Abstract
To circumvent the difficulty of isolating specific cell populations by MACS from dissociated complex animal tissue, when their proportions reached levels similar to that of the background, we developed the “Three-step MACS” strategy. Cells of interest are defined by their expression of a particular gene(s) of interest rather by than their natural cell surface markers or size. A two-component transgenic cell surface protein, for two sequential rounds of MACS, is expressed under the promoter control of the endogenous gene of interest by means of gene targeting and the generation of transgenic tissue. An initial step to remove dead cells is also used. Here, we describe proof-of-concept experiments, using the biotin acceptor peptide (BAP)-low-affinity nerve growth factor receptor as the two-component protein. The first component, the BAP, can be biotinylated in specific subsets of cells expressing a particular gene by expressing the biotinylating enzyme, hBirA = humanized BirA (hBirA), under the promoter control of another gene defining the specific subpopulation. We showed that a rare population of cells (1.1% of the 13.5 days postcoital mouse embryo) could be enriched to a sufficiently high purity (84.4%). From another sample with 0.1% of our cells of interest, we achieved a 40.3% pure sample. The low cost, speed, and technical ease of the Three-step MACS also make it scalable and hence, an ideal method for preparing sufficient quantities of biological samples for sensitive, high-throughput assays.
Keywords: magnetic-activated cell sorting, Lngfr, biotin-acceptor peptide, cell enrichment, cell isolation, cell purification, tissue dissociation
INTRODUCTION
Sensitive assays for transcriptome profiling, proteomics, and other biological processes require starting cell samples of high purity, as the data are easily distorted by contaminating, irrelevant cell types. This need is most keenly felt when developmentally regulated pathways are being investigated.1–4 Such studies require samples representative of spatiotemporal, cell-specific events occurring in vivo. Samples must be of sufficient quantity and quality for the sensitive downstream assays to produce reliable data. Current technologies commonly used fall short. Analyzing whole organs or tissues often introduces irrelevant cell types into the sample, which distorts the data (e.g., gene expression profiles) of all of the cell types, which, on average, are obtained, rather than that of any particular cell type.5–8 Microdissected samples are purer, but they are laborious and technically challenging to prepare; hence, sample quantities are small.9 Nucleic acid amplification is often needed downstream, a step that introduces bias.10–14 Cultured cells are homogenous, cheap, and convenient to use, but cells are taken out of their in vivo microenvironment, which greatly affects spatiotemporally regulated, developmental processes and gene expression.3,4,15–17 FACS has thus far yielded the most suitable samples, in terms of its sample purity, quality, and ease of preparation.18–21 The drawback of FACS, however, is its high start-up cost, which is prohibitively high, and its flow rate, which can be prohibitively low when scaling up is required. MACS is very similar to FACS, except that the start-up costs are 100-fold lower, and it is easily scalable to large input volumes.22 It has routinely been used in the clinical setting to isolate cells from large volumes of blood, based on the cell surface markers of the desired cells.23–26 However, single-step MACS does not sort cells as purely as FACS.27–30
When using MACS to isolate rare cell populations from dissociated animal tissue, even a low level of background renders the isolated cells insufficiently pure for sensitive assays. Three main sources contribute to the background of MACS: cells that express endogenous proteins with regions identical or similar to the epitope tag, dead cells, and cells expressing proteins on their extracellular surface that bind nonspecifically to the antibody. However, the proportion of these background cells in the purified fraction is not just determined by the background level in the starting cell sample; the percentage of desired cells of interest within the starting cell sample is a major determinant too. For example, if the background cells make up 1% of the starting cell sample, and the desired cell population also makes up 1% of the starting cell sample, then the final eluted fraction will consist of 50% background cells and 50% desired cells. The effects of the background level become very pronounced when the proportion of the cells of interest approximates or drops below the level of the background.
To address these needs and challenges, we developed a cell isolation strategy, termed “Three-step MACS”, where two consecutive rounds of MACS can be applied, each against a different epitope tag. In the first round, dead cells are removed. In the second round, rare cells are enriched to levels well above the background level of the antibody to be used in the third round. In that third round, the purity of the final sample can then be enriched to sufficient levels of purity. In our hands, the purity of isolated, rare cells (1.1% of the starting cells) from dissociated, solid animal tissue was doubled from 37.9% after the second round to 84.4% after the third round of MACS. Our sorting strategy also enables the isolation of cells defined by the expression of one or two genes of interest, thus eliminating the need to identify existing cell surface markers for antibody recognition, as is the limitation of current MACS strategies. This is a generic isolation method that can be applied to all cell types of interest defined this way, e.g., to study any transcription factor of interest within its in vivo environment. In this paper, we describe our protocol for obtaining highly purified cell samples using Three-step MACS.
MATERIALS AND METHODS
Transient Expression in Cells
Human embryo kidney (HEK)293 cells (ATCC CRL-1573) were transfected with circular plasmid expression constructs for transient protein expression, using FuGENE 6 with Plus reagent (Invitrogen, Carlsbad, CA, USA) or FuGENE HD (Roche Applied Science, Indianapolis, IN, USA). Optimem (Invitrogen) was used as the carrier medium. Cells were maintained in high-glucose DMEM/10% FBS/40 μg/ml gentamycin sulfate. Cells were harvested between 24 and 48 h.
