Abstract
The conserved oligomeric Golgi complex (COG) is a hetero-octomeric peripheral membrane protein required for retrograde vesicular transport and glycoconjugate biosynthesis within the Golgi. Mutations in subunits 1, 4, 5, 6, 7 and 8 are the basis for a rare inheritable human disease termed congenital disorders of glycosylation type-II. Defects to COG complex function result in aberrant glycosylation, protein trafficking and Golgi structure. The cellular function of the COG complex and its role in protein glycosylation are not completely understood. In this study, we report the first detailed structural analysis of N-glycans from a COG complex-deficient organism. We employed sequential ion trap mass spectrometry of permethylated N-glycans to demonstrate that the COG complex is essential for the formation of fucose-rich N-glycans, specifically antennae fucosylated structures in Caenorhabditis elegans. Our results support the supposition that disruption to the COG complex interferes with normal protein glycosylation in the medial and/or trans-Golgi.
Keywords: C. elegans, conserved oligomeric Golgi complex, fucosylation, N-glycosylation, sequential ion trap mass spectrometry
Introduction
The Golgi serves not only as the core of the secretory pathway, but it is essential for the glycosylation and trafficking of glycoproteins, glycolipids and proteoglycans. Glycosylation of proteins in the Golgi accounts for the diversification of N- and O-linked glycans and glycosaminoglycans. The Golgi acts by precisely sorting glycosylation enzymes, nascent glycoproteins and substrate transporters throughout the Golgi cisternal stacks in a non-uniform steady-state distribution (Nilsson et al. 1993; Rabouille et al. 1995; Opat et al. 2001). Transcriptional control of glycosylation enzymes involved in synthesis, modification and catabolism is believed to be the driving force in glycan diversity and abundance (Nairn et al. 2008). However, the fine details and changes in glycan structure cannot be accounted for by transcriptional regulation of glycosyltransferases or glycosidases alone. It is known that sequential modifications of glycoproteins by glycosyltransferases also depend on the precise localization and abundance of resident glycosylation enzymes within the Golgi (Rabouille et al. 1995) as well as the duration a glycoprotein traverses through the Golgi stack (Wang et al. 1991). To this end, an understanding of the secretory pathway and Golgi-associated functions is fundamental to fully characterize the diverse structural products.
As glycoproteins rapidly traverse the Golgi, resident enzymes, substrates and substrate transporters migrate from the cis- or medial-Golgi toward the trans-Golgi network. The outflow or mislocalization of resident enzymes is rectified by retrograde transport of specific material back toward the correct Golgi cisternae. This is facilitated by Golgi-specific fusion and tethering factors such as the conserved oligomeric Golgi complex (COGC), COPI and COPII coat proteins, Rab GTPases, soluble N-ethylmaleimide-sensitive factor attachment protein receptors and SM proteins (Smith and Lupashin 2008). Of these proteins, the COG complex is of considerable interest because of its association with a group of inherited autosomal recessive human diseases called congenital disorders of glycosylation (CDG). Mutations of the COG complex subunits 1, 4, 5, 6, 7 and 8 cause CDGs in humans (Wu et al. 2004; Spaapen et al. 2005; Foulquier et al. 2006, 2007; Kranz et al. 2007; Paesold-Burda et al. 2009; Reynders et al. 2009; Lubbehusen et al.). The severity of the disease varies and symptoms that include mental retardation, perinatal asphyxia, acute encephalopathy, alternating esotropia, pseudoptosis, hypotonia, hepatosplenomegaly, ventricular hypertrophy, growth retardation, progressive microcephaly, epileptic seizures and failure to thrive.
The COG complex is an eight subunit protein consisting of two lobes: COG 1–4 form lobe A and COG 5–8 form lobe B (Ungar et al. 2002; Loh and Hong 2004). Deletions of any subunit in lobe A lead to severe growth defects in yeast, whereas subunit deletions in lobe B have only mild consequences (Whyte and Munro 2001; Ram et al. 2002). Mutations of COG subunits 1 and 2 in Chinese hampster ovary (CHO) cells damage the integrity of the Golgi complex, causing intracellular abnormalities in protein sorting and secretion, with subsequent defects in cell growth, disruption of glycoprotein trafficking and protein glycosylation (Kingsley et al. 1986; Ungar et al. 2002; Oka et al. 2004, 2005; Shestakova et al. 2006). Interestingly, all eight mammalian COG components are conserved in Caenorhabditis elegans (Kubota and Nishiwaki 2006).
Investigations of the COG complex began with the identification of low-density lipoprotein receptor (LDLR)-deficient CHO mutants (Krieger et al. 1981). In this study, the mutant CHO cells showed changes in LDLR electrophoretic mobility that was attributed to the absence of N-glycans. The mutation in the LDLR-deficient CHO cells (IdlB and IdlC) was later found to be defects that occurred in subunits 1 and 2 in lobe A of the COG complex and resulted in alterations of cell surface glycoconjugates, namely the absence of galactose and sialic acid residues on the N-glycan terminus and heterogeneity of sialic acid content on O-glycans (Kingsley et al. 1986; Reddy and Krieger 1989). Abnormal glycosylation attributed to Golgi function have also been reported in yeast mutants with COG defects (Whyte and Munro 2001; Conde et al. 2003; Corbacho et al. 2004), but the specific glycan structural defects were not reported. A recent report by Peanne et al. (2011) demonstrated a depletion of lobe A of the COG complex (subunit 3 RNAi knockdown) in HeLa cells resulted in Golgi structural instability, whereas a decrease in lobe B (subunit 7 knockdown) resulted in a decrease in β1,4-galactosyltransferase 1 and α2,6-sialyltransferase expression. Interestingly, in the lobe A defective experiments, the glycosyltransferase expression levels were not decreased. In a short lobe, lobe A is required for the precise localization of terminal glycosyltransferases, but lobe B is required for the availability of terminal glycosylation enzymes in the Golgi.
Recently, a C. elegans strain (NF299) containing a mutation in the subunit 1 of the COG complex was created using a Tc1 transposon (Kubota et al. 2006). In C. elegans, the COG complex acts in the glycosylation of MIG-17, a member of the ADAM family of proteases, that is required for gonadogenesis. Worms lacking the cogc-1 gene show altered gonad morphology due to misdirection of the distal tip (Kubota et al. 2006). Kubota et al. prepared a model for the role of the COG complex-dependent (CCD) glycosylation of the MIG-17 ADAM protease. In brief, the COG complex is required for sorting or stabilizing MIG-23, a Golgi nucleoside phosphatase, along the body wall muscles and converts uridine diphosphate (UDP) to uridine monophosphate (UMP). UMP export from the Golgi is crucial for the higher energy UDP-sugar to be transported into the Golgi. The UDP-sugar is required for MIG-17 glycosylation. Therefore, the COG complex is indirectly required for the glycosylation of MIG-17.
The COG complex has been studied considerably at the cellular level, but its role in N-glycosylation during animal development remains unknown. To expand the current understanding of how the COG complex regulates N-glycosylation in the Golgi, we performed a detailed approach using ion trap mass spectrometry (ITMSn). A comparative N-glycan analysis was performed between the single gene mutant of the COG complex subunit 1 and N2 Bristol wild-type C. elegans strains. Our results showed that the COG complex is required for the formation of fucose-rich N-glycans and the addition of fucose to terminal residues on N-glycan antennae.
Results
Matrix-assisted laser desorption ionization-time of flight mass spectrometry of reduced/permethylted N-glycans
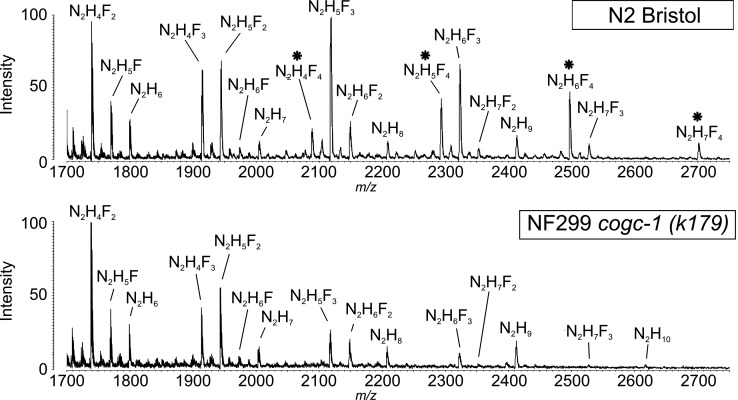
Mass spectrometry (MS) spectra of hydrazine-released reduced and permethylated N-linked glycans from N2 and NF299 illustrate the differences in compositions from whole cell lysates (Figure 1). Most notable was the absence of compositions containing four fucose residues in the NF299 strain. These compositions included N2H4F4 (m/z 2088.1), N2H5F4 (m/z 2292.2), N2H6F4 (m/z 2496.2) and N2H7F4 (m/z 2700.3), where N = HexNAc [e.g. N-acetylglucosamine (GlcNAc), N-acetylgalactosamine], H = Hexose (e.g. galactose, mannose, glucose), F = Deoxyhexose (e.g. fucose). Present in all compositions were the trimannosyl-chitobiose core (Manα1-3,Manα1,6)Manβ1-4GlcNAcβ1-4GlcNAc. In addition to the presence of mannose residues on the N-glycan core, the remaining hexose residues were presumably mannose added to the trimannosyl-chitobiose core in an α1-3, α1-4 or α1-6 linkage or galactose residues linked β1-4 to the Fucα1-6GlcNAc or Fucα1-3GlcNAc on each of the two core GlcNAcs. Also, compositions with three fucose residues are decreased in the NF299 strain. Moreover, N2H10 was detected in the NF299 strain, but was not present in the N2 spectrum.
