997). 2-APB significantly depressed maximal Ca2+ elevations to OA and GW9508 (Fig. 3B) and increased the transient nature of the responses measured (Supplemental Fig. 2).
Fig. 3.
The effects of PTX, 2-APB, and R2.64A or R4.65A mutation on GPR120S-stimulated intracellular calcium mobilization. Tetracycline-induced 293TR cells expressing SNAP-GPR120S were pretreated with 100 nM PTX for 18 h (A) or 50 μM 2-APB for 20 min (B), before Fluo4 loading and stimulation with OA or GW9508 (pooled data n = 4). In C, agonist responses were compared with cells expressing SNAP-GPR120S R2.64A or SNAP-GPR120SA R4.65A receptors (n = 4). The insets show receptor labeling obtained with membrane-impermeant SNAP label BG-AF488, imaged with the IX Ultra plate reader (see legend to Fig. 6 for full details). Vehicle or 300 μM OA treatment was for 30 min in these representative images.
Two basic amino acids within the transmembrane domains (TMD) of GPR120S were also investigated as potential counterions that might interact with the FFA and GW9508 carboxylate groups. Arg 2.64 (Ballesteros-Weinstein numbering or R99), at the top of TMD II (Fig. 1), has previously been implicated in agonist binding (Suzuki et al., 2008), and R2.64A substitution in the SNAP-GPR120S abolished both OA- and GW9508-mediated calcium responses (Fig. 3C). However, R2.64A also reduced SNAP-GPR120S receptor expression in 293TR cells, and surface SNAP-GPR120S R2.64A receptors labeled with cell-impermeant BG fluorophores underwent substantial basal internalization in otherwise unstimulated cells (Fig. 3). Alanine mutation of Arg 4.65 (R178), at the top of TMD IV (Fig. 1), did not alter SNAP-GPR120S expression or Ca2+ responses to GW9508 or OA (Fig. 3C).
Label-Free DMR Measurements Also Reflect Differences in Signaling between GPR120S and GPR120L Isoforms.
We used DMR technology (Corning EPIC biosensor) to probe integrated functional responses to the GPR120 receptor splice variants in the same cells. This label-free method measures signaling-induced changes in the distribution of proteins and other constituents within cells grown on a specialized optical grating biosensor. Such changes produce small alterations in refractive index and thus in the wavelength of reflected light after broad-spectrum polarized illumination (Schröder et al., 2010). In addition to the amplitude of DMR measurements, their profiles can indicate different types of agonist-stimulated GPCR signaling events, in particular by discriminating different Gα protein pathways (Schröder et al., 2010).
Figure 4 summarizes DMR responses obtained from 293TR cells expressing SNAP-GPR120S or SNAP-GPR120L receptors in comparison with those of nontransfected cells or cells expressing the SNAP-GPR120S R2.64A mutant. ATP responses, mediated by endogenous P2Y receptors (predominantly Gq/11 coupled) (Wirkner et al., 2004), were used as consistent positive controls. In tet-treated 293TR SNAP-GPR120S cells, the agonists OA, Myr, and GW9508 all produced monophasic positive deflections in reflected wavelength (Fig. 4A), with potencies equivalent to that of the Ca2+ mobilization assay (Fig. 4B; Table 1). Maximal responses to GW9508 were somewhat greater than those for the FFA agonists, although not significantly so. The R2.64A mutation substantially reduced agonist responses, without altering their relative potencies (Fig. 4C), and only small agonist responses (relative to ATP) were observed in SNAP-GPR120L cells, although these were greater than those for the vehicle controls (Fig. 4, D and E; Table 1). Similar low-potency effects of GW9508 and OA were observed in nontransfected 293TR cells (Fig. 4F). Neither PTX (100 nM) nor 2-APB (50 μM) preincubation altered DMR responses to ATP or GPR120 FFA agonists in either GPR120S- or GPR120L-expressing cells (Fig. 5, Table 2). However, a small (3-fold) but significant reduction in GW9508 potency was observed after treatment with either inhibitor (Table 2).
Fig. 4.
GPR120-mediated DMR responses. The effects of OA, Myr, and GW9508 in comparison with those of the endogenous agonist ATP were measured in cells expressing SNAP-GPR120S (A and C), SNAP-GPR120L (B and D), SNAP-GPR120S R2.64A (E), and nontransfected 293TR cells (F). A and B show 50-min time courses of the DMR changes observed (26°C) for agonists added at the indicated concentration. Concentration-response curves (n = 6–9, in quadruplicate) constructed from the pooled data are shown in C to F, yielding pEC50 and Rmax values quoted in Table 1.
TABLE 1.