Expression Plasmids Construction
The constructs for transient transfection and expression in cell cultures were based on the pDisplay (Invitrogen) vector. Three consecutive biotin acceptor peptide (BAP) epitopes were cloned after and in-frame with the HA tags on the vector. The base vector was modified such that three versions were made: one with IRES-hBirA-IRES-EGFP, one with an IRES-hBirA, and another one with EGFP in-frame and hence, fused to the transmembrane region on the intracellular side. The IRES-hBirA or IRES-hBirA-IRES-EGFP was cloned in after the Stop codon of the transmembrane protein but before the BghPolyA tail. Low-affinity nerve growth factor receptor (Lngfr; Miltenyi Biotec, Germany) was cloned in between the BAP tags and myc tag. hBirA is a humanized sequence of the Escherichia coli endogenous BirA.31
Confocal Imaging
Cells were grown on Lab-Tek chambered coverglass (Nunc, Thermo Scientific, Rochester, NY, USA) until 80% confluency for confocal imaging. Cells were fixed in 4% PFA for 10 min at 4°C and permeabilized with 0.1% Triton X-100/PBS for 3 min with ProLong Gold (Invitrogen) and mounted with DAPI. If antibodies were used, cells were blocked in 1% BSA/PBS after permeablization and then incubated with antibodies in 1% BSA/PBS. Imaging was done using the Carl Zeiss LSM 5 DUO inverted microscope.
Flow Cytometry (FC)
FC analysis was performed on a Becton Dickinson LSR II three-laser, using DPBS/0.5% BSA/5 mM EDTA, pH 8.0 (PBE), as a carrier medium, with each sample containing between 104 and 107 of single cells, resuspended in 1 ml PBE. Dissociated, WT mouse embryo cells were used for gating and to set the fluorescence baseline level. PI (Invitrogen) was used to stain dead cells.
Dissociation and MACS of Cells from Tissues
WT C57BL/6J mouse embryonic tissue [12.5 days postcoital (d.p.c.)] were used. Dissociation of tissue was carried out by a combination of enzymatic and mechanical dissociation. Embryo tissue was cut into small pieces of ∼3 mm × 3 mm × 3 mm. The pieces were immersed in 5–10× tissue volume of liver digest media (Invitrogen) at room temperature for enzymatic dissociation. Mechanical dissociation was used by pipetting up and down with a wide-bore, 1 ml pipette tip, followed by a narrow-bore, 1 ml pipette tip, until tissue is completely dissociated to single cells. Cold calcium-, magnesium-, and phenol red-free HBSS (5 vol; Gibco, Life Technologies, Carlsbad, CA, USA) was added to dilute out the liver digest media and reduce the enzymatic activity. Dissociated cells were pelleted, resuspended in cold HBSS, and passed through a 40-μ single cell filter and collected on ice. The entire process took ∼15 min.
MACS
Up to 107 cells/0.5 ml PBE were nutated for 20 min at room temperature with a magnetic bead-conjugated antibody (Miltenyi Biotec) or a primary antibody followed by a secondary magnetic bead-conjugated antibody. Table 1 provides details of the different antibodies used and their working concentration. Cold PBE (1 ml) was then added; the cells pelleted at 200 g at 4°C. Up to 108 cells were resuspended in 1 ml PBE. The cells were then put over a 40-μm single cell filter and through a magnetic separation (MS) column (Miltenyi Biotec), which was freshly equilibrated with 0.5 ml cold PBE. The column was then washed with a total of 2 ml cold PBE. Cells were eluted with 1 ml cold PBE using the plunger, into a second freshly equilibrated MS column. They were washed again and finally eluted into a 1.7-ml tube with 1 ml cold PBE and kept on ice. All subsequent analysis was done immediately. MACS (from applying the input cells to the MACS column to obtaining the desired washed and eluted fraction) took ∼20 min to complete.
TABLE 1.
Antibodies Used for MACS
| Epitope | Epitope seq | Antibody | Concentration (supplier, catalog number) |
|---|---|---|---|
| Antibodies against common epitope tags | |||
| HA | YPTDVPDYA | Mouse anti-HA-FITC Mouse anti-HA-PE | 1:11 (Miltenyi Biotec, 130-092-256, 7) |
| S-tag | KETAAAKFERQHMDS | Rabbit anti-S-tag | 1:20 (Delta Biolabs, Gilroy, CA, USA; DB115S-tag) |
| Lngfr | Lngfr | Mouse anti-Lngfr | 1:4 (Miltenyi Biotec, 130-091-330) |
| Myc | EQKLISEEDL | Mouse anti-c-myc-FITC | 1:100 (Invitrogen, R953-25) |
| V5 | GKPIPNPLLGLDST | Mouse anti-V5 | 1:100 (Invitrogen, R963-25) |
| His | HHHHHH | Mouse anti-His | 1:20 (BD PharMingen, San Diego, CA, USA; 552565) |
| CBP | KRRWKKNFIAVSAANRFKKISSSGAL | Rabbit anti-CBP | 1:50 (Upstate Biotechnology,Waltham, MA, USA; 07-482) |
| FLAG | DYKDDDDK | Mouse anti-FLAG | 1:500 (Sigma, St. Louis, MO, USA; F 3165) |
| Biotin | Biotin | Mouse anti-biotin | 1:4 (Miltenyi Biotec, 130-090-485) |
| Commonly used secondary antibodies | |||
| Rabbit | Rabbit IgG | Goat anti-rabbit | 1:5 (Miltenyi Biotec, 130-048-602) |
| Rat | Rat IgG | Goat anti-rat | 1:5 (Miltenyi Biotec, 130-048-501) |
| Mouse | Mouse IgG | Goat anti-mouse | 1:5 (Miltenyi Biotec, 130-048-401) |
| FITC | FITC | Mouse anti-FITC | 1:10 (Miltenyi Biotec, 130-048-701) |
| PE | PE | Mouse anti-PE | 1:5 (Miltenyi Biotec, 130-048-801) |
CBP, Calmodulin-binding peptide.
