Abstract
The formation of mucin-type O-glycans is initiated by an evolutionarily conserved family of enzymes, the UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyltransferases (GalNAc-Ts). The human genome encodes 20 transferases; 17 of which have been characterized functionally. The complexity of the GalNAc-T family reflects the differential patterns of expression among the individual enzyme isoforms and the unique substrate specificities which are required to form the dense arrays of glycans that are essential for mucin function. We report the expression patterns and enzymatic activity of the remaining three members of the family and the further characterization of a recently reported isoform, GalNAc-T17. One isoform, GalNAcT-16 that is most homologous to GalNAc-T14, is widely expressed (abundantly in the heart) and has robust polypeptide transferase activity. The second isoform GalNAc-T18, most similar to GalNAc-T8, -T9 and -T19, completes a discrete subfamily of GalNAc-Ts. It is widely expressed and has low, albeit detectable, activity. The final isoform, GalNAc-T20, is most homologous to GalNAc-T11 but lacks a lectin domain and has no detectable transferase activity with the panel of substrates tested. We have also identified and characterized enzymatically active splice variants of GalNAc-T13 that differ in the sequence of their lectin domain. The variants differ in their affinities for glycopeptide substrates. Our findings provide a comprehensive view of the complexities of mucin-type O-glycan formation and provide insight into the underlying mechanisms employed to heavily decorate mucins and mucin-like domains with carbohydrate.
Keywords: GaNAcT, splice variants, enzyme activity, expression analysis
Introduction
Mucin glycoproteins are heavily decorated with carbohydrate side chains, termed O-glycans, which are often clustered within repeating amino acid sequences of the protein (tandem repeats). Functionally, membrane-bound mucins are involved in signal transduction events (Singh and Hollingsworth 2006; Carraway et al. 2007; Cullen 2007) and targeted sorting into apical and basolateral domains of the plasma membrane (Singh and Hollingsworth 2006), whereas secreted mucins contribute to the formation of the extracellular matrix or to the gel-like mucus coat which envelopes mucosal surfaces of the body thereby forming the most exterior face of the innate immune system (Tabak 1995; Sheehan et al. 2006; Linden et al. 2008). Although it is known that O-glycans are ubiquitous among proteins, the precise extent of the “O-glycome” remains to be defined.
The first committed step in the formation of mucin-type O-glycans is catalyzed by a family of evolutionarily conserved UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyltransferases (GalNAc-Ts) (EC 2.4.1.41), which transfer α-N-acetylgalactosamine (α-GalNAc) from the sugar donor UDP-GalNAc to Ser or Thr residues of acceptor protein substrates. [The acronym for UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase has been variable in the literature including ppGaNTase-T, polypeptide GalNAc-T, ppGalNAc-T, ppGalNAc-T, ppGalNAc-T and GalNAc-T. To avoid future confusion, we and Bennett et al. (2012; this volume) suggest the adoption of the common abbreviation of GalNAc-T for all mammalian forms of this enzyme family.] Although no consensus amino acid sequence has emerged that is both necessary and sufficient for O-glycan formation, two classes of GalNAc-Ts have been identified—those with preference for peptide (“naked” or unglycosylated peptide) substrates and those which favor glycopeptide (peptides previously decorated with α-GalNAc) substrates (Ten Hagen et al. 1999, 2001, 2003; Pratt et al. 2004). Thus, the concerted action of the GalNAc-T family enables the formation of the dense arrays of O-glycans that typify mucins and mucin-like domains and underlie many of their functions.
The human GalNAc-T family has 20 distinct members; 17 of which have been characterized and shown to have distinct expression and substrate specificity (Ten Hagen et al. 2003; Peng et al. 2010). Of these, no activity has been detected for two isoforms, GalNAc-T8 which is ubiquitously expressed (White et al. 2000) and GalNAc-T19 which is expressed in the cerebellum and cerebral cortex (Nakamura et al. 2005). In the present study, we have sought to validate whether or not the remaining three of these putative isoforms (GalNAc-T16, -T18 and -T19) are indeed enzymatically active and have begun characterization of their expression patterns to better understand their biological roles. In addition, we demonstrate distinct kinetic properties of two splice variants of GalNAc-T13. This adds an additional dimension to the complexity of this large family of enzymes and potentially represents a unique mechanism for fine tuning glycosylation site selection.
Results
Conceptual translation of each of the GalNAc-Ts characterized here reveals a type-II transmembrane protein containing a short, N-terminal cytoplasmic tail, a transmembrane region, a stem region and a catalytic domain highly homologous to previously characterized GalNAc transferases [see Figure S1 in Bennett et al., 2012 (this issue)]. Like all previously characterized transferases (GalNAc-T1–T15 and GalNAc-T17), the isoforms GalNAc-T16 and -T18 also contain a ricin-like lectin domain following the catalytic domain. GaNAc-T20, which is closely related to GalNAc-T11, lacks this characteristic domain.
As detailed below (Table I), two of these previously uncharacterized gene products, GalNAc-T16 and -T18 display enzymatic activity. GalNAc-T20 was not active on any of the peptides assayed in this study.
Table I.
Sequences of peptides used and activitiesa of GalNAc-Ts
| Peptide | Sequence | GalNAc-T1 | GalNAc-T16 | GalNAc-T18 | GalNAc-T13 | GalNAc-T13V1 |
|---|---|---|---|---|---|---|
| EA2 | PTTDSTTPAPTTK | 1167 ± 59 | 203 ± 28 | ND | 298 ± 31 | 198 ± 25 |
| MUC5AC | GTTPSPVPTTSTTSAP | 807 ± 65 | 195 ± 21 | ND | 335 ± 40 | 309 ± 32 |
| MUC5AC-3 | GTTPSPVPTTSTTSAPb | 1326 ± 145 | 326 ± 45 | ND | 752 ± 80 | 501 ± 62 |
| MUC5AC-13 | GTTPSPVPTTSTTSAPb | 989 ± 93 | 319 ± 35 | ND | 270 ± 21 | 155 ± 18 |
| MUC5AC-3,13 | GTTPSPVPTTSTTSAPb | 752 ± 85 | ND | ND | 70 ± 9 | 67 ± 8 |
| Muc1B | PDTRPAPGSTAPPAC | 62 ± 11 | 13 ± 2 | ND | 13 ± 3 | 6 ± 2 |
| P7 | GTTAKPTTLKPTE | 23 ± 5 | 11 ± 3 | 12 ± 1.2 | 13 ± 2 | 16 ± 3 |
| IgAh | PSTPPTPSPSTPPTPSPS | 308 ± 12 | 45 ± 7 | ND | 22 ± 2 | 18 ± 0.6 |
| rMuc2 | SPTTSTPISSTPQPTS | 32 ± 4 | 14 ± 4 | ND | 56 ± 7 | 24 ± 4 |
| Muc1a | AHGVTSAPDTR | 114 ± 12 | 34 ± 2 | ND | ND | ND |
| PSGL-1b | Ac-QATEYEYLDYDFLPETEPPEM | 18 ± 3 | 8 ± 2 | ND | 11 ± 2 | 14 ± 3 |
| gp120 | Ac-CIRIQRGPGRAFVTIGKIGNMR | 3 ± 1 | ND | ND | ND | ND |
| hT1B | GAGAEAPTPAPAGAGK | 428 ± 37 | 34 ± 4 | 12 ± 1.5 | ND | ND |
ND, no activity detected. GalNAc-T17, GalNAC-T19 and GalNAc-T20 were not active on any of the peptides listed in the table.
aNumbers represent pmol of product formed in 1 h by enzyme quantities equivalent to GalNAc-T1 at a peptide concentration of 500 μM.
bEmboldened residues indicate the position of preexisting GalNAc.
