Abstract
The human genome encodes thousands of long noncoding RNAs (lncRNAs). Although most remain functionally uncharacterized biological “dark matter,” lncRNAs have garnered considerable attention for their diverse roles in human biology, including developmental programs and tumor suppressor gene networks. As the number of lncRNAs associated with human disease grows, ongoing research efforts are focusing on their regulatory mechanisms. New technologies that enable enumeration of lncRNA interaction partners and determination of lncRNA structure are well positioned to drive deeper understanding of their functions and involvement in pathogenesis. In turn, lncRNAs may become targets for therapeutic intervention or new tools for biotechnology.
Introduction
RNA is now recognized as an important regulator of biological systems. While its primary sequence can encode protein, RNA can also fold into non–protein-coding structural motifs that perform catalysis (1), bind small molecules (2), or serve as protein scaffolds (3). Noncoding RNAs can conditionally govern gene expression (4) and have impressive regulatory capacity, as small noncoding RNAs may modulate the expression of greater than 60% of human coding genes (5). Built upon the growing number of well-characterized regulatory RNAs, novel RNA-based control systems are now being applied to problems in microbial (6, 7) and mammalian biotechnology. Gene networks have been programmed to recognize and respond to cancer-associated miRNA profiles (8), shRNA-based genetic switches may support gene therapy applications (9), and drug-responsive RNA sensors have been developed for T cell therapy (10).
Despite the remarkable progress in characterizing RNA-based regulation and the promise of RNA biotechnology, much of the human transcriptome remains functionally uncharacterized. Although less than 1.5% of the human genome codes for proteins, a much larger fraction of the human genome appears to code for RNA. Some studies indicate that greater than 90% of the human genome is transcribed (11), although others have argued that technical artifacts or biological noise explain much of this pervasive transcription (12). Increasingly sensitive RNA sequencing approaches (13) are helping to address this controversy and have confirmed the existence of many novel transcripts (14).
Ongoing efforts are focused on classifying these presumed noncoding transcripts, although care and additional experiments are necessary to exclude their protein coding potential (15). Considerable attention has focused on long noncoding RNAs (lncRNAs), which are distinguished from short regulatory RNAs by having a length greater than 200 nucleotides. Recent studies have classified more than 8,000 intergenic lncRNAs (16), which are often transcribed at lower abundance than mRNAs by RNA polymerase II in a tissue-specific manner. Many lncRNAs map to regions associated with disease by genome-wide association studies (GWAS) (16), and the number of papers discussing lncRNA disease associations grows each year (17).
Here, we explore the rapidly expanding literature by discussing lncRNAs that exert epigenetic, transcriptional, and posttranscriptional regulation in gene networks that are relevant to human health, including tumor suppression and development. At each level of control, we highlight lncRNA regulatory mechanisms and putative roles in human disease. Finally, we discuss new technologies that are well-positioned to drive deeper mechanistic understanding of lncRNA function.
Epigenetic control
Despite having identical genomes, the different cell types within an individual exhibit unique and heritable gene expression patterns. Heritable variation (-genetic) must be encoded in molecular signatures beyond (epi-) the DNA sequence itself (18). These epigenetic signatures can be written to chromatin, the structural housing of genetic information in which DNA is wrapped around repeating octamers of histone proteins. Methylation of cytosine residues in DNA and posttranslational histone modifications can specify the state of chromatin, resulting in transcriptional activation or silencing. In mammalian systems, the chromatin-remodeling components that write and erase these epigenetic signatures generally lack domains to specify DNA localization (19) and depend upon ancillary factors in order to target specific loci. lncRNAs can serve as these factors, recruiting chromatin-remodeling components to their point of synthesis as nascent transcripts in cis or acting as diffuse scaffolds that guide these components to distant loci in trans. cis- and trans-acting lncRNAs facilitate epigenetic control over diverse processes, including tumor suppression and development.
Cell cycle control and tumor suppression.
Senescence is a state of cell-cycle arrest that guards the cell against unrestrained proliferation and tumor progression. Spanning an approximately 42-kb region on human chromosome 9p21, the INK4b (p15)–ARF (p14)–INK4a (p16) locus is an important regulator of cellular senescence, as it codes for three tumor suppressors: p15 and p16 promote retinoblastoma protein (pRB) function and cell-cycle arrest by inhibiting cyclin-dependent kinases (CDK4/6), and p14 increases the functionality of p53 signaling (20). Coordinated regulation of this locus is governed by polycomb repressive complex–2 (PRC-2), which trimethylates histone H3 on lysine 27 (H3K27) in transcriptionally silent heterochromatin, and PRC-1, which recognizes methylated H3K27 as a signal of heterochromatin maintenance. cis- and trans-acting lncRNAs recruit these complexes and help govern heterochromatin establishment.