Removal of Cell-Bound Magnetic Beads
Cells, which were bound to the column by magnetic bead-conjugated antibodies, were given a final wash with calcium-, magnesium-, and phenol red-free HBSS (Gibco, Life Technologies) and then eluted from the column with phenol red-free 1 ml 1× TrypLE Express (Gibco, Life Technologies). Cells were nutated with TrypLE Express at room temperature for 5 min, and then 100 μl FBS was added to inhibit the reaction. Cells were then pelleted and washed with PBE. Dead cells were removed, and the live cell fraction was then subjected to a second round of MACS. The Dead Cell Removal Kit (Miltenyi Biotec) was used to remove dead cells from cell samples, following the manufacturer's instructions. Briefly, MACS was performed on the dissociated cells, using magnetic bead-bound antibodies against apoptotic and necrotic cells.
Ethics Statement
All animal procedures were performed according to the Singapore Agency for Science Technology and Research (A*STAR) Biopolis Biological Resource Center Institutional Animal Care and Use Committee (IACUC) guidelines, and the IACUC protocols used were reviewed and approved by the aforementioned committee before any animal procedures were undertaken for the study described here (IACUC Protocol Nos. 080348 and 080377).
RESULTS
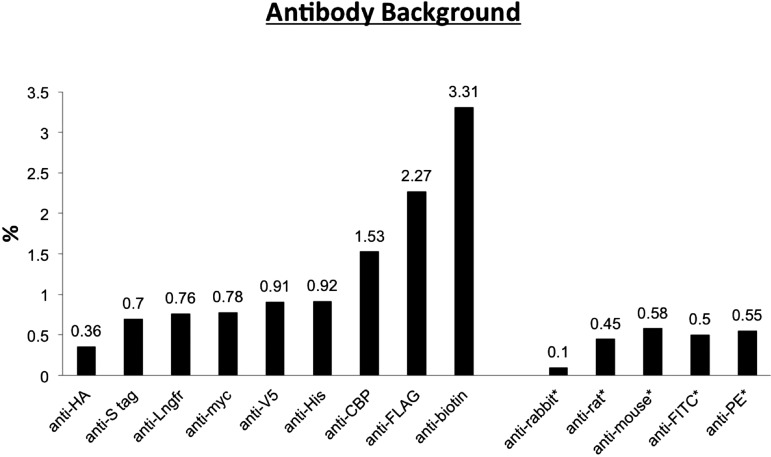
The difficulty of achieving high purities using MACS when desired cells were 1% or less of the starting population was acknowledged by Miltenyi et al.32 and others.33–36 Most of the antibodies against commonly used epitope tags (His, FLAG, myc, V5, S-tag, CBP, HA, Lngfr, or biotin tags), which we tested, resulted in 0.36% and 3.13% of the starting cells, inappropriately ending up into the eluted fraction (Fig. 1) when a single round of MACS was performed. When the proportion of the cells of interest within a complex mixture of cells approaches this background level, the resultant MACS eluted fraction would have a purity of ∼50% only.
FIGURE 1.
Antibody background: the proportion of the starting cells inappropriately ending up into the eluted fraction after a single round of MACS. Single cell populations dissociated from 13.5 d.p.c. WT mouse embryos were incubated with the antibodies to these epitopes standardly used in purification. The antibodies and their concentrations used are listed in Table 1, following the manufacturer's recommendations for FACS. Antibodies against commonly used epitopes and commonly used secondary antibodies (*) were tested. Ideally, none of these cells should be labeled, but in practice, a small percentage of these WT cells were labeled nonspecifically, and the percentage was determined by this experiment. The labeled cells were then subjected to MACS, and the background was defined as the number of cells in the eluted fraction as a percentage of the number of starting cells put through the MACS column.
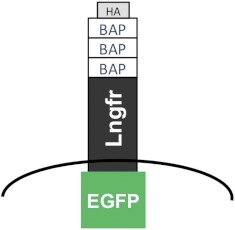
To isolate rare cells from a complex mix of cell types, we expressed a two-component cell surface marker on the cells of interest for two sequential rounds of MACS. Briefly, the first-round MACS targets the first component, biotinylated BAP, and the second round targets the second component, Lngfr (Fig. 2). In between both rounds, the two-component protein is cleaved at its trypsin sites to remove the beads labeling the cells.
FIGURE 2.
The design of the two-component cell surface protein that enables two sequential rounds of MACS, the Three-step MACS. Following removal of dead cells, the second MACS round targets the first component at the N-terminus, the biotinylated BAP. The third round targets the second component, the Lngfr. Three BAP tags were used to increase the number of epitopes and thus, improve retention on the column. In between both rounds, the two-component protein is cleaved at its trypsin sites to remove the beads labeling the cells. In the proof-of-concept experiments, EGFP is fused to the intracellular C-terminus and HA at the N-terminus to enable confocal imaging and FC analysis.