Summary of DMR response data for 293TR cells expressing SNAP-GPR120S or SNAPGPR120L receptors
pEC50 and Rmax values (300 μM FFA and 100 μM GW9508 normalized to 100 μM ATP) were obtained from the pooled concentration response curves in Fig. 7. Data represent the mean ± S.E.M. of six to nine experiments performed in quadruplicate.
| Agonist | 293TR Nontransfected |
293TR SNAP-GPR120S |
293TR SNAP-GPR120L |
293TR SNAP GPR120S R2.64A |
||||
|---|---|---|---|---|---|---|---|---|
| pEC50 | Rmax | pEC50 | Rmax | pEC50 | Rmax | pEC50 | Rmax | |
| % | % | % | % | |||||
| ATP | 5.8 ± 0.2 | 100 | 5.0 ± 0.1 | 100 | 5.2 ± 0.1 | 100 | 5.4 ± 0.1 | 100 |
| OA | 17 ± 4 | 4.6 ± 0.2 | 58 ± 6 | 21 ± 5 | 4.5 ± 0.3 | 20 ± 4 | ||
| Myr | 2 ± 1 | 4.9 ± 0.2 | 65 ± 5 | 16 ± 4 | 5.1 ± 0.2 | 19 ± 3 | ||
| GW9508 | 6 ± 1 | 5.7 ± 0.1 | 104 ± 6 | 27 ± 12 | 5.4 ± 0.2 | 20 ± 2 | ||
Fig. 5.
PTX or 2-APB pretreatment does not alter DMR responses. 293TR SNAP-GPR120S or SNAP-GPR120L cells were pretreated with PTX (100 nM) (A, C, and D) or 2-APB (50 mM (B, E, and F), and peak DMR changes were then measured to OA, Myr, and GW9508, together with the reference agonist ATP. The 50-min time courses in A and B are shown in comparison to the equivalent OA and GW 9508 data in the absence of inhibitors (dotted lines, shown in full in Fig. 4A). Pooled data represent the mean ± S.E.M. of three to six experiments performed in quadruplicate, with agonist potency and maximal responses indicated in Table 2.
TABLE 2.
Influence of 2-APB or PTX preincubation on GPR120 DMR responses
Pooled concentration response curves (Fig. 8, n = 3) yielded pEC50 and Rmax values (300 μM FFA and 100 μM GW9508 normalized to 100 μM ATP in control cells without inhibitor). Data represent the means ± S.E.M. of two to nine experiments performed in quadruplicate.
| Agonist | + 50 μM 2-APB |
+100 nM PTX |
||||||
|---|---|---|---|---|---|---|---|---|
| 293TR SNAP-GPR120S |
293TR SNAP-GPR120L |
293TR SNAP-GPR120S |
293TR SNAP GPR120L |
|||||
| pEC50 | Rmax | pEC50 | Rmax | pEC50 | Rmax | pEC50 | Rmax | |
| % | % | % | % | |||||
| ATP | 5.3 ± 0.2 | 89 ± 5 | 5.2 ± 0.1 | 81 ± 10 | 5.2 ± 0.1 | 92 ± 2 | 5.4 ± 0.1 | 94 ± 8 |
| OA | 4.5 ± 0.2 | 50 ± 13 | 20 ± 4 | 4.6 ± 0.2 | 49 ± 3 | 10 ± 2 | ||
| Myr | 5.0 ± 0.2 | 57 ± 9 | 9 ± 5 | 4.8 ± 0.2 | 53 ± 9 | 7 ± 2 | ||
| GW9508 | 5.2 ± 0.2** | 107 ± 11 | 37 ± 17 | 5.2 ± 0.1* | 107 ± 5 | 30 ± 14 | ||
Both GPR120 Splice Variants Undergo Agonist-Stimulated Endocytosis.
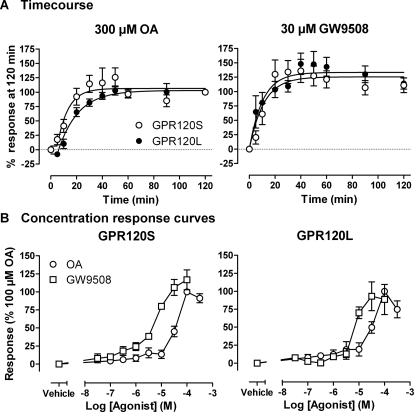
As a further alternative readout of GPR120 activation, SNAP-tagged receptor internalization in response to different agonists was quantified by automated imaging and analysis. Figure 6 shows representative images, acquired on an IX Ultra confocal plate reader, of the localization of BG-AF488-labeled SNAP-GPR120 receptors. In contrast to the Ca2+ and DMR assay findings, endocytosis of both SNAP-GPR120S and SNAP-GPR120L receptors was clearly observed after 300 μM OA pretreatment for 30 min. The fluorescence intensity within punctate compartments, predominantly intracellular, was quantified by granularity analysis to provide a measure of internalization on a cell-by-cell basis. As Fig. 7 indicates, 100 μM GW9508 stimulated internalization of SNAP-GPR120L (t1/2 21 ± 3 min, n = 4; after 2 min latency) and SNAP-GPR120S (t1/2 13 ± 1 min, n = 4) (Fig. 7A) receptors; 300 μM OA responses increased internalization with t1/2 of 20 ± 6 min for 293TR SNAP-GPR120S cells and 21 ± 3 min for SNAP-GPR120L cells (both n = 4, calculated after a 5-min latency) (Fig. 7A). Concentration-response relationships, after a 60-min agonist treatment (Fig. 7B), demonstrated a threshold of 30 μM for both OA- and Myr-stimulated internalization, and an approximate 3- to 10-fold increased potency for GW9508 (pEC50 4.4 ± 0.3 and 4.0 ± 0.4 for GPR120S and GPR120L, respectively; n = 4) compared with both FFAs. Internalization of SNAP-GPR120S receptors in response to GW9508 or OA was unaltered by PTX pretreatment or by R4.65A mutation (both n = 4; data not shown). We also observed that TZDs elicited SNAP-GPR120S receptor responses (10–30 μM threshold concentrations) in both intracellular calcium mobilization (rosiglitazone, ciglitazone, and troglitazone) and endocytosis assays (ciglitazone and troglitazone). A fourth TZD, pioglitazone, was only active at 100 μM in the calcium mobilization assay (Fig. 8).