PRC-1/2 complexes are recruited to this locus by ANRIL, a lncRNA expressed antisense to p14 and p15. ANRIL binds SUZ12, a subunit of the PRC-2 complex, (21) and CBX7, a subunit of the PRC-1 complex (22). ANRIL knockdown or deletion leads to upregulation of p15 (21) and p16 (22), which suggests that PRC-1/2 are recruited in cis to the locus gene through association with nascent ANRIL transcripts. Moreover, changes in ANRIL expression can affect the transcriptional state of the locus, which is frequently deleted or silenced in cancers (23). Specifically, ANRIL is upregulated in prostate cancer tissues (22), and risk-associated SNPs for type 2 diabetes, heart disease, and cancers overlap with the ANRIL genomic region (24). Many of these SNPs are found within enhancer elements, and one — associated with coronary artery disease (CAD) — disrupts the binding site for STAT1, a transcription factor that represses ANRIL expression (25). Because it abrogates STAT1 binding and leads to upregulation of ANRIL, this SNP may contribute to CAD through ANRIL-mediated silencing of p15.
trans-acting lncRNAs also play a role in tumor suppression through regulation of this locus. A screen for lncRNAs upregulated in hepatocellular carcinoma (HCC) indentified high expression in hepatocellular carcinoma (HEIH), a lncRNA that binds EZH2, the methyltransferase subunit of PRC-2 (26). HEIH knockdown resulted in derepression of PRC-2 target genes, including p16, and HEIH overexpression increased H3K27 levels at the p16 promoter, suggesting a trans model of regulation in which HEIH recruits PRC-2 to silence p16 and other genes involved in cell cycle control. As a result, HEIH recruitment of PRC-2 to tumor suppressors may facilitate HCC tumorigenesis.
Development.
Paralleling their role in regulation of the INK4b-ARF-INK4a locus, lncRNAs also govern the epigenetic state of HOX genes, a set of 39 transcription factors that are integral to normal temporospatial limb and organ development along the anatomical anterior-posterior axis. Expressed from the HOXC locus, the lncRNA HOTAIR drives transcriptional repression of HOXD in trans through recruitment of PRC-2 and LSD1, a complex that removes a chromatin modification (H3K4me2) associated with transcriptional activation (27). Like HEIH, HOTAIR overexpression leads to genome-wide retargeting of the PRC-2 complex, resulting in a PRC-2 occupancy and gene expression pattern that promotes metastasis in breast (28) and colorectal cancers (29).
Countering the transcriptional repression of PRC-1/2 complexes, the trithorax group (TrXG) proteins trimethylate lysine 4 of histone H3 (H3K4) in order to establish and maintain HOX gene transcriptional activation. LncRNAs recruit TrXG proteins, such as the MLL-1 complex, to chromatin for activation of specific HOX genes. The lncRNA HOTTIP binds WDR5, an adapter protein for MLL-1, and recruits the MLL-1/WDR5 complex to 5ι HOXA genes (30). HOTTIP-mediated recruitment of MLL-1 occurs in cis and is facilitated by chromosomal looping, which bring the nascent HOTTIP transcript into close spatial proximity with its 5ι HOX target genes. This interaction is disrupted by HOTTIP knockdown, which results in a loss of H3K4me3 across the HOXA locus and a notable shortening and bending of distal bony elements, including the radius, ulna, and third digit. Whereas HOTTIP knockdown most strongly reduced expression of 5ι HOXA genes (HOXA10–HOXA13), knockdown of Mira, another lncRNA transcribed within the HOXA locus, specifically reduced expression of HOXA6/7 (31). Unlike HOTTIP, Mira binds directly to MLL-1 and recruits the complex to the HOXA6/7 promoters. Through the activation of HOXA6/7, Mira indirectly triggers expression of 15 germ-layer marker genes during early mouse ES cell differentiation, further underscoring the importance of lncRNAs in the HOX developmental program.