Sequence analysis and enzymatic activity
Epitope-tagged transmembrane deletion constructs of all transferases described were expressed in COS7 cells and purified as described in Methods. A similar construct of GalNAc-T1 was also purified and used as a control for quantitation and activity. Purified enzymes were detected by chemiluminescence and quantified by densitometry on western blots probed with rabbit anti-FLAG™ antibody followed by anti-rabbit antibody conjugated to horseradish peroxidase (HRP; Figure 1). Activity was primarily measured on the peptides EA2 (PTTDSTTPAPTTK), MUC5AC (GTTPSPVPTTSTTSAP) peptide and glycosylated derivatives of MUC5AC carrying a GalNAc at Thr-3 or Thr-13 or Thr-3 and Thr-13. The peptides were chosen as the activities of the well-characterized transferases GalNAc-T1, -T2 and -T10 have been extensively studied on these peptides (Fritz et al. 2004, 2006; Kubota et al. 2006; Raman et al. 2008). Activity was also measured on a larger panel of peptides and these are listed in Table I, along with activities if observed. All activities presented are for quantities of enzyme equivalent to GalNAc-T1.
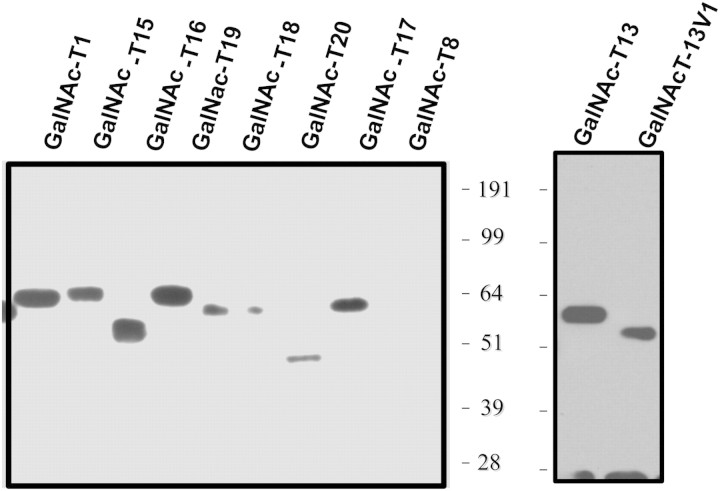
Fig. 1.
Quantitation of recombinantly expressed GalNAc-T isoforms by western blotting. GalNAc-T isoforms were expressed and analyzed by western blotting as described in Experimental procedures. Size markers are shown on the side of each blot. GalNAc-T isoform is denoted at the top of each lane. Quantitation was performed as described in Experimental procedures.
GalNAc-T16. This isoform forms a phylogenetic subfamily with GalNAc-T2 and -T14 sharing a sequence identity of 49 and 53%, respectively [see Figure 3 in Bennett et al., 2012 (this issue), for the phylogenetic tree]. GalNAc-T16 transfers GalNAc to a wide range of peptide substrates (Tables I and II). Table II compares the kinetic parameters of GalNAc-T16 and -T1 on the EA2 and MUC5AC-derived peptide and glycopeptide substrates. Although the Km for these peptides for the two enzymes is comparable, its rates of catalysis are less than one-fifth of that of GalNAc-T1. Both GalNAc-T16 and -T1 add GalNAc to an MUC5AC glycopeptide preglycosylated at either Thr-3 or Thr-13. However, GalNAc-T16 does not add GalNAc to the MUC5AC peptide glycosylated at both Thr-3 and Thr-13, whereas both GalNAc-T1 and GalNAc-T2 do (Raman et al. 2008). Sequencing of the peptides glycosylated by GalNAc-T16 with UDP-[1-14C]GalNAc showed glycosylation at Thr-3 and Ser-5 on the MUC5AC peptide, at Thr-2 and to a lesser extent at Thr-13 on the MUC5AC-3 peptide and at Thr-3 and Ser-5 on the MUC5AC-13 peptide (Figure 2). GalNAc-T1 glycosylates Thr-3 and Thr-13 on the MUC5AC peptide and Thr-12 on the MUC5AC-3,13 diglycopeptide (Ten Hagen et al. 2001), a peptide for which GalNAc-T16 displays no activity. The specificity is also different from that of GalNAc-T2 which glycosylates Thr-9 on MUC5AC, Thr-13 on MUC5AC-13, Thr-3 on MUC5AC-13 and Ser-5 on MUC5AC-3,13 (Raman et al. 2008).
Table II.
Kinetic parameters for peptide glycosylation by GalNAc-T1 and -T16
| GalNAc-T1a |
GalNAc-T16a |
|||
|---|---|---|---|---|
| Vmax (pmol/h) | Km (µM) | Vmax (pmol/h) | Km (µM) | |
| Muc5AC | 966 ± 101 | 171 ± 18 | 251 ± 28 | 344 ± 29 |
| Muc5AC-3 | 1464 ± 114 | 332 ± 38 | 310 ± 25 | 299 ± 30 |
| Muc5AC-13 | 1305 ± 143 | 460 ± 51 | 390 ± 32 | 115 ± 10 |
| Muc5AC-3,13 | 1031 ± 178 | 206 ± 19 | No activity | |
| EA2 | 1900 ± 185 | 75 ± 9 | 332 ± 37 | 172 ± 18 |
aEquivalent enzyme amounts as determined by western blot were used for the assays.
Fig. 2.
