Abstract
The constitutional t(11;22)(q23;q11) is the most common recurrent non-Robertsonian translocation in humans. The breakpoint sequences of both chromosomes are characterized by several hundred base pairs of palindromic AT-rich repeats (PATRRs). Similar PATRRs have also been identified at the breakpoints of other nonrecurrent translocations, suggesting that PATRR-mediated chromosomal translocation represents one of the universal pathways for gross chromosomal rearrangement in the human genome. We propose that PATRRs have the potential to form cruciform structures through intrastrand-base pairing in single-stranded DNA, creating a source of genomic instability and leading to translocations. Indeed, de novo examples of the t(11;22) are detected at a high frequency in sperm from normal healthy males. This review synthesizes recent data illustrating a novel paradigm for an apparent spermatogenesis-specific translocation mechanism. This observation has important implications pertaining to the predominantly paternal origin of de novo gross chromosomal rearrangements in humans.
Keywords: cruciform, gross chromosomal rearrangements, non-B DNA, palindrome, translocation
The constitutional t(11;22)(q23;q11) is the most frequent recurrent non-Robertsonian translocation in humans (Fig. 1a). Similar to Robertsonian translocations or many other nonrecurrent constitutional translocations, balanced carriers of the t(11;22) usually manifest no clinical symptoms, because the rearrangement does not disrupt functional genes. However, translocation carriers often have reproductive problems, such as male infertility, recurrent spontaneous abortions, and the birth of offspring with a chromosomal imbalance. Severely affected offspring often have supernumerary-der(22)t(11;22) syndrome (Emanuel syndrome, MIM# 609029), as a result of 3:1 meiotic malsegregation of the der(22) (1). The syndrome is characterized by severe mental retardation, preauricular tag or sinus, ear anomalies, cleft or high-arched palate, micrognathia, microcephaly, kidney abnormalities, heart defects, and genital abnormalities in males (2–4). Most of the t(11;22) carrier individuals are identified subsequent to the birth of an individual with Emanuel syndrome.
Fig. 1.
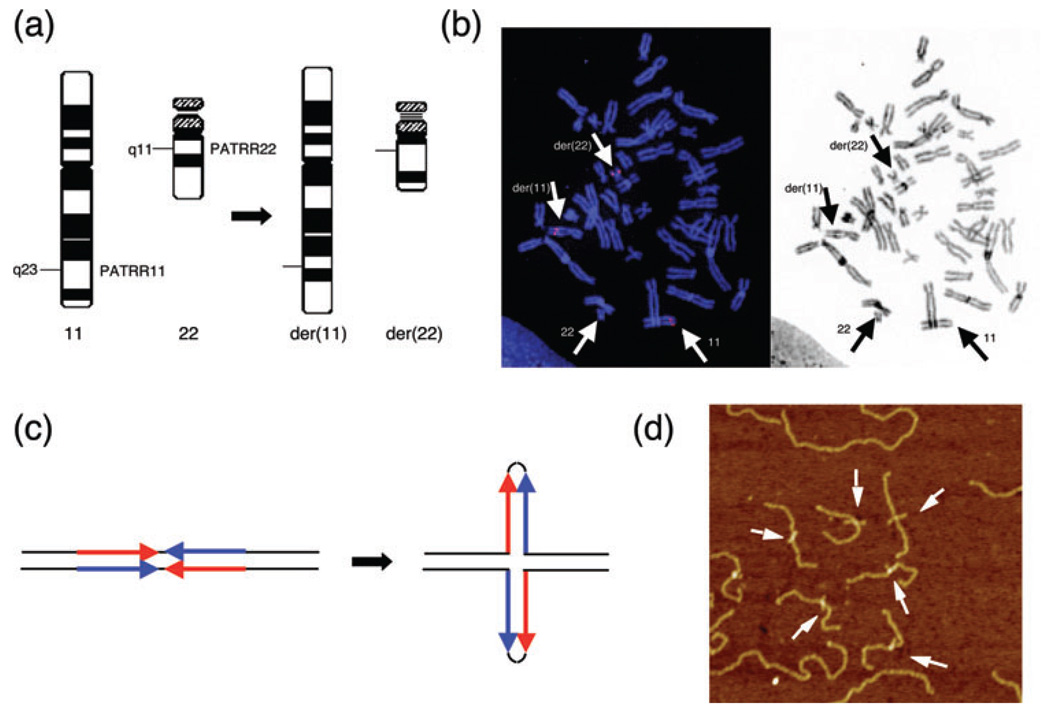
Palindrome-mediated translocation in humans. (a) Schematic representation of the t(11;22)(q23;q11). The PATRR11 and PATRR22 are located at the breakpoints on 11q23 and 22q11, respectively. (b) FISH analysis of metaphase chromosomes derived from a balanced t(11;22) carrier using a BAC spanning the breakpoint on chromosome 11. The signal from the chromosome 11 BAC is split by the translocation and appears on both the der(11) and the der(22) (left). The same FISH image was inverted to greyscale to show the location of the relevant chromosomes in the metaphase spreads (right). (c) Predicted secondary structure for the palindromic sequence. Short palindromic sequences have the potential to form double-stranded cruciform structures by intrastrand-base pairing in single-stranded DNA. DNA sequences indicated by blue arrows are complementary to those indicated by red arrows. (d) Cruciform extrusion of the plasmid harboring the PATRR11. A plasmid bearing the PATRR11 insert was fixed with psoralen treatment followed by ultraviolet exposure. The PATRR11 fragments were released with restriction enzyme digestion and were visualized using AFM. AFM, atomic force microscopy; BAC, bacterial artifical chromosome; FISH, fluorescent in situ hybridization; PATRR, palindromic AT-rich repeat.
Translocation is one of the most frequently occurring gross chromosomal rearrangements in humans. Translocation can potentially cause devastating disorders when it disrupts an important genomic element. The somatic translocations that are commonly identified in cancers or leukemias result in the disruption of proto-oncogenes, leading to the production of chimeric transcripts with oncogenic potential. Likewise, although constitutional translocations occasionally cause specific genetic diseases, most of them are harmless and do not disrupt essential genes. However, the offspring of individuals harboring a balanced translocation have a potential risk of unbalanced translocation, resulting in pregnancy loss or birth of a child with a congenital anomaly syndrome.
The formation of translocation is essentially dependent on two distinct processes; double-strand-breaks (DSBs) and an error in DSB repair (5). DSBs can result from exogenous agents such as ionizing radiation and chemotherapeutic drugs, and also from endogenously generated reactive oxygen species and mechanical stresses on the chromosomes (6, 7). DSBs can impair cellular function and eventually cause cell death by triggering apoptosis. To counteract such deleterious effects, DSBs are usually repaired through the activity of error-free repair systems, such as homologous recombination (8). However, translocations occasionally result from the activity of an error-prone repair system, such as non-homologous end joining (NHEJ), with or without microhomology (9, 10). Experimental induction of two DSBs is sufficient to produce translocations in mammalian cells (11). Indeed, NHEJ is suggested to be involved in translocation junction formation both in this system and in translocations identified in human disease patients as nonrecurrent translocations (12, 13).
