Abstract
Mutations in the E3 ubiquitin ligase tripartite motif-containing 32 (TRIM32) are responsible for the disease limb-girdle muscular dystrophy 2H (LGMD2H). Previously, we generated Trim32 knockout mice (Trim32–/– mice) and showed that they display a myopathic phenotype accompanied by neurogenic features. Here, we used these mice to investigate the muscle-specific defects arising from the absence of TRIM32, which underlie the myopathic phenotype. Using 2 models of induced atrophy, we showed that TRIM32 is dispensable for muscle atrophy. Conversely, TRIM32 was necessary for muscle regrowth after atrophy. Furthermore, TRIM32-deficient primary myoblasts underwent premature senescence and impaired myogenesis due to accumulation of PIAS4, an E3 SUMO ligase and TRIM32 substrate that was previously shown to be associated with senescence. Premature senescence of myoblasts was also observed in vivo in an atrophy/regrowth model. Trim32–/– muscles had substantially fewer activated satellite cells, increased PIAS4 levels, and growth failure compared with wild-type muscles. Moreover, Trim32–/– muscles exhibited features of premature sarcopenia, such as selective type II fast fiber atrophy. These results imply that premature senescence of muscle satellite cells is an underlying pathogenic feature of LGMD2H and reveal what we believe to be a new mechanism of muscular dystrophy associated with reductions in available satellite cells and premature sarcopenia.
Introduction
Tripartite motif-containing 32 (TRIM32) is a member of the tripartite motif (TRIM) family of proteins (1), which share the common features of a RING finger, a B-box, and a coiled-coil region. TRIM32 is an E3 ubiquitin ligase, the activity of which is contained in the RING finger domain (2). TRIM32 differs from most other TRIM family members because its C terminus contains a 6-bladed β-propeller NHL domain, which is known to mediate protein-protein interactions (3). Although the TRIM32 protein is ubiquitously expressed, different mutations in the Trim32 gene result in several different inherited diseases, with disparate phenotypes that affect both muscle and nonmuscle tissues. For example, all mutations that result in muscle phenotypes (limb-girdle muscular dystrophy 2H [LGMD2H] and sarcotubular myopathy [STM]) occur in the C-terminal NHL domain of TRIM32 (4–8). On the other hand, a missense mutation in the B-box of TRIM32 results in another genetically inherited disorder called Bardet-Biedl syndrome type 11 (BBS11). BBS11 is a multisystemic disorder that has no skeletal muscle involvement (9). These diseases are rare, and their incidence is still not known. Several muscle-specific substrates and/or interacting partners of TRIM32 have been identified, including myosin, actin (2), and dysbindin (10), yet the biological role of these interactions is not understood.
Previously, we created Trim32 knockout mice (Trim32–/– mice), which proved to be a good model for studying pathogenic mechanisms of LGMD2H (11). LGMD2H/STM (MIM 254110) is a neuromuscular disorder with a highly variable and slowly progressive clinical course (12). Phenotypic outcomes of LGMD2H and its more severe form STM are similar to other muscular dystrophies and are primarily characterized by proximal muscle weakness and wasting. LGMD2H biopsies show myopathic features of central nucleation, fiber splitting, Z-line streaming, and a dilated sarcotubular system with vacuoles (4, 5, 8, 13–16). In the Trim32–/– mouse model, the absence of TRIM32 resulted in a similar myopathic phenotype to that described in LGMD2H/STM biopsies, demonstrating similar muscle morphology and muscle weakness on grip strength and wire hang testing. In addition, our studies revealed that the Trim32–/– myopathy included a neurogenic component. The observation of reduced neurofilament protein concentration led to the discovery that Trim32–/– mice had reduced motor axon diameters, resulting in a shift to a slow myosin fiber-type composition (11). In agreement with our findings in the mouse, neuronal involvement was also confirmed in some patients with LGMD2H/STM. These LGMD2H-associated neurogenic features included mixed neuropathic and myopathic elements revealed by electromyography, muscle weakness, paresis, paresthesia, hypoactive or absent tendon reflexes, a slight dominance of type I slow muscle fibers, and decreased motor and sensory nerve conduction velocities (5, 7, 8, 14). Therefore, the Trim32–/– mouse replicates the LGMD2H phenotype and provides an excellent model system for elucidating functions of TRIM32 in vivo.
In this study, we sought to investigate the myogenic component of the Trim32–/– myopathy to lend insight into LGMD2H pathogenesis. We found that TRIM32 has a unique role in skeletal muscle, which is different from known roles of other E3 ubiquitin ligases involved in skeletal muscle remodeling. In contrast to the muscle-specific E3 ubiquitin ligases TRIM63 (also known as MURF1) and F-box protein 32 (FBXO32; also known as MAFbx/atrogin-1), which are key regulators of atrophy (17, 18), our data demonstrated that TRIM32 is not necessary for muscle atrophy but instead participates in muscle regrowth after atrophy. In this investigation, we present evidence that, in the absence of TRIM32, primary myogenic cells demonstrated impaired myogenesis and underwent premature senescence mediated by accumulation of an E3 small ubiquitin-related modifier (SUMO) ligase called protein inhibitor of activated STAT (PIAS4, also known as PIASy). PIAS4 was previously identified as a substrate of TRIM32 in keratinocytes (19). SUMO ligases are enzymes involved in the process of sumoylation, which is a cascade of SUMO activation, and transfer reactions that conjugate a SUMO moiety to a protein substrate. The sumoylation pathway regulates a wide range of cellular events, similar to ubiquitination (20). Recently, it was demonstrated that overexpression of PIAS4 or SUMO-2/3 led to senescence in several different cell models, thus implicating PIAS4 and sumoylation in regulation of cellular senescence (21–23). Senescence was first described as irreversible proliferation arrest that limits the replicative ability of cultured cells, a process termed “replicative senescence” (24). However, senescence can also be triggered in different cell types in response to certain kinds of stress, such as DNA damage or oncogenic or oxidative stress, and is referred to as “premature senescence” (25).
Since adult skeletal muscle contains a population of resident, self-renewable muscle stem cells called “satellite cells,” the premature senescence of these cells may lead to a decline in the satellite cell population, which in turn results in reduced growth potential of skeletal muscle and premature sarcopenia in vivo. Sarcopenia is defined as loss of muscle mass and function associated with aging; its pathogenesis involves multiple factors, including a reduction of satellite cell number and selective type II fast fiber atrophy (26–28). In this study, we identified premature senescence of myogenic cells as a mechanism that can cause adaptive growth deficiency and subsequent muscular dystrophy in adult muscles. In an animal model of induced muscle growth, we demonstrated that Trim32–/– muscles do indeed have substantially fewer satellite cells and reduced muscle growth in vivo. We also present evidence of premature sarcopenia in Trim32–/– mice. Collectively, these data identified premature myoblast senescence and sarcopenia as an underlying pathogenic feature of TRIM32 deficiency and, thus, revealed what we believe to be a novel mechanism of muscular dystrophy due to reduced replicative potential of satellite cells resulting in impaired growth capability of skeletal muscle.
Results
Skeletal muscles are able to atrophy in the absence of TRIM32.
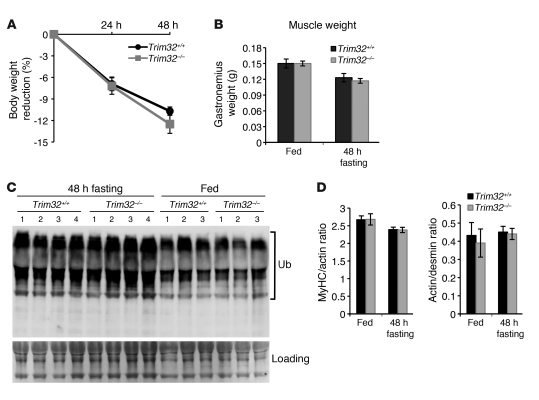
Previous studies demonstrated an involvement of the muscle-specific E3 ubiquitin ligases TRIM63 and FBXO32 in muscle atrophy (17, 18). FBXO32- or TRIM63-knockout mice are resistant to atrophy in multiple models of muscle wasting, whereas overexpression of FBXO32 induces atrophy (17). It was thus logical to hypothesize that TRIM32 might possess similar biological functions as TRIM63 in promoting muscle atrophy. To test the role of TRIM32 in the process of skeletal muscle atrophy, we first used a fasting model previously shown to induce significant atrophy after 48 hours of fasting (29). Food deprivation leads to muscle mass loss by ubiquitin-dependent proteolysis of muscle proteins (30); therefore, rates of atrophy and levels of ubiquitination were assessed in fasted Trim32–/– muscles and wild-type (Trim32+/+) muscles to test the involvement of TRIM32. During 48 hours of fasting, Trim32–/– mice showed the same loss of body weight and muscle mass as Trim32+/+ mice (Figure 1, A and B). Moreover, accumulation of ubiquitinated proteins induced by fasting occurred similarly in Trim32–/– and Trim32+/+ muscles (Figure 1C). TRIM32 is an E3 ubiquitin ligase, which is capable of ubiquitinating actin and interacting with myosin, at least in vitro (2). Since actin is a potential substrate of TRIM32, loss of TRIM32 would be predicted to lead to actin accumulation in skeletal muscle. To assess the rate of myofibrillar protein loss in mice undergoing atrophy in the presence and absence of TRIM32, we analyzed the ratio of myosin heavy chain to actin in isolated myofibrils from the Trim32+/+ and Trim32–/– muscles. Analyzing the ratio rather than the density of individual bands allowed us to neglect fluctuations in sample loading. If actin were an in vivo target of TRIM32, it would be expected to accumulate in the Trim32–/– muscles, and a decrease in the ratio of myosin to actin during atrophy would be observed; however, during fasting, the myosin-to-actin ratio in Trim32–/– muscles was indistinguishable from that of Trim32+/+ muscles (Figure 1D). In addition, the ratio of actin to desmin, another myofibrillar protein, was unchanged (Figure 1D). Thus, TRIM32 is not required for skeletal muscle atrophy induced by fasting, unlike the muscle-specific E3 ubiquitin ligases TRIM63 and FBXO32.
Figure 1. Fasting-induced atrophy occurs normally in Trim32–/– mice.