Although the HOX cluster is a good model for understanding lncRNA-mediated repression and activation (Figure 1A), the full scope of epigenetic regulation by lncRNAs in development appears to be much broader. A recent report identified 137 lncRNAs that globally affect gene expression levels in mouse ES cells, demonstrating that lncRNAs play roles in early development and stem cell biology. Many of these lncRNAs (approximately 30%) interact with chromatin-modifying proteins and affect gene expression patterns that maintain ES cell pluripotency or repress lineage-specific differentiation (32). Beyond early development, lncRNA scaffolds for chromatin-remodeling components maintain developmental programs in specific tissues, such as the retina (33). Despite the apparent diversity of target genes regulated by these lncRNAs, several questions underpin all cases of epigenetic control: it will be important to determine the RNA structural motifs that interact with chromatin-remodeling components, identify cases in which multiple complexes are recruited to a single lncRNA, and discover how lncRNAs target these complexes to specific regions of chromatin. In addition, a recent study in fission yeast showed that transcription of protein-coding mRNAs, along with their degradation factors, nucleates heterochromatin formation at specific loci in response to developmental signals (34). Moreover, it will be interesting to explore whether lncRNAs and protein-coding transcripts share mechanisms for exerting epigenetic control over genomic loci.
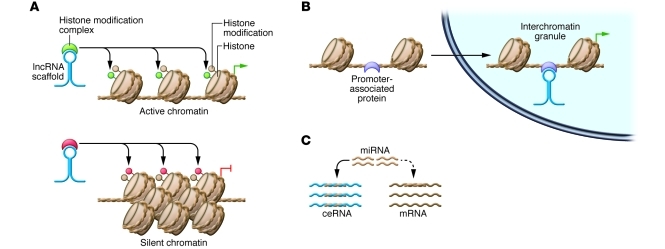
Figure 1. Models for lncRNA-mediated epigenetic, transcriptional, and posttranscriptional regulation.
(A) lncRNAs can recruit chromatin-remodeling components to specific genomic loci, reprogramming the state of chromatin in order to silence or activate transcription. (B) lncRNAs housed within nuclear subcompartments can bind promoter-associated proteins, recruiting specific genomic loci to the subcompartment and driving transcription of the localized genes. (C) lncRNAs can act as ceRNAs, which harbor miRNA binding sites and therefore reduce miRNA levels available for targeting mRNAs. In all cases, lncRNAs are shown in blue.
Transcriptional and cotranscriptional control
While the examples of epigenetic control involve recruitment of chromatin-remodeling components, transcriptional control often governs recruitment of RNA polymerase-II (PolII), transcription factors, and coregulators to the promoter regions of specific genes. Just as they recruit chromatin-modifying proteins, lncRNAs also nucleate assembly of transcription factors in diverse signaling pathways, including lncRNA-mediated repression of target genes in the p53 pathway (reviewed in ref. 35) and lncRNA coactivation of nuclear receptor signaling (reviewed in ref. 36). Yet the mechanisms employed by lncRNAs in transcription control extend well beyond scaffolds, as lncRNAs can also serve as decoys that bind and repress the activity of transcription factors or repress target genes by forming a triple helix with promoter DNA, blocking polII binding and transcription initiation (reviewed in ref. 37). Although mechanisms and disease associations (38) for lncRNA transcriptional regulators have been reviewed, recent reports extend both the mechanistic scope of lncRNA-mediated transcriptional control and the clinical relevance of these pathways.
Cell cycle control.
Cell cycle progression is intimately associated with expression of specific sets of lncRNAs (39). It is also well-established that some lncRNAs abundantly localize to nuclear bodies, which can house protein factors involved in transcriptional activation (e.g., interchromatin granules [ICGs]) or repression (e.g., Pc group [PcG] bodies) (reviewed in ref. 40). A recent report shows that these lncRNAs play a role in cell cycle control by recruiting specific genomic loci to each nuclear compartment, resulting in transcriptional activation or repression. The lncRNAs TUG1 and NEAT2 associate with PcG bodies and ICGs, respectively, and serve as scaffolds for protein factors involved in transcriptional repression and activation within their respective compartments (Figure 1B). These two lncRNAs also bind polycomb 2 protein (Pc2), which is associated with the promoters of cell cycle regulator genes and also contains a chromodomain that binds histone modifications. TUG1 binds the methylated form of Pc2, recruits the protein to PcG bodies, and increases its affinity for repressive heterochromatin marks, reinforcing localization within the PcG-repressed compartment. In contrast, NEAT2 binds the demethylated form of the protein, recruiting it to ICGs and increasing its affinity for active chromatin modifications. As a result, Pc2 methylation and demethylation govern the physical relocation and transcription of the cell cycle regulator genes to which it is bound (41). It will be interesting to learn whether other lncRNA scaffolds can recognize covalent modifications on promoter-associated signaling proteins and explore whether this is a common mechanism for conditional gene localization and transcriptional control.
Stress response.