Chromosomal translocations can be random events with nonrecurrent breakpoints, indicating the occurrence of random DSBs followed by an error in the DSB repair pathway. However, a subset of translocations shows recurrent manifestation, suggesting increased susceptibility of one of these two elements at specific loci. Regarding constitutional translocation, the recurrent translocation observed most frequently is the Robertsonian translocation. Breakpoint analyses indicated homologous recombination between pericentromeric repeats that commonly occur on the short arm of acrocentric chromosomes (14). This mechanism is known as non-allelic homologous recombination (NAHR) and is indicative of not only translocations, but also other recurrent gross chromosomal rearrangements, such as deletions and inversions (15–18).
Another example of recurrent translocation is t(4;8)(p16;p23). Both translocation breakpoints have been mapped within the olfactory receptor gene clusters at 4p16 and 8p23, suggesting that NAHR between olfactory receptor genes on different chromosomes is also responsible for the translocation (19). Interestingly, both de novo Robertsonian translocations and de novo t(4;8) arise preferentially during maternal gametogenesis (20, 21). Although rare, t(4;11)(p16.2;p15.4) is also an example of recurrent translocation characterized by breakpoint homology (22). In sharp contrast is t(11;22)(q23;q11), which is an example of recurrent constitutional translocation that may be caused by susceptibility to DSBs at specific loci.
Identification of the PATRR sequences at the breakpoints of constitutional t(11;22)
Flurorescent in situ hybridization (FISH) studies using multiple probes on chromosomes 11q23 and 22q11 have shown that in individuals with t(11;22), including de novo cases, the t(11;22) breakpoints are confined to same narrow intervals on both chromosomes (1, 23, 24) (Fig. 1b). The recurrent nature of the t(11;22) prompted us to examine the translocation breakpoints in detail to identify the specific genomic structure associated with the chromosome 11 and 22 breakpoints.
A conventional positional cloning strategy allowed us to identify both constitutional t(11;22) breakpoints, although the work was challenging due to the inherent genomic instability in the breakpoint regions. We first identified a breakpoint cluster region on 11q23, which was approximately 450 bp in length with a high AT content (93%). The breakpoint on 11q23 constitutes a nearly perfect palindromic structure, with 98% identity between its proximal and distal arms (Fig. 1c). We designated this configuration as a PATRR on 11q23 (PATRR11) (25, 26). The cloning of the 22q11 breakpoint was also an extraordinarily challenging. Eventually, a similar PATRR was identified within one of the unclonable gaps unresolved by the human genome project (27). The size of the PATRR on 22q11 (PATRR22) was approximately 590 bp, which was slightly larger than PATRR11. In spite of their similarity with regard to AT-richness, no substantial homology was observed between PATRR11 and PATRR22 (58% identity).
In addition, we examined junction fragments originating from more than 50 independent balanced t(11;22) carriers to localize the breakpoints precisely (28). The breakpoints on both chromosomes were located at the center of the PATRRs. Only a small number of identical nucleotides were found at the point where the original two sequences were joined, and there were always small deletions (no greater than 50 nucleotides) at the breakpoint regions of both PATRRs (26, 27). Collectively, these data suggested that the mechanism for this recurrent chromosomal translocation involved the occurrence of DSBs at the center of the two PATRRs, followed by repair via the NHEJ pathway.
These findings were confirmed by two other research groups who tested different subsets of translocation carriers and showed similar breakpoints (29, 30).
PATRR-mediated chromosomal translocation as one of the universal pathways for gross chromosomal rearrangements in humans
In the human genome, the PATRR22 site appears to be extremely susceptible to breakage. The chromosome 22q11 has been designated as a hotspot for translocation breakpoints, because cytogenetic studies have shown that a large number of translocation breakpoints cluster at 22q11. FISH mapping has indicated that the breakpoints of numerous translocations involving 22q11 cluster within the same interval that includes the PATRR22 (31–35). These data suggest the involvement of PATRR22 in the etiology of all 22q11-related translocations.
We analyzed two unrelated individuals with constitutional t(17;22)(q11;q11) that presented with neurofibromatosis type 1 (NF1) (32, 36). Each translocation disrupted the NF1 gene on 17q11, resulting in patients displaying the NF1 phenotype. As was expected, FISH analysis localized the 22q11 breakpoints within the same interval where the breakpoint of t(11;22) resides. Further analyses of the 17q11 breakpoints within the NF1 gene revealed the presence of an approximately 200 bp PATRR sequence within intron 31 of the NF1 gene (PATRR17) (36, 37). Subsequent molecular cloning of other translocation breakpoints has shown similar palindromic, and often AT-rich, sequences on partner chromosomes, such as 4q35.1, 1p21.2 and 8q24.1 (38–40). Hence, palindrome-mediated chromosomal translocation appears to be one of the possible universal pathways for creating human genomic rearrangements. This subset of translocations appears to occur in a nonrandom fashion and to be possibly mediated by the genomic instability of palindromic DNA.
Small palindromic DNAs like the PATRRs have the potential to form stem-loop structures through intrastrand-base pairing within single-stranded DNA. As a consequence, they form a specific structure consisting of a single-stranded hairpin or a double-stranded cruciform (Fig. 1c). We analyzed the tertiary structure of the cloned PATRR11 in vitro (41). A plasmid containing the PATRR11 adopts a non-B DNA conformation under negative superhelicity. Using atomic force microscopy (AFM), we were able to visualize the cruciform extrusion from the PATRR11 plasmid directly (Fig. 1d). Thus, we propose that the secondary structure of the PATRRs induces genomic instability leading to both recurrent and nonrecurrent chromosomal translocations in humans.
Translocation-specific polymerase chain reaction (PCR) detects de novo t(11;22)s in sperm from normal healthy males
We established a t(11;22)-specific PCR system utilizing sequence data from the junction fragments. PATRR-flanking primers were designed both on 11q23 and 22q11 to amplify the der(11) and the der(22) junction fragments (Fig. 2a). This PCR approach successfully amplified the der(11) and der(22) junction fragments from balanced t(11;22) carriers as well as the der(22) junction fragments of patients with Emanuel syndrome (28).
Fig. 2.