(A) A statistically significant (P < 0.05) body weight reduction due to 24 or 48 hours of fasting was observed in mice of both genotypes. The rate of body weight reduction in Trim32–/– fasting mice is indistinguishable from the Trim32+/+ rate (n = 4). Error bars represent sem. (B) After 48 hours of food deprivation, gastrocnemius muscle weight was reduced by 20%, similarly in Trim32–/– and Trim32+/+ mice. (C) Accumulation of high-molecular weight ubiquitin-protein conjugates occurs similarly in Trim32–/– and Trim32+/+ myofibrils, as demonstrated by anti-ubiquitin (Ub) Western blot staining. A representative blot is shown. Ponceau S staining is shown as a loading control. (D) Ratios of major myofibrillar proteins, myosin heavy chain (MyHC) to actin and actin to desmin, were assessed in myofibrils isolated from Trim32+/+ and Trim32–/– fed mice and 48 hour–fasted mice by densitometry of Coomassie-stained SDS-PAGE. No difference was observed between Trim32–/– and Trim32+/+ samples.
TRIM32 is essential for skeletal muscle regrowth after disuse atrophy.
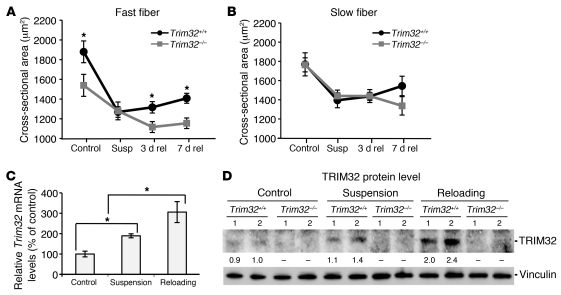
The unexpected finding that skeletal muscles are able to atrophy during fasting in the absence of TRIM32 led us to further examine whether TRIM32 participates in remodeling of skeletal muscles using a physiological model of hind limb unloading followed by reloading. Suspension of a mouse by its tail leads to disuse atrophy of the hind limb muscles, in particular the soleus, which can lose up to 30% of its mass (31). Return to normal cage activity (reloading) results in muscle regrowth (32). We subjected the Trim32–/– and Trim32+/+ mice to the procedure of 5-day hind limb unloading followed by 7 days of reloading to induce atrophy followed by regrowth. Measurement of the cross-sectional area of the soleus revealed that fast fibers in ambulatory Trim32–/– mice were smaller than fast fibers in ambulatory Trim32+/+ mice (Figure 2A), indicating type II fiber atrophy in Trim32–/– muscles. After 5 days of unloading, both fast and slow muscle fibers showed similar loss of cross-sectional area in both genotypes of mice, suggesting similar rates of atrophy. On the contrary, while Trim32+/+ mice started to regain muscle fiber size after 7 days of reloading, muscles from Trim32–/– mice, particularly fast muscle fibers, did not grow (Figure 2, A and B). Analysis of Trim32 expression by qPCR and Western blotting in Trim32+/+ skeletal muscles demonstrated an increase in both mRNA and protein concentration during unloading. Moreover, reloading induced an even greater increase in expression of Trim32 (Figure 2, C and D). Therefore, these data demonstrate that, unlike TRIM63 and FBXO32, TRIM32 is not essential for muscle atrophy in 2 different models, but rather, it plays a substantial role in regrowth of muscle mass after atrophy.
Figure 2. Trim32–/– muscles fail to regrow during reloading after hind limb suspension.
Measurements of cross-sectional areas of (A) fast and (B) slow fibers in Trim32–/– and Trim32+/+ soleus muscles were performed on frozen sections immunostained with anti-fast and anti-slow myosin antibodies. Four groups of mice for each genotype (n = 6) were analyzed as follows: ambulatory control mice (control); mice subjected to 5 days of suspension (susp); mice subjected to 5 days of suspension followed by 3 days (3d rel) or 7 days (7d rel) of reloading. Values represent the mean of measurements in several independent fields of view for each of 6 slides (>100 fibers were measured per each slide). Error bars represent SEM (*P < 0.05). (C) qPCR analysis using RNA harvested from Trim32+/+ control, suspended, and reloaded muscles (n = 3) shows elevated Trim32 expression during suspension and reloading phases. Results from 3 samples in each group were run in triplicate, normalized to Gapdh, and expressed as percentage relative to ambulatory control (*P < 0.05). Data are shown as mean ± SEM. (D) Anti-TRIM32 Western blot demonstrates absence of TRIM32 in Trim32–/– muscles and increase of TRIM32 protein level in Trim32+/+ muscles during suspension and reloading phases. An anti-vinculin blot is shown as a loading control. Numbers in each lane under the TRIM32 blot represent fold change in TRIM32 normalized to vinculin relative to ambulatory control. Representative blots are shown.
PI3K/AKT signaling is not impaired in Trim32–/– muscles.
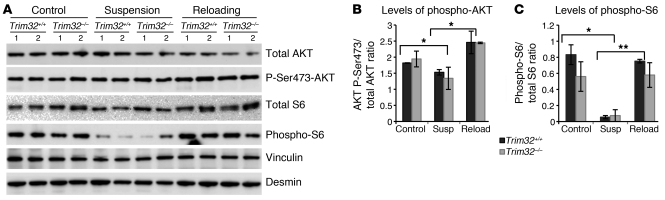
The PI3K/AKT signaling pathway is a key regulator of skeletal muscle growth (33, 34). Phosphorylation of the serine/threonine protein kinase AKT results in both increased protein synthesis (35) and transcriptional inhibition of the atrophy-specific genes TRIM63 and FBXO32 (36). We examined PI3K/AKT pathway activation in Trim32–/– mice during hind limb suspension/reloading to test whether insufficient muscle growth in Trim32–/– muscles resulted from inadequate AKT signaling. Using Western blot analysis, we assessed phosphorylation levels of AKT and one of its downstream targets, ribosomal protein S6, in soleus muscles (Figure 3A). As expected, muscle inactivity resulted in dephosphorylation of S6 protein after 5 days of hind limb suspension, and phosphorylation status of S6 protein was restored after 3 days of reloading in Trim32+/+ mice (Figure 3, A and C). This process was not impaired in Trim32–/– mice. Similarly, no difference in AKT phosphorylation was observed between Trim32–/– and Trim32+/+ muscles during hind limb suspension/reloading (Figure 3, A and B). Therefore, activation of the PI3K/AKT pathway occurs normally in Trim32–/– skeletal muscle, and failure of Trim32–/– muscle to regrow after disuse atrophy is not due to improper PI3K/AKT signaling.
Figure 3. AKT signaling is not impaired in Trim32–/– muscles.
(A) Representative Western blots of soleus muscle extracts from ambulatory control mice, mice suspended for 5 days (suspension), or mice reloaded for 3 days after suspension (reloading) were stained with total AKT, activated AKT phospho-specific (P-Ser473-AKT), total ribosomal protein S6, and phospho-S6 antibodies. Anti-vinculin and anti-desmin blots are shown as loading controls. Densitometry data of (B) phospho-AKT and (C) phospho-S6 blots were normalized to total AKT or total S6, respectively. Both phospho-AKT and phospho-S6 levels decline during inactivity after 5 days of hind limb suspension (*P < 0.05) and restore during reloading (*P < 0.05, **P < 0.03) similarly in Trim32–/– and Trim32+/+ muscles. Data (n = 4) are shown as mean ± SEM.
Apoptosis is not impaired in Trim32–/– muscles.
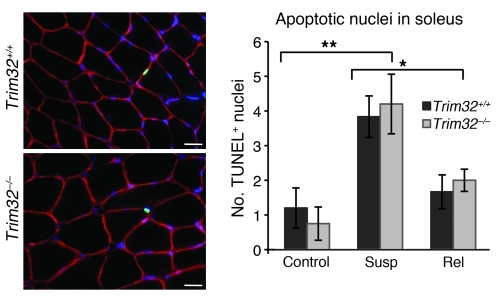
Apoptosis contributes to skeletal muscle remodeling to maintain a constant nuclear-to-cytoplasm ratio during hind limb suspension or immobilization (37). Moreover, it’s been previously shown that TRIM32 may play an antiapoptotic role in epidermal keratinocytes (38) and squamous epithelial cells (39) but a proapoptotic role in some established cell culture lines (40). Therefore, we examined whether apoptosis is altered in skeletal muscle during hind limb suspension/reloading in the absence of TRIM32. TUNEL labeling revealed low levels of apoptotic nuclei in both ambulatory Trim32+/+ and Trim32–/– solei (Figure 4). The rate of apoptosis in hind limb–suspended muscles was substantially increased, whereas reloading caused a decrease of apoptosis to the same extent in both genotypes of mice (Figure 4). Therefore, apoptosis was not altered in the TRIM32-deficient skeletal muscles, and this process is not an underlying mechanism of the Trim32–/– myopathy.
Figure 4. Apoptosis is not impaired in Trim32–/– skeletal muscles.
TUNEL labeling was performed, and TUNEL-positive apoptotic nuclei (green) were quantified in entire soleus muscle cross sections. To visualize muscle fibers and nuclei, sections were counterstained with anti-dystrophin (red) and DAPI (blue). Representative examples of the staining are shown. Scale bar: 25 μm. Four to six samples were analyzed in each group for Trim32–/– and Trim32+/+ genotype: ambulatory control; mice subjected to 5 days of suspension; and mice subjected to 5 days of suspension followed by 3 days of reloading (rel). The number of TUNEL-positive nuclei increased during inactivity after 5 days of hind limb suspension in both genotypes compared with ambulatory controls of the same genotype (**P < 0.02) and declined during reloading compared with the value observed in the suspended mice of the same genotype (*P < 0.05). There was no statistical difference between the 2 genotypes at any of the time points tested. Data are shown as mean ± SEM.
Because autophagic vesicles were previously observed in the Trim32–/– muscles and because autophagy increases during muscle atrophy (41), we analyzed the autophagy marker LC3B by Western blotting. Our data show that levels of LC3B did not change during suspension in either Trim32+/+ or Trim32–/– muscles, and both genotypes expressed LC3B to the same extent throughout the experiment (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI59581DS1).
Primary myoblasts from Trim32–/– muscles have impaired differentiation in vitro.