Fig. 1.
MALDI-TOF profiles of reduced and permethylated N-glycans following hydrazinolysis from N2 Bristol and NF299 cogc-1 (k179) C. elegans strains. Fucose-rich glycans, which are absent in NF299, are marked with an asterisk and represent compositions with four fucose residues. N, N-acetylhexosamine (e.g. GlcNAc, N-acetylgalactosamine); H, hexose (e.g. galactose, mannose, glucose); F, deoxyhexose (e.g. fucose).
Relative abundance of N-glycan compositions
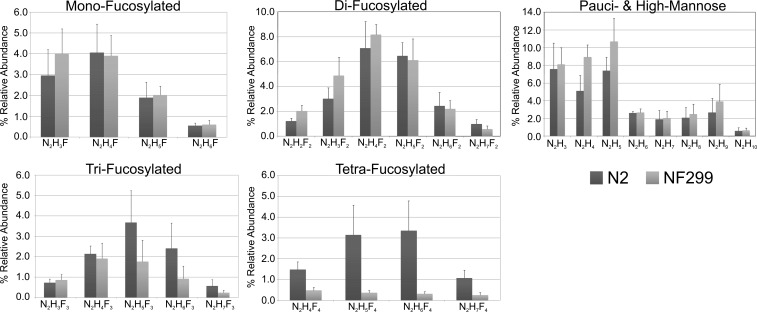
The presence of N-glycan compositions was supported by three independent analysis (Table I). The replicates confirmed the molecular compositions of N-glycans from N2 and NF299 strains including the decrease in fucosylation for the NF299 strain (Figure 2). The relative abundances of fucosylated structures were calculated by dividing the intensity of each molecular composition in the matrix-assisted laser desorption ionization (MALDI)-time of flight (TOF) spectra by the intensity sum of all the glycans. Phosphorylcholine-rich and truncated complex structures were not included in the comparative because they were not detected in significant amounts by previous hydrazinolysis-based reports (Hanneman et al. 2006) and only represent a minority of molecular compositions in the C. elegans glycome (Paschinger et al. 2008). Comparatively, the frequency of mono- and difucosylated compositions did not differ significantly between strains. Tetra-fucosylated compositions consisted of 1.43 ± 0.10% of the total NF299 N-glycome. Alternatively, the N2 N-glycome was 9.07 ± 1.15% tetra-fucosylated compositions. The amount of trifucosylated structures was 11.39 ± 1.39% in NF229 and 19.07 ± 1.15% in N2. Pauci- and high-mannose glycans were increased in the NF299 strain (39.61 ± 3.74%) compared with N2 (29.96 ± 2.63%).
Table I.
Relative abundance of permethylated/reduced N-linked glycans from N2 and NF299 strians via MALDI-MS
| N-Glycan Composition | [M + Na]+ | N2 % Composition | NF299 % Composition |
|---|---|---|---|
| N2H3 | 1187.62 | 7.58 ± 2.96 | 8.12 ± 1.87 |
| N2H2F2 | 1331.69 | 1.21 ± 0.24 | 2.03 ± 0.46 |
| N2H3F | 1361.70 | 5.92 ± 2.50 | 8.06 ± 2.36 |
| N2H4 | 1391.71 | 5.10 ± 1.75 | 8.95 ± 1.35 |
| N3H3 | 1432.74 | 1.75 ± 0.27 | 2.46 ± 0.75 |
| N2H3F2 | 1535.79 | 3.01 ± 0.88 | 4.89 ± 1.48 |
| N2H4F | 1565.80 | 8.13 ± 2.75 | 7.82 ± 1.95 |
| N2H5 | 1595.81 | 7.41 ± 1.51 | 10.70 ± 2.61 |
| N2H3F3 | 1709.88 | 1.47 ± 0.36 | 1.72 ± 0.54 |
| N2H4F2 | 1739.89 | 7.09 ± 2.14 | 8.18 ± 0.82 |
| N2H5F | 1769.90 | 3.80 ± 1.48 | 4.04 ± 0.85 |
| N2H6 | 1799.91 | 2.62 ± 0.18 | 2.69 ± 0.41 |
| N2H4F3 | 1913.98 | 4.28 ± 0.78 | 3.82 ± 1.52 |
| N2H5F2 | 1943.99 | 6.47 ± 1.07 | 6.12 ± 1.73 |
| N2H6F | 2147.99 | 1.09 ± 0.27 | 1.21 ± 0.40 |
| N2H7 | 2004.10 | 1.89 ± 1.02 | 2.06 ± 0.77 |
| N2H4F4 | 2088.07 | 1.48 ± 0.37 | 0.49 ± 0.13 |
| N2H5F3 | 2118.08 | 7.37 ± 3.11 | 3.53 ± 2.10 |
| N2H6F2 | 2148.09 | 2.44 ± 1.10 | 2.20 ± 0.67 |
| N2H8 | 2208.11 | 2.08 ± 1.17 | 2.51 ± 1.10 |
| N2H5F4 | 2292.17 | 3.15 ± 1.43 | 0.37 ± 0.10 |
| N2H6F3 | 2322.18 | 4.83 ± 2.48 | 1.85 ± 1.24 |
| N2H7F2 | 2352.28 | 0.99 ± 0.36 | 0.58 ± 0.26 |
| N2H9 | 2412.21 | 2.67 ± 1.60 | 3.92 ± 1.92 |
| N2H6F4 | 2496.27 | 3.36 ± 1.43 | 0.31 ± 0.11 |
| N2H7F3 | 2526.28 | 1.13 ± 0.62 | 0.47 ± 0.25 |
| N2H10 | 2616.31 | 0.60 ± 0.36 | 0.65 ± 0.26 |
| N2H7F4 | 2700.37 | 1.08 ± 0.38 | 0.26 ± 0.12 |
N-glycan compositions along with the theoretical mass of the singly charged sodium adducts are shown. The values represent the relative intensity of each composition to the sum of all measured glycan intensities (with the standard deviation). N = N-acetylhexosamine (e.g. N-acetylglucosamine, N-acetylgalactosamine), H = hexose (e.g. galactose, mannose, glucose), F = deoxyhexose (e.g. fucose)
Fig. 2.
Relative abundance of N-linked glycans from N2 and NF299 strains by type. The y-axis represents the percent relative abundance of each composition, and the x-axis represents the molecular compositions (N, N-acetylhexosamine; H, hexose; F, fucose).
Structural characterization of fucosylated N-glycans in NF299 via MSn
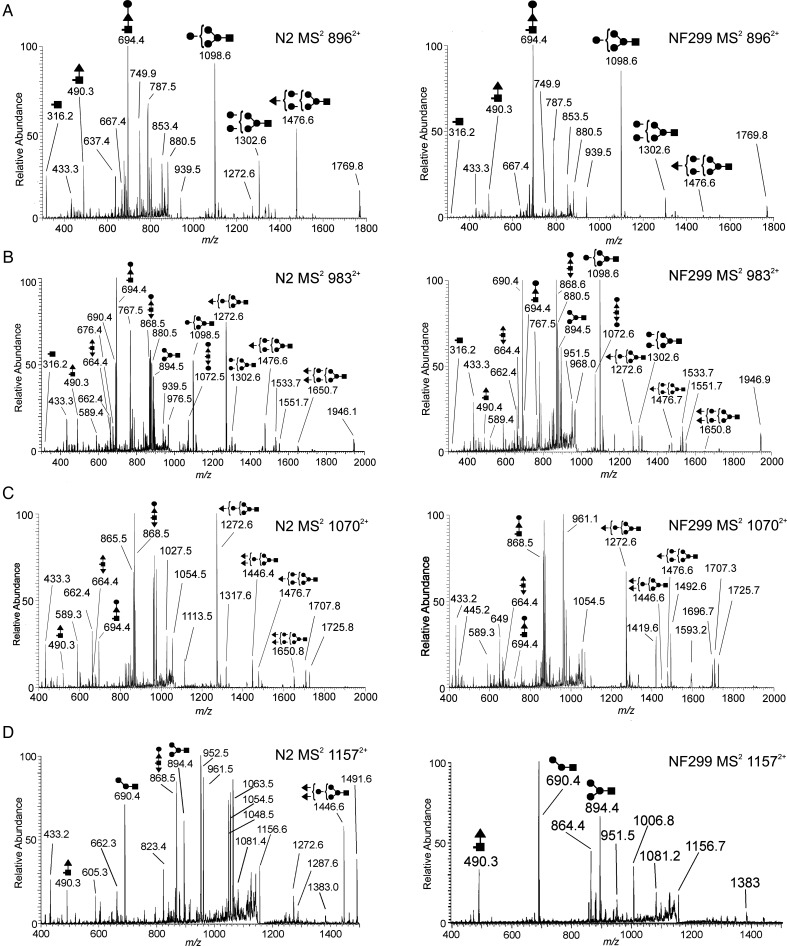
The compositions GlcNAc2Hex5F(1-4) were selected for MSn analysis due to their increased abundance compared with other compositions with up to four fucose residues (GlcNAc2Man4Fuc1-4, GlcNAc2Man5Fuc1-4, GlcNAc2Man6 Fuc1-4 and GlcNAc2Man7Fuc1-4). MS2 profiles of N2H5F (m/z 8962+), N2H5F2 (m/z 9832+), N2H5F3 (m/z 10702+) and N2H5F4 (m/z 11572+) provide topological information with regard to the fucose location (Figure 3). A common fragmentation pattern observed in the MS2 profiles was a result of labile glycosidic bond cleavage between the penultimate and reducing-end GlcNAc residues in the chitobiose core, resulting in B- and Y-complementary ions that partially identify the location of fucose residue(s) (i.e. on the reducing-end GlcNAc or antennae). It is important to note that the linkages shown in Figure 3 are not exact and the cartoon structures merely illustrate the potential topology. MS3 and MS4 spectra were generated to show the correct location of fucose in the structures mentioned above as similarly demonstrated by Hanneman et al. (2006). B/Y-ion complements in Figure 3 demonstrate the decrease in terminally fucosylated structures in the NF299 strain, most noticeably by the m/z 1650 in the MS2 profiles of N2H5F2 (m/z 9832+) and N2H5F3 (m/z 10702+) and m/z 1446 B-ion in the m/z 10702+ spectrum (Figure 3B and C). There were not any detectable B/Y-ions that corresponded to tetrafucosylated N-glycans in the NF299 strain (Figure 4).