The N-terminal first component is the biotinylated BAP tag for the first-round MACS. We incorporated three BAP tags in succession, rather than a single one, to increase the number of epitopes and thus, improve retention on the column. The BAP tag is a 13-aa sequence, to which biotin protein ligases catalyze the attachment of biotin. This requires the trans expression of BirA, the E. coli biotin protein ligase (35.5 kD).37–39 Biotinylated proteins can be affinity-purified using avidin.40
The second component of our two-component cell surface marker is the truncated human Lngfr, developed and optimized by Miltenyi Biotec's MACSelect Systems for MACSorting by their accompanying magnetic bead-conjugated anti-Lngfr antibody. It consists of only the extracellular domain of the human LNGFR so that the resultant cell surface protein is incapable of transducing further intracellular signals.
As both rounds of sorting depended on MACS, there was a need to remove the magnetic bead-conjugated antibodies from the first round of sorting. Lngfr has 14 trypsin sites; these were used to remove the N-terminal 3× BAP and the attached beads from the cells, between the two rounds of MACS. As the anti-Lngfr antibody was polyclonal, even after trypsin cleavage, there was still a sufficient length of the polypeptide (46 aa) exposed on the cell surface to provide epitopes for antibody binding in the second round of MACS. This method of bead removal meant that trypsin could not be used as one of the enzymes for tissue dissociation to single cell suspensions before the first round of MACS.
As proof of concept to rapidly ascertain the effectiveness of our rare cell isolation strategy, we made expression plasmids to express the two-component cell surface BAP-Lngfr proteins in HEK293 cells, spiked the transfected cells into a suspension of cells from dissociated WT embryos in predetermined quantities and ratios, and performed MACS to recover the transfected cells. Conceptually, to isolate a particular cell type from dissociated animal tissue, the BAP-Lngfr cell surface marker needs to be expressed together with the gene that has been selected to define the cells of interest within the animal tissue. This can be achieved, for example, by expressing BAP-Lngfr under the promoter of the gene defining the desired cells through homologous recombination in embryonic stem cells. Transgenic animals can then be created, which express the BAP-Lngfr on the surface of desired cells at the appropriate spatiotemporal developmental stage. Harvested tissue can be dissociated and MACS performed, as we describe in our proof-of-concept experiments, to obtain the desired cells for downstream expression-profiling assays. We chose to use BAP for the first component, as BAP requires the expression of BirA in trans, thus adding another layer of control and specification. By expressing the biotinylating enzyme, hBirA, under the promoter control of another gene, a smaller subset of cells, defined by the expression of two different genes, can be isolated.
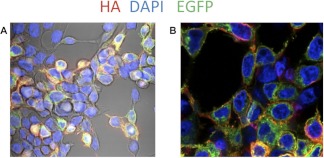
Our expression plasmids, used for the proof-of-concept experiments, drive the expression of the BAP-Lngfr through the CMV promoter. We expressed the humanized BirA31,41 under the same promoter using the IRES. The expression constructs also encoded EGFP, which was fused to the C-terminus intracellular side of BAP-Lngfr so that the various fractions of the MACS could be analysed by FC (Fig. 2). A HA tag was also included N-terminal of the three BAP epitopes for confocal imaging prior to MACSorting To determine if the cell surface molecule was properly translocated to the extracellular side of the cell surface, we performed anti-HA, PE-conjugated antibody staining without prior cell permeabalization (Fig. 3). Antibody staining of the cell membrane (red) confirmed that our two-component extracellular cell surface BAP-Lngfr was indeed exposed on the cell surface for the cell to be labeled by bead-conjugated anti-biotin or anti-Lngfr for MACS. BAP-Lngfr-EGFP, located in the cytoplasm (green) and the nuclei (blue), were also seen.
FIGURE 3.
The BAP-Lngfr-EGFP fusion cell surface protein is translocated to the extracellular cell surface. Confocal images show the subcellular distribution of the BAP-Lngfr-EGFP protein. Nonpermeabilized, transfected HEK293 cells were probed with PE-conjugated anti-HA, staining only the HA epitopes on the extracellular cell surface (red). BAP-Lngfr-EGFP (green) protein can be seen to be evenly distributed across the cytoplasm but not nucleus (DAPI, blue). (A) Fluorescent imaging is superimposed with the phase contrast image. (B) Fluorescent imaging only.
The Three-step MACS Protocol
WT C57BL/6J mouse embryos (12.5 d.p.c.) were dissociated using liver digest (Invitrogen) media and passed through a single cell filter. The single cell suspension was spiked with the BAP-Lngfr-EGFP-expressing HEK293 cells at 1% by cell count (1.1% by FC analysis), forming the starting input cell population. They were incubated with bead-conjugated anti-biotin and the first round of MACS performed. FC analysis showed that the resulting eluted fraction had a purity of 37.9% (Fig. 4). The unbound fraction had 0.2% GFP+ cells.
FIGURE 4.
The second round of the Three-step MACS doubles the purity of the first round. HEK293 cells expressing the BAP-Lngfr-EGFP fusion protein on their cell surface were added into dissociated, 12.5 d.p.c. mouse embryo cells. In the first round, they were sorted with anti-biotin; in the second round, with anti-Lngfr. The purity of the second and third round of sorting was 37.9% (B) and 84.4% (C), respectively. The first panel is a FC plot of the baseline control using dissociated mouse embryo cells without any BAP-Lngfr-EGFP-transfected HEK293 cells spiked in.