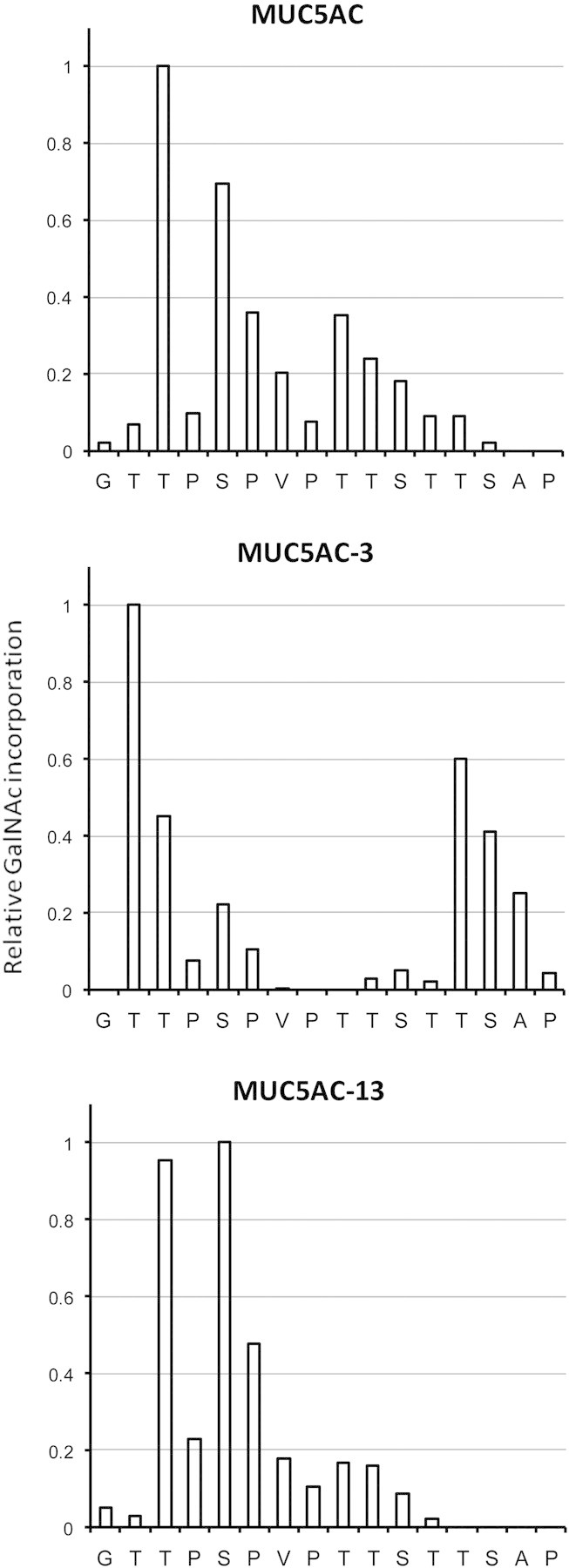
Site of glycosylation of MUC5AC peptides by GalNAc-T16. Peptides were glycosylated with UDP-[1-14C]GalNAc, subjected to the Edman degradation and radioactivity at each position determined to measure GalNAc addition. Data are expressed relative to the site of maximum incorporation, Thr-3 for MUC5AC, Thr-2 for MUC5AC-3 and Ser-5 for MUC5AC-13. Incorporation seen at sites of extant glycosylation in the glycopeptides and at residues lacking an acceptor hydroxyl group result from carryover counts from the previous position during the Edman sequencing.
However, preliminary experiments with a random unglycosylated peptide library, GAGAXXXXXTXXXXXAGAGK (where X = G,A,P,V,L,Y,E,Q,R,H; Gerken et al. 2008) indicate that the specificity of GalNAcT-16 is very similar to that of GalNAc-T2 (TA Gerken, unpublished data). Thus, the substrate specificity of GalNAc-T16 on glycopeptide substrates is quite distinct from both GalNAc-T1 and GalNAc-T2 while being similar on peptide substrates. The catalytic domains of GalNAc-T2 and -T16 share a sequence identity of ∼56%, whereas the lectin domains are significantly different, with an identity of only 16%. The differences in the specificity for glycopeptides might result from a contribution of the lectin domain to the recognition of glycopeptide substrates. Previous studies with GalNAc-T2 and -T10 have demonstrated the contribution of the lectin domain in determining the site of glycosylation on glycopeptides (Raman et al. 2008).
GalNAc-T18. GalNAc-T18 forms a phylogenetic branch together with GalNAc-T8, -T9 and -T19. Members of this subfamily differ significantly in sequence from other GalNAc-Ts with a number of otherwise conserved residues altered. In addition, the only other member of this branch for which activity has been demonstrated is GalNAc-T9 (Zhang et al. 2003). Therefore, GalNAc-T8 and -T17 were also cloned and expressed as recombinant proteins in COS7 cells. Weak activity of GalNAc-T18 was observed with only 2 of the 12 peptides used in this study (Table I) with no activity against the remaining substrates. Reactions required extended assay times for activity to be detected. GalNAc-T19 could be expressed and purified but no expression could be detected for the gene product encoded by GalNAc-T8 (Figure 1) and no activity could be detected for GalNAc-T19 or -T8. A previous study using recombinant GalNAc-T19 expressed in insect cells also failed to detect activity (Nakamura et al. 2005).
Examination of a sequence alignment of all 20 members of the GalNAc-T family shows that several residues conserved among other members of the GalNAc-T family are altered in this phylogenetic branch. Many of these residues are involved in a binding acceptor peptide as revealed by the crystal structure of the ternary complex of GalNAc-T2 with UDP and the acceptor peptide EA2 (Fritz et al. 2006). This suggests that the GalNAc-T members of this branch may glycosylated very specific substrates in vivo and that the low activities we observed could be the consequence of a lack of appropriate acceptor peptides among the substrate pool used in this study.
GalNAc-T20. This transcript is unique among human isoforms in that it does not encode a lectin domain. The catalytic domain of this putative transferase is closest to GalNAc-T11 on the phylogenetic tree. Although the expression of recombinant protein could be detected (Figure 1), no transfer of GalNAc to the peptides used in this study was observed. It has been demonstrated previously that the lectin domain is not necessary for catalytic activity of either GalNAc-T2 or -T10 (Fritz et al. 2006; Raman et al. 2008). The lectin domain deletion construct of mouse GalNAc-T11 is also active (unpublished observations), suggesting that the lectin domain may not be critical for the function of this isoform. However, GalNAc-T20 also lacks one of four invariant cysteine residues that form two disulfide bonds within the catalytic domain of all known, active GalNAc-Ts and which, when mutated to alanine, have been shown to abolish activity in rat GalNAc-T1 (Tenno et al. 2002). In addition, Gln152 replaces an invariant Arg and Arg334 replaces the invariant Gly of the WGGEN motif. Nevertheless, it is possible that detecting activity for this isoform will depend on the identification of a unique substrate related to its restricted expression in human sperm (see section of transcript expression).
Splice variants of GalNAc-T13. GalNAc-T13 was previously cloned and the activity of the recombinant enzyme demonstrated (Zhang et al. 2003). It is most closely related to the ubiquitously expressed isoform, GalNAc-T1 with a sequence identity of 84%. Examination of the expression of various GalNAc-T isoforms in cultured human embryonic kidney (HEK) cells showed that the GalNAc-T13 was expressed at high levels. HEK cell-derived cDNA was therefore used to clone this enzyme using polymerase chain reaction (PCR) amplification with gene-specific primers (Bennett et al. 2012, Table S1). When clones were screened, three distinct insert sizes were observed. Sequencing of the inserts showed that one was identical to the sequence reported for GalNAc-T13, whereas the other two were splice variants. The coding region of GalNAc-T13 comprises 11 exons. In splice variant 1 (GalNAc-T13-V1), a 198-bp intron between exons 10 and 11 is not spliced out resulting in alteration of one half of the lectin domain. This variant has been reported from a library of human cDNA pooled from various tissues (BC101032.2) sequenced as part of an effort to sequence all full-length expressed sequence tags in humans and mouse (Strausberg et al. 2002). Variant 2 (GalNAc-T13-V2) is truncated at exon 6 resulting in an incomplete catalytic domain.
Figure 3A shows the sequence alignment of the cloned sequences of GalNAc-T13 and GalNAc-T13-V1. As can be seen, the latter half of the lectin domain is altered. However, disulphide-bonded cysteine residues are conserved as is the QXW motif in the third carbohydrate-binding subdomain within the lectin domain. Variant 2 showed no activity for the peptides tested, whereas both GalNAc-T13 and GalNAc-T13-V1 were active on a wide range of peptides with little difference between the activity of the two for the peptide and glycopeptide substrates assayed (Table I). To exclude the possibility that GalNAc-T13-V1 protein expressed in COS7 cells resulted from a transcript spliced to form GalNAc-T13, we also expressed GalNAc-T13-V1 from a transcript that could not be spliced into GalNAc-T13. Recombinant enzyme produced from the modified transcript exhibited activity similar to that produced from the non-modified transcript suggesting that the activity observed was indeed from the protein encoded by the splice variant.