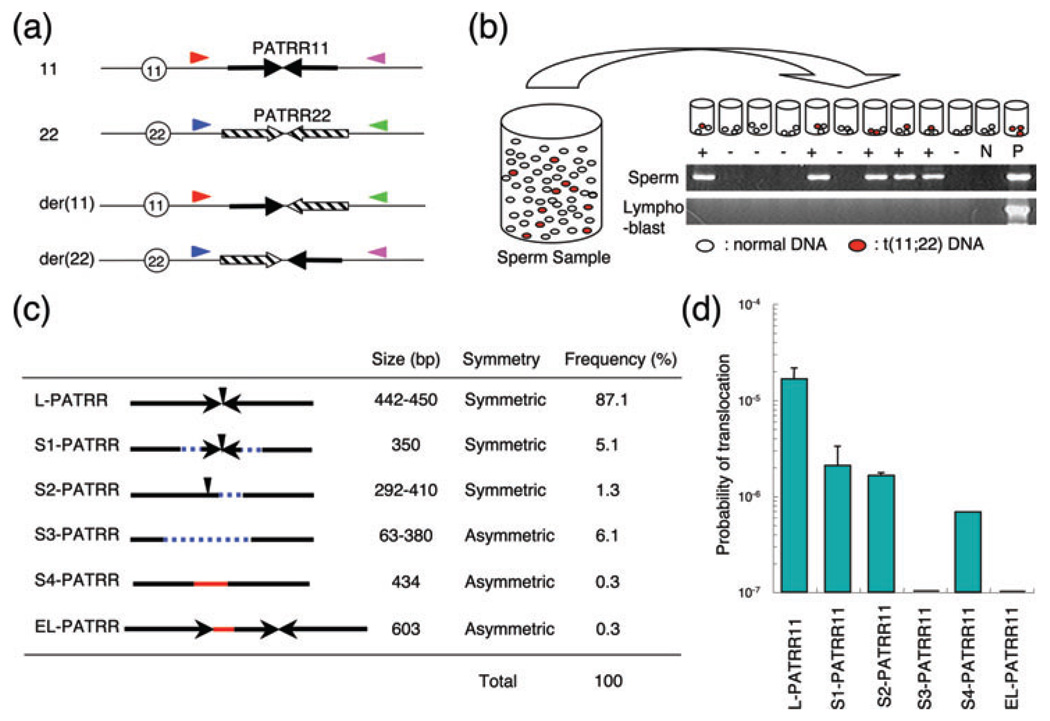
Polymorphisms of the palindrome affect the de novo translocation frequency. (a) Location of PCR primers. Arrows indicate each arm of the PATRR11 (solid arrows) and PATRR22 (hatched arrows). Size polymorphisms of the PATRR11 were examined by PCR using primers indicated by red and pink triangles. Translocations were detected using one of the primers flanking the PATRR11 (red or pink triangles) and with one of the primers flanking the PATRR22 (blue or green triangles). Centromeres are represented by circles. (b) Strategy for estimation of translocation frequency by PCR. Genomic DNA was extracted from sperm samples. Translocation-specific PCR was performed using multiple batches of template DNA. The translocation frequency was calculated using the equation, q = 1 − (1 − p)1/n; with n = number of haploid genomes per aliquot, p = the probability that an aliquot contains a translocation, product and q = the probability that one randomly selected haploid genome in a given aliquot sustained a translocation. The gel images show representative PCR results. The upper panel shows results derived from sperm DNA, whereas the lower panel presents results from lymphoblast DNA. Lane M, size marker; lane N, negative control; lane P, genomic DNA from a t(11;22) balanced carrier serving as a positive control. (c) Characterization of the PATRR11 variants. Arrows indicate each unit of inverted repeats. Vertical arrowheads indicate the center of the palindromic sequence. Dotted blue lines show the deleted region, while red lines indicate the insertion of sequences of unknown origin. Other characteristics and the frequency in the general population are shown on the right. (d) A histogram showing the probability of de novo translocation by allele types. Each bar represents the mean value and the vertical bar indicates standard deviation on a log scale. The allele types are abbreviated. PCR, polymerase chain reaction; PATRR, palindromic AT-rich repeat.
To determine the prevalence of t(11;22), we examined the frequency of de novo translocations in DNA derived from sperm samples obtained from normal, healthy male volunteers (42). Translocation-specific PCR was performed using conditions that would allow for the detection of a single molecule of target DNA. Multiple aliquots were amplified from each sperm sample (Fig. 2b). When 100 ng aliquots of sperm DNA, each containing 33,000 haploids as templates, were amplified, translocation-specific PCR products were detected in a substantial number of reactions (42). The presence of both positive and negative PCRs clearly indicates the de novo origin of the translocation (Fig. 2b). Translocation-specific PCR products were never detected in DNA from blood or cheek swab DNA from the same donors, essentially excluding the possibility that the positive PCR results seen in sperm were the result of contamination or PCR artifacts.
The frequency of sperm de novo translocation events was calculated based on the presence of positive PCRs. Using the number of positive PCRs expressed relative to the total number of PCRs performed, the frequency was calculated on the basis that the probability of observing a positive PCR corresponded to the total sum of a binomial series of the translocation frequency calculated as described previously (42). For an initial analysis, we examined sperm samples from four randomly selected healthy male volunteers, and the estimated frequency of the translocation was approximately 1 × 10−5, which is unexpectedly high. The frequencies for der(11) and der(22) were approximately equal, suggesting that de novo translocation occurs as a reciprocal rearrangement. This observation is in agreement with the observation that the majority of patients with the supernumerary-der(22) syndrome are the offspring of a balanced translocation carrier, rather than representing a de novo event (2–4).
PATRR polymorphisms affect the de novo translocation frequency
In the course of our PATRR analyses, we identified that the PATRRs on several chromosome 22 partner chromosomes are hypervariable among individuals (37, 43). Size polymorphisms due to deletion, insertion and duplication, including the center region, are common, supporting the idea that the center of a PATRR is fragile. The central modifications often resulted in asymmetric palindromic structures (Fig. 2c). This observation prompted us to test the hypothesis that PATRR polymorphism might affect the frequency of de novo t(11;22)s.
We examined sperm samples from normal healthy males with various PATRR11 genotypes (43). Homozygotes for the long symmetric PATRR11 (L-PATRR11), the most frequent allele, produced de novo translocations at a frequency ranging between 1.52 × 10−5 and 1.57 × 10−4. Heterozygotes for the L-PATRR11 and symmetric short PATRR11 alleles (S1, S2) produced de novo translocations at an overall frequency similar to that detected for L-PATRR11 homozygotes. However, the translocation products derived from the symmetric short PATRR were observed less frequently (~10−6) suggesting that the frequency is dependent on the size of the PATRR (Fig. 2d). However, although individuals heterozygous for an L-PATRR11 and an asymmetric short PATRR11 (S3) produced de novo translocations at a similar overall frequency, the asymmetric short PATRR did not produce any de novo translocations. These observations clearly show that PATRR polymorphisms affect the frequency of translocations. Collectively, these data, including the data for rare genotypes, indicate that the size and symmetry of the center of the PATRR11 determines the frequency of de novo t(11;22)s (Fig. 2d). Indeed, the translocation frequency reflects the propensity of each polymorphic allele for secondary structure formation (44). PATRR22 is slightly longer than other PATRRs, which might explain why all of the known PATRR-mediated translocations involve PATRR22 (27). Thus, it is reasonable to propose that the potential for adopting a secondary structure is probably to contribute to the susceptibility to translocation development.