Following this first round, the bead antibody-bound BAP epitopes had to be cleaved off. We used TrypLE (Invitrogen), which cleaves proteins at the trypsin-cleavage sites but produces higher cell viability than trypsin and works at room temperature. As the carrier solution used to resuspend and wash cells for MACS contained 0.5% BSA, which can potentially compromise the TrypLE cleavage, we used HBSS solution for the third and final wash before eluting the cells bound to the MACS column. The cells were then eluted using TrypLE and left to incubate with gentle rotation for 10 min at room temperature. FBS was added to 20% concentration to stop the reaction. The Dead Cell Removal Kit was then applied. Finally, the cells were washed in PBE and then incubated with the bead-conjugated, anti-Lngfr antibody for the second round of MACS. The purity of the cells after the second round was increased to 84.4% (Fig. 4), and the wash had 0.2% GFP+ cells. The enitre process, between harvest to tissue dissociation (15 min), and the completion of the two rounds of MACS (about 2 h) took ∼2¼ h in total. Starting with ∼4 × 107 cells pooled from five dissociated embryos inititally, our final purified sample had ∼5 × 104 cells, of which ∼70% were viable, by trypan blue staining on a hemocytometer. In the second and third rounds of MACS, the recovery was nearly 100% by cell count. Approximately 25% of cells were recovered after applying the Dead Cell Removal Kit.
We also made an attempt, using this same protocol, to isolate cells expressing BAP-Lngfr-EGFP when the WT embryo single cell suspensions were spiked at an ultra-low level of 0.15% by cell count (0.1% GFP+ by FC analysis). Purity of the eluted cells following the first round of MACS was 17% and 40.3% after the second round. We did a negative control MACS using this same protocol, where we used HEK293 cells expressing EGFP intracellularly without BAP-Lngfr-EGFP at the cell surface. We spiked them into the WT embryo-dissociated cells at 1% by cell count (0.7% GFP+ by FC analysis). Purity of the eluted cells following the second round of MACS was 0.6% and 2.2% after the third round.
DISCUSSION
Our experiments described here showed that a 1.1% rare cell population could be enriched to over 84.4% purity through our Three-step MACS, which is then acceptable for sensitive downstream assays. From a 0.1% starting cell sample, we achieved an enriched fraction of 40.3% purity. Spike-in experiments offer two advantages. Firstly, as the positive cells have already been precharacterized and enumerated, we had a basis for evaluating the performance of MACS. If transgenic tissue samples were used, the uncertainty of the exact proportion and fluorescence would make it difficult to evaluate and optimize each step of MACS. The second advantage is that this is a much faster way of optimizing the MACS, as long durations may be required for wholly transgenic animals or tissue to be obtained.
In the course of developing the Three-step MACS protocol, we optimized each of the following parameters to produce the optimized protocol described earlier: the Two-component BAP-Lngfr extracellular surface molecule design for MACS, the embryonic tissue dissociation conditions, the BAP-Lngfr cleavage conditions, and the conditions for bead-conjugated antibody labeling of the target cells. For example, we tested an alternative option for the removal of magnetic beads after the first round of MACS: Miltenyi Biotec's Anti-Biotin Multisort Kit. The bead-conjugated Anti-Biotin Multisort Kit antibody was identical to the anti-biotin antibody that we used for the Three-step MACS, except that had been modified to contain a proprietary cleavage site, which would be cleaved to remove the conjugated bead when treated with the kit's accompanying multisort release reagent. We chose to use TrypLE, however, as in our hands, TrypLE seemed more effective at removing the beads (unpublished results). The modified multisort anti-biotin antibody also seemed to bind with lower affinity than the unmodified antibody (unpublished results).
A critical issue that we dealt with during optimization was the amount of dead cells in the dissociated tissue input sample. Dead cells tend to bind antibodies nonspecifically and form a significant source of background cells, which has great impact on the eventual purity of the eluted, desired cells. Dead cells tend to be sticky as well and will form clumps with each other, which might, in turn, trap live cells. These clumps tend to be retained on the column and get eluted out with the desired cells of interest in the eluted fraction. Embryos and animal tissues start to die the moment they are removed from the mother or animal, respectively. During the tissue dissociation process required to obtain single cell suspensions, the mechanical force and enzymes will further kill a proportion of the cells. We addressed the issue of dead cells in two ways. Firstly, we minimized cell death as much as possible by minimizing the time spent from the sacrifice of the host animal until the cells are completely sorted and dissolved in Trizol for RNA extraction. Cells were kept on ice wherever possible. Secondly, we introduced a dead cell removal step using Miltenyi Biotec's Dead Cell Removal Kit between the cell dissociation steps and the MACS procedure. Here, magnetic bead-conjugated antibodies against various apoptotic and necrotic cell surface markers bind to dead or dying cells, which themselves tend to bind easily and nonspecifically to antibodies anyway. When the cell suspension is put through the magnetized column, the dead and dying cells are retained, allowing only live cells to pass through.