Fig. 3.
MS2 spectra of N2H5F(1-4) compositions from N2 and NF299. The decrease in terminal fucosylation (the absence in N2H5F4) in NF299 is illustrated in the MS2 spectrum. B-type ions with increased fucosylation are noticeable less in NF299 than N2. The presence in multiple structural isomers is also evident in both strains, but the extent of structural diversity cannot be resolved without further fragmentation of B- and Y-ions (i.e. MS3) (filled triangle, fucose; filled square, N-acetylhexosamine; filled circle, hexose).
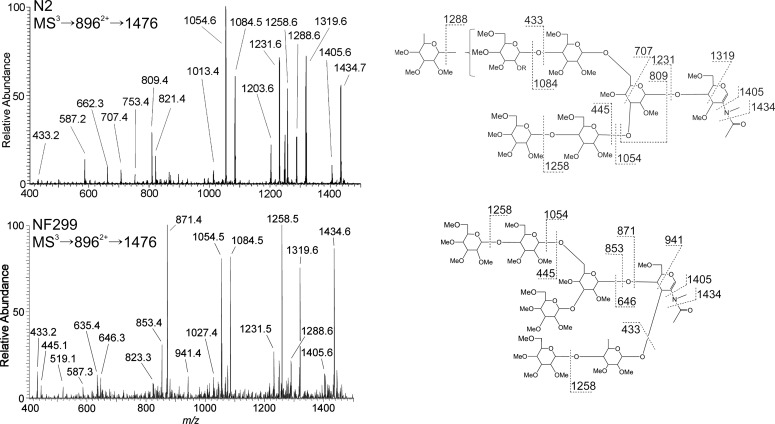
Fig. 4.
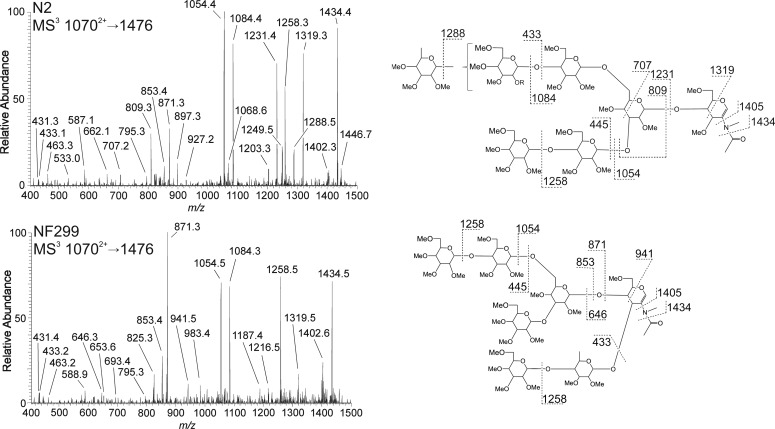
MS3 spectra and fragmentation assignments of the m/z 1476 B-ion from GlcNAc2Man5Fuc (8962+) in N2 and NF299. Fragmentation profiles differ between the wild-type and COG-deficient strain illustrating changes in fucosylation. The wild-type N2 strain added a terminal fucose residue to the antenna and is evident from the m/z 1288, 1231, 707 and 433 ions. Fucose position in the NF299 strain was at the non-reducing core GlcNAc with an additional terminal hexose. This motif was apparent from the m/z 941, 871 and 646 ions as well as the absence of m/z 1288 ion, corresponding to the neutral loss of a terminal fucose.
Structural characterization of GlcNAc2Hex5Fuc
The relative abundance of mono-fucosylated N-glycans in N2 and NF299 glycome were 18.94 ± 3.0 and 21.12 ± 3.28%, respectively. The MS2 profiles of N2 and NF299 differed with regard to the intensity of B-ions containing outer arm fucose residues. The B/Y-ion complementation in N2H5F (m/z 8962+) confirmed three locations where fucose was present (Figure 3A). The three positions are based on ion complements: m/z 316/1476, 490/1302 and 694/1098. The NF299 strain contained trace amounts of the m/z 316 and 1476 ions, demonstrating that outer arm fucosylation was decreased. From the N2 spectrum, it can be inferred that fucose was added either on a terminal hexose, the reducing-end GlcNAc or the reducing-end GlcNAc with an additional hexose residue. The m/z 1476 contained an outer arm fucose, the only such B-ion fragment in the GlcNAc2Man5Fuc MS2 spectrum.
MS3 of the m/z 1476 B-ion from N2 and NF299 were markedly different (Figure 4). The NF299 strain contained an N-glycan structure that is largely absent in N2. In this structure, the penultimate GlcNAc residue in the chitobiose core was modified with a Galβ(1-4)Fuc linked to the 3-position. This was evident from the m/z 646, 853, 871 and 941 ions. N2H5F in N2 was principally a branched structure with fucose present on either antennae. Ion fragments m/z 707, 809, 1231, 1288 and 1319 are specific to this structure. The presence of the m/z 1231, 1288 and 1319 ions in the NF299 spectrum indicated that this structure was also expressed in the COG mutant, but to a lesser extent than the aforementioned isomer.
Structural characterization of GlcNAc2Hex5Fuc2
The relative abundance of difucosylated structures in the N2 strain was 21.21 ± 2.63%. Similarly, the N-glycome of the NF299 strain was 23.99 ± 2.89% difucosylated compositions. Of the four GlcNAc2Man5(Fuc1-4) compositions analyzed, GlcNAc2Man5Fuc2 contained the most isomeric diversity (Figure 3B). The MS2 spectra of the doubly charged GlcNAc2Hex5Fuc2 composition (m/z 9832+) indicated the presence of six B/Y-complement ion fragments in both strains. These compositions ranged from GlcNAcMan4/GlcNAcMan2Fuc2 B/Y-complements (m/z 894 and 1072) to a GlcNAc2Man5Fuc2 B-ion with a single reducing-end GlcNAc Y-ion (m/z 316 and 1650). Despite the presence of these ions in the MS2 profiles, the abundance of these ions differed, specifically m/z 1650 and 1476 in the NF299 strain. These B-ions had one and two outer arm fucose residues, respectively.
Of the six B/Y-ion complement fragments generated in the MS2 profile from N2, all but one (m/z 1650) were present in sufficient amounts to generate an MS3 spectrum in the NF299 strain. Three B-ions (m/z 894, 1098 and 1302) did not contain fucose residues and were the most abundant structures in the NF299 strain. MS3 spectra of these non-fucosylated B-ions were identical in both the N2 and NF299 strains (Supplementary data, Figure 1).