The t(11;22) is independent of DNA replication
Curiously, translocation-specific PCR has never detected a de novo translocation event in any tissues other than sperm. Diverse human tissues such as peripheral leukocytes, lymphoblasts, and skin fibroblasts were consistently negative for de novo translocation by PCR analysis (42). We also tested various cultured somatic cell lines derived from human cells (HEK293, HeLa, HepG2 and THP-1), but all were negative (0 positive PCR in 40 reactions, <7.67 × 10−7) (45). The fact that only sperm samples produce de novo t(11;22)s supports the idea that PATRR-mediated translocations occur primarily during gametogenesis. These findings are unusual and appear to be inconsistent with the established mechanisms pertaining to the instability of palindromic DNA sequences.
The instability of palindromic DNA has been extensively investigated over the last 20 years. Susceptibility to deletion within palindromic regions has been consistently shown in many experimental organisms, including Escherichia coli (46–48), Saccharomyces cerevisiae (49, 50), and mice (51, 52). Such deletions appear to be primarily mediated by the stalling of DNA replication at a region that has formed a secondary structure. This blockade appears to be resolved by ‘slippage’ or ‘strand switch’. Another important pathway for palindromic DNA deletions is endonuclease cleavage of such a secondary structure during replication. The PATRR11 was found to be deleted from the BAC encompassing the breakpoint region (25, 26). Furthermore, the fact that PATRR22 is underrepresented in BAC/PAC/YAC libraries also suggests that PATRRs are highly unstable in bacteria and yeast. Indeed, translocations have been induced in regions containing palindromic or inverted repeats in vegetative yeast when DNA replication is compromised (53).
Combined with data supporting the sperm-specific occurrence of de novo translocations, PATRR-mediated translocations may result from palindrome instability facilitated by the high number of cell divisions and DNA replications during spermatogenesis. Paradoxically, we analyzed sperm samples from 10 male donors and found no age-dependent increase in the frequency of de novo t(11;22)s (54). We also obtained samples from the same donors after a 6-year interval, and no age-dependent increase in translocation frequency was observed (54). If the translocation occurs during DNA replication, the samples from older donors that have undergone greater number of germline divisions should theoretically include greater number of translocations. Thus, these findings may invoke a novel paradigm regarding palindrome instability that is independent of DNA replication.
The consequence of replication-related palindromic instability is often a deletion. Indeed, PATRRs often manifest themselves as size polymorphisms among individuals due to deletions, insertions and duplications, as mentioned in the previous section (43). However, our recent breakpoint analyses clearly distinguished the mechanisms of deletion from those of translocation (55). Microhomology identified at the deletion endpoints is significantly longer than that at translocation breakpoints, suggesting that PATRR-mediated deletions develop in a homology-directed manner. Another interesting finding is that all of the insertions that were identified within the deletion junctions were AT-rich sequences, whereas insertions found at translocation junctions were non-AT-rich DNA sequences. Thus, it could be hypothesized that rearrangements within the PATRR, such as deletions, insertions and duplications, are induced by secondary structure formation during DNA replication followed by repair through a homology-dependent pathway. In contrast, PATRR-mediated translocation may be driven by a completely different mechanism. Indeed, inhibition of DNA replication in cultured human cells induces deletions within the PATRR11, but not translocations between different PATRRs (45). Thus, these findings lend support to the possibility of replication-independent translocations.
Establishment of a model system
To gain a better understanding of how DNA secondary structure contributes to palindrome-mediated genomic instability, there have been ongoing attempts to establish a model system that recapitulates PATRR-mediated translocation. Establishment of such a model system would also provide information on the underlying mechanisms of PATRR-mediated gross chromosomal rearrangements.
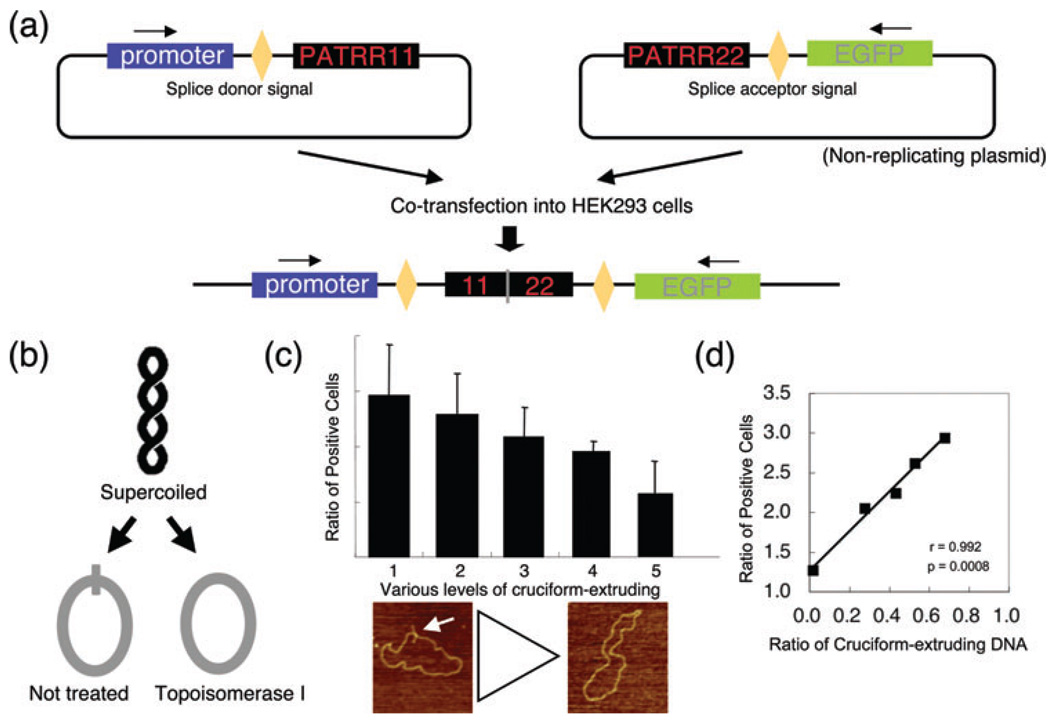
We have recently developed a plasmid-based model system of PATRR-mediated translocation (56). This system utilizes two plasmids harboring either PATRR11 or PATRR22 sequences, which act as substrates for rearrangement within mammalian nuclei. The first plasmid includes a promoter and a splice donor sequence upstream of PATRR11, whereas the second plasmid contains the PATRR22 sequence followed by a splice acceptor sequence and then the green fluorescence protein (GFP) gene coding sequence (Fig. 3a). Translocation-like rearrangements between the plasmids were detected by PCR 24 h after co-transfection into human cell lines. The junction sequences were found to be analogous to the human t(11;22) junction sequences, suggesting that this model system recapitulates the PATRR-mediated t(11;22) translocations, which occur in humans.
Fig. 3.