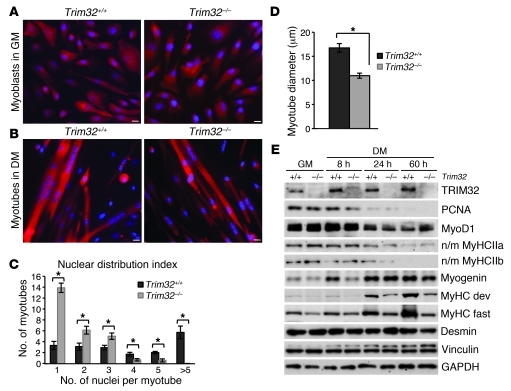
Atrophy induced by hind limb suspension results in loss of both myonuclei and satellite cells, whereas muscle regrowth due to reloading is associated with activation and proliferation of satellite cells (42). Upon growth stimulation, quiescent skeletal muscle precursor (satellite) cells are activated, enter the cell cycle, and eventually undergo differentiation. This process involves either the fusion of satellite cells to the existing myofibers or the alignment and fusion of satellite cells together to produce new myofibers (43). Some of the satellite cells reestablish a quiescent precursor cell pool through a process of self-renewal (44). Therefore, a healthy population of satellite cells is necessary for proper muscle growth. To gain insight into the role of TRIM32 in myogenic cell growth and differentiation, we isolated primary myoblasts from skeletal muscles of Trim32–/– and Trim32+/+ mice. These cells were cultured in media containing high levels of fetal bovine serum and bFGF to allow for proper cell growth and proliferation. To initiate differentiation, serum was removed from the media, which induced myoblasts to withdraw from the cell cycle, undergo myogenic differentiation, and fuse into multinucleated myotubes. Trim32–/– myoblasts were able to align and fuse like Trim32+/+ myoblasts (Figure 5, A and B); however, Trim32–/– myotubes were thinner and contained fewer nuclei than Trim32+/+ myotubes (Figure 5, C and D). Downregulation of proliferating cell nuclear antigen (PCNA) in response to mitogen withdrawal and the initial steps of myogenic differentiation, such as downregulation of early myogenic regulatory factor MyoD and nonmuscle myosin heavy chains IIa and IIb, occurred normally in Trim32–/– cells (Figure 5E). However, late markers of differentiation (such as muscle-specific myosin heavy chain developmental and fast isotypes and myogenin) showed deferred expression in Trim32–/– cultures, suggesting impaired differentiation in the absence of TRIM32 (Figure 5E). It is noteworthy that the level of TRIM32 in Trim32+/+ cells was increased upon differentiation, as early as 8 hours after serum withdrawal (Figure 5E), which is in agreement with our previously published data on TRIM32 expression in C2C12 myogenic cell cultures (2). Therefore, TRIM32 is required for proper myogenic differentiation.
Figure 5. Trim32–/– primary myoblasts have impaired differentiation in vitro.
(A and B) Primary myogenic cells isolated from Trim32+/+ and Trim32–/– muscles immunostained with the myogenic cell marker desmin to show purity and enrichment of the myogenic cell cultures. Cells were counterstained with Hoechst dye to visualize the nuclei. Scale bar: 10 μm. (A) Primary myoblasts in growth medium (GM). (B) Fusion of myoblasts and formation of myotubes is evident in both cultures after 48 hours of switching to differentiation medium (DM). (C) The nuclear distribution index (number of nuclei per myotube) was quantified in 10 independent fields of view. Data are shown as mean ± SEM (*P < 0.05). (D) Diameters of 30 myotubes were measured for each culture. Data are shown as mean ± SEM (*P < 0.05). (E) Western blot analysis of Trim32+/+ and Trim32–/– primary myoblast cultures in growth medium and 8, 24, or 60 hours after switching to differentiation medium. TRIM32 is undetectable in Trim32–/– cells. Vinculin and GAPDH blots are included to show the relative concentration of proteins in the cell lysates. MyoD1, early marker of myogenic commitment; n/m MyHCIIa, nonmuscle myosin heavy chain IIa; n/m MyHCIIb, nonmuscle myosin heavy chain IIb; myogenin, marker of myogenic differentiation; MyHC dev, muscle-specific myosin heavy chain developmental; MyHC fast, muscle-specific myosin heavy chain fast.
Cell cycle analysis of primary myoblasts.
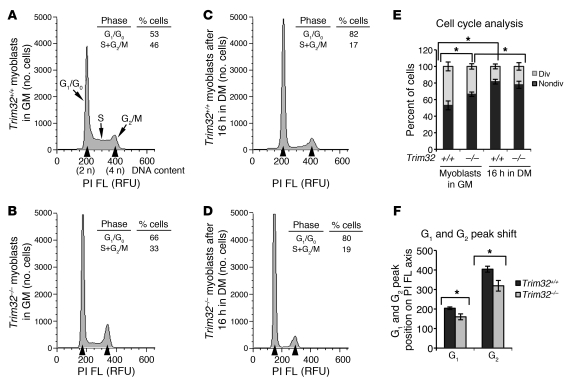
It’s been shown that TRIM32 affects cell growth rates in NIH 3T3 cultured cells in which overexpression of TRIM32 accelerates cell growth, whereas TRIM32-specific siRNA attenuates it (39). Therefore, to examine whether loss of TRIM32 affects the cell cycle in primary myogenic cultures, we used the widely used method of FACS analysis of propidium iodide–labeled (PI-labeled) cells. This analysis demonstrated that both Trim32–/– and Trim32+/+ cell cultures exhibited normal cell cycle withdrawal upon differentiation, as evidenced by the increased nondividing G1/G0 population after 16 hours in differentiation medium (Figure 6, A–E). However, under normal growth conditions in media with high levels of serum, the myoblast population in the G1/G0 phase was significantly larger in Trim32–/– cultures, compared with that in Trim32+/+ cultures, suggesting growth arrest of Trim32–/– myoblasts (Figure 6, A, B, and E).
Figure 6. Cell cycle analysis of primary myoblasts.
Primary myoblasts were stained with PI and analyzed by flow cytometry, as described in the Methods. (A–D) Representative flow cytometric histograms showing the cell cycle distribution. (A) Trim32+/+ and (B) Trim32–/– myoblasts in growth medium. (C) Trim32+/+ and (D) Trim32–/– cells cultured for 16 hours in differentiation medium. G1/G0, S, and G2/M peaks are shown. Positions of G1 and G2 peaks are indicated with the first and second arrowheads, respectively. FL, fluorescence; RFU, relative fluorescence units. (E) Percentages of dividing (div) (S+G2/M) and nondividing (nondiv.) (G1/G0) cells were calculated. Values represent the mean of at least 3 independent experiments ± SEM (*P < 0.05). (F) Positions of G1 and G2 peaks (shown with arrowheads on A–D) are shifted to the left on the PI fluorescence axis in Trim32–/– cultures. Data are summarized and presented as the mean of 3 independent experiments ± SEM (*P < 0.05).
Unexpectedly, we also noticed that Trim32–/– cells consistently showed a leftward shift of the G1/G0 and G2/M peak positions on the PI fluorescence axis (Figure 6, A–D and F). We used equal numbers of cells in our FACS experiments and aligned the position of the peaks using Trim32+/+ myoblasts. As one would expect, the position of these peaks should remain unchanged during the experiment if diploid cells of the same origin were used. Indeed, the peaks were preserved in Trim32+/+ cells before and after the induction of differentiation (Figure 6, A and C). However, these peaks were noticeably shifted to the left in both Trim32–/– myoblasts and Trim32–/– differentiating cultures (Figure 6, B, D, and F). Since the total genomic DNA content isolated from an equal amount of cells was similar for both genotypes, as revealed by spectrophotometric analysis (Supplemental Figure 2), such reduced PI fluorescence may indicate a decreased amount of DNA available for PI binding. This situation may occur with heterochromatin formation, when accessibility of DNA to the fluorophore is diminished due to chromatin condensation. Characteristic chromatin condensation occurs during senescence (45). In support of this hypothesis, a similar change in PI fluorescence intensity (and a shift of G1/G0 peak to the left) was reported during senescence of plant cells (46). This observation, together with the finding of growth arrest in Trim32–/– myoblasts, led us to further investigate whether senescence is a characteristic process in Trim32–/– primary myoblast cultures.
Premature senescence in Trim32–/– myoblasts.
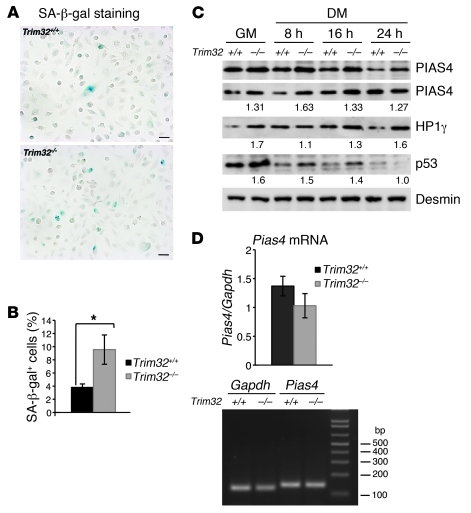
Cellular senescence is described as irreversible cell cycle arrest (24). Aside from growth arrest, senescent cells demonstrate characteristic metabolic changes, such as elevated senescence-associated β-galactosidase (SA-β-gal) (47) and formation of transcriptionally silent foci of heterochromatin that recruit specific heterochromatin proteins (45). A higher percentage of SA-β-gal–positive cells was observed in Trim32–/– myoblast cultures compared with that in Trim32+/+ cultures of the same passage number (Figure 7, A and B). Furthermore, levels of heterochromatin protein 1 (HP1γ) were also increased in Trim32–/– myoblasts and myotubes compared with those in Trim32+/+ cells (Figure 7C). Therefore, senescence markers were present in Trim32–/– cells in an increased concentration compared with those in Trim32+/+ cells, suggesting that premature senescence results from the absence of TRIM32.
Figure 7. Premature senescence in Trim32–/– myoblasts.
(A) Micrographs of Trim32+/+ and Trim32–/– primary myoblast cultures stained for SA-β-gal activity (blue staining). Scale bar: 20 μm. (B) SA-β-gal–positive cells were counted in 20 independent fields of view. Data are represented as percentage of total cell number and shown as mean ± SEM (*P < 0.05). (C) Western blot analysis demonstrates expression of PIAS4 (2 blots are shown), HP1γ, and p53 in myogenic cells in growth medium and at different indicated times after switching to differentiation medium. The desmin blot is shown as a loading control. The numbers under the blots are densitometry data normalized to loading and represent a fold difference of the indicated protein expression in Trim32–/– cells relative to the value in the corresponding Trim32+/+ culture. (D) Real-time PCR analysis of Pias4 mRNA expression using RNA harvested from Trim32+/+ and Trim32–/– myoblasts in growth medium. Data are shown as mean ± SEM. A representative agarose gel of Gapdh and Pias4 PCR using qPCR primers is shown.