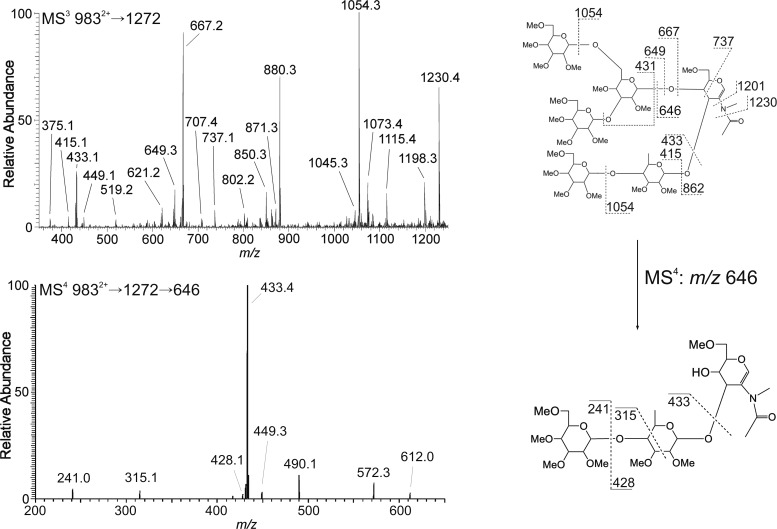
Two fucosylated B-ions (m/z 1272 and 1476) were present in both strains. The MS3 spectra of the m/z 1272 B-ion (Man4FucGlcNAc) differed considerably between strains (Figures 5 and 6). Most noticeable was the m/z 646 fragment which is a FucHex disaccharide linked to the non-reducing-end GlcNAc on the chitobiose core (Figure 5). The MS4 spectrum of m/z 646 demonstrated that the linkage is consistent with a 1-4 linkage (presumably a β1-4 galactose), which has been demonstrated previously (Hanneman et al. 2006). An additional diagnostic ion showing the composition FucHexGlcNAc core modification was the decrease in the m/z 1027 and 1045 ion fragments, which were the B- and C-type ions from the neutral loss of an internal GlcNAc. This confirmed that the GlcNAc residue in NF299 was more commonly modified with a Hex-Fuc disaccharide. A second diagnostic ion in the NF299 strain was m/z 667 that is a trimannosyl C-type ion of the antennae. The m/z 709, 621, 593, 449 and 431 ions were more prevalent in NF299 and could only be derived from the structure in Figure 5. In the NF299 strain, the location of fucose remained linked to the chitobiose core and not the antennae. Also the m/z 1084 ion generated from the neutral loss of a terminal fucose was significantly reduced in the NF299 strain, further suggesting the prevalence of the complex core structure and decrease in outer arm fucose additions.
Fig. 5.
N-Glycan structure and core fucose linkage assignment of GlcNAc2Man5Fuc2 (9832+) from NF299. The MS3 profile of the 1272 B-type ion from the difucosylated precursor illustrates that fucosylation is at the non-reducing-end GlcNAc of the chitobiose core and not at the antenna. The MS4 spectrum of the 646 ion (9832+ → 1272 → 646) shows that the topology is Hex-Fuc-GlcNAc and the fragmentation assignment is consistent with a 1-4 linkage.
Fig. 6.
N-Glycan structure and antenna fucose linkage assignment of GlcNAc2Man5Fuc2 (9832+) from N2. The wild-type structural isomer of the 1272 B-type ion differs from the NF299, where fucose is a terminal residue on the antenna and not at the chitobiose core. MS4 of the 433 ion, which is the Fuc-Hex C-type disaccharide, shows the linkage to be both 1-2 and 1-4 with fucose at the terminus.
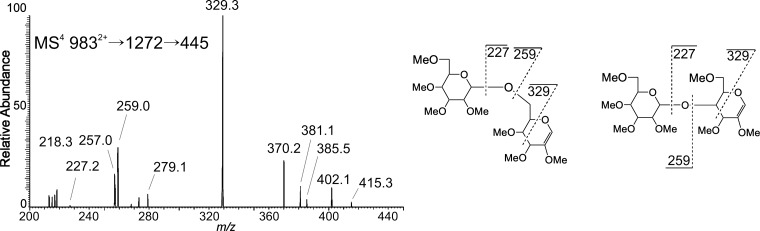
The MS3 spectrum of the m/z 1272 ion from N2 showed that the antenna is modified with a terminal fucose (Figure 6). The m/z 433 fragment ion is a Fuc-Hex C-type disaccharide and the MS4 fragmentation spectrum of this ion contained two linkage isomers. The m/z 271 ion results from a 0,2A cross-ring cleavage of the internal hexose residue and points to the fucose being linked at the 2-position. This is further supported by the m/z 359 0,4A fragment; however, this ion is not unique to a 1–2 linkage. The second diagnostic ion is m/z 299, a 3,5A cross-ring fragment of the hexose. This ion is specific for a 1-6 linked fucose. Therefore, wild-type fucosylation in N2 is principally on the antenna in a 1-2 and a 1-6 linkage to a hexose. The linkage of Hex-Hex disaccharides on the antenna was found to be either 1-6 or 1-4 from the MS4 fragmentation of the m/z 445 ion (9832+ → 1272 → 445; Figure 7).
Fig. 7.
Assignment of the Hex-Hex disaccharide on the antenna from N2 GlcNAc2Man5Fuc2 (9832+). The topology and linkage of the Hex-Hex on the opposite arm of the Fuc-Hex disaccharide from that illustrated in Figure 6 was determined to be 1-6 and 1-4.
The spectra of the m/z 1476 B-ion in N2H5F2 from N2 and NF299 (Supplementary data, Figure S2) had similar profiles to N2H5F (Figure 4). The NF299 strain maintains the m/z 646 Gal-Fuc-GlcNAc ion that was mostly absent in N2. Additionally, the ion that correlates with a neutral loss of a terminal fucose (m/z 1288) was scarcely detectable in the NF299 spectrum. The majority of ions in the NF299 profile indicate the expression of core-modified N-glycans over biantennary structures with terminal fucosylation found in N2.
Structural characterization of GlcNAc2Hex5Fuc3
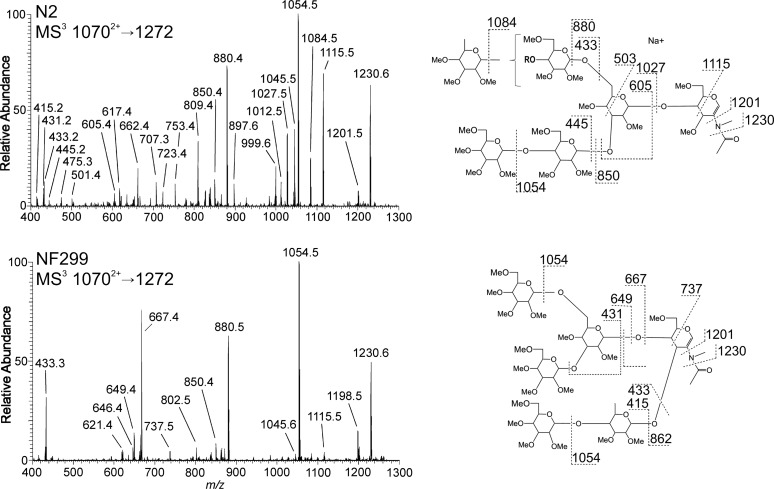
The N-glycome of N2 features 19.07 ± 1.15% trifucosylated compositions, whereas NF299 contain 11.39 ± 1.39%. The MS2 profile of this composition showed four B/Y-ion compliments (m/z 490/1651, 664/1476, 694/1446 and 868/1272; Figure 3C). The m/z 1651 B-ion peak was absent in NF299 and the m/z 1446 B-ion peak was notably reduced, both of which contain two fucose residues.
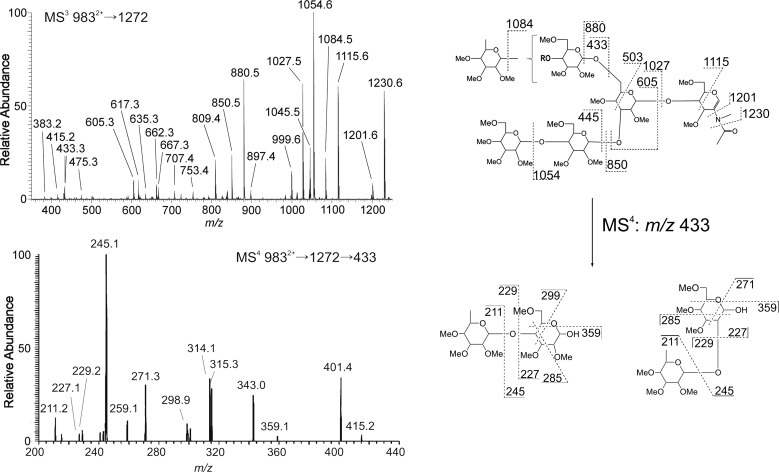
The MS3 spectra of the m/z 1272 B-ion (GlcNAcMan4Fuc) had noticeable differences between strains (Figure 8). The trend of the NF299 to either lack or exhibit decreased terminal fucosylation was evident from the m/z 723, 1027, 1045, 1084 and 1115 ions. Also, the presence of other ions (m/z 646, 737, 649 and 667), which were largely absent in the N2 N-glycan spectrum, point to an increase in fucosylation at the non-reducing core GlcNAc. Isolation and fragmentation of the m/z 1476 B-ion (GlcNAcMan5Fuc) in both strains (Figure 9) showed fucose on the antenna in N2 and the core in NF299. Again ions were present in NF299 that were not in N2, namely m/z 646 and 911. The NF299 strain lacked higher m/z ions that correspond to the neutral loss of an internal GlcNAc (245 amu), which is a non-modified GlcNAc from the chitobiose core.