Plasmid-based model system of palindrome-mediated translocation. (a) Schematic representation of the detection system. When the two plasmids harboring PATRR11 and PATRR22 are rearranged within each PATRR region, the transcript from the fusion product splices out the intervening junction sequence and expresses the downstream GFP gene product. (b) Strategy for preparing PATRR plasmids with or without cruciform extrusion. Plasmid purified from Escherichia coli is negatively supercoiled (upper). The PATRR plasmid with negative superhelicity energetically favors cruciform extrusion forming intrastrand-base pairing. The positive free energy of cruciform formation is offset by relaxation of the negative superhelical density (left). If the negative superhelicity is abrogated by incubation with topoisomerase I prior to cruciform extrusion, a relaxed plasmid without the extruded cruciform can be obtained (right). (c) The incidence of translocation-like rearrangements following the use of different preparative techniques for PATRR plasmids. GFP-positive cells were counted after co-transfection with various levels of cruciform-extruding PATRR plasmids. The plasmids were prepared by the alkaline-SDS method (1) and by the Triton method followed by either incubation in various NaCl concentrations (2, 10 mM; 3, 50 mM; 4, 200 mM) or topoisomerase I treatment (5). Representative AFM images of the PATRR plasmid are indicated as follows: arrow indicates cruciform extrusion of the PATRR plasmids. (d) The degree of cruciform in the transfected plasmids affects the levels of rearrangement. The degrees of cruciform extrusion observed by AFM correlate with the ratio of GFP-positive cells (right; r = 0.992, p = 0.0008). AFM, atomic force microscopy; GFP, green fluorescence protein; PATRR, palindromic AT-rich repeat; SDS, sodium dodecyl sulfate.
To examine the contribution of DNA secondary structure to this translocation-like rearrangement, we prepared plasmid samples that contained differing proportions of cruciform-extruding plasmids (Fig. 3b). The plasmids were then transfected into human cell lines and the rearrangement frequency was quantified by GFP reporter gene expression as detected using flow cytometry. The plasmids isolated using the alkali-method, which contained an abundance of cruciform-extruding plasmid DNA, showed the greatest number of GFP-positive cells (Fig. 3c). In contrast, plasmids treated with topoisomerase I, which had fewer cruciform-extruding plasmids, resulted in smaller numbers of GFP-positive cells. The proportion of cruciform-extruding plasmids monitored by AFM was correlated with the number of GFP-positive cells (Fig. 3c,d). These data show that cruciform conformation of the PATRR contributes to this translocation-like event.
Furthermore, our data provide indirect evidence for the presence of DNA secondary structures in living cells, and also indirectly but strongly support the hypothesis that the PATRR sequences adopt a cruciform conformation that induces genomic instability, leading to the translocation. In this model system, no translocation was detected between endogenous PATRRs or between PATRR plasmids and endogenous PATRRs. These results indicate that all the enzymatic activity that is required for translocation to occur is present in human somatic cell lines with the exception of the capacity for cruciform extrusion of the PATRR. These results also support the hypothesis described in the next section.
Hypothesis: underlying mechanism dictating spermatogenesis-specific translocation
As mentioned previously, PATRR-mediated translocation appears to be specific to meiotic cells. During mammalian meiosis, there are two types of physiological DNA breakage. A substantial number of DSBs occur as an initiating step for meiotic recombination. SPO11, a meiosis-specific endonuclease, mainly functions in this step (57). Subsequently, repair of the DSBs by homologous recombination progresses by a RAD52-mediated homology detection and RAD51/DMC1-mediated strand invasion. Holliday junctions (HJs), which form as intermediates during this process, are finally resolved by endonuclease activity. The four-way junction of a cruciform DNA structure is analogous to this HJ structure and could represent a substrate for the HJ resolvase, although such an enzyme has not been definitely identified in higher organisms such as yeast or mammals (58). Either of these steps might provide a good ‘candidate process’ for the generation of the DSBs leading to translocation events (59, 60).
To prove the presence of such meiosis-specific cruciform resolution, it is necessary to determine whether female germ cells can also produce de novo t(11;22) translocations. Toward this end, it might be reasonable to survey female germ cells for the presence of de novo t(11;22)s using similar translocation-specific single molecule detection PCR. However, performing this experiment is limited by the finite number of human oocytes available for examination. Even with the existing evidence of a high frequency of de novo translocations in sperm, female germ cells cannot be analyzed using a similar strategy.
As an alternative, we obtained samples from eight individuals with a de novo t(11;22) translocation together with samples from their parents to determine the parental origin of the translocation. Because the PATRR shows sequence variation among individuals, we theorized that determination of a parental origin for an individual translocation was possible through comparison of the sequence of translocation junction fragments on the der(11) and the der(22) with the PATRR11 and PATRR22 sequences on normal chromosomes 11 and 22 in the parental samples. Indeed, segregation analysis has shown that the de novo events are exclusively of paternal origin in our patient population, although to date only a finite number of samples have been examined (61). This finding implies that it is not necessarily a meiotic mechanism, but rather a spermatogenesis-specific mechanism that permits the development of the t(11;22) translocation.
DNA breakage might occur during late spermatogenesis when DNA is packaged into dense chromatin. The successive transition of chromatin components from histones to protamines causes dynamic changes in chromatin structure (62, 63). During this process, release from nucleosomes may contribute to the release of free negative supercoiling. With nucleosome withdrawal, temporary and local accumulation of an excess of negative superhelicity is possible, potentially exacerbating secondary structure formation within PATRR sequences with subsequent formation of strand breaks. A similar mechanism has been proposed for triplet-repeat expansion, which has been shown to be of postmeiotic origin (64).
All of the hypotheses described above relate to the causes of frequent DSBs at the PATRR sequences. However, the factors that facilitate such high efficiency DSB repair and aberrant joining between two PATRRs are still an enigma that remains to be elucidated. The role of spatial proximity between the two PATRRs in meiotic and postmeiotic cells deserves further investigation (65). Thus, there are still numerous factors surrounding the mechanism of translocation formation that remain to be elucidated.
What the t(11;22) research tells us
Chromosomal abnormalities in humans often manifest a gender bias with respect to parental origin. Aneuploidy, or a numerical abnormality, is more probably to arise in maternal gametogenesis (66). In contrast, 80% of known structural chromosomal abnormalities are of paternal origin (67). This is not surprising because late spermatids and sperm, which are non-dividing haploid cells, cannot undergo homologous recombination. However, components of NHEJ repair are expressed at low levels in these cells (68). It is formally possible that such structural abnormalities arise when DSBs induced by endogeneous or exogeneous mutagens are repaired by error-prone NHEJ system. However, our current hypothesis points to the importance of postmeiotic physiological events during spermatogenesis for the generation of the translocation. This hypothesis has important implications of the predominantly paternal origin of de novo gross chromosomal rearrangements in humans.
To elucidate the precise mechanism of palindrome-mediated translocation, it will be necessary to establish a mouse model system for t(11;22) research. Because the mouse genome does not possess PATRR-like sequences, one could postulate that the introduction of two copies of PATRRs will be sufficient for the creation of palindrome-mediated translocations in a mouse. Further characterization of the PATRR dynamics in a mammalian germline should aid the future elucidation of the complex biology of non-B DNA-mediated translocations.