Fig. 3.
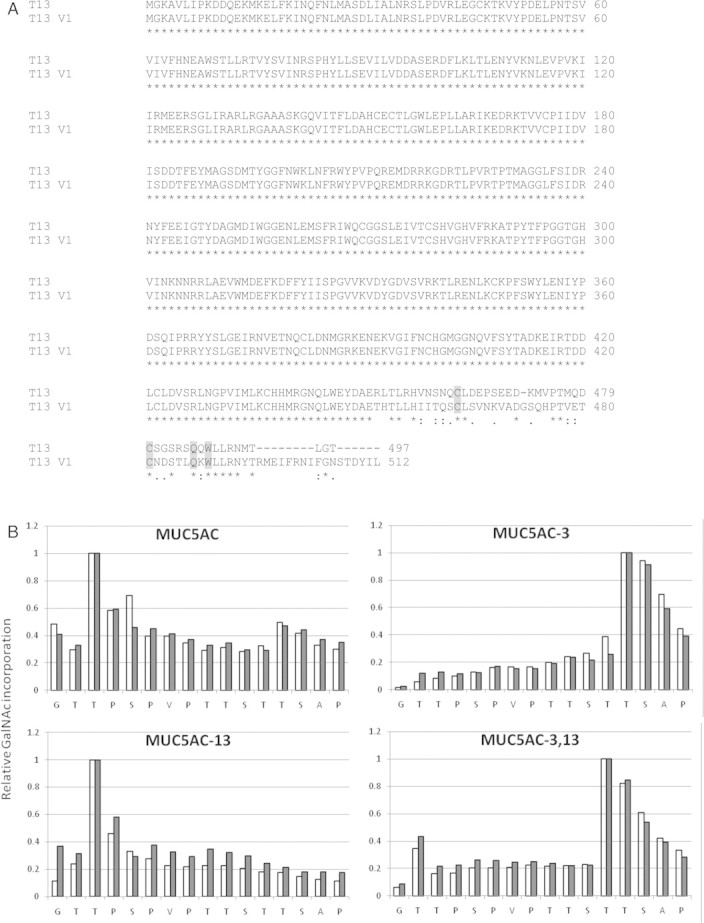
(A) Sequence alignment of the two active GalNAc-T13 splice variants (sequence accession nos BC101033.1-GalNAc-T13 and BC101032.2-GalNAc-T13 V1, the N-terminal-truncated versions used in this study are shown). Residues important for the structure and function of the lectin domain are shaded. “*”. “:” and “.” indicate identity, conserved substitution and semi-conserved substitution respectively. (B) Site of glycosylation of MUC5AC peptides by GalNAc-T13 splice variants. Open bars, GalNAc-T13; closed bars, GalNAc-T13V1. Peptides were glycosylated with UDP-[1-14C]GalNAc, subjected to the Edman degradation and radioactivity at each position determined to measure GalNAc addition. Data are expressed relative to the site of maximum incorporation, Thr-3 for MUC5AC, Thr-13 for MUC5AC-3, Thr-3 for MUC5AC-13 and Thr-12 for MUC5AC-3,13. Incorporation seen at sites of extant glycosylation in the glycopeptides and at residues lacking an acceptor hydroxyl group result from carryover counts from the previous position during the Edman sequencing.
The GalNAc-T lectin domain has been shown to influence activity on glycopeptide substrates and direct glycosylation site selection (Raman et al. 2008). We, therefore, compared the kinetics for glycosylation of MUC5AC-derived glycopeptides and determined the preferred site of glycosylation by the Edman sequencing. Table III lists the Vmax and Km values for GalNAc-T13 and GalNAc-T13-V1. Although the reaction velocities on both peptide and glycopeptide substrates and the Km values for the peptide substrates are comparable for the two variants, there is significant variation in the Km values for the glycopeptide substrates. The Km of V1 for MUC5AC-3 and MUC5AC-13 is ∼8- and 3-fold lower, respectively, compared with GalNAc-T13. The Edman sequencing of the products glycosylated with UDP-[1-14C]GalNAc, however, did not show any pronounced difference in the glycosylation site preference of the two GalNAc-T13 splice forms. Both forms glycosylate Thr-3 on MUC5AC, Thr-13 and Thr-12 on MUC5AC-3, Thr-3 on MUC5AC-13 and Thr-12 on MUC5AC-3,13 (Figure 3B). This is similar to the site preference of GalNAc-T1 (Ten Hagen et al. 2001) with which GalNAc-T13 shares the maximum sequence identity.
Table III.
Kinetic parameters for peptide glycosylation by GalNAc-T13 splice variants
| GalNAc-T13a |
GalNAc-T13-V1a |
|||
|---|---|---|---|---|
| Vmax (pmol/h) | Km (µM) | Vmax (pmol/h) | Km (µM) | |
| Muc5AC | 357 ±53 | 111 ± 12 | 326 ± 57 | 67 ± 10 |
| Muc5AC-3 | 815 ± 76 | 263 ± 39 | 522 ± 80 | 35 ± 6 |
| Muc5AC-13 | 309 ± 44 | 353 ± 21 | 458 ± 46 | 130 ± 15 |
| Muc5AC-3,13 | 97 ± 14c | >2000b | 215 ± 25c | >2000b |
| EA2 | 375 ± 38 | 124 ± 30 | 157 ± 25 | 108 ± 15 |
aEnzyme amount equivalent to GalNAc-T1 in Table II, as determined by western blot, was used for the assays. Significant differences are highlighted in bold.
bThe Km was higher than the maximum peptide concentrations used in the assay.
cValue represents activity at a peptide concentration of 2 mM.
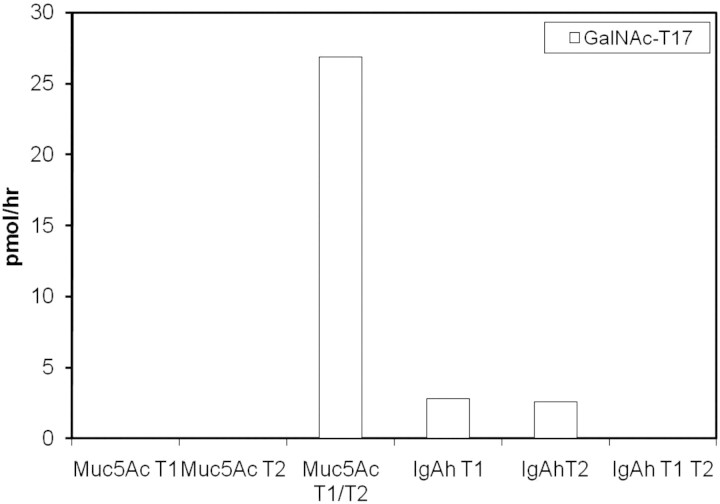
GalNAc-T17. Although this manuscript was being prepared, Peng et al. (2010) published a preliminary characterization of GalNAc-T17 (referred to as “GalNAc-T20” in their work) and our work corroborates their findings. This isoform is most closely related to GalNAc-T10 (70% sequence identity). These two isoforms form a branch on the phylogenetic tree along with GalNAc-T7. GalNAc-T7 and -T10 are the only transferases with a restricted preference for glycopeptides, i.e. substrates that are already decorated with GalNAc added by other isoforms (Bennett et al. 1999; Ten Hagen et al. 1999, 2001; Perrine et al. 2009). Therefore, we challenged GalNAc-T17 with several glycopeptides to gain additional insight into its substrate preferences. Like its nearest sequence neighbors, GalNAc-T17 was most active with the MUC5AC peptide that had been partially glycosylated by a combination of GalNAc-T1 and -T2. However, no activity was observed with MUC5AC glycosylated with either GalNAc-T1 or -T2 alone (Figure 4) or with the synthetic MUC5AC glycopeptides used in this study. This indicates that GalNAc-T17 has a substrate specificity distinct from both GalNAc-T7 and -T10, each of which can glycosylate MUC5AC glycopeptides generated by the action of either GalNAc-T1 or -T2 (Tetaert et al. 2001). The specific glycosylated species that forms the substrate for GalNAc-T17 could not be purified and identified due to the heterogeneity of the peptide preparation obtained by partial glycosylation of MUC5AC by GalNAc-T1 and -T2.