Fig. 8.
MS3 spectra and fragmentation assignments of the m/z 1272 B-ions from the MS2 of GlcNAc2Man5Fuc3 (10702+) in N2 and NF299. The structures detected in both spectra from NF299 contain fucose at the core and the location of fucose in N2 was terminal and on the antenna. The NF299 strain did not add fucose to the antenna.
Fig. 9.
MS3 spectra and fragmentation assignments of the m/z 1476 B-ions from the MS2 of GlcNAc2Man5Fuc3 (10702+) in N2 and NF299. Similar to the m/z 1272 structure in Figure 8, the structures from NF299 contain fucose at the core. The location of fucose in N2 was terminal and on the antenna.
The m/z 1446 and 1650 ions, which were mostly absent in the NF299 strain, had fucose residues linked to the penultimate GlcNAc residue or at the terminus of the structure in the N2 strain. The MS3 spectra from NF299 were too poor to make any accurate inferences, and therefore, it could be concluded that these structures are expressed at low levels in the NF299 strain. In the N2 strain, the penultimate GlcNAc was mostly modified with a Hex-Fuc disaccharide (m/z 646), established from the absence of an m/z 1201 ion and m/z 1405 (245 neutral loss of an internal GlcNAc) in the m/z 1446 and 1650 profiles, respectively. That is to say that N-glycans in N2 do not have two terminal fucose residues. One fucose residue was attached to the penultimate GlcNAc in the chitobiose core and the second was on the terminus of one antennae. The NF299 strain did not have fucose on the antennal terminus. Specifically, NF299 did not readily generate N-glycans with two fucose residues distal from the reducing-end GlcNAc.
Structural characterization of GlcNAc2Hex5Fuc4
The MS2 spectra of the 11572+ precursor ion from NF299 did not produce any B/Y-ion complements which verified the absence of N2H5F4 structures. The three ions that corresponded to a carbohydrate m/z composition in the NF299 MS2 spectrum of the 11572+ parent ion were m/z 490, 690 and 894. The MS3 spectra demonstrated that the fragments of m/z 490 and 690 were specific to carbohydrates. The m/z 490 ion corresponded to a fucosylated reducing-end GlcNAc and the m/z 690 was a Man2GlcNAc composition. Interestingly, the sum of the m/z 490 and 690 ions was 1157, which is the sodium adduct of the N2H2F glycan that has been identified in C. elegans. The detection of m/z 490 and 690 in Figure 3D were most likely a result from the isolation window parameter specified in the mass spectrometer. The N2H5F4 sodium adduct was observed and consequently selected as a double-sodiated adduct with an m/z of 11572+ in the MS1 profile, but the singly charged 1157+ ion would also be selected, fragmented and present in the MS2 spectrum. The presence of a 1157+ ion in the MALDI profile (corresponding to the parent ion of B/Y-fragments m/z 490 and 690) was detected (data not shown) and has been reported previously (Paschinger et al. 2004; Hanneman et al. 2006). Most important was the absence of the B/Y-ions 1446/868 that represents the only wild-type N-glycan with the composition N2H5F4, which was not present in the NF299 strain. Therefore, the NF299 does not express any N-glycans with the composition GlcNAc2Hex5Fuc4.
Discussion
Glycosylation is a complex process that involves a substantial fraction of cellular components. The correlation between glycan-related gene transcription, biosynthesis and glycan structure is one aspect of glycobiology that remains poorly understood. The conventional belief has been that the regulation of fucosylation depends on fucosyltransferases, GDP-fucose biosynthetic enzymes and GDP-fucose transporters alone. Here, we demonstrate that fucosylation in C. elegans is also controlled by the expression of the COG complex.
The comparative analysis of N2 (Bristol) and NF299 (k179) has led to two principle findings of the COG complex in C. elegans glycosylation, both of which involve fucosylation. Most notable was the absence of tetrafucosylated compositions N2H4F4, N2H5F4, N2H6F4 and N2H7F4. This was evident from the MALDI-TOF spectra (Figure 1) and further supported by the electrospray ionization MS2 spectra of N2H5F4 in Figure 3D [MS2 of tetrafucosylated compositions in NF299 illustrated the absence of tetrafucosylated N-glycans (Supplementary data, Figure S3)]. The N2H4F4, N2H5F4, N2H6F4 and N2H7F4 compositions constitute the fucose-rich glycans in C. elegans, which have been reported in the glycome of the N2 Bristol strain (Hanneman et al. 2006; Paschinger et al. 2008). Moreover, these structures constitute the core chitobiose modifications, where fucose residues are found extensively on the core as well as on the antennae and were only reported using hydrazinolysis (Hanneman et al. 2006).
The second finding in the COG-deficient strain was the inability to modify N-glycans with terminal fucosyl residues, as supported in Figures 4 and 6 and illustrated in Figure 10. N-Glycans in the NF299 strain were more readily fucosylated at the reducing-end GlcNAc or penultimate GlcNAc chitobiose core residues thereby increasing the level of complex core N-glycans. The level of pauci- and high-mannose glycans was increased in the NF299 strain and may arise from a block in the glycosylation pathway caused by the disruption of the trans-Golgi from COG defects. Interestingly, the NF299 strain, which lacks fucose-rich glycans and is deficient in antennal fucosylation, exhibits only slowed growth and protruding vulva phenotypes. Therefore, tetrafucosylated fucose-rich N-glycans and antennal fucosylation are not vital for C. elegans survival in laboratory conditions. It is valuable to note that α1,2-fucosyltransferase knockout mice are viable and fertile, further suggesting that α1,2-fucosylation is not required for animal survival (Domino et al. 2001).
Fig. 10.
Changes in fucosylation of the Hex5Fuc(1-4) N-glycan isomers. Glycan structures are annotated using the Consortium for Functional Glycomics representation. ✓ represents a detected structure, an ↓ or ↑ indicates a decrease or increase and “nd” denotes the glycans that were not detected. The corresponding m/z is also shown.
Fucosylation in C. elegans is relatively intricate having two characterized α1,2-fucosyltransferases CE2FT-1 and CE2FT-2 (from 25 putative genes), five α1,3/4 fucosyltransferases (including the reducing-end GlcNAc α1,3-fucosyltransferase) and one reducing-end GlcNAc α1,6-fucosyltransferase. α1,2-, α1,3/4- and α1,6-fucosyltranserases are type II membrane proteins and are localized in the medial and/or trans-Golgi. The linkage of terminal fucosylation has been shown to be Fucα1-2Galβ-R (Zheng et al. 2002; Cipollo et al. 2004; Paschinger et al. 2004, 2008; Schachter 2004). Although Zheng et al. (2008) showed that the CE2FT-2 fucosyltransferase in C. elegans generates H-type 3 glycan epitopes (Fucα1-2Galβ1-3GalNAcα1-R), we, however, did not find an Fuc-Hex-HexNAc trisaccharides from the N-glycan antennae in either strain. The fucose linkage in N2 was determined to be 1-2 and 1-4 on the antenna and the fucose linkage at the non-reducing core GlcNAc in NF299 was established as 1-4.
The function of terminal fucose on N-glycans and the role of fucose-rich N-glycans are not completely understood in C. elegans. The lack of sialic acid in the worm may be compensated by the complexity and degree of fucosylation of N-glycans. The expression of CE2FT-1 and therefore α1,2-fucosylated N-glycans in C. elegans is localized to the intestines and has been shown to act in defense toward bacterium, parasites and pathogens, but may also be vital as a substrate for sought after lectins.
There are several examples in C. elegans that illustrate the importance of fucosylation in bacterial and toxin defense. The C. elegans srf-3 mutant, which lacks a UDP-galactose and UDP-GlcNAc Golgi transporter, does not express terminal fucosylated N-glycans because the required galactose is absent for binding. These animals were resistant to Microbacterium nematophilum infection and binding of the biofilm formed by Yersinia pseudotuberculosis and Yersinia pestis (Cipollo et al. 2004). Also, the C. elegans bre-1 mutant, which lacks a GDP-mannose 4,6-dehdratase, exhibited a decrease in fucosylated N- and O-glycans, as well as glycolipids. The bre-1 mutants were resistant to the Bacillus thuringiensis crystal protein Cry5B and were attributed to the absence of fucose on glycoconjugates, namely glycolipids. Fucosylated N-glycans were not detected in the bre-1 mutant by MALDI-MS; however, the whole range of N-glycans (especially fucose-rich) was not reported in the N2 stain control, possibly due to the use of enzymes in the N-glycan release procedure (Barrows et al. 2007). Third, the CGL2 galectin produced by the fungus Coprinopsis cinerea binds to the reducing-end core Galβ1,4Fucα1,6 on N-glycans expressed in the intestine of C. elegans and results in decrease brood size, delayed larval development and lethality in the N2 strain. Interestingly, the bre-1 mutant showed nearly complete resistance to CGL2, demonstrating the role of core fucosylation in toxin binding (Butschi et al. 2010). In all the above-mentioned studies, the NF299 strain would provide a valuable tool to dissect the complex nature of bacteria, toxin and biofilm interactions where fucose is central.