Fig. 7.
Agonist-stimulated internalization of GPR120S and GPR120L receptors. Endocytosis was assessed by granularity analysis of SNAP-tagged receptors expressed in 293TR cells and imaged using the IX Ultra plate reader, as described in the legend to Fig. 6. A, internalization time courses for 300 μM OA or 100 μM GW9508, pooled from experiments in which 293TR GPR120S and GPR120L cells were simultaneously treated with agonist on the same plate (n = 4). B, a 60-min agonist treatment period, after SNAP fluorophore labeling, was used to construct the concentration-response relationships to OA, Myr, and GW9508. In each case, data represent the mean ± S.E.M. of 4 experiments performed in triplicate.
Fig. 8.
GPR120S receptor responses to TZD ligands. Pooled data illustrate peak intracellular calcium mobilization responses (A, n = 4–5) or internalization after 60 min agonist treatment (B, n = 4, measured as in the legend to Fig. 6) in tetracycline-induced 293TR SNAP-120S cells. In each case, responses were normalized to 300 μM OA reference controls (100%). Troglitazone and pioglitazone caused changes in cell morphology at 100 μM, and so this concentration has been excluded from the concentration-response curves.
Both GPR120S and GPR120L Receptors Recruit β-Arrestin2.
GPCR endocytosis is often a downstream indicator of their interaction with β-arrestins. As a consequence, the molecular association between GPR120 and β-arrestin2 was measured directly using a BiFC assay that we and others have described previously (Auld et al., 2006; Kilpatrick et al., 2010). Its basis is that receptor and arrestin interaction brings complementary Yn and Yc fragments of yellow fluorescent protein together to allow refolding and reconstitution of a fluorescent signal. In HEK293 cells stably cotransfected with GPR120-Yc and β-arrestin2-Yn (GPR120 A2), OA or GW9508 stimulation resulted in the development of receptor-arrestin BiFC in intracellular compartments (Supplemental Fig. 3), and the resulting fluorescence was then quantified by granularity analysis (Fig. 9). Time courses for BiFC development in GPR120S A2 and GPR120L A2 cells were comparable in response to 30 μM GW9508 [t1/2 7 ± 1 and 8 ± 1 min, respectively (n = 5–6), no latency] (Fig. 9, A and B). Somewhat slower development of the GPR120L A2 300 μM OA response was observed [t1/2 18 ± 2 min (n = 5) compared with 9 ± 1 min (n = 6) for GPR120S, both after a 5-min latency] (Fig. 9, A and B). Concentration-response relationships demonstrated agonist potencies for GPR120S recruitment of β-arrestin2 that were equivalent to intracellular Ca2+ responses (GW9508 pEC50 5.1 ± 0.1 and OA pEC50 4.4 ± 0.1; n = 4) (Fig. 9C). Moreover, GW9508 (pEC50 5.1 ± 0.2, n = 4) and OA (pEC50 4.6 ± 0.2, n = 4) also stimulated GPR120L β-arrestin2 interaction with potencies similar to those for GPR120S in this assay (Fig. 9D).
Fig. 9.
Comparison of GPR120S and GPR120L association with β-arrestin2 using bimolecular fluorescence complementation. Stable transfected HEK293T cells coexpressed β-arrestin2-Yn and either GPR120S-Yc or GPR120L-Yc. This allowed agonist-stimulated receptor-arrestin BiFC to be quantified in intracellular compartments by granularity analysis of the IX Ultra plate reader images (Supplemental Fig. 3). A, response time courses to 30 μM GW9508 or 300 μM OA (n = 5–6), over 120 min at 37°C. B, pooled OA and GW9508 concentration-response curves (30 min treatment, n = 4) with pEC50 values quoted in the text.
GPR120 Receptors Are Internalized to Nonrecycling Pathways Associated with Acidic Late Endosomes and Lysosomes.