The E3 SUMO ligase PIAS4 is a regulator of cellular senescence; its overexpression in fibroblasts led to a senescence arrest through activation of the p53 and Rb tumor suppressor pathways (21). It was previously demonstrated that PIAS4 is a TRIM32 substrate in keratinocytes (19). Since TRIM32 ubiquitinates PIAS4 and targets it for proteasomal degradation (19), we examined PIAS4 protein levels in myoblast cultures. As demonstrated by Western blotting, PIAS4 accumulated in Trim32–/– myoblasts and myotubes (Figure 7C). This increase in PIAS4 protein levels in Trim32–/– myoblasts is not due to a change in Pias4 mRNA levels, as revealed by qPCR analysis of RNA harvested from Trim32+/+ and Trim32–/– myoblasts (Figure 7D). In addition, the concentration of the tumor suppressor p53, a PIAS4 substrate essential for implementing senescence (21), was also increased in Trim32–/– cells (Figure 7C).
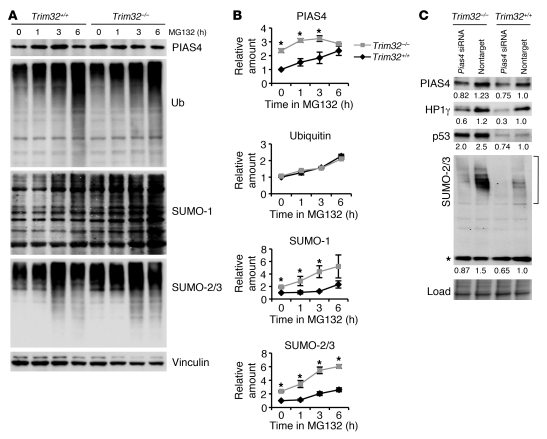
To investigate whether accumulation of PIAS4 occurred due to its decreased ubiquitination and proteasomal degradation, we treated primary myoblast cultures with the proteosome inhibitor MG132. We found that over 3 hours of incubation with MG132, PIAS4 accumulated in Trim32+/+ myoblasts (Figure 8, A and B). These studies suggest a role for the ubiquitin/proteasomal pathway in degradation of PIAS4. In the Trim32–/– cells, however, PIAS4 did not accumulate above baseline values and was thus independent of the presence of MG132. These results suggest a role for TRIM32 in degradation of PIAS4 in vivo, in accordance with the previously published data that indicate that PIAS4 is a TRIM32 E3 ubiquitin ligase substrate (19). The concentration of ubiquitin-conjugated proteins was similar in Trim32+/+ and Trim32–/– cells (Figure 8, A and B), implying that TRIM32 does not participate in widespread protein degradation but rather is likely involved in the ubiquitination of a limited number of substrates.
Figure 8. Accumulation of PIAS4 and sumoylated proteins in Trim32–/– cells.
(A and B) Primary myogenic cells were treated with 50 μM of the inhibitor of proteasome activity, MG132, for 1, 3, or 6 hours. Cells represented by the lane labeled “0” were treated with DMSO (vehicle) alone for 6 hours. MG132 treatment for 6 hours had a toxic effect on the cells, which showed marked signs of cell death (the majority of the cells were rounded and detached). (A) Western blot analysis of the treated cells using anti-PIAS4, anti-ubiquitin, anti–SUMO-1, and anti–SUMO-2/3 antibody. Vinculin is shown as a loading control. Note the reduction of band densities in lanes representing 6 hours in MG132 due to cellular toxicity. (B) Densitometry data normalized to vinculin relative to vehicle-treated Trim32+/+ values and shown as mean ± sem (*P < 0.05). (C) RNAi using an siRNA targeting Pias4 or nontargeting control in myoblast cultures. Western blot analysis demonstrates expression of PIAS4, HP1γ, p53, and SUMO-2/3 in myogenic cells. A Ponceau S–stained blot is shown as a loading control. Numbers under the blots are densitometry data normalized to loading and represent a fold difference of the indicated protein expression in myoblasts relative to the value in the Trim32+/+ culture treated with nontargeting control siRNA. Note that the asterisk denotes unconjugated free SUMO-2/3; only conjugated SUMO species (shown in bracket) were used for densitometry of SUMO-2/3 blots.
Given the known role of PIAS4 as an E3 SUMO ligase, we investigated the effect of PIAS4 accumulation on levels of sumoylation. This analysis revealed that the concentration of SUMO-conjugated proteins was considerably higher in Trim32–/– cells, even in the absence of MG132 (Figure 8, A and B). Though the exact role of the SUMO pathway in implementing senescence is obscure, increased levels of sumoylation in senescent cells, as compared with presenescent or quiescent cells, has been reported (22). Finally, suppression of PIAS4 by siRNA resulted in a reduction of sumoylation levels as well as the levels of p53 and HP1γ in primary myoblasts (Figure 8C). Therefore, in the absence of TRIM32 myoblasts accumulate PIAS4 and undergo premature senescence.
Premature sarcopenia in Trim32–/– muscles.
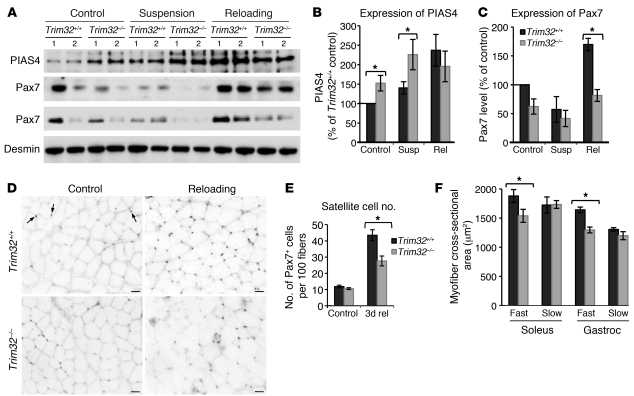
The observation of premature senescence in Trim32–/– myogenic cells suggests that these cells have a reduced capability for self-renewal, which may result in reduced growth potential of muscle tissue and a premature sarcopenic phenotype in vivo. Sarcopenia is characterized by degenerative loss of muscle mass and strength, usually associated with aging. A reduction in satellite cell number and selective type II (fast) fiber atrophy contribute to progression of sarcopenia (26–28). We used Trim32–/– and Trim32+/+ skeletal muscles obtained from hind limb suspension and reloading experiments to examine whether the defect observed in Trim32–/– myoblasts in vitro impacts skeletal muscle in vivo. Since our data demonstrate that myoblast senescence is accompanied by accumulation of PIAS4, we examined the concentration of PIAS4 in Trim32–/– and Trim32+/+ whole muscle extracts. Western blot analysis revealed increased PIAS4 concentration in Trim32–/– muscles from both ambulatory control and hind limb–suspended mice compared with PIAS4 concentration in Trim32+/+ muscles (Figure 9, A and B). Therefore, in the absence of TRIM32, PIAS4 accumulates not only in primary myoblast cultures, but also in skeletal muscles in vivo. To assess satellite cell content in skeletal muscle during hind limb suspension/reloading, we used Western blotting and immunohistochemical analysis of Pax7, a satellite cell marker, which is specifically expressed in quiescent and newly activated satellite cells (48–50). As expected, expression of Pax7 was induced after 3 days of reloading in Trim32+/+ muscles (Figure 9, A and C). In contrast, the level of Pax7 in reloaded Trim32–/– muscles was significantly lower than that in reloaded Trim32+/+ muscles. Similarly, immunohistochemical staining for Pax7 demonstrated that Trim32–/– ambulatory control muscles had the same number of satellite cells as Trim32+/+ muscles; however, satellite cell induction upon reloading was diminished in Trim32–/– muscles compared with that in Trim32+/+ muscles (Figure 9, D and E). These data suggest that Trim32–/– satellite cells have impaired replicative potential in vivo. Moreover, both soleus and gastrocnemius muscles of Trim32–/– ambulatory control mice demonstrated selective fast fiber atrophy (Figure 2A and Figure 9F). Therefore, Trim32–/– muscles demonstrate both key features of sarcopenia, a reduction in satellite cell number and type II fast fiber atrophy.
Figure 9. Satellite cell number is reduced in Trim32–/– muscles during muscle reloading.
(A) Representative Western blots of soleus muscle extracts from ambulatory control mice, mice suspended for 5 days (suspension), or mice reloaded for 3 days after suspension (reloading) were stained with Pax7 (2 blots are shown) and PIAS4. An anti-desmin blot is shown as a loading control. Densitometry data (n = 4) of (B) PIAS4 and (C) Pax7 blots were normalized to desmin and shown as mean ± SEM (*P < 0.05). (D) Frozen sections of Trim32+/+ and Trim32–/– soleus muscles from control mice and mice subjected to reloading for 3 days after suspension were stained with Pax7. Pax7-positive satellite cells appear on the micrographs as black dots (examples are indicated by arrows). Scale bar: 50 μm. (E) The amount of Pax7-positive cells was quantified in 3 independent fields of view per each slide. Data (n = 6) are shown as mean ± SEM (*P < 0.05). (F) Frozen sections of ambulatory control Trim32–/– and Trim32+/+ mice were immunostained with anti-fast and anti-slow myosin antibodies. Fiber area measurements show a significant decrease in the average cross-sectional area for fast fibers in soleus (n = 6) and gastrocnemius (gastroc) (n = 3) muscles of the Trim32–/– animals (18.1% and 21.04%, correspondingly). 100–300 fibers were measured per each slide. Data are shown as mean ± SEM (*P < 0.05).
We also assayed canonical (via phospho-SMAD) and noncanonical (via phospho-ERK1/2 and phospho-p38) TGF-β signaling pathways (51) in muscles of both genotypes undergoing hind limb suspension and reloading. These results showed subtle but statistically significant differences between Trim32–/– and Trim32+/+ muscles subjected to reloading after unloading (Supplemental Figure 3). Even though it is difficult to interpret whether such a small difference is biologically meaningful, it is in agreement with previously published observations. Phosphorylation of p38 has been shown to increase in atrophic muscles (52). Phosphorylated SMAD is upregulated and phospho-ERK1/2 is downregulated in aged skeletal muscles (53, 54). Therefore, these findings most likely reflect both the atrophic changes and premature aging that occur in Trim32–/– muscles and are consistent with the hypothesis of premature senescence caused by the absence of TRIM32.