Future prospects
PATRR-mediated translocation provides the opportunity for novel insight into mechanisms of chromosomal translocations. This concept could also be applied to translocations that develop in somatic cells. The recurrent nature of translocations observed in cancers or leukemias has been long thought to be random events resulting from a selective growth advantage. However, some translocation breakpoints have the potential for non-B DNA conformation (69). For example, the breakpoint regions of translocation partners in immunoglobulin gene-related translocation are reported to form Z-DNA or triplex DNA (70–72). Increasing research efforts are being applied toward elucidation of the molecular components leading to the DSB formation at these non-B DNAs, such as structure-directed nucleases.
Considerable progress has been made toward understanding the molecular mechanisms underlying the generation of recurrent translocations. However, questions still remain pertaining to the mechanisms of nonrecurrent translocations, which have also been long thought to be random events. A novel molecular mechanism has also been proposed to explain the occurrence of another subset of gross chromosomal rearrangements, deletions and duplications. This proposed mechanism explaining nonrecurrent gross chromosomal rearrangements is not based on the occurrence of DSBs, but is associated with DNA replication (73, 74). In brief, a free DNA end is generated by a stall during DNA synthesis, which restarts after switching to the template DNA on a different chromosomal site, resulting in gross chromosomal rearrangement. This mechanism, called replication Fork Stalling and Template Switching (FoSTeS) and microhomology-mediated break-induced replication (MMBIR), might be involved in some nonrecurrent chromosomal translocations.
A recent report confirmed the preferential paternal origin of de novo nonrecurrent translocations using flow-sorted derivative translocation chromosomes followed by segregation analysis. Interestingly, development of the translocation appeared to be associated with increased paternal age (75). Based on the fact that male gametogenesis undergoes much higher number of mitotic divisions than that in female, this observation is in agreement with a replication-based mechanism of the translocation formation. This result is in clear contrast with the existing data on t(11;22) translocations and the possible PATRR-mediated mechanism (54). Further detailed studies and accumulated breakpoint information for nonrecurrent translocations are needed to elucidate the molecular mechanisms leading to gross chromosomal rearrangements.
Acknowledgements
The author wishes to thank Drs. T.H. Shaikh and M. Taniguchi for helpful discussion, and Ms. A.M. Hacker, H. Kowa, K. Nagaoka, T. Mori, and E. Hosoba for technical assistance. These studies were supported by a grant-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (16390102), the Ministry of Health, Labour and Welfare and Daiko Foundation to H.K. Support was also provided by a grant from the National Institutes of Health (CA39926) and funds from the Charles E. H. Upham endowed chair to B. S. E.
Footnotes
Conflict of interest
We declare no conflict of interest.
References
- 1.Shaikh TH, Budarf ML, Celle L, et al. Clustered 11q23 and 22q11 breakpoints and 3:1 meiotic malsegregation in multiple unrelated t(11;22) families. Am J Hum Genet. 1999;65:1595–1607. doi: 10.1086/302666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zackai EH, Emanuel BS. Site-specific reciprocal translocation, t(11;22) (q23;q11), in several unrelated families with 3:1 meiotic disjunction. Am J Med Genet. 1980;7:507–521. doi: 10.1002/ajmg.1320070412. [DOI] [PubMed] [Google Scholar]
- 3.Fraccaro M, Lindsten J, Ford CE, et al. The 11q;22q translocation: a European collaborative analysis of 43 cases. Hum Genet. 1980;56:21–51. doi: 10.1007/BF00281567. [DOI] [PubMed] [Google Scholar]
- 4.Carter MT, St. Pierre SA, Zackai EH, et al. Phenotypic delineation of Emanuel syndrome (supernumerary derivative 22 syndrome): clinical features of 63 individuals. Am J Med Genet A. 2009;149A:1712–1721. doi: 10.1002/ajmg.a.32957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kurahashi H, Bolor H, Kato T, et al. Recent advance in our understanding of the molecular nature of chromosomal abnormalities. J Hum Genet. 2009;54:253–260. doi: 10.1038/jhg.2009.35. [DOI] [PubMed] [Google Scholar]
- 6.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 7.van Gent DC, Hoeijmakers JH, Kanaar R. Chromosomal stability and the DNA double-stranded break connection. Nat Rev Genet. 2001;2:196–206. doi: 10.1038/35056049. [DOI] [PubMed] [Google Scholar]
- 8.Cromie GA, Connelly JC, Leach DR. Recombination at double-strand breaks and DNA ends: conserved mechanisms from phage to humans. Mol Cell. 2001;8:1163–1174. doi: 10.1016/s1097-2765(01)00419-1. [DOI] [PubMed] [Google Scholar]
- 9.Lieber MR, Ma Y, Pannicke U, et al. Mechanism and regulation of human non-homologous DNA end-joining. Nat Rev Mol Cell Biol. 2003;4:712–720. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- 10.Elliott B, Richardson C, Jasin M. Chromosomal translocation mechanisms at intronic alu elements in mammalian cells. Mol Cell. 2005;17:885–894. doi: 10.1016/j.molcel.2005.02.028. [DOI] [PubMed] [Google Scholar]
- 11.Richardson C, Jasin M. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature. 2000;405:697–700. doi: 10.1038/35015097. [DOI] [PubMed] [Google Scholar]
- 12.Kurahashi H, Sakamoto M, Ono J, et al. Molecular cloning of the chromosomal breakpoint in the LIS1 gene of a patient with isolated lissencephaly and balanced t(8;17) Hum Genet. 1998;103:189–192. doi: 10.1007/s004390050805. [DOI] [PubMed] [Google Scholar]
- 13.D’Angelo CS, Gajecka M, Kim CA, et al. Further delineation of nonhomologous-based recombination and evidence for subtelomeric segmental duplications in 1p36 rearrangements. Hum Genet. 2009;125:551–563. doi: 10.1007/s00439-009-0650-9. [DOI] [PubMed] [Google Scholar]
- 14.Page SL, Shin JC, Han JY, et al. Breakpoint diversity illustrates distinct mechanisms for Robertsonian translocation formation. Hum Mol Genet. 1996;5:1279–1288. doi: 10.1093/hmg/5.9.1279. [DOI] [PubMed] [Google Scholar]
- 15.Shaffer LG, Lupski JR. Molecular mechanisms for constitutional chromosomal rearrangements in humans. Annu Rev Genet. 2000;34:297–329. doi: 10.1146/annurev.genet.34.1.297. [DOI] [PubMed] [Google Scholar]