The fate of internalized GPR120 receptors was investigated by confocal microscopy. After a 60-min vehicle or 300 μM OA treatment, SNAP-labeled receptors were imaged in live cells in the presence of either 10 nM LysoTracker Red or 5 μg/ml transferrin-AF647. LysoTracker Red is a marker of acidic organelles such as lysosomes, whereas transferrin-labeled receptors constitutively engage the clathrin-mediated endocytosis and recycling pathway. As illustrated in the representative images presented in Fig. 10, agonist-stimulated SNAP-GPR120S and SNAP-GPR120L receptors showed extensive colocalization with LysoTracker Red-positive compartments and also some distribution to compartments labeled by transferrin.
Fig. 10.
Colocalization of GPR120 with markers for the clathrin-mediated endocytic and late endosomal/lysosomal pathways after internalization. 293TR cells expressing SNAP-GPR120S (A) or SNAP-GPR120L (B) receptors were visualized by live cell confocal microscopy after labeling with SNAP-Surface AF488 (Receptor) and incubation with vehicle or 300 μM OA (37°C, 60 min). Simultaneous images of the markers transferrin-AF633 (Tf) (5 μg/ml), or LysoTracker Red (Lyso) (10 nM) were acquired. Representative images of four independent experiments are shown, together with the overlay of the two channels. White arrows highlight examples of colocalized receptor and marker fluorescence (yellow).
The pattern of distribution suggested a nonrecycling trafficking phenotype for both receptors, and this was investigated using internalization analysis on the plate reader. Cells were stimulated for 30 min with 300 μM OA, followed by two rinse washes and a final wash step of 5 to 60 min in HBSS containing an increased concentration (0.1%) of fatty acid-free BSA. Figure 11 shows that there was a less than 25% reduction in OA-stimulated internalization over the 60-min wash period, for either 293TR SNAP-GPR120S or 293TR SNAP-GPR120L cells.
Fig. 11.
Internalized GPR120 receptors do not recycle rapidly to the cell surface. SNAP-GPR120S (A) and SNAP-GPR120L (B) recycling was quantified by granularity analysis of IX Ultra plate reader images, as for Fig. 7. 293TR SNAP-GPR120S or SNAP-GPR120L cells were labeled with 0.1 μM SNAP-Surface BG-AF488, followed by addition of vehicle or 300 μM OA (in HBSS-0.02% BSA) for 30 min at 37°C. Cells were washed twice with HBSS-0.1% BSA and then were incubated in this medium for 5 to 60 min at 37°C (the “wash period”) before fixation. Pooled granularity data are normalized to 300 μM OA control responses without a wash step and represent the mean ± S.E.M. of five experiments performed in triplicate.
Discussion
GPR120 is a long-chain FFA receptor that regulates incretin hormone secretion and has important modulatory roles for adipocyte and macrophage function (Hirasawa et al., 2005; Oh et al., 2010; Talukdar et al., 2011). Thus, GPR120 synthetic ligands offer a multipronged approach to the treatment of obesity and type II diabetes. Achieving an understanding of the pharmacology of GPR120 is complicated by the existence of two isoforms in the coding region of the human receptor, generated by alternative splicing, which are not apparent in lower primates and rodents (Moore et al., 2009). This study provides the first direct comparison between these splice variants in the same defined cell system, using a variety of approaches to detect G protein- and β-arrestin-dependent events. We demonstrated that the insertion of 16 amino acids in the third intracellular loop of GPR120L (Fig. 1) results in a receptor that fails to activate G proteins in HEK293 cells, while preserving arrestin recruitment and downstream internalization and trafficking to lysosomal compartments. This characterization of a GPR120L as an arrestin-biased receptor has relevance for future studies on the basis of the recent critical role discovered for GPR120 arrestin-mediated pathways in immune cells (Oh et al., 2010).
GPR120S G Protein Coupling and Pharmacology.
GPR120S activation stimulated intracellular calcium mobilization and changes in DMR global cell responses through a PTX-insensitive pathway, indicating the involvement of Gq/11α proteins. The time profile observed for GPR120S DMR responses also showed a signature appropriate for this type of G protein (Schröder et al., 2010). The absence of a PTX effect on GPR120S DMR measurements (other than a small reduction in GW9508 potency) contrasts with results of studies using FFA1, in which the potential for Gi/oα as well as predominant Gq/11α coupling has been suggested (Itoh et al., 2003; Kotarsky et al., 2003), most convincingly by DMR (Schröder et al., 2010). However, PTX insensitivity is characteristic of GPR120 signaling (Hirasawa et al., 2005; Oh et al., 2010), and small interfering RNA knockdown of Gq/11α abolished GPR120-mediated glucose uptake in adipocytes (Oh et al., 2010). 2-APB had a partial effect on GPR120S Ca2+ responses, consistent with its potency for inositol 1,4,5-trisphosphate receptor inhibition (Maruyama et al., 1997). DMR measurements were largely unaffected, suggesting involvement of additional arms of the Gq/11-dependent pathway, for example, activation of protein kinase C.