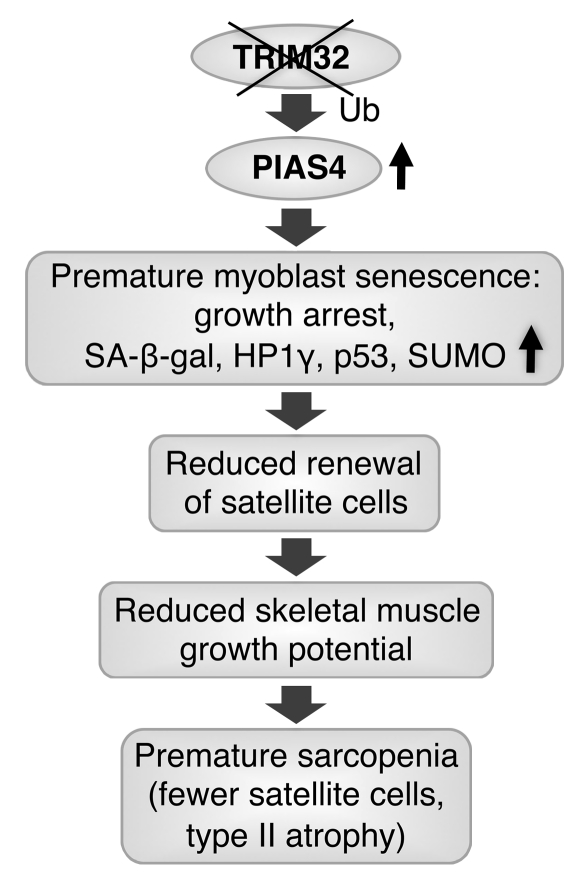
In conclusion, our data suggest that lack of TRIM32 leads to accumulation of PIAS4 and premature senescence of myoblasts. Trim32–/– satellite cells with reduced replicative capacity are unable to provide sufficient numbers of muscle precursor cells to achieve adequate muscle growth, resulting in a premature sarcopenic phenotype in response to growth-demanding stimuli (Figure 10). Our study implies that premature senescence of myoblasts is an underlying mechanism of Trim32–/– myopathy and LGMD2H.
Figure 10. Proposed role for TRIM32 in muscular dystrophy pathogenesis.
Lack of E3 ubiquitin ligase TRIM32 leads to accumulation of its substrate E3 SUMO ligase PIAS4 in skeletal muscle. Myoblasts undergo premature senescence, as evidenced by growth arrest and increased concentrations of senescence markers, such as the PIAS4 substrate p53, SA-β-gal, HP1γ, and elevated levels of global sumoylation. Trim32–/– satellite cells with reduced replicative capacity are unable to provide adequate adaptive muscle growth, resulting in a premature sarcopenic phenotype.
Discussion
Skeletal muscle adapts to physiological and pathological signals by undergoing remodeling. To sustain muscle performance, myofibers hypertrophy (for example, due to exercise training) and increase muscle bulk through elevated protein synthesis and addition of nuclei by satellite cell-mediated fusion. Muscle protein loss or atrophy in response to cancer cachexia, food deprivation, denervation, or inactivity is accomplished primarily through the ubiquitin proteasome pathway. Two muscle-specific E3 ligases, TRIM63 and FBXO32, have been shown to be major contributors to skeletal muscle atrophy through the ubiquitin proteasome pathway (17, 18). Since TRIM63 is a TRIM family member, it was logical to presume that TRIM32 might perform a similar biological role as TRIM63, especially since TRIM32 was shown to bind to myosin and ubiquitinate actin (2). In this investigation, we examined the role of TRIM32 in muscle atrophy and growth. Our data demonstrate that, unexpectedly, TRIM32 does not play a role in protein degradation processes that take place during muscle atrophy caused by either food deprivation or inactivity. Rather, TRIM32 appears to play an important role in muscle growth after disuse atrophy. The process of muscle growth requires activation, proliferation, and differentiation of muscle satellite cells. Our data demonstrate that TRIM32 deficiency leads to premature senescence of myogenic cells, which become unable to provide adequate muscle growth in response to growth-demanding stimuli. Premature senescence contributes to satellite cell proliferation and differentiation defects in Trim32–/– myogenic cultures, leading to impaired myotube formation. These findings are in agreement with the observations that senescent myoblasts are able to fuse less well and more slowly, forming thinner myotubes with fewer nuclei (55, 56). Whether aberrant differentiation in Trim32–/– myoblasts is caused solely by senescence or involves additional mechanisms is yet to be explored; however, these results provide what we believe to be the first suggestion that senescence in myoblasts could underlie pathogenic mechanisms in LGMD2H.
Our data link the E3 ubiquitin ligase TRIM32 and myoblast senescence via the E3 SUMO ligase PIAS4, which accumulates in myoblasts and skeletal muscle tissue in the absence of TRIM32. Similar to fibroblasts overexpressing PIAS4 (21), accumulation of PIAS4 in Trim32–/– myoblasts also leads to premature senescence. TRIM32 and PIAS4 have been shown to occupy different cellular compartments under basal conditions: TRIM32 is mainly a cytoplasmic protein, while PIAS4 is primarily nuclear (19). However, both proteins can shuttle between the cytoplasm and nucleus: while PIAS4 translocation from nucleus to cytoplasm seems to occur only upon cellular stress conditions (19), recently published data ascertain that TRIM32 can be relocated from the cytoplasm to the nucleus upon interaction with defined types of E2 ubiquitin-conjugating enzymes, such as UBE2N and UBE2V2 (57). Strikingly, these E2 enzymes have been implicated in the cell cycle progression and cellular responses to DNA damage upon genotoxic stress (58, 59). These data not only open up the possibility for an in vivo interaction between TRIM32 and PIAS4, but also suggest a fine adjustment for such an interaction in the cell.
Interestingly, our data demonstrated that PIAS4 concentration increases during unloading and increases even further after reloading of Trim32+/+ murine skeletal muscles. This observation is in agreement with a recent report of global gene expression patterns after 48 hours of unloading and 24 hours of reloading in humans (60). It was observed that human PIAS gene expression was upregulated in both unloaded and reloaded muscles; however, this finding is yet to be addressed with regard to its role in skeletal muscle remodeling.
The sumoylation pathway has been implicated in the process of cellular senescence (61). There are several SUMO isoforms in mammals that differ in structure and functional activity. While SUMO-2 and SUMO-3 are 95% identical to each other (and therefore are referred to as SUMO-2/3), SUMO-1 shares only 50% sequence identity with SUMO-2/3. Moreover, in contrast to SUMO-1, SUMO-2/3 can form poly-SUMO chains on a target protein (62). Under normal conditions SUMO-1 is mainly conjugated to target proteins, while free unconjugated pools of SUMO-2/3 are abundant and ready to be used under stress conditions. The E3 SUMO ligase PIAS4 is able to use all SUMO isoforms in sumoylation reactions (63–66). Interestingly, global levels of both SUMO-1 and SUMO-2/3 are increased in Trim32–/– myoblasts, though only SUMO-2/3 has been implicated in cellular senescence (23, 67). Despite the structure/functional dissimilarities between the isoforms, sumoylation is widely regarded as a protective mechanism against cellular stresses such as heat shock, hypoxia, and osmotic and high oxidative stress, upon which global levels of SUMO-1 (68) and SUMO-2/3 (69) are elevated. Thus, elevated levels of both SUMO-1 and SUMO-2/3 conjugation suggest that Trim32–/– cells may experience additional cellular stresses besides senescence.
It is noteworthy that, in contrast to sumoylation, total ubiquitination levels are not perturbed in the absence of TRIM32, neither under normal nor under pathological conditions (Figure 1C and Figure 8A), suggesting a highly selective role for TRIM32’s E3 ubiquitin ligase activity, which has been implicated in a number of diverse biological processes and most likely functions in a cell type–dependent manner. A controversial role for TRIM32 in apoptosis has recently emerged in studies using different cell models. TRIM32 has been shown to be elevated in epidermal and epithelial tumorigenic cells, where its antiapoptotic and carcinogenic roles have been discovered (19, 38, 39). However, TRIM32 overexpression/knockdown experiments in established cell culture lines have suggested a proapoptotic, tumor suppressive role for TRIM32 (40). Our data demonstrate that apoptosis is not perturbed in TRIM32-deficient skeletal muscle and that it is not an essential pathogenic feature of Trim32–/– myopathy. In support of our finding, no signs of apoptosis were found in the muscle biopsies from patients with LGMD2H/STM (8).
Skeletal muscle is a dynamic tissue capable of regeneration and remodeling. Its plasticity depends on satellite cell function. Loss of skeletal muscle mass (sarcopenia) progresses with age at a rate of 0.5%–1% a year and is associated with failure of the regeneration machinery to replace damaged muscle fibers (70–72). Factors other than aging that lower the ability of satellite cells to replicate and/or differentiate will contribute to premature sarcopenia. The importance of normal replicative capacity of satellite cells for effective skeletal muscle maintenance has recently emerged. The abrogation of age-related atrophy by satellite cell transplantation was demonstrated to arise from an increased regenerative capacity of donor stem cells (73). Moreover, the introduction of a null mutation in the telomerase gene dramatically exacerbates muscular dystrophy in the mdx mouse, a model for Duchenne muscular dystrophy (74). Lack of telomerase activity in this model results in decreased proliferative ability of satellite cells with shortened telomeres, leading to their reduced regenerative capacity. These data further emphasize the importance of replicative senescence of satellite cells in the pathogenesis of muscular dystrophy. In Duchenne muscular dystrophy, however, sarcopenia represents a secondary mechanism of satellite cell exhaustion, as it occurs due to the large number of cycles of degeneration/regeneration. In contrast, sarcopenia seems to play a primary role in the pathogenesis of LGMD2H due to TRIM32 deficiency, through accumulation of PIAS4, leading to premature myoblast senescence and resulting in inadequate adaptive muscle growth. Even though the majority of the LGMD2H pathogenic mutations in TRIM32 are point mutations, at least some of these mutations result in TRIM32 loss in vivo (7, 19). Concordantly, our recently published data analyzing a knock-in mouse carrying the common D489N TRIM32 mutation revealed a dramatic reduction of mutated TRIM32 at the protein level, which leads to a muscle pathology similar to the myopathy described for the Trim32-null mouse (75). Therefore, at least in some cases, LGMD2H can arise from insufficiency of TRIM32 due to protein instability caused by pathogenic mutations. Thus, our data not only reveal a role for TRIM32 in implementing myoblast senescence in skeletal muscles but also suggest a pathogenic mechanism of muscular dystrophy that involves accumulation of PIAS4 and satellite cell senescence in vivo, leading to premature sarcopenia.