Fig. 4.
Activity of GalNAc-T17 on glycopeptides. MUC5AC and IgA hinge region peptides were first glycosylated with either or both GalNAc-T1 and GalNAc-T2 in the presence of a 2-fold excess of a donor substrate, purified on a Sep-Pak C-18 cartridge and used as a substrate for GalNAc-T17. No activity was detected on any other peptides listed in Table I.
Expression of the previously uncharacterized GalNAc-T transcripts
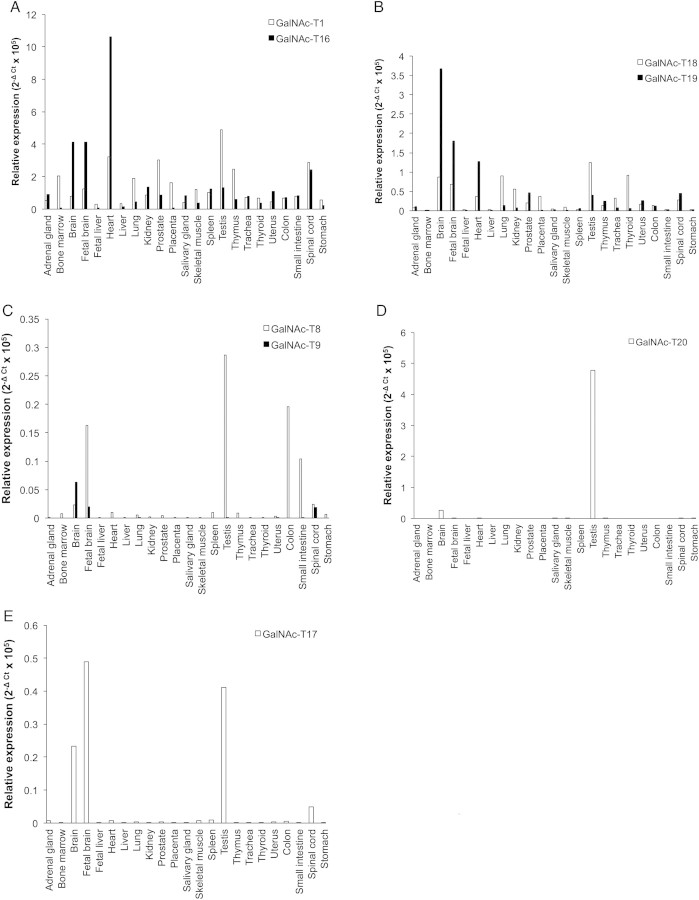
The expression of the transcripts encoding the isoforms described in this manuscript was examined by real-time PCR using two human multiple tissue RNA arrays with the expression of GalNAc-T1 examined for comparison (Figure 5). It should be noted that these expression levels represent a specific time during development and may vary with parameters such as developmental stage and gender. Of the putative isoforms characterized here, GalNAc-T16 and GalNAc-T18 transcripts were found to be expressed in all tissues examined. In the adult heart and adult and fetal brain, transcripts of GalNAc-T16 were more than twice as abundant as those of GalNAc-T1 (Figure 5A) making it one of the most highly expressed isoforms in these tissues. The expression of GalNAc-T18 message was much lower than GalNAc-T16, with the testis, thyroid and adult brain containing the highest levels of transcript (Figure 5B). This is in contrast to the expression of the mouse orthologs. Mouse GalNAc-T16 (GalNAc-Ta in Young et al. 2003) was shown to have maximum expression in the brain with lower levels in the lung. GalNAc-T18 (GalNAc-T8 in Young et al. 2003) was detected only in the lung in mice (Young et al. 2003). These two isoforms constitute the most strongly expressed isoforms after GalNAc-T1 and -T2 in these mouse tissues, respectively (Young et al. 2003).
Fig. 5.
Gene expression of GalNAc-Ts in human tissues as determined by quantitative real-time PCR. Shown are relative expression levels for (A) GalNAc-T1 and GalNAc-T16, (B) GalNAc-T18 and GalNAc-T19, (C) GalNAc-T8 and GalNAc-T9, (D) GalNAc-T20 and (E) GalNAc-T17.
Transcript levels of the remaining isoforms were much more modest. Transcripts of GalNAc-T19 were detected at low levels in a wide variety of tissue with highest expression observed in the adult and fetal brain and heart in agreement with previous reports (Figure 5B; Nakamura et al. 2005). This isoform has been reported to be expressed in neuronal but not glial cells of the rat cerebellum, hippocampus, thalamus and cerebral cortex. GalNAc-T8, without detectable activity, was found to have low levels of expression in the testis, colon and fetal brain (Figure 5C). This is in contrast to the studies by White et al. (2000) who reported expression in a wide variety of tissue including the skeletal muscle, heart, kidney and liver. The close homolog of this isoform, GalNAc-T9, was expressed at very low levels with transcript detected in the brain (Figure 5C), in agreement with earlier reports that this isoform is brain-specific (Toba et al. 2000). GalNAc-T20 was expressed exclusively in the testis (Figure 5D), a pattern similar to the expression of the mouse ortholog designated GalNAc-TB in Young et al. (2003). GalNAc-T17 showed limited expression in the adult and fetal brain, spinal cord and testis (Figure 5E), in agreement with that reported by Peng et al. (2010).
Discussion
In this study, we demonstrate that two additional putative transferases, GalNAc-T16 and -T18 are active. With this, 17 of the 20 human GalNAc-Ts are shown to be bona fide transferases. We were unable to detect activity for GalNAc-T19 and -T20 and no expression was detected for GalNAc-T8. The classification of these three genes as GalNAc-Ts remains putative, pending demonstration of their ability to glycosylate Ser/Thr residues of proteins.
The members of the GalNAc-T family differ in the tissue-specific expression of their transcripts and in their enzymatic substrate specificity. Previous studies demonstrated that the transcripts of GalNAc-T1, -T2 and -T10 are the most widely expressed, whereas the messages for other isoforms have a more restricted expression profile. GalNAc-Ts also differ in their in vitro substrate preferences and have been classified as early, middle and late transferases based on their preference for peptide or glycopeptide substrates (Ten Hagen et al. 1999, 2003; Pratt et al. 2004).