The potencies of oleic acid and myristic acid and the synthetic agonist GW 9508 in the GPR120S calcium and DMR assays were consistent with previous reports (Hirasawa et al., 2005; Briscoe et al., 2006; Suzuki et al., 2008; Moore et al., 2009; Smith et al., 2009; Galindo et al., 2012) and indicated that receptor modification with FLAG or SNAP epitope tags did not alter agonist activity. A 3- to 10-fold lower potency of FFAs, compared with some studies (Hirasawa et al., 2005; Briscoe et al., 2006), is likely to result from our inclusion of BSA (3 μM) in all assay media, which aids fatty acid solubility and limits the use of DMSO as a solvent but also reduces FFA concentration (Hirasawa et al., 2005). An inducible expression system, as previously used for a number of studies on FFA1 (Smith et al., 2009), demonstrated the specificity of GPR120 agonist responses. Furthermore, we confirmed that mutation of TMD II Arg99 eliminated GPR120S receptor signaling but not substitution of the second basic residue (Arg178) located at the top of TMD IV. This may support a role for Arg99 in recognizing FFA carboxylate anions in an orthosteric binding site otherwise poorly defined by mutagenesis (Suzuki et al., 2008). However, this mutant also substantially reduced cell surface expression and led to extensive constitutive internalization of surface-labeled SNAP GPR120S receptors. Thus, some caution is necessary in interpreting the functional effects of R2.64A substitution only in the context of changes in agonist activity.
TZD PPARγ agonists stimulated GPR120S receptor signaling and internalization with low potency (threshold >10 μM for calcium mobilization). The demonstration that TZDs containing the 2,4-dione rather than a carboxylic acid group can show GPR120 agonism parallels their additional actions as FFA1 agonists (Kotarsky et al., 2003; Hara et al., 2009; Smith et al., 2009). In contrast with other TZDs tested, including troglitazone, pioglitazone was without effect at 30 μM, and this ligand also exhibits the lowest potency among TZDs in activating FFA1 (Hara et al., 2009; Smith et al., 2009). Because pioglitazone retains PPARγ agonist activity equivalent to or greater than that of troglitazone (Sakamoto et al., 2000), its relative position in experimental orders of potency should discriminate PPAR and FFA GPCR-mediated actions of TZD ligands.
GPR120L as a β-Arrestin-Coupled GPCR.
Poor G protein-coupling efficiency of GPR120L receptors was indicated by the absence of intracellular calcium and DMR responses, despite levels of cell surface expression similar to that of the GPR120S isoform. It is possible that HEK293 cells lack critical G protein subunits for GPR120L coupling, although Gq/11α, the mediator of GPR120 signaling in adipocytes, is present (Oh et al., 2010). A specific impairment of G protein coupling is indirectly supported by previous investigations using the GPR120L splice variant. These required receptor fusion to the promiscuous G protein Gα16 (Hirasawa et al., 2005), viral receptor overexpression (Briscoe et al., 2006), or overexpression of chimeric Gα subunits (Galindo et al., 2012) to detect intracellular calcium signaling after activation. However, both GPR120S and GPR120L recruited β-arrestin2 equally and underwent endocytosis in response to agonists. Quantitative imaging approaches demonstrated that both isoforms exhibited similar agonist potencies for each assay. Thus, the perceived selectivity of GPR120L for arrestin signaling pathways is unlikely to result from a general impairment of GPR120L signaling, which becomes apparent only in the calcium assay format (Kenakin and Miller, 2010), given, in particular, an increased receptor reserve here, compared with internalization and arrestin-binding assays measured at the receptor. We conclude that GPR120L represents a GPCR whose signaling exhibits functional selectivity for arrestin-mediated pathways.
Molecular Basis for GPR120 Isoform Coupling Selectivity.
The effect of the GPR120L splice insert might be predicted from the critical role that ICL3 plays for rhodopsin-like GPCRs, in forming part of the cytoplasmic cleft involved in Gα recognition (Rosenbaum et al., 2009). Subtle alterations in G protein coupling specificity occur between dopamine D2S and D2L receptors (Senogles et al., 2004; Lane et al., 2008), whereas a splice insert in the central ICL3 of the histamine H3 receptor influences its constitutive activity (Bongers et al., 2007). However, the positioning of the GPR120L insert in the key juxtamembrane region between TMD V and ICL3 may account for more extensive changes in G protein signaling than have previously been observed for other GPCR splice variants. Mutagenesis studies in other GPCRs have indicated the critical importance of this region for G coupling specificity and activation (Cotecchia et al., 1992; Kostenis et al., 1997).