It may be possible to apply the conclusions made here to the existing model of the COG complex in C. elegans. Kubota et al. concluded that COG complex-1 and COG complex-3 were required for the glycosylation and localization of MIG-17 along the body wall muscles. In addition, MIG-17 is underglycosylated in mig-23 mutants. Distal tip migration was defective in cogc-1(k179), cogc-3(k181) and mig-17(k174) mutants and presumably functions along a common pathway. It may be plausible that a refined simpler model of distal tip migration exists where COG complex-1 and COG complex-3 function to synthesize fucose-rich or terminal fucosylated N-glycans on MIG-17. Moreover, the cogc-1(k179) and cogc-3(k181) phenotypes are identical and more severe than mig-17 alone, most likely from the complete lack of fucose-rich and/or terminal fucosylation on all glycoproteins throughout the animal. The double mutant cogc-1(k179);mig-17(k174) or cogc-3(k181);mig-17(k174) does not exhibit an enhanced distal tip migration phenotype, most likely due to the absence of the MIG-17 protein component, the lack of N-glycans is inconsequential. It is also possible that a corresponding lectin(s) along the body wall muscles binds fucose-rich or terminal fucosylated residues (including N-glycans on MIG-17) to direct distal tip cells during embryogenesis.
This study clearly shows that the COG complex acts as a non-direct factor of fucosylation in C. elegans. The COG complex is required for proper function of the glycosylation machinery via intracellular trafficking and in the absence of the COG complex terminal fucosylation and tetrafucosylated N-glycans are absent. It is known that the COG complex is compulsory for retrograde trafficking of COPI vesicles within the Golgi (Cavanaugh et al. 2007) and that COG defects give rise to Golgi fragmentation and the formation of CCD vesicles (Zolov and Lupashin 2005; Pokrovskaya et al. 2011). Also, changes in Golgi morphology from COG complex disruption are lobe-dependent and affect the stability of two glycosyltransferases (Peanne et al. 2011). Depletion of COG subunits 1–4 (lobe A) results in normal β1,4-galactosyltransferase and α2,6-sialyltransferase expression but the enzymes are incorrectly localized and accumulated in CCD vesicles. Conversely, defects in subunits 5–8 (lobe B) caused a decreased expression of the same enzymes due to a rapid relocalization to the endoplasmic reticulum and ultimately the proteosome. Decreases in COG complex-3 also result in a reduction in COG complex-1, -2 and -4 (Zolov and Lupashin 2005). Therefore, the NF299 strain does not form a functional lobe A of the COG complex and the decrease in terminal fucosylation is from the mislocalization of the α1,2-fucosyltransferases CE2FT-1 from the trans-Golgi into CCD vesicles and is unable to actively glycosylate proteins. Furthermore, α1,2-mannosidase II has been shown to accumulate in CCD vesicles (Zolov and Lupashin 2005) and this observation would explain the increase in high-mannose structures in NF299.
As the discovery of glycan structural variations increase and the number of enzymes enlisted to create these structures remains consistent, it is critical to consider other molecular mechanisms by which the Golgi functions and localizes resident enzymes that give rise to the glycan diversity observed in all eukaryotes. Furthermore, the pathogenic mechanisms linking glycosylation defects to clinical presentations in CDG cases remain to be elucidated. This approach not only supports the trend of glycosylation in CDG-II/COG-1-like conditions, but also presents an opportunity to study several disorders or functions where fucosylation is fundamental. These include leukocyte adhesion deficiency, bacterial toxicology, as well as providing a target for pharmacological modulation where a disruption in sialylation, galactosylation and/or outer arm fucosylation is modulated, such as tumor metastasis, inflammation and graft rejection.
Material and methods
N2 (Bristol) and NF299 cogc-1 (k179) strains were obtained from the Caenorhabditis Genetic Center at the University of Minnesota. Mixed stage N2 and NF299 C. elegans strains were grown separately on large trays (six trays per strain) containing peptone-rich nematode growth media and a bacterial food source of the E. coli OP50 strain as described (Hope 1999). Animals were rinsed from the trays with three aliquots of 250 mL of cold distilled de-ionized (DDI) water. The worm solutions were centrifuged at 800 RCF for 4 min at 4°C. The supernatant was removed and the worms were washed with cold DDI water; spun at 1000 RCF for 60 s at 4°C until the supernatant was clear. The worm pellet was resuspended in 20 mL of cold DDI water. To this solution, 30 mL of cold 50% sucrose solution was added and mixed by inversion. Each tube was centrifuged at 50 × g for 5 min at 4°C. The top layer of the solution, containing live animals, were aspirated and collected in a fresh tube containing 20 mL of cold DDI water. Tubes containing live sample were centrifuged at 1000 × g for 2 min at 4°C. This step was repeated once with DDI water and once with M9 solution to remove excess sucrose. The worm solution was centrifuged again at 1000 × g for 2 min at 4°C and the supernatant was removed. The pellet was resuspended in 5 volumes of M9, agitated at room temperature to allow the gut contents to be digested. A final series of three DDI water washes was performed to remove the gut contents.
Approximately, 1 g of wet weight worms (∼1.5 mL), thawed from a frozen pellet, was added to 1.5-mL CHAPS lysis buffer containing 4% CHAPS, 8 M urea, 40 mM Tris–HCl, pH 8, 65 mM dithiothreitol. Worms were lysed using a Pressure Biosciences Barocycler NEP2320 (South Easton, MA) with 20 cycles, each consisting of 40 s at 35,000 PSI, followed by 10 s at ambient pressure. Whole lysate protein concentration was quantified via Bradford assay (Pierce, Rockford, IL) using a UV-Viz spectrophotometer (Shimadzu Corporation, Kyoto, Japan). Sufficient lysate to yield 5 mg of protein was transferred to a glass culture tube (13 × 100 mm), and protein was precipitated using 15% trichloroacetic acid for 1 h on ice. Following precipitation, the sample was centrifuged at low speed for 15 min. After the supernatant was discarded, the protein pellet was washed and vortexed with 2 × 1 mL of cold acetone, and 2 × 1 mL of chloroform:methanol:water (10:10:1), with centrifugation between each wash. The protein pellet was dried under N2 flow and lyophilized overnight.
Prior to the N-glycan release, the protein pellet was dried over P2O5 for 48 h. About 500 µL of anhydrous hydrazine (Sigma-Aldrich, St Louis, MO) was added to the dried protein pellet and heated at 100°C for 6 h. Samples were cooled to room temperature at which time the hydrazine was removed under N2 flow. Samples were then lyophilized overnight. The released glycans were re-N-acetylated using 150 µL of acetic anhydride in 300 µL of saturated sodium bicarbonate on ice for 30 min, followed by 1 h at room temperature. The samples were desalted on a DOWEX AG 50 W X8-400 cation-exchange resin (Sigma-Aldrich). Free N-glycans were purified using a hand-packed column containing 500 µL of medium fibrous cellulose. The column was washed with 4:1:1 n-butanol:methanol:water to remove the peptide components. N-Glycans were eluted with 1:1 ethanol:water. N-Linked glycans were reduced with a 200-µL solution of 10 mg/mL of sodium borohydride (NaBH4) in 0.01 M NaOH for 1 h on ice followed by room temperature overnight. To stop the reduction reaction, glacial acetic acid was added drop-wise until the sample pH reaches 5. Borate was removed by co-evaporation of 3 mL ethanol, 2 × 2 mL 1% acetic acid in methanol and 2 × 2 mL toluene. The reduced N-glycans were dissolved in water and desalted on a pre-conditioned 1.5-mL column of porous graphitized carbon (Alltech Associates, Deerfield, IL). After washing the column with 3 mL of water, N-glycans were eluted using 3 mL of 25% acetonitrile/water. Purified N-glycans in solution were dried in a SpeedVac® Concentrator (Savant Instruments, Holbrook, NY).
After reduction, N-glycans were permethylated as described (Kerek 1984). Briefly, dried glycans were dissolved in 500 µL of dimethyl sulfoxide (HPLC grade, Sigma-Aldrich), followed by addition of powdered sodium hydroxide (99.999%, Sigma-Aldrich). After vortexing, 100 mL of iodomethane (99.9%, Sigma-Aldrich) was added. Sample tubes were flushed with argon, capped and wrapped in aluminum foil and vortexed for 1 h. Samples were placed on ice and 1 mL of water was added to each sample to stop the reactions followed by 1 mL of HPLC grade dichloromethane. The sample mixtures were vortexed and the top organic phase (containing N-glycan) was removed and put into a clean test tube. The dichloromethane addition was repeated twice and the extracts pooled. The new tube containing the organic phase was washed five times with HPLC grade water, removing the water layer after each wash. The dichloromethane was evaporated under N2 flow, lyophilized overnight and stored at −20°C.