Of the isoforms characterized in the present study, the GalNAc-T16 transcript shows the most widespread expression and the recombinant enzyme the highest activity on a wide variety of substrates. Surprisingly, this isoform was expressed at higher levels than GalNAc-T1 in some tissues including the heart and brain. This suggests that it can be classified, together with GalNAc-T1 and -T2 as an early GalNAc-T, catalyzing the initial addition of GalNAc to nascent proteins. GalNAc-T17, on the other hand, appears to be a very late transferase, catalyzing the transfer of GalNAc to proteins that have already been extensively glycosylated by multiple other GalNAc-Ts. The substrate specificity of GalNAc-T16 is distinct from that of both GalNAc-T1 and -T2 suggesting a unique cellular function. The Xenopus ortholog of GalNAc-T16 has been suggested to play a role in early development with the expression of the isoform correlating with suppression of transforming growth factor-β signaling. Inhibition of expression of this enzyme in Xenopus results in developmental defects of the anterior neural crest, anterior notochord and spinal cord (Herr et al. 2008).
GalNAc-T18 along with GalNAc-T8, -T9 and -T19 constitutes a phylogenetic subfamily that differs significantly in sequence from the other members of the GalNAc-T family. A number of catalytic domain residues conserved across other GalNAc-Ts are altered in these transferases. The most striking alteration is the substitution of Tyr for Trp in the conserved WGGE motif. This Trp, which is conserved not only in the GalNAc-T family but also in other related glycosyltransferases like the β1-4 galactosyltransferase, appears to “flip” during an important loop movement during catalysis (Gunasekaran et al. 2003; Milac et al. 2007). Evidently, a Tyr at this position is able to fulfill this role during catalysis. Three of the changes map to residues that contact the acceptor substrate in the structure suggesting unique substrate specificity. In spite of these differences, weak activity could also be detected for GalNAc-T18, as for GalNAc-T9, indicating that the various amino acid substitutions compensate each other resulting in an active enzyme. The sequence changes may be necessary for the glycosylation of as yet unidentified brain-specific substrates where these GalNAc-Ts are expressed.
The demonstration of an active splice variant of GalNAc-T13 in which the lectin domain is altered adds another dimension to the complexity that already exists due to the large number of functional GalNAc-T genes. We have previously shown that the lectin domain directs site selection on glycopeptide substrates by GalNAc-T2 (Raman et al. 2008). This raises the possibility that the alternate splicing might result in variation in the carbohydrate specificity of the lectin domain and represents a novel mechanism of generating additional complexity in this already large family. The different affinities exhibited by the splice variants for glycopeptides substrates support this premise. Conversely, the presence of a splice variant that lacks a complete catalytic domain may also represent a post-transcriptional mechanism to diminish enzyme activity.
This comprehensive description of the GalNAc transferase family underscores the complexity of mucin-type O-glycosylation regulation. In vitro substrate preferences suggest that the different family members play unique roles by adding GalNAc early, mid and late in the post-translational modification of substrates. A number of transferases including GalNAc-T16 characterized in this study prefer “naked” peptides, suggesting their role earlier in the glycosylation process. Recently, GalNAc-T17 was shown to exclusively glycosylate glycopeptides, like its close homolog of GalNAc-T10 (Peng et al. 2010). We have also observed activity of this isoform on the MUC5AC peptide after partial glycosylation with a combination of GalNAc-T1 and -T2 but not either one alone (Figure 4). GalNAc-T10 along with -T7 and -T17 thus constitute late transferases, coming into play only after the early and mid-transferases. Such a hierarchical strategy would yield a fine level of control.
Pulse chase analysis of rat submandibular gland mucin biosynthesis indicates that the addition of core GalNAc residues does not occur simultaneously (Nehrke and Tabak 1997). Temporally, it is unknown whether all GalNAcs are first added to the protein core and then subsequently elongated by the addition of galactose, N-acetylglucosamine and/or sialic acid, or whether GalNAc elongation can begin prior to final GalNAc addition. However, there is some evidence that GalNAc-Ts are localized throughout the Golgi and thus initiation of O-glycosylation may not be restricted to the cis-Golgi supporting the latter possibility (Rottger et al. 1998). If there is simultaneous addition and elongation, it would seem plausible that the resultant structures may influence acquisition of late GalNAcs. The substrate preferences of “late acting” GalNAc-Ts have not yet included more completed forms of the O-glycan chains. While significant challenges remain, the completion of the GalNAc-T “family tree” represents an important first step in understanding the functional role(s) played by each GalNAc-T isoform and in decoding the O-glycome.
Methods
Cloning and expression of GalNAc transferases
cDNA clones of the human isoforms used in this study were purchased from commercial sources. RNA isolated from HEK cells was used to amplify isoform GalNAc-T8 and -T13 by reverse transcriptase–PCR. Transmembrane and partial stem region deletions of these genes were PCR amplified and cloned into the Mlu1/Age1 restriction sites of the expression vector pIMKF-4 (Ten Hagen et al. 2001) using the primers listed in Bennett et al. 2012, Table S1. Expression vectors were transfected into COS7 cells using Lipofectamine 2000 (Life Technologies, Grand Island, NY) according to the manufacturer's protocol. Proteins were expressed for 36 h at 37°C and recombinant transferases were purified using anti-FLAG™ m2 agarose (Sigma, St. Louis, MO) according to the manufacturer's protocol. Recombinant proteins were eluted with 100 μM FLAG peptide. FLAG peptide was removed by dialysis against Tris-buffered saline, pH 7.5. Purified enzymes were detected by chemiluminescence and quantified by densitometry on western blots probed with rabbit anti-FLAG™ antibody followed by anti-rabbit antibody conjugated to HRP.
Enzyme assays
Enzyme assays were done as previously described (Hagen et al. 1997) using the panel of peptides and glycopeptides. Kinetic analysis was carried out primarily on the peptides EA2 (PTTDSTTPAPTTK), MUC5AC (GTTPSPVPTTSTTSAP) peptide and glycosylated derivatives of MUC5AC carrying a GalNAc at Thr-3 or Thr-13 or Thr-3 and Thr-13. The MUC5AC peptide used for this study was synthesized by the Facility for Biotechnology Resources, Center for Biologics Evaluation and Research at the National Institutes of Health and glycopeptides MUC5AC-3, MUC5AC-13 and MUC5AC-3,13 were synthesized by Anaspec (San Jose, CA). The complete list of peptides on which the transferases were assayed is presented in Table I. Assays contained 500 µM peptide and 50 µM UDP-[1-14C]GalNAc (0.18 µCi/mmol). Reactions were initiated by the addition of enzyme and allowed to proceed at 37°C for 1–3 h. For the kinetic analysis, peptide concentrations were varied between 30 and 500 µM or between 2 mM and 125 µM with a UDP-GalNAc concentration of 100 µM (0.18 µCi/mmol). Reactions were allowed to proceed for 3 h and stopped by the addition of 0.1% trifluoroacetic acid (TFA). The reactions were then loaded onto Sep-pak C-18 reverse phase cartridges (Waters, NJ) equilibrated with 0.1% TFA in water, the column washed with 0.1% TFA and 5% methanol three times to remove unreacted UDP-GalNAc and hydrolyzed GalNAc and the bound peptide/glycopeptide eluted with 0.1% TFA and 70% methanol. Incorporated radiolabel was determined by liquid scintillation counting. Activities presented are for quantities of enzyme equivalent to that of GalNAc-T1 as determined by densitometric scanning of western blots probed with anti-FLAG™ antibody. The activity of this amount of GalNAc-T1 on the panel of substrates is shown in Tables I and II.