Direct repression of PolII is another form of transcriptional control exerted by Alu lncRNAs, which are approximately 300-bp products of PolIII transcription from prevalent genomic repeats known as Alu elements (42). These lncRNAs are often upregulated by stress induction, such as heat shock, and in cancers, including hepatocellular carcinoma (43). Alu knockdown in heat shock–induced kidney cells lead to derepression of four genes, prompting biochemical studies identifying specific Alu regions that bind RNA PolII (44) and demonstrating that Alu lncRNAs exert transcriptional repression by blocking contacts between PolII and promoter DNA (45). Identifying the regulatory factors that restrict Alu repression to specific target genes remains an important area of research and may complement efforts focused on the clinical implications of this regulatory pathway.
A recent report provides an intriguing link between Alu expression and geographic atrophy (GE), a form of macular degeneration that is a common cause of blindness. Retinal tissues affected by GE exhibit reduced levels of Dicer1, an enzyme that cleaves long double-stranded RNA and is a core component of small RNA gene silencing pathways. Surprisingly, knockdown of other enzymes required for miRNA processing did not lead to the GE phenotype, and Dicer1 knockdown caused accumulation of Alu lncRNAs (46). Overexpression and direct injection of Alu lncRNAs into the retina resulted in GE, and the condition could be ameliorated by increased Dicer1 expression. Although it appears that Dicer1 is needed to cleave toxic long double-stranded RNA Alu transcripts, failure to produce the smaller Alu products may also contribute to GE. Moreover, it will be important to learn whether the cleaved Alu products are functional in the retina and whether downregulation of Dicer1 in other diseased tissues, such as cancers (47), alters Alu expression levels. Furthermore, using antisense oligonucleotides to reduce toxic Alu lncRNA levels might be a useful therapeutic strategy to combat GE.
Posttranscriptional control
Epigenetic and transcriptional control by lncRNAs share common themes: lncRNA-protein interactions and lncRNA-DNA/chromatin interactions establish a considerable diversity of linkages between transcriptional activator or repressor proteins (e.g., transcription factors or chromatin-remodeling components) and the genomic loci to which these proteins are recruited. Although effort continues to focus on these two areas, lncRNAs have an increasingly appreciated role in posttranscriptional regulatory networks. Many lncRNAs that exert posttranscriptional control fall within the broader class of competitive endogenous RNAs (ceRNAs), which harbor miRNA binding sites (miRNA response elements; MREs) and reduce miRNA levels available for targeting mRNAs (Figure 1C and refs. 48, 49). As a result, ceRNA upregulation is typically followed by symmetric upregulation of transcripts that the sequestered miRNA normally targets. Because miRNAs regulate thousands of human genes and each miRNA may have many targets, the scope of ceRNA regulation may be significant, and recent reports highlight its importance in development as well as tumor suppression.
Cell cycle control and tumor suppression.
Evidence for lncRNAs acting as ceRNAs emerged from a study on phosphatase and tensin homolog (PTEN), a tumor suppressor for which gene dosage changes are linked to cancer susceptibility (50). PTEN was in part chosen for investigation because it is subject to miRNA regulation and has a noncoding pseudogene, PTEN-P1, that harbors a 3′ untranslated region (UTR) with the same MRE. Overexpression of the PTEN-P1 3′ UTR regulated PTEN levels in trans, but only in the presence of Dicer1, which suggests that miRNAs must be present in order to establish the regulatory link (51). Because it exerts trans regulation by sequestering PTEN-targeting miRNAs miR-19b and miR-20a, PTEN-P1 is a tumor suppressor, and copy number losses at PTEN-P1 are observed in colon cancer cells. The same study showed that a similar relationship exists between v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS), which is amplified in numerous human tumors, and its pseudogene, KRAS1P, suggestive of a broad role for lncRNA ceRNAs in tumorigenesis. Two recent reports have extended this initial study by showing that PTEN is also affected by coding ceRNAs, including the Zeb2 transcript in melanoma (52) and the RB1 transcript in glioblastoma (53). Collectively, these pioneering efforts on ceRNAs establish a framework for understanding the role of lncRNAs as posttranscriptional regulators in tumor suppressor or oncogenic miRNA networks. It will be interesting to understand the sensitivity of these networks to changes in ceRNA abundance relative the total miRNA pool.
Development.