- 16.Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- 17.McDermid HE, Morrow BE. Genomic disorders on 22q11. Am J Hum Genet. 2002;70:1077–1088. doi: 10.1086/340363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bailey JA, Eichler EE. Primate segmental duplications: crucibles of evolution, diversity and disease. Nat Rev Genet. 2006;7:552–564. doi: 10.1038/nrg1895. [DOI] [PubMed] [Google Scholar]
- 19.Giglio S, Broman KW, Matsumoto N, et al. Olfactory receptorgene clusters, genomic-inversion polymorphisms, and common chromosome rearrangements. Am J Hum Genet. 2001;68:874–883. doi: 10.1086/319506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Page SL, Shaffer LG. Nonhomologous Robertsonian translocations form predominantly during female meiosis. Nat Genet. 1997;15:231–232. doi: 10.1038/ng0397-231. [DOI] [PubMed] [Google Scholar]
- 21.Giglio S, Calvari V, Gregato G, et al. Heterozygous submicroscopic inversions involving olfactory receptor-gene clusters mediate the recurrent t(4;8)(p16;p23) translocation. Am J Hum Genet. 2002;71:276–285. doi: 10.1086/341610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas NS, Maloney V, Bryant V, et al. Breakpoint mapping and haplotype analysis of three reciprocal translocations identify a novel recurrent translocation in two unrelated families: t(4;11)(p16.2;p15.4) Hum Genet. 2009;125:181–188. doi: 10.1007/s00439-008-0611-8. [DOI] [PubMed] [Google Scholar]
- 23.Edelmann L, Spiteri E, McCain N, et al. A common breakpoint on 11q23 in carriers of the constitutional t(11;22) translocation. Am J Hum Genet. 1999;65:1608–1616. doi: 10.1086/302689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tapia-Paez I, O’Brien KP, Kost-Alimova M, et al. Fine mapping of the constitutional translocation t(11;22)(q23;q11) Hum Genet. 2000;106:506–516. doi: 10.1007/s004390000287. [DOI] [PubMed] [Google Scholar]
- 25.Kurahashi H, Shaikh TH, Hu P, et al. Regions of genomic instability on 22q11 and 11q23 as the etiology for the recurrent constitutional t(11;22) Hum Mol Genet. 2000;9:1665–1670. doi: 10.1093/hmg/9.11.1665. [DOI] [PubMed] [Google Scholar]
- 26.Kurahashi H, Emanuel BS. Long AT-rich palindromes and the constitutional t(11;22) breakpoint. Hum Mol Genet. 2001;10:2605–2617. doi: 10.1093/hmg/10.23.2605. [DOI] [PubMed] [Google Scholar]
- 27.Kurahashi H, Inagaki H, Hosoba E, et al. Molecular cloning of a translocation breakpoint hotspot in 22q11. Genome Res. 2007;17:461–469. doi: 10.1101/gr.5769507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurahashi H, Shaikh TH, Zackai EH, et al. Tightly clustered 11q23 and 22q11 breakpoints permit PCR-based detection of the recurrent constitutional t(11;22) Am J Hum Genet. 2000;67:763–768. doi: 10.1086/303054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edelmann L, Spiteri E, Koren K, et al. AT-rich palindromes mediate the constitutional t(11;22) translocation. Am J Hum Genet. 2001;68:1–13. doi: 10.1086/316952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tapia-Paez I, Kost-Alimova M, Hu P, et al. The position of t(11;22)(q23;q11) constitutional translocation breakpoint is conserved among its carriers. Hum Genet. 2001;109:167–177. doi: 10.1007/s004390100560. [DOI] [PubMed] [Google Scholar]
- 31.Budarf ML, Eckman B, Michaud D, et al. Regional localization of over 300 loci on human chromosome 22 using a somatic cell hybrid mapping panel. Genomics. 1996;35:275–288. doi: 10.1006/geno.1996.0358. [DOI] [PubMed] [Google Scholar]
- 32.Kehrer-Sawatzki H, Haussler J, Krone W, et al. The second case of a t(17;22) in a family with neurofibromatosis type 1: sequence analysis of the breakpoint regions. Hum Genet. 1997;99:237–247. doi: 10.1007/s004390050346. [DOI] [PubMed] [Google Scholar]
- 33.Rhodes CH, Call KM, Budarf ML, et al. Molecular studies of an ependymoma-associated constitutional t(1;22)(p22;q11.2) Cytogenet Cell Genet. 1997;78:247–252. doi: 10.1159/000134667. [DOI] [PubMed] [Google Scholar]
- 34.Debeer P, Mols R, Huysmans C, et al. Involvement of a palindromic chromosome 22-specific low-copy repeat in a constitutional t(X; 22)(q27;q11) Clin Genet. 2002;62:410–414. doi: 10.1034/j.1399-0004.2002.620510.x. [DOI] [PubMed] [Google Scholar]
- 35.Spiteri E, Babcock M, Kashork CD, et al. Frequent translocations occur between low copy repeats on chromosome 22q11.2 (LCR22s) and telomeric bands of partner chromosomes. Hum Mol Genet. 2003;12:1823–1837. doi: 10.1093/hmg/ddg203. [DOI] [PubMed] [Google Scholar]
- 36.Kurahashi H, Shaikh T, Takata M, et al. The constitutional t(17;22): another translocation mediated by palindromic AT-rich repeats. Am J Hum Genet. 2003;72:733–738. doi: 10.1086/368062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inagaki H, Ohye T, Kogo H, et al. A palindromic AT-rich repeat in the NF1 gene is hypervariable in humans and evolutionarily conserved among primates. Hum Mutat. 2005;26:332–342. doi: 10.1002/humu.20228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nimmakayalu MA, Gotter AL, Shaikh TH, et al. A novel sequence-based approach to localize translocation breakpoints identifies the molecular basis of a t(4;22) Hum Mol Genet. 2003;12:2817–2825. doi: 10.1093/hmg/ddg301. [DOI] [PubMed] [Google Scholar]
- 39.Gotter AL, Shaikh TH, Budarf ML, et al. A palindrome-mediated mechanism distinguishes translocations involving LCR-B of chromosome 22q11.2. Hum Mol Genet. 2004;13:103–115. doi: 10.1093/hmg/ddh004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gotter AL, Nimmakayalu MA, Jalali GR, et al. A palindrome-driven complex rearrangement of 22q11.2 and 8q24.1 elucidated using novel technologies. Genome Res. 2007;17:470–481. doi: 10.1101/gr.6130907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kurahashi H, Inagaki H, Yamada K, et al. Cruciform DNA structure underlies the etiology for palindrome-mediated human chromosomal translocations. J Biol Chem. 2004;279:35377–35383. doi: 10.1074/jbc.M400354200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurahashi H, Emanuel BS. Unexpectedly high rate of de novo constitutional t(11;22) translocations in sperm from normal males. Nat Genet. 2001;29:139–140. doi: 10.1038/ng1001-139. [DOI] [PubMed] [Google Scholar]