Determination of sites of GalNAc incorporation
Reactions were carried out with 500 µM peptide and ∼7 µM UDP-[1-14C]GalNAc (2.25 µCi/mmol) for 16 h at 37°C. Peptides were purified using Sep-pak C-18 cartridges as described above and subjected to the Edman degradation as described previously (Gerken et al. 1998). Radiolabel incorporation at each position was determined by liquid scintillation counting as described (Perrine et al. 2009).
Quantitative analysis by real-time PCR
Two human RNA panels were purchased from Clontech (Mountain View, CA) and Agilent Technologies (Santa Clara, CA). Complimentary DNA was generated using the High Capacity cDNA Reverse Transcription Kit with RNase Inhibitor (Applied Biosystems, Foster City, CA). TaqMan gene expression assays used are listed in Bennett et al. 2012, Table S2. Thermal cycling conditions used followed the manufacturer's recommendations (50°C for 2 min; 95°C for 10 min; 40 cycles of 95°C for 15 s and 60°C for 1 min). Relative expression levels were calculated using the comparative computed tomography method and normalized to 18S rRNA endogenous control. The data, averaged over the two RNA sources, are presented as 1-fold difference to 18S RNA so that higher values indicate stronger expression.
Supplementary data
Supplementary data for this article is available online at http://glycob.oxfordjournals.org/.
Funding
Supported by funds of the intramural program of NIDDK, NIH to LAT and the National Cancer Institute grant R01-CA-78834 to TAG.
Note added in proof
ppGalNAc-T18 described by Li et al, 2011, doi: 10.1093/glycob/cwr179, published while this manuscript was under review, corresponds to GalNAc-T18 described here.
Conflict of interest
None declared.
Abbreviations
GalNAc-T, polypeptide N-acetylgalactosaminyltransferase; HEK, human embryonic kidney; HRP, horseradish peroxidase; PCR, polymerase chain reaction; TFA, trifluoroacetic acid; α-GalNAc, α-N-acetylgalactosamine.
Supplementary Material
Acknowledgements
We thank Dr. Kelly Ten Hagen and Dr. Timothy A. Fritz for helpful suggestions.
References
- Bennett EP, Mandel U, Clausen H, Gerken TA, Fritz TA, Tabak LA. Control of mucin-type O-glycosylation: A classification of the polypeptide GalNAc-transferase gene family. Glycobiology. 2012;22:736–756. doi: 10.1093/glycob/cwr182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett EP, Hassan H, Hollingsworth MA, Clausen H. A novel human UDP-N-acetyl-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase, GalNAc-T7, with specificity for partial GalNAc-glycosylated acceptor substrates. FEBS Lett. 1999;460:226–230. doi: 10.1016/s0014-5793(99)01268-5. doi:10.1016/S0014-5793(99)01268-5. [DOI] [PubMed] [Google Scholar]
- Carraway KL, 3rd, Funes M, Workman HC, Sweeney C. Contribution of membrane mucins to tumor progression through modulation of cellular growth signaling pathways. Curr Top Dev Biol. 2007;78:1–22. doi: 10.1016/S0070-2153(06)78001-2. doi:10.1016/S0070-2153(06)78001-2. [DOI] [PubMed] [Google Scholar]
- Cullen PJ. Signaling mucins: The new kids on the MAPK block. Crit Rev Eukaryot Gene Expr. 2007;17:241–257. doi: 10.1615/critreveukargeneexpr.v17.i3.50. [DOI] [PubMed] [Google Scholar]
- Fritz TA, Hurley JH, Trinh LB, Shiloach J, Tabak LA. The beginnings of mucin biosynthesis: The crystal structure of UDP-GalNAc:polypeptide α-N-acetylgalactosaminyltransferase-T1. Proc Natl Acad Sci USA. 2004;101:15307–15312. doi: 10.1073/pnas.0405657101. doi:10.1073/pnas.0405657101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz TA, Raman J, Tabak LA. Dynamic association between the catalytic and lectin domains of human UDP-GalNAc:polypeptide α-N-acetylgalactosaminyltransferase-2. J Biol Chem. 2006;281:8613–8619. doi: 10.1074/jbc.M513590200. doi:10.1074/jbc.M513590200. [DOI] [PubMed] [Google Scholar]
- Gerken TA, Owens CL, Pasumarthy M. Site-specific core 1 O-glycosylation pattern of the porcine submaxillary gland mucin tandem repeat. J Biol Chem. 1998;273:26580–26588. doi: 10.1074/jbc.273.41.26580. doi:10.1074/jbc.273.41.26580. [DOI] [PubMed] [Google Scholar]
- Gerken TA, Ten Hagen KG, Jamison O. Conservation of peptide acceptor preferences between Drosophila and mammalian polypeptide-GalNAc transferase ortholog pairs. Glycobiology. 2008;18:861–870. doi: 10.1093/glycob/cwn073. doi:10.1093/glycob/cwn073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasekaran K, Ma B, Ramakrishnan B, Qasba PK, Nussinov R. Interdependence of backbone flexibility, residue conservation, and enzyme function: A case study on β1,4-galactosyltransferase-I. Biochemistry. 2003;42:3674–3687. doi: 10.1021/bi034046r. doi:10.1021/bi034046r. [DOI] [PubMed] [Google Scholar]
- Hagen FK, Ten Hagen KG, Beres TM, Balys MM, VanWuyckhuyse BC, Tabak LA. cDNA cloning and expression of a novel UDP-N-acetyl-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase. J Biol Chem. 1997;272:13843–13848. doi: 10.1074/jbc.272.21.13843. doi:10.1074/jbc.272.21.13843. [DOI] [PubMed] [Google Scholar]
- Herr P, Korniychuk G, Yamamoto Y, Grubisic K, Oelgeschlager M. Regulation of TGF-β signalling by N-acetylgalactosaminyltransferase-like 1. Development. 2008;135:1813–1822. doi: 10.1242/dev.019323. doi:10.1242/dev.019323. [DOI] [PubMed] [Google Scholar]
- Kubota T, Shiba T, Sugioka S, Furukawa S, Sawaki H, Kato R, Wakatsuki S, Narimatsu H. Structural basis of carbohydrate transfer activity by human UDP-GalNAc:polypeptide α-N-acetylgalactosaminyltransferase (pp-GalNAc-T10) J Mol Biol. 2006;359:708–727. doi: 10.1016/j.jmb.2006.03.061. doi:10.1016/j.jmb.2006.03.061. [DOI] [PubMed] [Google Scholar]
- Li X, Wang J, Li W, Xu Y, Shao D, Xie Y, Xie W, Kubota T, Narimatsu H, Zhang Y. Characterization of ppGalNAc-T18, a member of the vertebrate-specific Y subfamily of UDP-N-acetyl-α-D-galactosamine:polypeptide N-acetylgalactosaminyltransferases. Glycobiology. 2011;22:602–615. doi: 10.1093/glycob/cwr179. [DOI] [PubMed] [Google Scholar]
- Linden SK, Sutton P, Karlsson NG, Korolik V, McGuckin MA. Mucins in the mucosal barrier to infection. Mucosal Immunol. 2008;1:183–197. doi: 10.1038/mi.2008.5. doi:10.1038/mi.2008.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milac AL, Buchete NV, Fritz TA, Hummer G, Tabak LA. Substrate-induced conformational changes and dynamics of UDP-N-acetylgalactosamine:polypeptide N-acetylgalactosaminyltransferase-2. J Mol Biol. 2007;373:439–451. doi: 10.1016/j.jmb.2007.08.028. doi:10.1016/j.jmb.2007.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Toba S, Hirai M, Morishita S, Mikami T, Konishi M, Itoh N, Kurosaka A. Cloning and expression of a brain-specific putative UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase gene. Biol Pharm Bull. 2005;28:429–433. doi: 10.1248/bpb.28.429. doi:10.1248/bpb.28.429. [DOI] [PubMed] [Google Scholar]