The absence of isoform-specific effects on β-arrestin association implies that the structural elements involved in its recruitment to active GPR120 receptors differ from those required by the G protein. The recognition of active rhodopsin-like GPCRs by arrestin has been suggested instead to involve ICL2 (Marion et al., 2006), but the specific proline-based motif implicated is absent in GPR120. A second arrestin sensor domain binds phosphorylated Ser/Thr residues in ICL3 or in the C tail of GPCRs (Gurevich and Gurevich, 2006). Although additional potential phosphorylation sites are introduced into GPR120L by the amino acid insertion (Fig. 1), our data, together with similar patterns of agonist-induced phosphorylation for both GPR120S and GPR120L (Burns and Moniri, 2010), indicate that they are not critical for initial β-arrestin association and internalization. The extent and duration of receptor phosphorylation could also influence subsequent intracellular trafficking of the receptor through recycling or degradative pathways, for example, by altering the stability of arrestin association (Gurevich and Gurevich, 2006). However, no differences were observed between GPR120 isoforms in this respect. Both variants were sorted to lysosomal compartments on internalization and were unable to recycle rapidly to the cell surface after removal of agonist.
DMR Assays Indicate G Protein but Not β-Arrestin Signaling by GPR120.
The absence of specific DMR responses mediated by SNAP-GPR120L receptors, beyond those also apparent in nontransfected 293TR cells, suggests that the DMR system is relatively insensitive to β-arrestin-mediated signaling pathways. Indeed, previous findings have indicated that GPCR DMR responses largely reflect G protein-mediated events whether these are assessed by blocking G protein activation or by considering the minor contribution of downstream arrestin pathways, such as extracellular signal-regulated kinase activation (Schröder et al., 2010). It should also be noted that DMR assays are performed at room temperature, and this would minimize any contribution that might be expected from GPR120 receptor clustering and endocytosis.
We have demonstrated that alternative splicing generates GPR120 receptors with altered coupling specificity between G protein and β-arrestin pathways. Isoform-specific expression in different cell types might thus tune GPR120 signaling, for example, to β-arrestin-dependent mechanisms in macrophages (Oh et al., 2010). Moreover, our observations on GPR120L, together with an initial investigation of the chemokine receptor CXCR7 (Rajagopal et al., 2010), provide early examples of native “G protein-coupled” receptors that in fact display significant selectivity for β-arrestin recruitment.
Supplementary Material
Acknowledgments
We thank Dr. Stephen Briddon for valuable discussions during preparation of this article.
 The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
This work was supported by AstraZeneca (Alderley Park, UK); the UK Medical Research Council [Grant G0700049] (to N.D.H.); and the UK Engineering and Physical Sciences Research Council (postgraduate studentship to S.J.W.).
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
- FFA
- free fatty acid
- PPAR
- peroxisome proliferator-activated receptor
- GPCR
- G protein-coupled receptor
- ICL
- intracellular loop
- HEK
- human embryonic kidney
- GW9508
- (4-[[(3-phenoxyphenyl)methyl]amino]benzenepropanoic acid)
- DMR
- dynamic mass redistribution
- DMSO
- dimethyl sulfoxide
- PTX
- pertussis toxin
- TR
- tetracycline repressor
- GFP
- green fluorescent protein
- BiFC
- bimolecular fluorescence complementation
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- HBSS
- HEPES buffered saline solution
- BSA
- bovine serum albumin
- BG
- benzyl guanine
- BG-AF488
- benzyl guanine-Alexa Fluor-488
- H33342
- Hoechst 33342
- BSA
- bovine serum albumin
- OA
- oleic acid
- Myr
- myristic acid
- 2-APB
- 2-aminoethoxydiphenyl borate
- TMD
- transmembrane domain
- TZD
- thiazolidinedione.
Authorship Contributions
Participated in research design: Brown and Holliday.
Conducted experiments: Watson.
Contributed new reagents or analytic tools: Brown.
Performed data analysis: Watson.
Wrote or contributed to the writing of the manuscript: Watson, Brown, and Holliday.