- 43.Kato T, Inagaki H, Yamada K, et al. Genetic variation affects de novo translocation frequency. Science. 2006;311:971. doi: 10.1126/science.1121452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kogo H, Inagaki H, Ohye T, et al. Cruciform extrusion propensity of human translocation-mediating palindromic AT-rich repeats. Nucleic Acids Res. 2007;35:1198–1208. doi: 10.1093/nar/gkm036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kurahashi H, Inagaki H, Kato T, et al. Impaired DNA replication prompts deletions within palindromic sequences, but does not induce translocations in human cells. Hum Mol Genet. 2009;18:3397–3406. doi: 10.1093/hmg/ddp279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trinh TQ, Sinden RR. Preferential DNA secondary structure mutagenesis in the lagging strand of replication in E. coli. Nature. 1991;352:544–547. doi: 10.1038/352544a0. [DOI] [PubMed] [Google Scholar]
- 47.Leach DR. Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. Bioessays. 1994;16:893–900. doi: 10.1002/bies.950161207. [DOI] [PubMed] [Google Scholar]
- 48.Bzymek M, Lovett ST. Evidence for two mechanisms of palindrome-stimulated deletion in Escherichia coli: single-strand annealing and replication slipped mispairing. Genetics. 2001;158:527–540. doi: 10.1093/genetics/158.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nag DK, Kurst A. A 140-bp-long palindromic sequence induces double-strand breaks during meiosis in the yeast Saccharomyces cerevisiae. Genetics. 1997;146:835–847. doi: 10.1093/genetics/146.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gordenin DA, Lobachev KS, Degtyareva NP, et al. Inverted DNA repeats: a source of eukaryotic genomic instability. Mol Cell Biol. 1993;13:5315–5322. doi: 10.1128/mcb.13.9.5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Collick A, Drew J, Penberth J, et al. Instability of long inverted repeats within mouse transgenes. EMBO J. 1996;15:1163–1171. [PMC free article] [PubMed] [Google Scholar]
- 52.Akgun E, Zahn J, Baumes S, et al. Palindrome resolution and recombination in the mammalian germ line. Mol Cell Biol. 1997;17:5559–5570. doi: 10.1128/mcb.17.9.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lemoine FJ, Degtyareva NP, Lobachev K, et al. Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell. 2005;120:587–598. doi: 10.1016/j.cell.2004.12.039. [DOI] [PubMed] [Google Scholar]
- 54.Kato T, Yamada K, Inagaki H, et al. Age has no effect on de novo constitutional t(11;22) translocation frequency in sperm. Fertil Steril. 2007;88:1446–1448. doi: 10.1016/j.fertnstert.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kato T, Inagaki H, Kogo H, et al. Two different forms of palindrome resolution in the human genome: deletion or translocation. Hum Mol Genet. 2008;17:1184–1191. doi: 10.1093/hmg/ddn008. [DOI] [PubMed] [Google Scholar]
- 56.Inagaki H, Ohye T, Kogo H, et al. Chromosomal instability mediated by non-B DNA: Cruciform conformation and not DNA sequence is responsible for recurrent translocation in humans. Genome Res. 2009;19:191–198. doi: 10.1101/gr.079244.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nasar F, Jankowski C, Nag DK. Long palindromic sequences induce double-strand breaks during meiosis in yeast. Mol Cell Biol. 2000;20:3449–3458. doi: 10.1128/mcb.20.10.3449-3458.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lobachev KS, Gordenin DA, Resnick MA. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell. 2002;108:183–193. doi: 10.1016/s0092-8674(02)00614-1. [DOI] [PubMed] [Google Scholar]
- 59.Kurahashi H, Inagaki H, Ohye T, et al. Palindrome-mediated chromosomal translocations in humans. DNA Repair (Amst) 2006;5:1136–1145. doi: 10.1016/j.dnarep.2006.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kurahashi H, Inagaki H, Ohye T, et al. Chromosomal translocations mediated by palindromic DNA. Cell Cycle. 2006;5:1297–1303. doi: 10.4161/cc.5.12.2809. [DOI] [PubMed] [Google Scholar]
- 61.Ohye T, Inagaki H, Kogo H, et al. Paternal origin of the de novo constitutional t(11;22)(q23;q11) Eur J Hum Genet. doi: 10.1038/ejhg.2010.20. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boissonneault G. Chromatin remodeling during spermiogenesis: a possible role for the transition proteins in DNA strand break repair. FEBS Lett. 2002;514:111–114. doi: 10.1016/s0014-5793(02)02380-3. [DOI] [PubMed] [Google Scholar]
- 63.Meistrich ML, Mohapatra B, Shirley CR, et al. Roles of transition nuclear proteins in spermiogenesis. Chromosoma. 2003;111:483–488. doi: 10.1007/s00412-002-0227-z. [DOI] [PubMed] [Google Scholar]
- 64.Kovtun IV, McMurray CT. Trinucleotide expansion in haploid germ cells by gap repair. Nat Genet. 2001;27:407–411. doi: 10.1038/86906. [DOI] [PubMed] [Google Scholar]
- 65.Ashley T, Gaeth AP, Inagaki H, et al. Meiotic recombination and spatial proximity in the etiology of the recurrent t(11;22) Am J Hum Genet. 2006;79:524–538. doi: 10.1086/507652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2001;2:280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- 67.Buwe A, Guttenbach M, Schmid M. Effect of paternal age on the frequency of cytogenetic abnormalities in human spermatozoa. Cytogenet Genome Res. 2005;111:213–228. doi: 10.1159/000086892. [DOI] [PubMed] [Google Scholar]
- 68.Leduc F, Nkoma GB, Boissonneault G. Spermiogenesis and DNA repair: a possible etiology of human infertility and genetic disorders. Syst Biol Reprod Med. 2008;54:3–10. doi: 10.1080/19396360701876823. [DOI] [PubMed] [Google Scholar]
- 69.Bacolla A, Wojciechowska M, Kosmider B, et al. The involvement of non-B DNA structures in gross chromosomal rearrangements. DNA Repair (Amst) 2006;5:1161–1170. doi: 10.1016/j.dnarep.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 70.Adachi M, Tsujimoto Y. Potential Z-DNA elements surround the breakpoints of chromosome translocation within the 5′ flanking region of bcl-2 gene. Oncogene. 1990;5:1653–1657. [PubMed] [Google Scholar]
- 71.Wang G, Christensen LA, Vasquez KM. Z-DNA-forming sequences generate large-scale deletions in mammalian cells. Proc Natl Acad Sci U S A. 2006;103:2677–2682. doi: 10.1073/pnas.0511084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Raghavan SC, Swanson PC, Wu X, et al. A non-B-DNA structure at the Bcl-2 major breakpoint region is cleaved by the RAG complex. Nature. 2004;428:88–93. doi: 10.1038/nature02355. [DOI] [PubMed] [Google Scholar]
- 73.Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 74.Zhang F, Khajavi M, Connolly AM, et al. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat Genet. 2009;41:849–853. doi: 10.1038/ng.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thomas NS, Morris JK, Baptista J, Ng BL, Crolla JA, Jacobs PA. De novo apparently balanced translocations in man are predominantly paternal in origin and associated with a significant increase in paternal age. J Med Genet. 2010;47:112–115. doi: 10.1136/jmg.2009.069716. [DOI] [PubMed] [Google Scholar]