- Nehrke K, Tabak LA. Biosynthesis of a low-molecular-mass rat submandibular gland mucin glycoprotein in COS7 cells. Biochem J. 1997;323(Pt 2):497–502. doi: 10.1042/bj3230497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C, Togayachi A, Kwon YD, Xie C, Wu G, Zou X, Sato T, Ito H, Tachibana K, Kubota T. Identification of a novel human UDP-GalNAc transferase with unique catalytic activity and expression profile. Biochem Biophys Res Commun. 2010;402:680–686. doi: 10.1016/j.bbrc.2010.10.084. doi:10.1016/j.bbrc.2010.10.084. [DOI] [PubMed] [Google Scholar]
- Perrine CL, Ganguli A, Wu P, Bertozzi CR, Fritz TA, Raman J, Tabak LA, Gerken TA. Glycopeptide-preferring polypeptide GalNAc transferase 10 (ppGalNAc T10), involved in mucin-type O-glycosylation, has a unique GalNAc-O-Ser/Thr-binding site in its catalytic domain not found in ppGalNAc T1 or T2. J Biol Chem. 2009;284:20387–20397. doi: 10.1074/jbc.M109.017236. doi:10.1074/jbc.M109.017236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt MR, Hang HC, Ten Hagen KG, Rarick J, Gerken TA, Tabak LA, Bertozzi CR. Deconvoluting the functions of polypeptide N-α-acetylgalactosaminyltransferase family members by glycopeptide substrate profiling. Chem Biol. 2004;11:1009–1016. doi: 10.1016/j.chembiol.2004.05.009. doi:10.1016/j.chembiol.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Raman J, Fritz TA, Gerken TA, Jamison O, Live D, Liu M, Tabak LA. The catalytic and lectin domains of UDP-GalNAc:polypeptide α-N-acetylgalactosaminyltransferase function in concert to direct glycosylation site selection. J Biol Chem. 2008;283:22942–22951. doi: 10.1074/jbc.M803387200. doi:10.1074/jbc.M803387200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottger S, White J, Wandall HH, Olivo JC, Stark A, Bennett EP, Whitehouse C, Berger EG, Clausen H, Nilsson T. Localization of three human polypeptide GalNAc-transferases in HeLa cells suggests initiation of O-linked glycosylation throughout the Golgi apparatus. J Cell Sci. 1998;111(Pt 1):45–60. doi: 10.1242/jcs.111.1.45. [DOI] [PubMed] [Google Scholar]
- Sheehan JK, Kesimer M, Pickles R. Innate immunity and mucus structure and function. Novartis Found Symp. 2006;279:155–166. discussion 167–159, 216–159 doi:10.1002/9780470035399.ch13. [PubMed] [Google Scholar]
- Singh PK, Hollingsworth MA. Cell surface-associated mucins in signal transduction. Trends Cell Biol. 2006;16:467–476. doi: 10.1016/j.tcb.2006.07.006. doi:10.1016/j.tcb.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler GD, Altschul SF, et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci USA. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. doi:10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabak LA. In defense of the oral cavity: Structure, biosynthesis, and function of salivary mucins. Annu Rev Physiol. 1995;57:547–564. doi: 10.1146/annurev.ph.57.030195.002555. doi:10.1146/annurev.ph.57.030195.002555. [DOI] [PubMed] [Google Scholar]
- Ten Hagen KG, Bedi GS, Tetaert D, Kingsley PD, Hagen FK, Balys MM, Beres TM, Degand P, Tabak LA. Cloning and characterization of a ninth member of the UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase family, ppGaNTase-T9. J Biol Chem. 2001;276:17395–17404. doi: 10.1074/jbc.M009638200. doi:10.1074/jbc.M009638200. [DOI] [PubMed] [Google Scholar]
- Ten Hagen KG, Fritz TA, Tabak LA. All in the family: The UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases. Glycobiology. 2003;13:1R–16R. doi: 10.1093/glycob/cwg007. doi:10.1093/glycob/cwg007. [DOI] [PubMed] [Google Scholar]
- Ten Hagen KG, Tetaert D, Hagen FK, Richet C, Beres TM, Gagnon J, Balys MM, VanWuyckhuyse B, Bedi GS, Degand P. Characterization of a UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase that displays glycopeptide N-acetylgalactosaminyltransferase activity. J Biol Chem. 1999;274:27867–27874. doi: 10.1074/jbc.274.39.27867. doi:10.1074/jbc.274.39.27867. [DOI] [PubMed] [Google Scholar]
- Tenno M, Toba S, Kezdy FJ, Elhammer AP, Kurosaka A. Identification of two cysteine residues involved in the binding of UDP-GalNAc to UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1) Eur J Biochem. 2002;269:4308–4316. doi: 10.1046/j.1432-1033.2002.03123.x. doi:10.1046/j.1432-1033.2002.03123.x. [DOI] [PubMed] [Google Scholar]
- Tetaert D, Ten Hagen KG, Richet C, Boersma A, Gagnon J, Degand P. Glycopeptide N-acetylgalactosaminyltransferase specificities for O-glycosylated sites on MUC5AC mucin motif peptides. Biochem J. 2001;357:313–320. doi: 10.1042/0264-6021:3570313. doi:10.1042/0264-6021:3570313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toba S, Tenno M, Konishi M, Mikami T, Itoh N, Kurosaka A. Brain-specific expression of a novel human UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase (GalNAc-T9) Biochim Biophys Acta. 2000;1493:264–268. doi: 10.1016/s0167-4781(00)00180-9. [DOI] [PubMed] [Google Scholar]
- White KE, Lorenz B, Evans WE, Meitinger T, Strom TM, Econs MJ. Molecular cloning of a novel human UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase, GalNAc-T8, and analysis as a candidate autosomal dominant hypophosphatemic rickets (ADHR) gene. Gene. 2000;246:347–356. doi: 10.1016/s0378-1119(00)00050-0. doi:10.1016/S0378-1119(00)00050-0. [DOI] [PubMed] [Google Scholar]
- Young WW, Jr, Holcomb DR, Ten Hagen KG, Tabak LA. Expression of UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase isoforms in murine tissues determined by real-time PCR: A new view of a large family. Glycobiology. 2003;13:549–557. doi: 10.1093/glycob/cwg062. doi:10.1093/glycob/cwg062. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Iwasaki H, Wang H, Kudo T, Kalka TB, Hennet T, Kubota T, Cheng L, Inaba N, Gotoh M, et al. Cloning and characterization of a new human UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase, designated pp-GalNAc-T13, that is specifically expressed in neurons and synthesizes GalNAc alpha-serine/threonine antigen. J Biol Chem. 2003;278:573–584. doi: 10.1074/jbc.M203094200. doi:10.1074/jbc.M203094200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.