The use of multistep cell isolation methods previously has been used successfully to increase the purity of the final cell sample. Busch et al.42 reported a double MACS protocol. Here, CD45+ leukocytes were first labeled and depleted from peripheral blood. The depleted fraction was then incubated with magnetic bead-conjugated anti-CD71, and this time, the CD71+ cells were positively selected—to a final purity of 62–87%. The first round of MACS depletion enriched for CD71+ by 9.4-fold, and the second round of MACS selection enriched it by a further 32.1-fold. Recovery rates for the positive selection ranged from 38% to 55%. In another example, a dual-step MACS procedure was used to isolate fetal cells from peripheral maternal blood for paternity testing. CD45+ and CD14+ cells were depleted by 780-fold from PBMCs. From this fraction, CD71 cells were enriched by 500-fold to 80% purity by MACS, with a recovery of 40–55%. PCR detected paternal DNA sequences from the purified fetal cell fraction but not from the unsorted maternal blood.43 The above cases of multiple rounds of cell isolation were possible, as several methods of sorting were already available for each cell type. For example, a lot of cell types from blood have multiple cell surface markers identified, or they have unique cell sizes and densities that separate them from other cell types. Without prior, further knowledge of these cells or prior purification to define their properties, it would be impossible to purify the cells by density or to know what kinds of cell surface markers are unique to those cells. Our Three-step MACS can circumvent this problem because of the introduction of a transgenic cell surface marker, which can be expressed under the same promoter control of the gene of interest to sort the cells. This is similar to other reports of successful isolations of specific cell populations by FACS, although these cells did not express any known, unique cell surface protein to which fluorescent antibodies could bind. They achieved this by expressing EGFP under the promoter control of a gene, which was uniquely expressed in the desired cell population.44–48
Another significant drawback to defining cells of interest by their endogenous cell surface markers is that these markers may not be expressed at the earliest stages of differentiation. Onset of stem cell differentiation down a particular lineage is often defined by a transcription factor functioning as a master regulator. Our MACS strategy allows the isolation of cells from the earliest stages of differentiation, as the expression of the cell surface molecule can be placed under the promoter control of the gene expressed at the earliest stage of a cell lineage's differentiation. Thus, gene expression and other events at the earliest stages of differentiation can be observed. At the earliest stages of differentiation, the numbers of precursors to a particular lineage are usually very small, before they enter the proliferative phases. This makes it important for the strategy to be able to isolate these rare cells of interest to sufficient purity and quantity. A single round of MACS does not yield samples of sufficient purity when the proportion of the cells of interest approaches the level of the background or false positives. This background tends to be high in a complex cell population. Doing two consecutive rounds of MACS significantly improves this purity.
The scalability of Three-step MACS means that sufficient quantities of the rare cell sample can be obtain simultaneously from multiple tissue samples. This reduces the need to depend on cell cultures or nucleic acid amplification. From the ∼5 × 104 cells, which we obtained from a single MS column, up to 50 ng RNA can be extracted, sufficient for downstream applications, such as an Illumina Beadchip Microarray (San Diego, CA, USA). By simultaneously using multiple MS columns (capacity to sort 2×108 cells in 20 min) or larger columns, such as the large separation (LS) columns (capacity to sort 2×109 cells in 20 min; Miltenyi Biotec), the amount of sample obtained can be scaled up proportionally, with minimal extra time and effort spent, at the cost of only the additional columns. MACS is scalable because of its low cost (compared with FACS, with a capacity to sort 108 cells in 20 min) and technical ease (compared with microdisections).
In conclusion, we have contributed to the available options for isolating rare cell populations from animal tissues by developing this Three-step MACS strategy. Compared with current available options, our strategy is fast, low-cost, technically simple, and scalable to produce quantities as required. The Three-step MACS strategy can potentially isolate any rare cells from dissociated animal tissue by ensuring that the transgenic cell surface marker is expressed in tandem with the gene of interest. It is capable of isolating specific cell populations or subpopulations, as defined by the expression of one or two genes of interest, thus eliminating the need to have prior information about the cells size, density, or surface markers available. The precaution that needs to be taken, however, is that not all cells may be compatible with the BAP-Lngfr two-component cell surface molecule design. Depending on the cell type and animal under study, the BAP or Lngfr component may need to be replaced. This requires empirical observation to know if the cells of interest within the tissues can express, fold, and translocate this protein appropriately and at suitable levels for sorting.
Beyond the mapping of genomes, spatiotemporally specific tissue transcriptomes and proteomes are now being mapped, with the advent of fast, high-throughput sequencing and array technologies. Protein interactions, protein-nucleic acid interactions, and micro-RNA expression profiles are other areas of interest enabled by technological advances in profiling methods. The development of a strategy to provide spatiotemporally specific animal tissue samples will cater to these downstream technologies so that information gleaned will be truly reflective of the in vivo processes.
ACKNOWLEDGMENTS
This work was supported by A*STAR (Singapore). We are grateful to the members of the Lufkin Lab for support and interesting discussions.
Footnotes
The authors declare no competing, conflicting, or financial interests.