Methods
Animals and experimental models.
Trim32–/– mice were created previously (11) and were backcrossed to BALB/cJ wild-type mice (The Jackson Laboratory) to generation F5.
For fasting-induced muscle atrophy, 1-year-old mice (n = 4 for each genotype) were deprived of food for 48 hours. All animals received water ad libitum. Body weight was assessed at the beginning of the experiment and after 24 and 48 hours of fasting. Gastrocnemius muscles from 48 hour–starved mice and normally fed control mice were harvested for myofibrillar preparation (see below).
For hind limb suspension and reloading, male mice of Trim32–/– or Trim32+/+ genotype (6–7 months of age; n = 6 in each group) were subjected to hind limb unloading for 5 days by placing an animal in a harness so that the feet of the hind limbs did not contact the cage floor, as described previously (76, 77). Care was taken to ensure that all animals received food and water ad libitum. Two other groups of animals were subjected to 5 days of hind limb suspension followed by reloading for 3 or 7 days by allowing them to return to their normal cage activity. For each group, there were age-, sex-, and genotype-matched ambulatory controls. At the end of the experiment, soleus muscles were dissected from each animal; one soleus was frozen for immunohistochemistry, and second one was placed in liquid nitrogen and used for Western blot analysis.
Tissue extract preparation and Western blot analysis.
For Western blot analysis, muscles were homogenized in reducing sample buffer (80 mM Tris [pH 6.8], 0.1 M dithiothreitol, 2% SDS, and 10% glycerol) and protease inhibitor cocktail (Sigma-Aldrich) using a Dounce homogenizer. An equal amount of total protein (40 μg per well) was loaded on SDS-PAGE followed by transfer to nitrocellulose membrane.
Myofibrillar preparations from gastrocnemius muscles was performed as described previously (78). Isolated myofibrils were separated on SDS-PAGE (2 μg per well) and stained with Coomassie blue.
Antibodies.
We generated specific rabbit affinity-purified antipeptide antibody against mouse TRIM32 peptide (240–372 aa) as described previously (10).
The following commercial antibodies were used for Western blot analysis: anti-ubiquitin (rabbit 1:200), anti-vinculin (mouse 1:1,000), and anti-PIAS4 (mouse 1:300) from Sigma-Aldrich; anti-desmin (mouse 1:250), anti–myosin developmental heavy chain (mouse 1:100), and anti–myosin fast heavy chain (mouse 1:100) from Leica Microsystems; anti-AKT total (rabbit 1:500), anti–phospho-AKT (Ser473) (rabbit 1:500), anti-S6 ribosomal protein (rabbit 1:1,000), anti–phospho-S6 (Ser240/244) (rabbit 1:1,000), anti–myosin heavy chain IIa nonmuscle (rabbit 1:1,000), anti-HP1γ (rabbit 1:500), anti-p53 (mouse 1:500), anti-LC3B (rabbit 1:500), anti-p38 (mouse 1:500), anti–phospho-p38 (mouse 1:500), and SMAD2/3 (rabbit 1:500) from Cell Signalling; anti-PCNA (mouse 1:500) and anti-MyoD1 (mouse 1:100) from DAKO; anti-GAPDH (mouse 1:500) from Millipore; anti–SUMO-1 (rabbit 1:500) from Boston Biochem; anti–SUMO-2/3 (rabbit 1:500) from Abcam; anti-ERK1/2 (rabbit) from Promega; anti–phospho-ERK1/2 (rabbit) from Chemicon; and anti–phospho-SMAD2/3 (goat) from R&D Systems.
Anti-myogenin, anti–myosin heavy chain IIb nonmuscle, anti–myosin embryonic heavy chain, and anti-Pax7 mAb were obtained from Developmental Studies Hybridoma Bank, developed under the auspices of the National Institute of Child Health and Human Development and maintained by the University of Iowa, Department of Biological Sciences, Iowa City, Iowa, USA.
Secondary antibodies conjugated with HRP were from Sigma-Aldrich. Specific signals were developed using ChemiGlow chemiluminescent substrate for HRP (Alpha Innotech). Images of the blots were acquired using FlourChem FC2 Imager (Alpha Innotech). Densitometry was performed using AlphaEase FC Software Version 6.0.2 (Alpha Innotech) and ImageJ 1.45s ( http://rsbweb.nih.gov/ij/).
Real-time PCR.
Analysis of relative Trim32 mRNA levels in Trim32+/+ gastrocnemius muscles was performed using real-time PCR with iQ SYBR Green Supermix (Bio-Rad) in an MyIQ Single Color Real-Time PCR Detection System (Bio-Rad), as described previously (11). Primers for mouse Pias4 qPCR were as follows: forward, 5′-CAAGAGAGCCCATGCATCTTCG; reverse, 5′-CCTCCTGCGGACAGCTGGTATC.
Immunohistochemistry.
Muscles were dissected from the mice, placed in O.C.T. Compound (Sakura Finetek), and frozen in isopentane cooled in liquid nitrogen. Frozen muscles were cross-sectioned mid-belly at 10 μm and stained with monoclonal antibodies to myosin fast-type or slow-type heavy chains (1:50; Leica Microsystems) using the M.O.M. Kit and AEC Peroxidase Substrate Kit (Vector Laboratories). Images were captured using an AxioImager microscope (Carl Zeiss). Cross-sectional areas of slow and fast fibers were measured in 3 independent fields of view for each slide using AxioVision software (Carl Zeiss). Satellite cell number was assessed using anti-Pax7 monoclonal antibodies (1:50; Developmental Studies Hybridoma Bank) and the M.O.M. Kit and DAB Peroxidase Substrate Kit with nickel (Vector Laboratories). Pax7-positive cells were counted in 3 independent fields of view for each slide. Images were captured and processed using AxioVision software (Carl Zeiss) and displayed using Adobe Photoshop CS4.
TUNEL labeling and immunofluorescence.
TUNEL was performed as described previously (79) using biotin-16-dUTP (Enzo Life Sciences) and TdT (GE Healthcare). The TUNEL reaction was detected with FITC-Avidin D (Vector Laboratories). After the TUNEL reaction, sections were washed in PBS; blocked in PBS with 0.2% gelatin, 0.5% Tween-20, and 3% BSA for 1 hour; and immunostained with rabbit anti-dystrophin antibody (Abcam) and anti–rabbit Texas Red (Vector Laboratories). Slides were mounted in Vectashield Mounting Medium with DAPI to counterstain nuclei (Vector Laboratories).
Primary muscle cell cultures.
Myogenic cells were isolated as previously described (80) with minor modifications. Briefly, single muscle fibers were prepared from skeletal muscles of Trim32–/– and Trim32+/+ mice under a stereo dissecting microscope. Isolated single fibers with associated satellite cells were incubated on plates coated with ECL Cell Attachment Matrix (Millipore). Proliferating cells were maintained as described previously (81). Myoblast fusion was induced by placing the cells in differentiation medium: DMEM supplemented with insulin-transferrin-selenium (1:100; Invitrogen) for indicated amount of time.
For immunostaining, cells were washed with PBS, permeabilized for 4 minutes at room temperature with 0.5% Triton X-100 and 2% paraformaldehyde in PBS, and postfixed for 25 minutes with 2% PFA. Anti-desmin antibodies (1:50; Leica Microsystems) were used to ensure myogenic origin of isolated cells.
Where indicated, cells were treated with 50 μg/ml MG132 (Z-Leu-Leu-Leu-al) in DMSO (Sigma-Aldrich) for the indicated amount of time. Control cells were incubated in DMSO alone.
For siRNA transfection, primary myoblasts were grown to 50% confluence and transfected with either a Mouse ON-TARGETplus SMARTpool PIAS4-specific siRNA (Thermo Scientific, Dharmacon) or a nontargeting pool siRNA as a negative control, using DharmaFECT 1 Transfection Reagent according to the manufacturer’s instructions (Thermo Scientific, Dharmacon).
Analysis of cellular DNA content by flow cytometry.
Cell cycle analysis was achieved by PI labeling of the nuclei and analyzing the fluorescence properties of the cells using FACS (82). Cells were trypsinized, collected by centrifugation, washed in PBS without calcium or magnesium, and resuspended at 1 × 106 cells in 1 ml of ice-cold FACS buffer (0.1% BSA in PBS). Three volumes of cold ethanol (to a final concentration of 70%) were forced added to the cell suspension. After overnight incubation at 4°C, fixed cells were washed in PBS and incubated with 20 μg/ml PI (Sigma-Aldrich), 0.2 mg/ml RNase A (Sigma-Aldrich), and 0.1% Triton X-100 in PBS for 30 minutes before analysis. Cells were acquired on a BD FACSCalibur flow cytometer (Becton Dickinson) available at the Center for Duchenne Muscular Dystrophy Core facility, and the data were analyzed using FlowJo 9.3.1 software (Tree Star).
SA-β-gal staining.
Cell senescence was assessed by detecting the SA-β-gal activity at pH 6.0 (47). Cells were fixed in 0.2% glutaraldehyde for 15 minutes at room temperature and stained in solution containing 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 2 mM magnesium chloride, 1 mg/ml bromo-chloro-indolyl-galactopyranoside (X-gal) (pH 6.0). SA-β-gal–positive cells were detected by phase contrast microscopy using Zeiss Axiovert 200M fluorescence microscope (Carl Zeiss). Positive (blue) cells were counted in 20 independent fields of view, and images were captured and processed using AxioVision software (Carl Zeiss) and displayed using Adobe Photoshop CS4.
Statistics.
Statistical analysis of all data was carried out by 2-tailed Student’s t test. Differences were considered statistically significant if the P value was less than 0.05. Error bars on all graphs are represented by SEM.
Study approval.
All experimental protocols and use of animals were conducted in accordance with the NIH Guide for Care and Use of Laboratory Animals and approved by the UCLA Institutional Animal Care and Use Committee.
Supplementary Material
Acknowledgments
We thank Jane Wen, Julia Overman, and Leonel Martinez for excellent technical support. This work was supported by funding from the National Institute of Arthritis, Musculoskeletal and Skin Diseases (NIAMS) (RO1 AR052693, RO1 AR/NS48177), NIAMS for a P30 Muscular Dystrophy Core Center (P30AR057230-01), and the Muscular Dystrophy Association.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(5):1764–1776. doi:10.1172/JCI59581.