Permethylated N-glycan spectra were generated using a MALDI-TOF mass spectrometer (Kratos-Shimadzu Analytical, Manchester, UK) equipped with an ultraviolet 337 nm wavelength nitrogen laser. For permethylated samples, 75 µL of methanol and 25 µL of HPLC grade H2O were added and vortexed; 1 µL of reconstituted sample solution was mixed with 1 µL of 2,5-dihydroxyl benzoic acid matrix (12 mg/mL in 50% (v/v) acetonitrile aqueous solution) on a stainless-steel MALDI target plate. All samples were ionized with 50–60% of the maximum power while rastering on the plate surface. Mass spectra were acquired by averaging ∼200 profiles generated by five laser shots per profile with the post-extraction parameter optimized at m/z 2500. Spectra processing was preformed with Kratos Launchpad software. Ions were observed as [M + Na]+adducts.
Spectra peak annotation was performed initially by using the online program Glycomod Tool, a package under the ExPASY (Export Protein Analysis System) proteomics server of the Swiss Institute of Bioinformatics (Gasteiger et al. 2003). GlycoMod can predict the possible oligosaccharide compositions from their experimentally determined masses and on compositional constraints applied by the user (i.e. mass tolerance). The search parameters were strict; the experimental values were compared with hypothetical mass values within a ±0.3-Da threshold.
ITMSn N-glycan mass spectra were obtained with a linear ion trap LTQ mass spectrometer (ThermoFinnigan, San Jose, CA) equipped with a TriVersa Nanomate (Advion Biosciences, Ithaca, NY) automated nanoelectrospray ion source supported with automation and related computational tools. Dried samples were diluted in 150 µL of methanol and 50 µL of water. Samples were infused (10 µL) at a low flow rate of 0.30 µL/min and the spectra were collected using Xcaliber 2.0 software (ThermoFinnigan). Signal averaging was accomplished by adjusting the number of microscans within each scan, generally ranging between 3 and 5. Collision parameters were left at default values with normalized collision energy set to 35%. Activation Q was set at 0.25, and activation time for 30 ms. Ions were observed as singly and/or doubly charged sodium adducts.
Supplementary data
Supplementary data for this article is available online at http://glycob.oxfordjournals.org/.
Funding
This work was supported by the NIGMS grant RO1 54045.
Conflict of interest
None declared.
Abbreviations
CCD, conserved oligomeric Golgi complex-dependent; CDG, congenital disorders of glycosylation; COG, conserved oligomeric Golgi; COGC, conserved oligomeric Golgi complex; DDI, distilled de-ionized; F, fucose; Fuc, fucose; GlcNAc, N-acetylglucosamine; H, hexose; Hex, hexose; ITMSn, sequential ion trap mass spectrometry; LDLR, low-density lipoprotein receptor; MALDI, matrix-assisted laser desorption ionization; Man, mannose; MS, mass spectrometry; MSn, sequential mass spectrometry; N, N-acetylhexosamine; TOF, time of flight; UDP, uridine diphosphate; UMP, uridine monophosphate.
Supplementary Material
Acknowledgements
We thank Theresa Stiernagle at the Caenorhabditis Genetics Center for the C. elegans strains used in this study.
References
- Barrows BD, Haslam SM, Bischof LJ, Morris HR, Dell A, Aroian RV. Resistance to Bacillus thuringiensis toxin in Caenorhabditis elegans from loss of fucose. J Biol Chem. 2007;282:3302–3311. doi: 10.1074/jbc.M606621200. doi:10.1074/jbc.M606621200. [DOI] [PubMed] [Google Scholar]
- Butschi A, Titz A, Walti MA, Olieric V, Paschinger K, Nobauer K, Guo X, Seeberger PH, Wilson IB, Aebi M, et al. Caenorhabditis elegans N-glycan core β-galactoside confers sensitivity towards nematotoxic fungal galectin CGL2. PLoS Pathog. 2010;6:e1000717. doi: 10.1371/journal.ppat.1000717. doi:10.1371/journal.ppat.1000717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh LF, Chen X, Richardson BC, Ungar D, Pelczer I, Rizo J, Hughson FM. Structural analysis of conserved oligomeric Golgi complex subunit 2. J Biol Chem. 2007;282:23418–23426. doi: 10.1074/jbc.M703716200. doi:10.1074/jbc.M703716200. [DOI] [PubMed] [Google Scholar]
- Cipollo JF, Awad AM, Costello CE, Hirschberg CB. srf-3, a mutant of Caenorhabditis elegans, resistant to bacterial infection and to biofilm binding, is deficient in glycoconjugates. J Biol Chem. 2004;279:52893–52903. doi: 10.1074/jbc.M409557200. doi:10.1074/jbc.M409557200. [DOI] [PubMed] [Google Scholar]
- Conde R, Pablo G, Cueva R, Larriba G. Screening for new yeast mutants affected in mannosylphosphorylation of cell wall mannoproteins. Yeast. 2003;20:1189–1211. doi: 10.1002/yea.1032. doi:10.1002/yea.1032. [DOI] [PubMed] [Google Scholar]
- Corbacho I, Olivero I, Hernandez LM. Identification of low-dye-binding (ldb) mutants of Saccharomyces cerevisiae. FEMS Yeast Res. 2004;4:437–444. doi: 10.1016/S1567-1356(03)00162-4. doi:10.1016/S1567-1356(03)00162-4. [DOI] [PubMed] [Google Scholar]
- Domino SE, Zhang L, Gillespie PJ, Saunders TL, Lowe JB. Deficiency of reproductive tract α(1,2)fucosylated glycans and normal fertility in mice with targeted deletions of the FUT1 or FUT2 α(1,2)fucosyltransferase locus. Mol Cell Biol. 2001;21:8336–8345. doi: 10.1128/MCB.21.24.8336-8345.2001. doi:10.1128/MCB.21.24.8336-8345.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulquier F, Ungar D, Reynders E, Zeevaert R, Mills P, Garcia-Silva MT, Briones P, Winchester B, Morelle W, Krieger M, et al. A new inborn error of glycosylation due to a Cog8 deficiency reveals a critical role for the Cog1–Cog8 interaction in COG complex formation. Hum Mol Genet. 2007;16:717–730. doi: 10.1093/hmg/ddl476. doi:10.1093/hmg/ddl476. [DOI] [PubMed] [Google Scholar]
- Foulquier F, Vasile E, Schollen E, Callewaert N, Raemaekers T, Quelhas D, Jaeken J, Mills P, Winchester B, Krieger M, et al. Conserved oligomeric Golgi complex subunit 1 deficiency reveals a previously uncharacterized congenital disorder of glycosylation type II. Proc Natl Acad Sci USA. 2006;103:3764–3769. doi: 10.1073/pnas.0507685103. doi:10.1073/pnas.0507685103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. doi:10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanneman AJ, Rosa JC, Ashline D, Reinhold VN. Isomer and glycomer complexities of core GlcNAcs in Caenorhabditis elegans. Glycobiology. 2006;16:874–890. doi: 10.1093/glycob/cwl011. doi:10.1093/glycob/cwl011. [DOI] [PubMed] [Google Scholar]
- Hope IA. C. elegans: A Practical Approach. Oxford NY: Oxford University Press; 1999. [Google Scholar]
- Kerek ICaF. A simple and rapid method for the permethylation of carbohydrates. Carbohydr Res. 1984;131:209–217. doi:10.1016/0008-6215(84)85242-8. [Google Scholar]
- Kingsley DM, Kozarsky KF, Segal M, Krieger M. Three types of low density lipoprotein receptor-deficient mutant have pleiotropic defects in the synthesis of N-linked, O-linked, and lipid-linked carbohydrate chains. J Cell Biol. 1986;102:1576–1585. doi: 10.1083/jcb.102.5.1576. doi:10.1083/jcb.102.5.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz C, Ng BG, Sun L, Sharma V, Eklund EA, Miura Y, Ungar D, Lupashin V, Winkel RD, Cipollo JF, et al. COG8 deficiency causes new congenital disorder of glycosylation type IIh. Hum Mol Genet. 2007;16:731–741. doi: 10.1093/hmg/ddm028. doi:10.1093/hmg/ddm028. [DOI] [PubMed] [Google Scholar]
- Krieger M, Brown MS, Goldstein JL. Isolation of Chinese hamster cell mutants defective in the receptor-mediated endocytosis of low density lipoprotein. J Mol Biol. 1981;150:167–184. doi: 10.1016/0022-2836(81)90447-2. doi:10.1016/0022-2836(81)90447-2. [DOI] [PubMed] [Google Scholar]
- Kubota Y, Nishiwaki K. C. elegans as a model system to study the function of the COG complex in animal development. Biol Chem. 2006;387:1031–1035. doi: 10.1515/BC.2006.127. doi:10.1515/BC.2006.127. [DOI] [PubMed] [Google Scholar]