REFERENCES
- 1.Kawauchi S, Takahashi S, Nakajima O, et al. Regulation of lens fiber cell differentiation by transcription factor c-Maf. J Biol Chem 1999;274:19254–19260 [DOI] [PubMed] [Google Scholar]
- 2.Wilding Crawford L, Tweedie Ables E, Oh YA, et al. Gene expression profiling of a mouse model of pancreatic islet dysmorphogenesis. PLoS ONE 2008;3:e1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aiba K, Sharov AA, Carter MG, et al. Defining a developmental path to neural fate by global expression profiling of mouse embryonic stem cells and adult neural stem/progenitor cells. Stem Cells 2006;24:889–895 [DOI] [PubMed] [Google Scholar]
- 4.Odom DT, Dowell RD, Jacobsen ES, et al. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat Genet 2007;39:730–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang WX, Wilfred BR, Baldwin DA, et al. Focus on RNA isolation: obtaining RNA for microRNA (miRNA) expression profiling analyses of neural tissue. Biochim Biophys Acta 2008;1779:749–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sugiyama Y, Sugiyama K, Hirai Y, Akiyama F, Hasumi K. Microdissection is essential for gene expression profiling of clinically resected cancer tissues. Am J Clin Pathol 2002;117:109–116 [DOI] [PubMed] [Google Scholar]
- 7.Capodieci P, Donovan M, Buchinsky H, et al. Gene expression profiling in single cells within tissue. Nat Methods 2005;2:663–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steffensen KR, Neo SY, Stulnig TM, et al. Genome-wide expression profiling; a panel of mouse tissues discloses novel biological functions of liver X receptors in adrenals. J Mol Endocrinol 2004;33:609–622 [DOI] [PubMed] [Google Scholar]
- 9.Croner RS, Guenther K, Foertsch T, et al. Tissue preparation for gene expression profiling of colorectal carcinoma: three alternatives to laser microdissection with preamplification. J Lab Clin Med 2004;143:344–351 [DOI] [PubMed] [Google Scholar]
- 10.Ma C, Lyons-Weiler M, Liang W, et al. In vitro transcription amplification and labeling methods contribute to the variability of gene expression profiling with DNA microarrays. J Mol Diagn 2006;8:183–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis JE, Eberwine JH, Hinkle DA, et al. Methodological considerations regarding single-cell gene expression profiling for brain injury. Neurochem Res 2004;29:1113–1121 [DOI] [PubMed] [Google Scholar]
- 12.Jensen KB, Watt FM. Single-cell expression profiling of human epidermal stem and transit-amplifying cells: Lrig1 is a regulator of stem cell quiescence. Proc Natl Acad Sci USA 2006;103:11958–11963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esumi S, Wu SX, Yanagawa Y, et al. Method for single-cell microarray analysis and application to gene-expression profiling of GABAergic neuron progenitors. Neurosci Res 2008;60:439–451 [DOI] [PubMed] [Google Scholar]
- 14.Sooriakumaran P, Henderson A, Denham P, Langley SE. A novel method of obtaining prostate tissue for gene expression profiling. Int J Surg Pathol 2009;17:238–243 [DOI] [PubMed] [Google Scholar]
- 15.Sievertzon M, Wirta V, Mercer A, et al. Transcriptome analysis in primary neural stem cells using a tag cDNA amplification method. BMC Neurosci 2005;6:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holley MC, Kneebone A, Milo M. Information for gene networks in inner ear development: a study centered on the transcription factor gata2. Hear Res 2007;227:32–40 [DOI] [PubMed] [Google Scholar]
- 17.Olsavsky KM, Page JL, Johnson MC, et al. Gene expression profiling and differentiation assessment in primary human hepatocyte cultures, established hepatoma cell lines, and human liver tissues. Toxicol Appl Pharmacol 2007;222:42–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Terahara K, Yoshida M, Igarashi O, et al. Comprehensive gene expression profiling of Peyer's patch M cells, villous M-like cells, and intestinal epithelial cells. J Immunol 2008;180:7840–7846 [DOI] [PubMed] [Google Scholar]
- 19.Kidd M, Modlin IM, Eick GN, Champaneria MC. Isolation, functional characterization, and transcriptome of Mastomys ileal enterochromaffin cells. Am J Physiol Gastrointest Liver Physiol 2006;291:G778–G791 [DOI] [PubMed] [Google Scholar]
- 20.Qian S, Ding JY, Xie R, et al. MicroRNA expression profile of bronchioalveolar stem cells from mouse lung. Biochem Biophys Res Commun 2008;377:668–673 [DOI] [PubMed] [Google Scholar]
- 21.Shigenobu S, Kitadate Y, Noda C, Kobayashi S. Molecular characterization of embryonic gonads by gene expression profiling in Drosophila melanogaster. Proc Natl Acad Sci USA 2006;103:13728–13733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.David R, Groebner M, Franz WM. Magnetic cell sorting purification of differentiated embryonic stem cells stably expressing truncated human CD4 as surface marker. Stem Cells 2005;23:477–482 [DOI] [PubMed] [Google Scholar]
- 23.Pongsritasana T, Wongratanacheewin S, Prasertcharoensuk V, Sermswan RW. Isolation of fetal nucleated red blood cell from maternal blood using immunomagnetic beads for prenatal diagnosis. Asian Pac J Allergy Immunol 2006;24:65–71 [PubMed] [Google Scholar]
- 24.Schindlbeck C, Stellwagen J, Jeschke U, et al. Immunomagnetic enrichment of disseminated tumor cells in bone marrow and blood of breast cancer patients by the Thomsen-Friedenreich-antigen. Clin Exp Metastasis 2008;25:233–240 [DOI] [PubMed] [Google Scholar]