Elena Kudryashova’s present address is: Department of Biochemistry, The Ohio State University, Columbus, Ohio, USA.
References
- 1.Reymond A, et al. The tripartite motif family identifies cell compartments. Embo J. 2001;20(9):2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kudryashova E, Kudryashov D, Kramerova I, Spencer MJ. Trim32 is a ubiquitin ligase mutated in limb girdle muscular dystrophy type 2H that binds to skeletal muscle myosin and ubiquitinates actin. J Mol Biol. 2005;354(2):413–424. doi: 10.1016/j.jmb.2005.09.068. [DOI] [PubMed] [Google Scholar]
- 3.Edwards TA, Wilkinson BD, Wharton RP, Aggarwal AK. Model of the brain tumor-Pumilio translation repressor complex. Genes Dev. 2003;17(20):2508–2513. doi: 10.1101/gad.1119403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frosk P, et al. Limb-girdle muscular dystrophy type 2H associated with mutation in TRIM32, a putative E3-ubiquitin-ligase gene. Am J Hum Genet. 2002;70(3):663–672. doi: 10.1086/339083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saccone V, et al. Mutations that impair interaction properties of TRIM32 associated with limb-girdle muscular dystrophy 2H. Hum Mutat. 2008;29(2):240–247. doi: 10.1002/humu.20633. [DOI] [PubMed] [Google Scholar]
- 6.Cossee M, et al. Use of SNP array analysis to identify a novel TRIM32 mutation in limb-girdle muscular dystrophy type 2H. Neuromuscul Disord. 2009;19(4):255–260. doi: 10.1016/j.nmd.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 7.Borg K, et al. Intragenic deletion of TRIM32 in compound heterozygotes with sarcotubular myopathy/LGMD2H. Hum Mutat. 2009;30(9):E831–E844. doi: 10.1002/humu.21063. [DOI] [PubMed] [Google Scholar]
- 8.Schoser BG, Frosk P, Engel AG, Klutzny U, Lochmuller H, Wrogemann K. Commonality of TRIM32 mutation in causing sarcotubular myopathy and LGMD2H. Ann Neurol. 2005;57(4):591–595. doi: 10.1002/ana.20441. [DOI] [PubMed] [Google Scholar]
- 9.Chiang AP, et al. Homozygosity mapping with SNP arrays identifies TRIM32, an E3 ubiquitin ligase, as a Bardet-Biedl syndrome gene (BBS11). Proc Natl Acad Sci U S A. 2006;103(16):6287–6292. doi: 10.1073/pnas.0600158103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Locke M, Tinsley CL, Benson MA, Blake DJ. TRIM32 is an E3 ubiquitin ligase for dysbindin. Hum Mol Genet. 2009;18(13):2344–2358. doi: 10.1093/hmg/ddp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kudryashova E, Wu J, Havton LA, Spencer MJ. Deficiency of the E3 ubiquitin ligase TRIM32 in mice leads to a myopathy with a neurogenic component. Hum Mol Genet. 2009;18(7):1353–1367. doi: 10.1093/hmg/ddp036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shieh PB, Kudryashova E, Spencer MJ. Limb-girdle muscular dystrophy 2H and the role of TRIM32. Handb Clin Neurol. 2011;101:125–133. doi: 10.1016/B978-0-08-045031-5.00009-8. [DOI] [PubMed] [Google Scholar]
- 13.Jerusalem F, Engel AG, Gomez MR. Sarcotubular myopathy. A newly recognized, benign, congenital, familial muscle disease. Neurology. 1973;23(9):897–906. doi: 10.1212/wnl.23.9.897. [DOI] [PubMed] [Google Scholar]
- 14.Muller-Felber W, Schlotter B, Topfer M, Ketelsen UP, Muller-Hocker J, Pongratz D. Phenotypic variability in two brothers with sarcotubular myopathy. J Neurol. 1999;246(5):408–411. doi: 10.1007/s004150050374. [DOI] [PubMed] [Google Scholar]
- 15.Shokeir MH, Kobrinsky NL. Autosomal recessive muscular dystrophy in Manitoba Hutterites. Clin Genet. 1976;9(2):197–202. doi: 10.1111/j.1399-0004.1976.tb01568.x. [DOI] [PubMed] [Google Scholar]
- 16.Shokeir MH, Rozdilsky B. Muscular dystrophy in Saskatchewan Hutterites. Am J Med Genet. 1985;22(3):487–493. doi: 10.1002/ajmg.1320220306. [DOI] [PubMed] [Google Scholar]
- 17.Bodine SC, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294(5547):1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 18.Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc Natl Acad Sci U S A. 2001;98(25):14440–14445. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Albor A, et al. The interaction of Piasy with Trim32, an E3-ubiquitin ligase mutated in limb-girdle muscular dystrophy type 2H, promotes Piasy degradation and regulates UVB-induced keratinocyte apoptosis through NFkappaB. J Biol Chem. 2006;281(35):25850–25866. doi: 10.1074/jbc.M601655200. [DOI] [PubMed] [Google Scholar]
- 20.Hannoun Z, Greenhough S, Jaffray E, Hay RT, Hay DC. Post-translational modification by SUMO. Toxicology. 2010;278(3):288–293. doi: 10.1016/j.tox.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 21.Bischof O, et al. The E3 SUMO ligase PIASy is a regulator of cellular senescence and apoptosis. Mol Cell. 2006;22(6):783–794. doi: 10.1016/j.molcel.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 22.Bischof O, Dejean A. SUMO is growing senescent. Cell Cycle. 2007;6(6):677–681. doi: 10.4161/cc.6.6.4021. [DOI] [PubMed] [Google Scholar]
- 23.Li T, et al. Expression of SUMO-2/3 induced senescence through p53- and pRB-mediated pathways. . J Biol Chem. 2006;281(47):36221–36227. doi: 10.1074/jbc.M608236200. [DOI] [PubMed] [Google Scholar]
- 24.Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 25.Serrano M, Blasco MA. Putting the stress on senescence. Curr Opin Cell Biol. 2001;13(6):748–753. doi: 10.1016/S0955-0674(00)00278-7. [DOI] [PubMed] [Google Scholar]
- 26.Snijders T, Verdijk LB, van Loon LJ. The impact of sarcopenia and exercise training on skeletal muscle satellite cells. Ageing Res Rev. 2009;8(4):328–338. doi: 10.1016/j.arr.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 27.Lexell J. Human aging, muscle mass, and fiber type composition. J Gerontol A Biol Sci Med Sci. 1995;50 spec no:11–16. doi: 10.1093/gerona/50a.special_issue.11. [DOI] [PubMed] [Google Scholar]
- 28.Balagopal P, Schimke JC, Ades P, Adey D, Nair KS. Age effect on transcript levels and synthesis rate of muscle MHC and response to resistance exercise. Am J Physiol Endocrinol Metab. 2001;280(2):E203–E208. doi: 10.1152/ajpendo.2001.280.2.E203. [DOI] [PubMed] [Google Scholar]
- 29.Jagoe RT, Lecker SH, Gomes M, Goldberg AL. Patterns of gene expression in atrophying skeletal muscles: response to food deprivation. FASEB J. 2002;16(13):1697–1712. doi: 10.1096/fj.02-0312com. [DOI] [PubMed] [Google Scholar]
- 30.Wing SS, Haas AL, Goldberg AL. Increase in ubiquitin-protein conjugates concomitant with the increase in proteolysis in rat skeletal muscle during starvation and atrophy denervation. Biochem J. 1995;307(pt 3):639–645. doi: 10.1042/bj3070639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomason DB, Booth FW. Atrophy of the soleus muscle by hindlimb unweighting. J Appl Physiol. 1990;68(1):1–12. doi: 10.1152/jappl.1990.68.1.1. [DOI] [PubMed] [Google Scholar]
- 32.Krippendorf BB, Riley DA. Distinguishing unloading- versus reloading-induced changes in rat soleus muscle. Muscle Nerve. 1993;16(1):99–108. doi: 10.1002/mus.880160116. [DOI] [PubMed] [Google Scholar]
- 33.Glass DJ. PI3 kinase regulation of skeletal muscle hypertrophy and atrophy. Curr Top Microbiol Immunol. 2010;346:267–278. doi: 10.1007/82_2010_78. [DOI] [PubMed] [Google Scholar]
- 34.Glass DJ. Signaling pathways perturbing muscle mass. Curr Opin Clin Nutr Metab Care. 2010;13(3):225–229. doi: 10.1097/MCO.0b013e32833862df. [DOI] [PubMed] [Google Scholar]
- 35.Rommel C, et al. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat Cell Biol. 2001;3(11):1009–1013. doi: 10.1038/ncb1101-1009. [DOI] [PubMed] [Google Scholar]
- 36.Stitt TN, et al. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol Cell. 2004;14(3):395–403. doi: 10.1016/S1097-2765(04)00211-4. [DOI] [PubMed] [Google Scholar]
- 37.Allen DL, et al. Apoptosis: a mechanism contributing to remodeling of skeletal muscle in response to hindlimb unweighting. Am J Physiol. 1997;273(2 pt 1):C579–C587. doi: 10.1152/ajpcell.1997.273.2.C579. [DOI] [PubMed] [Google Scholar]
- 38.Horn EJ, et al. RING protein Trim32 associated with skin carcinogenesis has anti-apoptotic and E3-ubiquitin ligase properties. Carcinogenesis. 2004;25(2):157–167. doi: 10.1093/carcin/bgh003. [DOI] [PubMed] [Google Scholar]
- 39.Kano S, Miyajima N, Fukuda S, Hatakeyama S. Tripartite motif protein 32 facilitates cell growth and migration via degradation of Abl-interactor 2. Cancer Res. 2008;68(14):5572–5580. doi: 10.1158/0008-5472.CAN-07-6231. [DOI] [PubMed] [Google Scholar]
- 40.Ryu YS, et al. TRIM32 sensitizes cells to TNF{alpha}-induced apoptosis via its RING domain-dependent E3 ligase activity against XIAP. J Biol Chem. 2011;286(29):25729–25738. doi: 10.1074/jbc.M111.241893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sandri M. Autophagy in health and disease. 3. Involvement of autophagy in muscle atrophy. Am J Physiol Cell Physiol. 2010;298(6):C1291–C1297. doi: 10.1152/ajpcell.00531.2009. [DOI] [PubMed] [Google Scholar]
- 42.Mitchell PO, Pavlath GK. Skeletal muscle atrophy leads to loss and dysfunction of muscle precursor cells. Am J Physiol Cell Physiol. 2004;287(6):C1753–1762. doi: 10.1152/ajpcell.00292.2004. [DOI] [PubMed] [Google Scholar]