Further support for the importance of lncRNAs acting as ceRNA is provided by a recent study on muscle cell differentiation, a process modulated by miRNAs and subject to miRNA dysregulation in diseases such as myocardial infarction and Duchenne muscular dystrophy (DMD). Expressed in skeletal muscles, the lncRNA linc-MD1 (a long intergenic noncoding RNA) contains MREs for miR-133 and miR-135, which regulate transcription factors involved in myogenic differentiation and muscle cell integrity, including MEF2C and MAML1 (54). Knockdown of linc-MD1 resulted in a symmetric reduction of these two myogenic markers, whereas overexpression led to the expected increase in protein levels. Although linc-MD1 levels, as well as those of MEF2C and MAML1, were strongly reduced in DMD myoblasts, linc-MD1 overexpression restored MEF2C and MAML1 synthesis and partially rescued timing of the differentiation program. Importantly, these results suggest that ceRNAs may be dysregulated in tissues that exhibit aberrant differentiation, such as DMD. They also highlight the potential of therapeutic strategies that restore dysregulated ceRNA networks (Figure 2B).
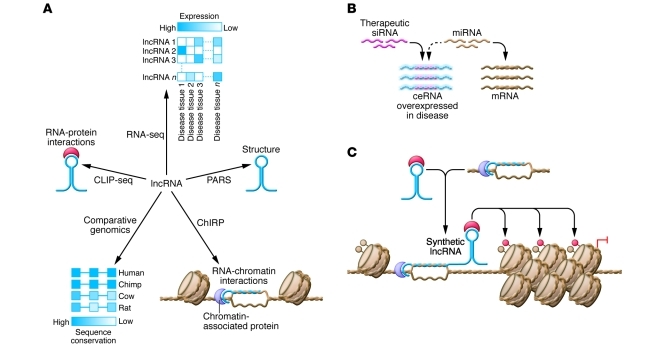
Figure 2. Technologies for discovering and studying lncRNAs may lead to the identification of new therapeutic targets or new tools for biotechnology.
(A) Differential expression profiling has been used to discover lncRNAs that are upregulated in specific tissues or diseases. Comparative genomics can be used to infer functional domains within these lncRNAs based upon conservation of RNA sequence or structure. New technologies provide complimentary information about the lncRNA interactome: PARS, ChIRP, and CLIP-seq enable genome-wide enumeration of lncRNA structure, genomic binding sites, and protein partners, respectively. RNA-seq, RNA deep sequencing. (B) Antisense oligo therapy may be used to target lncRNAs that are dysregulated in disease, such as overexpressed ceRNAs that lead to pathologic miRNA sequestration. (C) Synthetic lncRNAs may be used to reprogram specific regions of the epigenome for control over stem cell differentiation or for regenerative medicine applications.
Future perspectives
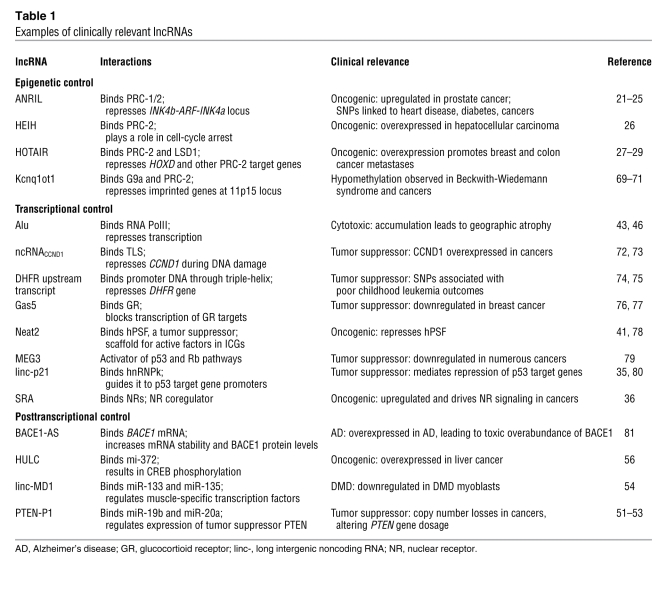
Beyond the examples discussed herein, recent advances in RNA sequencing technology have led to the discovery of thousands of lncRNAs that are upregulated in diseases, such as prostate (55), liver (56), and hepatocellular (26) cancers. A subset of these disease-associated lncRNAs as well as other clinically relevant lncRNAs is shown in Table 1. Although the ability to identify lncRNAs that correlate with disease far outpaces the elucidation of mechanistic links, sequencing technologies also provide ways to help close this gap. Because lncRNAs exert regulatory function through molecular interactions, sequencing technologies have been adapted to support high-throughput mapping of the lncRNA interactome. RNA immunoprecipitation followed by sequencing (RIP-seq) identified thousands of cis- and trans-acting lncRNAs that associate with PRC-2 (57), and direct cross-linking of RNA-protein (CLIP-seq) interactions in vivo is a promising strategy to improve stringency of such protein-RNA interactome measurements (58). Just as these methods take a protein-centric view, chromatin isolation by RNA purification (ChIRP) takes an RNA-centric view, as a lncRNA can be isolated from a cross-linked pool of chromatin to retrieve and enumerate associated DNA sequences and protein (59). Emerging technologies for transcriptome-wide determination of RNA structure (e.g., parallel analysis of RNA structure [PARS]) provide complimentary information (60), allowing researchers to associate interaction domains with the underlying RNA structures. Collectively, these methods should help identify structural domains that allow lncRNAs to associate specifically with other proteins and chromatin (Figure 2A). It may soon be possible to test phenotypic consequences of these interactions in model organisms, such as zebrafish, using morpholino oligonucleotides that specifically target and suppress lncRNA domains (61). Such studies may help inform whether molecular interactions or cryptic small peptide coding capacity (15) give rise to lncRNA function.