- Kubota Y, Sano M, Goda S, Suzuki N, Nishiwaki K. The conserved oligomeric Golgi complex acts in organ morphogenesis via glycosylation of an ADAM protease in C. elegans. Development. 2006;133:263–273. doi: 10.1242/dev.02195. doi:10.1242/dev.02195. [DOI] [PubMed] [Google Scholar]
- Loh E, Hong W. The binary interacting network of the conserved oligomeric Golgi tethering complex. J Biol Chem. 2004;279:24640–24648. doi: 10.1074/jbc.M400662200. doi:10.1074/jbc.M400662200. [DOI] [PubMed] [Google Scholar]
- Lubbehusen J, Thiel C, Rind N, Ungar D, Prinsen BH, de Koning TJ, van Hasselt PM, Korner C. Fatal outcome due to deficiency of subunit 6 of the conserved oligomeric Golgi complex leading to a new type of congenital disorders of glycosylation. Hum Mol Genet. 2010;19:3623–3633. doi: 10.1093/hmg/ddq278. [DOI] [PubMed] [Google Scholar]
- Nairn AV, York WS, Harris K, Hall EM, Pierce JM, Moremen KW. Regulation of glycan structures in animal tissues: Transcript profiling of glycan-related genes. J Biol Chem. 2008;283:17298–17313. doi: 10.1074/jbc.M801964200. doi:10.1074/jbc.M801964200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T, Pypaert M, Hoe MH, Slusarewicz P, Berger EG, Warren G. Overlapping distribution of two glycosyltransferases in the Golgi apparatus of HeLa cells. J Cell Biol. 1993;120:5–13. doi: 10.1083/jcb.120.1.5. doi:10.1083/jcb.120.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T, Ungar D, Hughson FM, Krieger M. The COG and COPI complexes interact to control the abundance of GEARs, a subset of Golgi integral membrane proteins. Mol Biol Cell. 2004;15:2423–2435. doi: 10.1091/mbc.E03-09-0699. doi:10.1091/mbc.E03-09-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T, Vasile E, Penman M, Novina CD, Dykxhoorn DM, Ungar D, Hughson FM, Krieger M. Genetic analysis of the subunit organization and function of the conserved oligomeric golgi (COG) complex: Studies of COG5- and COG7-deficient mammalian cells. J Biol Chem. 2005;280:32736–32745. doi: 10.1074/jbc.M505558200. doi:10.1074/jbc.M505558200. [DOI] [PubMed] [Google Scholar]
- Opat AS, van Vliet C, Gleeson PA. Trafficking and localisation of resident Golgi glycosylation enzymes. Biochimie. 2001;83:763–773. doi: 10.1016/s0300-9084(01)01312-8. doi:10.1016/S0300-9084(01)01312-8. [DOI] [PubMed] [Google Scholar]
- Paesold-Burda P, Maag C, Troxler H, Foulquier F, Kleinert P, Schnabel S, Baumgartner M, Hennet T. Deficiency in COG5 causes a moderate form of congenital disorders of glycosylation. Hum Mol Genet. 2009;18:4350–4356. doi: 10.1093/hmg/ddp389. doi:10.1093/hmg/ddp389. [DOI] [PubMed] [Google Scholar]
- Paschinger K, Gutternigg M, Rendic D, Wilson IB. The N-glycosylation pattern of Caenorhabditis elegans. Carbohydr Res. 2008;343:2041–2049. doi: 10.1016/j.carres.2007.12.018. doi:10.1016/j.carres.2007.12.018. [DOI] [PubMed] [Google Scholar]
- Paschinger K, Rendic D, Lochnit G, Jantsch V, Wilson IB. Molecular basis of anti-horseradish peroxidase staining in Caenorhabditis elegans. J Biol Chem. 2004;279:49588–49598. doi: 10.1074/jbc.M408978200. doi:10.1074/jbc.M408978200. [DOI] [PubMed] [Google Scholar]
- Peanne R, Legrand D, Duvet S, Mir AM, Matthijs G, Rohrer J, Foulquier F. Differential effects of lobe A and lobe B of the conserved oligomeric Golgi complex on the stability of β1,4-galactosyltransferase 1 and α2,6-sialyltransferase 1. Glycobiology. 2011;21:864–876. doi: 10.1093/glycob/cwq176. doi:10.1093/glycob/cwq176. [DOI] [PubMed] [Google Scholar]
- Pokrovskaya ID, Willett R, Smith RD, Morelle W, Kudlyk T, Lupashin VV. Conserved oligomeric Golgi complex specifically regulates the maintenance of Golgi glycosylation machinery. Glycobiology. 2011;21:1554–1569. doi: 10.1093/glycob/cwr028. doi:10.1093/glycob/cwr028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouille C, Hui N, Hunte F, Kieckbusch R, Berger EG, Warren G, Nilsson T. Mapping the distribution of Golgi enzymes involved in the construction of complex oligosaccharides. J Cell Sci. 1995;108(Pt 4):1617–1627. doi: 10.1242/jcs.108.4.1617. [DOI] [PubMed] [Google Scholar]
- Ram RJ, Li B, Kaiser CA. Identification of Sec36p, Sec37p, and Sec38p: Components of yeast complex that contains Sec34p and Sec35p. Mol Biol Cell. 2002;13:1484–1500. doi: 10.1091/mbc.01-10-0495. doi:10.1091/mbc.01-10-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy P, Krieger M. Isolation and characterization of an extragenic suppressor of the low-density lipoprotein receptor-deficient phenotype of a Chinese hamster ovary cell mutant. Mol Cell Biol. 1989;9:4799–4806. doi: 10.1128/mcb.9.11.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynders E, Foulquier F, Leao Teles E, Quelhas D, Morelle W, Rabouille C, Annaert W, Matthijs G. Golgi function and dysfunction in the first COG4-deficient CDG type II patient. Hum Mol Genet. 2009;18:3244–3256. doi: 10.1093/hmg/ddp262. doi:10.1093/hmg/ddp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachter H. Protein glycosylation lessons from Caenorhabditis elegans. Curr Opin Struct Biol. 2004;14:607–616. doi: 10.1016/j.sbi.2004.09.005. doi:10.1016/j.sbi.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Shestakova A, Zolov S, Lupashin V. COG complex-mediated recycling of Golgi glycosyltransferases is essential for normal protein glycosylation. Traffic. 2006;7:191–204. doi: 10.1111/j.1600-0854.2005.00376.x. doi:10.1111/j.1600-0854.2005.00376.x. [DOI] [PubMed] [Google Scholar]
- Smith RD, Lupashin VV. Role of the conserved oligomeric Golgi (COG) complex in protein glycosylation. Carbohydr Res. 2008;343:2024–2031. doi: 10.1016/j.carres.2008.01.034. doi:10.1016/j.carres.2008.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaapen LJ, Bakker JA, van der Meer SB, Sijstermans HJ, Steet RA, Wevers RA, Jaeken J. Clinical and biochemical presentation of siblings with COG-7 deficiency, a lethal multiple O- and N-glycosylation disorder. J Inherit Metab Dis. 2005;28:707–714. doi: 10.1007/s10545-005-0015-z. doi:10.1007/s10545-005-0015-z. [DOI] [PubMed] [Google Scholar]
- Ungar D, Oka T, Brittle EE, Vasile E, Lupashin VV, Chatterton JE, Heuser JE, Krieger M, Waters MG. Characterization of a mammalian Golgi-localized protein complex, COG, that is required for normal Golgi morphology and function. J Cell Biol. 2002;157:405–415. doi: 10.1083/jcb.200202016. doi:10.1083/jcb.200202016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WC, Lee N, Aoki D, Fukuda MN, Fukuda M. The poly-N-acetyllactosamines attached to lysosomal membrane glycoproteins are increased by the prolonged association with the Golgi complex. J Biol Chem. 1991;266:23185–23190. [PubMed] [Google Scholar]
- Whyte JR, Munro S. The Sec34/35 Golgi transport complex is related to the exocyst, defining a family of complexes involved in multiple steps of membrane traffic. Dev Cell. 2001;1:527–537. doi: 10.1016/s1534-5807(01)00063-6. doi:10.1016/S1534-5807(01)00063-6. [DOI] [PubMed] [Google Scholar]
- Wu X, Steet RA, Bohorov O, Bakker J, Newell J, Krieger M, Spaapen L, Kornfeld S, Freeze HH. Mutation of the COG complex subunit gene COG7 causes a lethal congenital disorder. Nat Med. 2004;10:518–523. doi: 10.1038/nm1041. doi:10.1038/nm1041. [DOI] [PubMed] [Google Scholar]
- Zheng Q, Van Die I, Cummings RD. Molecular cloning and characterization of a novel α1,2-fucosyltransferase (CE2FT-1) from Caenorhabditis elegans. J Biol Chem. 2002;277:39823–39832. doi: 10.1074/jbc.M207487200. doi:10.1074/jbc.M207487200. [DOI] [PubMed] [Google Scholar]
- Zheng Q, Van Die I, Cummings RD. A novel α1,2-fucosyltransferase (CE2FT-2) in Caenorhabditis elegans generates H-type 3 glycan structures. Glycobiology. 2008;18:290–302. doi: 10.1093/glycob/cwn007. doi:10.1093/glycob/cwn007. [DOI] [PubMed] [Google Scholar]
- Zolov SN, Lupashin VV. Cog3p depletion blocks vesicle-mediated Golgi retrograde trafficking in HeLa cells. J Cell Biol. 2005;168:747–759. doi: 10.1083/jcb.200412003. doi:10.1083/jcb.200412003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.