- 25.Okegawa T, Nutahara K, Higashihara E. Immunomagnetic quantification of circulating tumor cells as a prognostic factor of androgen deprivation responsiveness in patients with hormone naive metastatic prostate cancer. J Urol 2008;180:1342–1347 [DOI] [PubMed] [Google Scholar]
- 26.Cools-Lartigue JJ, McCauley CS, Marshall JC, et al. Immunomagnetic isolation and in vitro expansion of human uveal melanoma cell lines. Mol Vis 2008;14:50–55 [PMC free article] [PubMed] [Google Scholar]
- 27.Schmitz B, Radbruch A, Kummel T, et al. Magnetic activated cell sorting (MACS)—a new immunomagnetic method for megakaryocytic cell isolation: comparison of different separation techniques. Eur J Haematol 1994;52:267–275 [DOI] [PubMed] [Google Scholar]
- 28.Wang JY, Zhen DK, Falco VM, et al. Fetal nucleated erythrocyte recovery: fluorescence activated cell sorting-based positive selection using anti-γ globin versus magnetic activated cell sorting using anti-CD45 depletion and anti-γ globin positive selection. Cytometry 2000;39:224–230 [PubMed] [Google Scholar]
- 29.Ordog T, Redelman D, Horowitz NN, Sanders KM. Immunomagnetic enrichment of interstitial cells of Cajal. Am J Physiol Gastrointest Liver Physiol 2004;286:G351–G360 [DOI] [PubMed] [Google Scholar]
- 30.Heng BC, Cao T. Immunoliposome-mediated delivery of neomycin phosphotransferase for the lineage-specific selection of differentiated/committed stem cell progenies: potential advantages over transfection with marker genes, fluorescence-activated and magnetic affinity cell-sorting. Med Hypotheses 2005;65:334–336 [DOI] [PubMed] [Google Scholar]
- 31.Mechold U, Gilbert C, Ogryzko V. Codon optimization of the BirA enzyme gene leads to higher expression and an improved efficiency of biotinylation of target proteins in mammalian cells. J Biotechnol 2005;116:245–249 [DOI] [PubMed] [Google Scholar]
- 32.Miltenyi S, Muller W, Weichel W, Radbruch A. High gradient magnetic cell separation with MACS. Cytometry 1990;11:231–238 [DOI] [PubMed] [Google Scholar]
- 33.Thomas TE, Abraham SJ, Otter AJ, Blackmore EW, Lansdorp PM High gradient magnetic separation of cells on the basis of expression levels of cell surface antigens. J Immunol Methods 1992;154:245–252 [DOI] [PubMed] [Google Scholar]
- 34.Owen CS, Sykes NL. Magnetic labeling and cell sorting. J Immunol Methods 1984;73:41–48 [DOI] [PubMed] [Google Scholar]
- 35.Owen CS, Lindsay JG. Ferritin as a label for high-gradient magnetic separation. Biophys J 1983;42:145–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Owen CS. High gradient magnetic separation of erythrocytes. Biophys J 1978;22:171–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beckett D, Kovaleva E, Schatz PJ. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci 1999;8:921–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schatz PJ. Use of peptide libraries to map the substrate specificity of a peptide-modifying enzyme: a 13 residue consensus peptide specifies biotinylation in Escherichia coli. Biotechnology (N Y) 1993;11:1138–1143 [DOI] [PubMed] [Google Scholar]
- 39.Barker DF, Campbell AM. Genetic and biochemical characterization of the BirA gene and its product: evidence for a direct role of biotin holoenzyme synthetase in repression of the biotin operon in Escherichia coli. J Mol Biol 1981;146:469–492 [DOI] [PubMed] [Google Scholar]
- 40.Lesley SA, Groskreutz DJ. Simple affinity purification of antibodies using in vivo biotinylation of a fusion protein. J Immunol Methods 1997;207:147–155 [DOI] [PubMed] [Google Scholar]
- 41.Sharp PM, Cowe E, Higgins DG, Shields DC, Wolfe KH, Wright F. Codon usage patterns in Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster and Homo sapiens; a review of the considerable within-species diversity. Nucleic Acids Res 1988;16:8207–8211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Busch J, Huber P, Pfluger E, et al. Enrichment of fetal cells from maternal blood by high gradient magnetic cell sorting (double MACS) for PCR-based genetic analysis. Prenat Diagn 1994;14:1129–1140 [DOI] [PubMed] [Google Scholar]
- 43.Zheng YL, Carter NP, Price CM, et al. Prenatal diagnosis from maternal blood: simultaneous immunophenotyping and FISH of fetal nucleated erythrocytes isolated by negative magnetic cell sorting. J Med Genet 1993;30:1051–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bouchard M, Grote D, Craven SE, et al. Identification of Pax2-regulated genes by expression profiling of the mid-hindbrain organizer region. Development 2005;132:2633–2643 [DOI] [PubMed] [Google Scholar]
- 45.Covassin L, Amigo JD, Suzuki K, et al. Global analysis of hematopoietic and vascular endothelial gene expression by tissue specific microarray profiling in zebrafish. Dev Biol 2006;299:551–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Von Stetina SE, Watson JD, Fox RM, et al. Cell-specific microarray profiling experiments reveal a comprehensive picture of gene expression in the C. elegans nervous system. Genome Biol 2007;8:R135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liadaki K, Kho AT, Sanoudou D, et al. Side population cells isolated from different tissues share transcriptome signatures and express tissue-specific markers. Exp Cell Res 2005;303:360–374 [DOI] [PubMed] [Google Scholar]
- 48.Cahoy JD, Emery B, Kaushal A, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci 2008;28:264–278 [DOI] [PMC free article] [PubMed] [Google Scholar]