Table 1 .
Examples of clinically relevant lncRNAs
Further characterization of the lncRNA interactome and follow-up functional studies may have at least two clinical implications. Initially, lncRNAs may serve as targets for clinical intervention. Although they may now be used as biomarkers for breast (28), hepatocellular (26, 62), liver (63), prostate (55), and lung (64) cancers, lncRNAs may also be targeted when dysregulated expression gives rise to disease or aberrant development. Antisense oligo therapy may be used to reduce toxic lncRNA overexpression, as in the case of Alu toxicity in the retina (Figure 2B). In addition, siRNA knockdown of Xist, a well-studied lncRNA that leads to epigenetic silencing of one female X-chromosome for gene dosage compensation (reviewed in ref. 65), can improve the developmental competence of embryos cloned via somatic cell nuclear transfer (66). Beyond its potential benefits for human-assisted reproductive technologies (67), this study suggests that lncRNA knockdown may be effective in cases where their overexpression leads to pathogenic epigenetic reprogramming.
lncRNAs may also find clinical application as tools for biotechnology. Characterization of the lncRNA interactome will reveal a wealth of linkages that may be rewired in order to obtain synthetic control over gene regulatory networks. Applying lncRNAs for control over chromatin modifications is a particularly interesting possibility, considering the importance of epigenetic memory in tumorigenesis, stem cell maintenance, and aging. LncRNAs that target chromatin-remodeling components to specific loci (Figure 2C) may be useful tools for controlling the state of cultured stem cells, as lncRNAs have already been shown to play important roles in maintaining pluripotency and driving differentiation programs. Furthermore, a challenge in regenerative medicine is to restore aged tissues to a young-adult state without resetting the differentiation program to embryonic or postnatal developmental stages. As current interventions for tissue rejuvenation may act by altering the epigenetic signature of aged cells (68), synthetic lncRNAs may compliment existing strategies by reprogramming specific regions of the epigenome. The potential benefits for disease diagnosis and treatment as well as stem cell therapy and regenerative medicine certainly justify ongoing efforts aimed at a deeper mechanistic understanding of lncRNA function.
Acknowledgments
We thank Robert Spitale and Jeff Quinn for comments on the manuscript. We thank Pedro Batista for assistance with Adobe Illustrator. We apologize to colleagues whose work could not be discussed and cited due to space limitations. The authors’ work is supported by a National Defense Science and Engineering Graduate Fellowship (to L. Martin), and by California Institute for Regenerative Medicine, NIH (to H.Y. Chang). H.Y. Chang is an Early Career Scientist of the Howard Hughes Medical Institute.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(5):1589–1595. doi:10.1172/JCI60020
References
- 1.Cech TR. Crawling out of the RNA world. Cell. 2009;136(4):599–602. doi: 10.1016/j.cell.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Caprara MG, Nilsen TW. RNA: versatility in form and function. Nat Struct Biol. 2000;7(10):831–833. doi: 10.1038/82816. [DOI] [PubMed] [Google Scholar]
- 3.Zappulla DC, Cech TR. RNA as a flexible scaffold for proteins: yeast telomerase and beyond. Cold Spring Harb Symp Quant Biol. 2006;71:217–224. doi: 10.1101/sqb.2006.71.011. [DOI] [PubMed] [Google Scholar]
- 4.Mandal M, Breaker RR. Gene regulation by riboswitches. Nat Rev Mol Cell Biol. 2004;5(6):451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- 5.Friedman RC, Farh KK-H, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delebecque CJ, Lindner AB, Silver PA, Aldaye FA. Organization of intracellular reactions with rationally designed RNA assemblies. Science. 2011;333(6041):470–474. doi: 10.1126/science.1206938. [DOI] [PubMed] [Google Scholar]
- 7.Sinha J, Reyes SJ, Gallivan JP. Reprogramming bacteria to seek and destroy an herbicide. Nat Chem Biol. 2010;6(6):464–470. doi: 10.1038/nchembio.369. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Xie Z, Wroblewska L, Prochazka L, Weiss R, Benenson Y. Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science. 2011;333(6047):1307–1311. doi: 10.1126/science.1205527. [DOI] [PubMed] [Google Scholar]
- 9.Deans TL, Cantor CR, Collins JJ. A tunable genetic switch based on RNAi and repressor proteins for regulating gene expression in mammalian cells. Cell. 2007;130(2):363–372. doi: 10.1016/j.cell.2007.05.045. [DOI] [PubMed] [Google Scholar]
- 10.Chen YY, Jensen MC, Smolke CD. Genetic control of mammalian T-cell proliferation with synthetic RNA regulatory systems. Proc Natl Acad Sci U S A. 2010;107(19):8531–8536. doi: 10.1073/pnas.1001721107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lander ES. Initial impact of the sequencing of the human genome. Nature. 2011;470(7333):187–197. doi: 10.1038/nature09792. [DOI] [PubMed] [Google Scholar]