- 43.Lipton BH, Schultz E. Developmental fate of skeletal muscle satellite cells. Science. 1979;205(4412):1292–1294. doi: 10.1126/science.472747. [DOI] [PubMed] [Google Scholar]
- 44.Conboy IM, Rando TA. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev Cell. 2002;3(3):397–409. doi: 10.1016/S1534-5807(02)00254-X. [DOI] [PubMed] [Google Scholar]
- 45.Narita M, et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113(6):703–716. doi: 10.1016/S0092-8674(03)00401-X. [DOI] [PubMed] [Google Scholar]
- 46.O’Brien IE, Murray BG, Baguley BC, Morris BA, Ferguson IB. Major changes in chromatin condensation suggest the presence of an apoptotic pathway in plant cells. Exp Cell Res. 1998;241(1):46–54. doi: 10.1006/excr.1998.4036. [DOI] [PubMed] [Google Scholar]
- 47.Dimri GP, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92(20):9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Halevy O, et al. Pattern of Pax7 expression during myogenesis in the posthatch chicken establishes a model for satellite cell differentiation and renewal. Dev Dyn. 2004;231(3):489–502. doi: 10.1002/dvdy.20151. [DOI] [PubMed] [Google Scholar]
- 49.Zammit PS, Golding JP, Nagata Y, Hudon V, Partridge TA, Beauchamp JR. Muscle satellite cells adopt divergent fates: a mechanism for self-renewal? J Cell Biol. 2004;166(3):347–357. doi: 10.1083/jcb.200312007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zammit PS, et al. Pax7 and myogenic progression in skeletal muscle satellite cells. J Cell Sci. 2006;119(pt 9):1824–1832. doi: 10.1242/jcs.02908. [DOI] [PubMed] [Google Scholar]
- 51.Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell Res. 2009;19(1):128–139. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Childs TE, Spangenburg EE, Vyas DR, Booth FW. Temporal alterations in protein signaling cascades during recovery from muscle atrophy. Am J Physiol Cell Physiol. 2003;285(2):C391–C398. doi: 10.1152/ajpcell.00478.2002. [DOI] [PubMed] [Google Scholar]
- 53.Burks TN, et al. Losartan restores skeletal muscle remodeling and protects against disuse atrophy in sarcopenia. Sci Transl Med. 2011;3(82):82ra37. doi: 10.1126/scitranslmed.3002227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carlson ME, et al. Molecular aging and rejuvenation of human muscle stem cells. EMBO Mol Med. 2009;1(8–9):381–391. doi: 10.1002/emmm.200900045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Renault V, et al. Skeletal muscle regeneration and the mitotic clock. Exp Gerontol. 2000;35(6–7):711–719. doi: 10.1016/S0531-5565(00)00151-0. [DOI] [PubMed] [Google Scholar]
- 56.Bigot A, et al. Replicative aging down-regulates the myogenic regulatory factors in human myoblasts. Biol Cell. 2008;100(3):189–199. doi: 10.1042/BC20070085. [DOI] [PubMed] [Google Scholar]
- 57.Napolitano LM, Jaffray EG, Hay RT, Meroni G. Functional interactions between ubiquitin E2 enzymes and TRIM proteins. Biochem J. 2011;434(2):309–319. doi: 10.1042/BJ20101487. [DOI] [PubMed] [Google Scholar]
- 58.Kobayashi J, et al. Current topics in DNA double-strand break repair. J Radiat Res. 2008;49(2):93–103. doi: 10.1269/jrr.07130. [DOI] [PubMed] [Google Scholar]
- 59.Huen MS, et al. Noncanonical E2 variant-independent function of UBC13 in promoting checkpoint protein assembly. Mol Cell Biol. 2008;28(19):6104–6112. doi: 10.1128/MCB.00987-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reich KA, Chen YW, Thompson PD, Hoffman EP, Clarkson PM. Forty-eight hours of unloading and 24 h of reloading lead to changes in global gene expression patterns related to ubiquitination and oxidative stress in humans. J Appl Physiol. 2010;109(5):1404–1415. doi: 10.1152/japplphysiol.00444.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Andreou AM, Tavernarakis N. SUMOylation and cell signalling. Biotechnol J. 2009;4(12):1740–1752. doi: 10.1002/biot.200900219. [DOI] [PubMed] [Google Scholar]
- 62.Tatham MH, et al. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem. 2001;276(38):35368–35374. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- 63.Azuma Y, Arnaoutov A, Anan T, Dasso M. PIASy mediates SUMO-2 conjugation of Topoisomerase-II on mitotic chromosomes. EMBO J. 2005;24(12):2172–2182. doi: 10.1038/sj.emboj.7600700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chun TH, Itoh H, Subramanian L, Iniguez-Lluhi JA, Nakao K. Modification of GATA-2 transcriptional activity in endothelial cells by the SUMO E3 ligase PIASy. Circ Res. 2003;92(11):1201–1208. doi: 10.1161/01.RES.0000076893.70898.36. [DOI] [PubMed] [Google Scholar]
- 65.Kang X, et al. PIASy stimulates HIF1alpha SUMOylation and negatively regulates HIF1alpha activity in response to hypoxia. Oncogene. 2010;29(41):5568–5578. doi: 10.1038/onc.2010.297. [DOI] [PubMed] [Google Scholar]
- 66.Subramanian L, Benson MD, Iniguez-Lluhi JA. A synergy control motif within the attenuator domain of CCAAT/enhancer-binding protein alpha inhibits transcriptional synergy through its PIASy-enhanced modification by SUMO-1 or SUMO-3. . J Biol Chem. 2003;278(11):9134–9141. doi: 10.1074/jbc.M210440200. [DOI] [PubMed] [Google Scholar]
- 67.Li T, et al. Sumoylation of heterogeneous nuclear ribonucleoproteins, zinc finger proteins, and nuclear pore complex proteins: a proteomic analysis. Proc Natl Acad Sci U S A. 2004;101(23):8551–8556. doi: 10.1073/pnas.0402889101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shao R, et al. Increase of SUMO-1 expression in response to hypoxia: direct interaction with HIF-1alpha in adult mouse brain and heart in vivo. FEBS Lett. 2004;569(1–3):293–300. doi: 10.1016/j.febslet.2004.05.079. [DOI] [PubMed] [Google Scholar]
- 69.Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem. 2000;275(9):6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- 70.Janssen I, Heymsfield SB, Wang ZM, Ross R. Skeletal muscle mass and distribution in 468 men and women aged 18-88 yr. J Appl Physiol. 2000;89(1):81–88. doi: 10.1152/jappl.2000.89.1.81. [DOI] [PubMed] [Google Scholar]
- 71.Frontera WR, Hughes VA, Fielding RA, Fiatarone MA, Evans WJ, Roubenoff R. Aging of skeletal muscle: a 12-yr longitudinal study. J Appl Physiol. 2000;88(4):1321–1326. doi: 10.1152/jappl.2000.88.4.1321. [DOI] [PubMed] [Google Scholar]
- 72.Brooks SV, Faulkner JA. Contraction-induced injury: recovery of skeletal muscles in young and old mice. Am J Physiol. 1990;258(3 pt 1):C436–C442. doi: 10.1152/ajpcell.1990.258.3.C436. [DOI] [PubMed] [Google Scholar]
- 73.Hall JK, Banks GB, Chamberlain JS, Olwin BB. Prevention of muscle aging by myofiber-associated satellite cell transplantation. Sci Transl Med. 2010;2(57):57ra83. doi: 10.1126/scitranslmed.3001081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sacco A, et al. Short telomeres and stem cell exhaustion model Duchenne muscular dystrophy in mdx/mTR mice. Cell. 2010;143(7):1059–1071. doi: 10.1016/j.cell.2010.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kudryashova E, Struyk A, Mokhonova E, Cannon SC, Spencer MJ. The common missense mutation D489N in TRIM32 causing limb girdle muscular dystrophy 2H leads to loss of the mutated protein in knock-in mice resulting in a Trim32-null phenotype. Hum Mol Genet. 2011;20(20):3925–3932. doi: 10.1093/hmg/ddr311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tidball JG, Spencer MJ. Expression of a calpastatin transgene slows muscle wasting and obviates changes in myosin isoform expression during murine muscle disuse. J Physiol. 2002;545(pt 3):819–828. doi: 10.1113/jphysiol.2002.024935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kramerova I, Kudryashova E, Venkatraman G, Spencer MJ. Calpain 3 participates in sarcomere remodeling by acting upstream of the ubiquitin-proteasome pathway. Hum Mol Genet. 2005;14(15):2125–2134. doi: 10.1093/hmg/ddi217. [DOI] [PubMed] [Google Scholar]
- 78.Cohen S, et al. During muscle atrophy, thick, but not thin, filament components are degraded by MuRF1-dependent ubiquitylation. J Cell Biol. 2009;185(6):1083–1095. doi: 10.1083/jcb.200901052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kramerova I, Kudryashova E, Tidball JG, Spencer MJ. Null mutation of calpain 3 (p94) in mice causes abnormal sarcomere formation in vivo and in vitro. Hum Mol Genet. 2004;13(13):1373–1388. doi: 10.1093/hmg/ddh153. [DOI] [PubMed] [Google Scholar]
- 80.Rosenblatt JD, Lunt AI, Parry DJ, Partridge TA. Culturing satellite cells from living single muscle fiber explants. In Vitro Cell Dev Biol Anim. 1995;31(10):773–779. doi: 10.1007/BF02634119. [DOI] [PubMed] [Google Scholar]
- 81.Kramerova I, Kudryashova E, Wu B, Spencer MJ. Regulation of the M-cadherin-beta-catenin complex by calpain 3 during terminal stages of myogenic differentiation. Mol Cell Biol. 2006;26(22):8437–8447. doi: 10.1128/MCB.01296-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Crissman HA, Steinkamp JA. Rapid, simultaneous measurement of DNA, protein, and cell volume in single cells from large mammalian cell populations. J Cell Biol. 1973;59(3):766–771. doi: 10.1083/jcb.59.3.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.