References
- Auld DS, Johnson RL, Zhang YQ, Veith H, Jadhav A, Yasgar A, Simeonov A, Zheng W, Martinez ED, Westwick JK, et al. (2006) Fluorescent protein-based cellular assays analyzed by laser-scanning microplate cytometry in 1536-well plate format. Methods Enzymol 414:566–589 [DOI] [PubMed] [Google Scholar]
- Bongers G, Krueger KM, Miller TR, Baranowski JL, Estvander BR, Witte DG, Strakhova MI, van Meer P, Bakker RA, Cowart MD, et al. (2007) An 80-amino acid deletion in the third intracellular loop of a naturally occurring human histamine H3 isoform confers pharmacological differences and constitutive activity. J Pharmacol Exp Ther 323:888–898 [DOI] [PubMed] [Google Scholar]
- Briscoe CP, Peat AJ, McKeown SC, Corbett DF, Goetz AS, Littleton TR, McCoy DC, Kenakin TP, Andrews JL, Ammala C, et al. (2006) Pharmacological regulation of insulin secretion in MIN6 cells through the fatty acid receptor GPR40: identification of agonist and antagonist small molecules. Br J Pharmacol 148:619–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe CP, Tadayyon M, Andrews JL, Benson WG, Chambers JK, Eilert MM, Ellis C, Elshourbagy NA, Goetz AS, Minnick DT, et al. (2003) The orphan G protein-coupled receptor GPR40 is activated by medium and long chain fatty acids. J Biol Chem 278:11303–11311 [DOI] [PubMed] [Google Scholar]
- Burns RN, Moniri NH. (2010) Agonism with the omega-3 fatty acids alpha-linolenic acid and docosahexaenoic acid mediates phosphorylation of both the short and long isoforms of the human GPR120 receptor. Biochem Biophys Res Commun 396:1030–1035 [DOI] [PubMed] [Google Scholar]
- Cartoni C, Yasumatsu K, Ohkuri T, Shigemura N, Yoshida R, Godinot N, le Coutre J, Ninomiya Y, Damak S. (2010) Taste preference for fatty acids is mediated by GPR40 and GPR120. J Neurosci 30:8376–8382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotecchia S, Ostrowski J, Kjelsberg MA, Caron MG, Lefkowitz RJ. (1992) Discrete amino acid sequences of the α1-adrenergic receptor determine the selectivity of coupling to phosphatidylinositol hydrolysis. J Biol Chem 267:1633–1639 [PubMed] [Google Scholar]
- Edfalk S, Steneberg P, Edlund H. (2008) Gpr40 is expressed in enteroendocrine cells and mediates free fatty acid stimulation of incretin secretion. Diabetes 57:2280–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksson R, Höglund PJ, Gloriam DE, Lagerström MC, Schiöth HB. (2003) Seven evolutionarily conserved human rhodopsin G protein-coupled receptors lacking close relatives. FEBS Lett 554:381–388 [DOI] [PubMed] [Google Scholar]
- Galindo MM, Voigt N, Stein J, van Lengerich J, Raguse JD, Hofmann T, Meyerhof W, Behrens M. (2012) G protein-coupled receptors in human fat taste perception. Chem Senses 37:123–139 [DOI] [PubMed] [Google Scholar]
- Gotoh C, Hong YH, Iga T, Hishikawa D, Suzuki Y, Song SH, Choi KC, Adachi T, Hirasawa A, Tsujimoto G, et al. (2007) The regulation of adipogenesis through GPR120. Biochem Biophys Res Commun 354:591–597 [DOI] [PubMed] [Google Scholar]
- Gurevich VV, Gurevich EV. (2006) The structural basis of arrestin-mediated regulation of G-protein-coupled receptors. Pharmacol Ther 110:465–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Hirasawa A, Sun Q, Koshimizu TA, Itsubo C, Sadakane K, Awaji T, Tsujimoto G. (2009) Flow cytometry-based binding assay for GPR40 (FFAR1; free fatty acid receptor 1). Mol Pharmacol 75:85–91 [DOI] [PubMed] [Google Scholar]
- Hirasawa A, Tsumaya K, Awaji T, Katsuma S, Adachi T, Yamada M, Sugimoto Y, Miyazaki S, Tsujimoto G. (2005) Free fatty acids regulate gut incretin glucagon-like peptide-1 secretion through GPR120. Nat Med 11:90–94 [DOI] [PubMed] [Google Scholar]
- Itoh Y, Kawamata Y, Harada M, Kobayashi M, Fujii R, Fukusumi S, Ogi K, Hosoya M, Tanaka Y, Uejima H, et al. (2003) Free fatty acids regulate insulin secretion from pancreatic β cells through GPR40. Nature 422:173–176 [DOI] [PubMed] [Google Scholar]
- Kenakin T, Miller LJ. (2010) Seven transmembrane receptors as shapeshifting proteins: the impact of allosteric modulation and functional selectivity on new drug discovery. Pharmacol Rev 62:265–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K. (2003) A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat Biotechnol 21:86–89 [DOI] [PubMed] [Google Scholar]
- Kilpatrick LE, Briddon SJ, Hill SJ, Holliday ND. (2010) Quantitative analysis of neuropeptide Y receptor association with β-arrestin2 measured by bimolecular fluorescence complementation. Br J Pharmacol 160:892–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostenis E, Conklin BR, Wess J. (1997) Molecular basis of receptor/G protein coupling selectivity studied by coexpression of wild type and mutant m2 muscarinic receptors with mutant Gαq subunits. Biochemistry 36:1487–1495 [DOI] [PubMed] [Google Scholar]
- Kotarsky K, Nilsson NE, Flodgren E, Owman C, Olde B. (2003) A human cell surface receptor activated by free fatty acids and thiazolidinedione drugs. Biochem Biophys Res Commun 301:406–410 [DOI] [PubMed] [Google Scholar]