- 12.van Bakel H, Nislow C, Blencowe BJ, Hughes TR. Response to “The Reality of Pervasive Transcription.”. PLoS Biol. 2011;9(7):e1001102. doi: 10.1371/journal.pbio.1000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mercer TR, et al. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat Biotechnol. 2011;30(1):99–104. doi: 10.1038/nbt.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark MB, et al. The reality of pervasive transcription. PLoS Biol. 2011;9(7):e1000625. doi: 10.1371/journal.pbio.1000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of Mammalian proteomes. Cell. 2011;147(4):789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cabili MN, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25(18):1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330(6004):612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469(7330):343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gil J, Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol. 2006;7(9):667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- 21.Kotake Y, et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15INK4B tumor suppressor gene. Oncogene. 2010;30(16):1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yap KL, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38(5):662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim WY, Sharpless NE. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127(2):265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 24.Pasmant E, Sabbagh A, Vidaud M, Bieche I. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J. 2011;25(2):444–448. doi: 10.1096/fj.10-172452. [DOI] [PubMed] [Google Scholar]
- 25.Harismendy O, et al. 9p21 DNA variants associated with coronary artery disease impair interferon-γ signalling response. Nature. 2011;470(7333):264–268. doi: 10.1038/nature09753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang F, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54(5):1679–1689. doi: 10.1002/hep.24563. [DOI] [PubMed] [Google Scholar]
- 27.Tsai M-C, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kogo R, et al. Long non–coding RNA HOTAIR regulates Polycomb–dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71(20):6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 30.Wang KC, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472(7341):120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bertani S, Sauer S, Bolotin E, Sauer F. The noncoding RNA mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Mol Cell. 2011;43(6):1040–1046. doi: 10.1016/j.molcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Guttman M, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rapicavoli NA, Poth EM, Zhu H, Blackshaw S. The long noncoding RNA Six3OS acts in trans to regulate retinal development by modulating Six3 activity. Neural Dev. 2011;6:32. doi: 10.1186/1749-8104-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Science. 2012;335(6064):96–100. doi: 10.1126/science.1211651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rinn JL, Huarte M. To repress or not to repress: This is the guardian’s question. Trends Cell Biol. 2011;21(6):344–353. doi: 10.1016/j.tcb.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 36.Colley SM, Leedman PJ. Steroid Receptor RNA Activator–A nuclear receptor coregulator with multiple partners: Insights and challenges. Biochimie. 2011;93(11):1966–1972. doi: 10.1016/j.biochi.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 37.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21(6):354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 39.Hung T, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43(7):621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ip JY, Nakagawa S. Long non–coding RNAs in nuclear bodies [published online ahead of print November 10, 2011].Dev Growth Differ . doi:10.1111/j.1440-169X.2011.01303.x. [DOI] [PubMed] [Google Scholar]
- 41.Yang L, et al. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147(4):773–788. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3(5):370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- 43.Tang R-B, et al. Increased level of polymerase III transcribed Alu RNA in hepatocellular carcinoma tissue. Mol Carcinog. 2005;42(2):93–96. doi: 10.1002/mc.20057. [DOI] [PubMed] [Google Scholar]
- 44.Mariner PD, et al. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell. 2008;29(4):499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 45.Yakovchuk P, Goodrich JA, Kugel JF. B2 RNA and Alu RNA repress transcription by disrupting contacts between RNA polymerase II and promoter DNA within assembled complexes. Proc Natl Acad Sci U S A. 2009;106(14):5569–5574. doi: 10.1073/pnas.0810738106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaneko H, et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011;471(7338):325–330. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lambertz I, et al. Monoallelic but not biallelic loss of Dicer1 promotes tumorigenesis in vivo. Cell Death Differ. 2009;17(4):633–641. doi: 10.1038/cdd.2009.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seitz H. Redefining microRNA targets. Curr Biol. 2009;19(10):870–873. doi: 10.1016/j.cub.2009.03.059. [DOI] [PubMed] [Google Scholar]
- 49.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alimonti A, et al. Subtle variations in Pten dose determine cancer susceptibility. Nat Genet. 2010;42(5):454–458. doi: 10.1038/ng.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465(7301):1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karreth FA, et al. In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell. 2011;147(2):382–395. doi: 10.1016/j.cell.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sumazin P, et al. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011;147(2):370–381. doi: 10.1016/j.cell.2011.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cesana M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prensner JR, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29(8):742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Panzitt K, et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132(1):330–342. doi: 10.1053/j.gastro.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 57.Zhao J, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40(6):939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460(7254):479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44(4):667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wan Y, Kertesz M, Spitale RC, Segal E, Chang HY. Understanding the transcriptome through RNA structure. Nat Rev Genet. 2011;12(9):641–655. doi: 10.1038/nrg3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147(7):1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Matouk IJ, et al. The H19 non-coding RNA is essential for human tumor growth. Plos One. 2007;2(9):e845. doi: 10.1371/journal.pone.0000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang Z, et al. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 2011;18(5):1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 64.Ji P, et al. MALAT-1, a novel noncoding RNA, and thymosin beta 4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22(39):8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 65.Augui S, Nora EP, Heard E. Regulation of X-chromosome inactivation by the X-inactivation centre. Nat Rev Genet. 2011;12(6):429–442. doi: 10.1038/nrg2987. [DOI] [PubMed] [Google Scholar]
- 66.Matoba S, et al. RNAi-mediated knockdown of Xist can rescue the impaired postimplantation development of cloned mouse embryos. Proc Natl Acad Sci U S A. 2011;108(51):20621–20626. doi: 10.1073/pnas.1112664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wells KD. More with less Xist. Proc Natl Acad Sci U S A. 2012;109(2):349–350. doi: 10.1073/pnas.1118081109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rando TA, Chang HY. Aging, rejuvenation, and epigenetic reprogramming: Resetting the aging clock. Cell. 2012;148(1–2):46–57. doi: 10.1016/j.cell.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pandey RR, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32(2):232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 70.Chiesa N, et al. The KCNQ1OT1 imprinting control region and non-coding RNA: new properties derived from the study of Beckwith-Wiedemann syndrome and Silver-Russell syndrome cases. Hum Mol Genet. 2012;21(1):10–25. doi: 10.1093/hmg/ddr419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wijnen M, Alders M, Zwaan CM, Wagner A, van den Heuvel-Eibrink MM. KCNQ1OT1 hypomethylation: A Novel disguised genetic predisposition in sporadic pediatric adrenocortical tumors [published online ahead of print December 11, 2011].Pediatr Blood Cancer . doi:10.1002/pbc.23398. [DOI] [PubMed] [Google Scholar]
- 72.Wang X, et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454(7200):126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Diehl JA. Cycling to cancer with cyclin D1. Cancer Biol Ther. 2002;1(3):226–231. doi: 10.4161/cbt.72. [DOI] [PubMed] [Google Scholar]
- 74.Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445(7128):666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 75.Al-Shakfa F, et al. DNA variants in region for noncoding interfering transcript of dihydrofolate reductase gene and outcome in childhood acute lymphoblastic leukemia. Clin Cancer Res. 2009;15(22):6931–6938. doi: 10.1158/1078-0432.CCR-09-0641. [DOI] [PubMed] [Google Scholar]
- 76.Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28(2):195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- 77.Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3(107):ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li L, Feng T, Lian Y, Zhang G, Garen A, Song X. Role of human noncoding RNAs in the control of tumorigenesis. Proc Natl Acad Sci U S A. 2009;106(31):12956–12961. doi: 10.1073/pnas.0906005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Benetatos L, Vartholomatos G, Hatzimichael E. MEG3 imprinted gene contribution in tumorigenesis. Int J Cancer. 2011;129(4):773–779. doi: 10.1002/ijc.26052. [DOI] [PubMed] [Google Scholar]
- 80.Huarte M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Coulson DTR, et al. BACE1 mRNA expression in Alzheimer’s disease postmortem brain tissue. . J Alzheimers Dis. 2010;22(4):1111–1122. doi: 10.3233/JAD-2010-101254. [DOI] [PubMed] [Google Scholar]