- Lane JR, Powney B, Wise A, Rees S, Milligan G. (2008) G protein coupling and ligand selectivity of the D2L and D3 dopamine receptors. J Pharmacol Exp Ther 325:319–330 [DOI] [PubMed] [Google Scholar]
- Marion S, Oakley RH, Kim KM, Caron MG, Barak LS. (2006) A β-arrestin binding determinant common to the second intracellular loops of rhodopsin family G protein-coupled receptors. J Biol Chem 281:2932–2938 [DOI] [PubMed] [Google Scholar]
- Maruyama T, Kanaji T, Nakade S, Kanno T, Mikoshiba K. (1997) 2APB, 2-aminoethoxydiphenyl borate, a membrane-penetrable modulator of Ins(1,4,5)P3-induced Ca2+ release. J Biochem 122:498–505 [DOI] [PubMed] [Google Scholar]
- Moore K, Zhang Q, Murgolo N, Hosted T, Duffy R. (2009) Cloning, expression, and pharmacological characterization of the GPR120 free fatty acid receptor from cynomolgus monkey: comparison with human GPR120 splice variants. Comp Biochem Physiol B Biochem Mol Biol 154:419–426 [DOI] [PubMed] [Google Scholar]
- Oh DY, Talukdar S, Bae EJ, Imamura T, Morinaga H, Fan W, Li P, Lu WJ, Watkins SM, Olefsky JM. (2010) GPR120 is an omega-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell 142:687–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pédelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. (2006) Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol 24:79–88 [DOI] [PubMed] [Google Scholar]
- Rajagopal S, Kim J, Ahn S, Craig S, Lam CM, Gerard NP, Gerard C, Lefkowitz RJ. (2010) Beta-arrestin- but not G protein-mediated signaling by the “decoy” receptor CXCR7. Proc Natl Acad Sci USA 107:628–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum DM, Rasmussen SG, Kobilka BK. (2009) The structure and function of G-protein-coupled receptors. Nature 459:356–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto J, Kimura H, Moriyama S, Odaka H, Momose Y, Sugiyama Y, Sawada H. (2000) Activation of human peroxisome proliferator-activated receptor (PPAR) subtypes by pioglitazone. Biochem Biophys Res Commun 278:704–711 [DOI] [PubMed] [Google Scholar]
- Schröder R, Janssen N, Schmidt J, Kebig A, Merten N, Hennen S, Müller A, Blättermann S, Mohr-Andrä M, Zahn S, et al. (2010) Deconvolution of complex G protein-coupled receptor signaling in live cells using dynamic mass redistribution measurements. Nat Biotechnol 28:943–949 [DOI] [PubMed] [Google Scholar]
- Senogles SE, Heimert TL, Odife ER, Quasney MW. (2004) A region of the third intracellular loop of the short form of the D2 dopamine receptor dictates Gi coupling specificity. J Biol Chem 279:1601–1606 [DOI] [PubMed] [Google Scholar]
- Smith NJ, Stoddart LA, Devine NM, Jenkins L, Milligan G. (2009) The action and mode of binding of thiazolidinedione ligands at free fatty acid receptor 1. J Biol Chem 284:17527–17539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steneberg P, Rubins N, Bartoov-Shifman R, Walker MD, Edlund H. (2005) The FFA receptor GPR40 links hyperinsulinemia, hepatic steatosis, and impaired glucose homeostasis in mouse. Cell Metab 1:245–258 [DOI] [PubMed] [Google Scholar]
- Stoddart LA, Smith NJ, Milligan G. (2008) International Union of Pharmacology. LXXI. Free fatty acid receptors FFA1, -2, and -3: pharmacology and pathophysiological functions. Pharmacol Rev 60:405–417 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Igari S, Hirasawa A, Hata M, Ishiguro M, Fujieda H, Itoh Y, Hirano T, Nakagawa H, Ogura M, et al. (2008) Identification of G protein-coupled receptor 120-selective agonists derived from PPARγ agonists. J Med Chem 51:7640–7644 [DOI] [PubMed] [Google Scholar]
- Talukdar S, Olefsky JM, Osborn O. (2011) Targeting GPR120 and other fatty acid-sensing GPCRs ameliorates insulin resistance and inflammatory diseases. Trends Pharmacol Sci 32:543–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Yano T, Adachi T, Koshimizu TA, Hirasawa A, Tsujimoto G. (2008) Cloning and characterization of the rat free fatty acid receptor GPR120: in vivo effect of the natural ligand on GLP-1 secretion and proliferation of pancreatic β cells. Naunyn Schmiedebergs Arch Pharmacol 377:515–522 [DOI] [PubMed] [Google Scholar]
- Varga T, Czimmerer Z, Nagy L. (2011) PPARs are a unique set of fatty acid regulated transcription factors controlling both lipid metabolism and inflammation. Biochim Biophys Acta 1812:1007–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirkner K, Schweigel J, Gerevich Z, Franke H, Allgaier C, Barsoumian EL, Draheim H, Illes P. (2004) Adenine nucleotides inhibit recombinant N-type calcium channels via G protein-coupled mechanisms in HEK 293 cells; involvement of the P2Y13 receptor-type. Br J Pharmacol 141:141–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaney GC, Corkey BE. (2003) Fatty acid metabolism and insulin secretion in pancreatic beta cells. Diabetologia 46:1297–1312 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.