Abstract
Aspergillus fumigatus is a model fungal pathogen and a common cause of infection in individuals with the primary immunodeficiency chronic granulomatous disease (CGD). Although primarily considered a deficiency of innate immunity, CGD is also linked to dysfunctional T cell reactivity. Both CD4+ and CD8+ T cells mediate vaccine-induced protection from experimental aspergillosis, but the molecular mechanisms leading to the generation of protective immunity and whether these mechanisms are dysregulated in individuals with CGD have not been determined. Here, we show that activation of either T cell subset in a mouse model of CGD is contingent upon the nature of the fungal vaccine, the involvement of distinct innate receptor signaling pathways, and the mode of antigen routing and presentation in DCs. Aspergillus conidia activated CD8+ T cells upon sorting to the Rab14+ endosomal compartment required for alternative MHC class I presentation. Cross-priming of CD8+ T cells failed to occur in mice with CGD due to defective DC endosomal alkalinization and autophagy. However, long-lasting antifungal protection and disease control were successfully achieved upon vaccination with purified fungal antigens that activated CD4+ T cells through the endosome/lysosome pathway. Our study thus indicates that distinct intracellular pathways are exploited for the priming of CD4+ and CD8+ T cells to A. fumigatus and suggests that CD4+ T cell vaccination may be able to overcome defective antifungal CD8+ T cell memory in individuals with CGD.
Introduction
Aspergillus fumigatus is a model fungal pathogen and a common cause of severe infections and diseases.
Humans inhale hundreds of conidia per day without adverse consequences (1), except for a small minority of persons who may develop life-threatening forms of aspergillosis. Despite the lack of evidence for susceptibility to aspergillosis in patients with congenital absence of T cells (2), CD4+ and CD8+ T cells are present in the human T cell repertoire to the fungus (3–5), and adoptive transfer of A. fumigatus–specific CD4+ T cells conferred protection against invasive aspergillosis (5, 6). As with other fungi (7–10), antigen-specific CD4+ and CD8+ T cell memory to A. fumigatus has been demonstrated in experimental aspergillosis (11). However, the molecular mechanisms leading to T cell memory are largely unknown.
It is becoming increasingly apparent that the optimization of vaccines depends crucially on fuller understanding of the mechanisms and processes that give rise to immunological memory, including adjuvant formulation, innate immunity, and antigen presentation by DCs (12). Although DCs process and present antigens efficiently irrespective of the route of antigen capture (13), the nature of the protein, perhaps through its mechanism of uptake, is important in determining the access to the endocytic pathway, the intersection of which with MHC-containing subcellular compartments determines crucially the course and types of antigen presentation (14). Owing to the ability of DCs to initiate and regulate fungus-specific adaptive immune responses and to improve fungal vaccines (15), defining fungal uptake and presentation by DCs may help in the understanding of immunological memory to Aspergillus and its dysregulation in diseases. The above consideration may be particularly relevant in chronic granulomatous disease (CGD), a primary immunodeficiency caused by the inherited disorder of the phagocyte NADPH oxidase (NOX2) (16, 17) that is associated with infections by Aspergillus spp (18). Although it is considered an “innate immunity” disease, increasing evidence links CGD with a higher risk of developing autoimmunity (19). It has indeed been shown that NOX2 activity participates in the regulation of the phagosomal and endosomal pH in DCs and is required for efficient antigen cross-presentation (20, 21). Thus, the defect in antigen presentation described in patients with CGD may reveal an unsuspected role for T cell and adaptive immunity in the pathogenesis of aspergillosis in this immunodeficiency.
In this study we have assessed the antigen presentation pathways in DCs underlying the generation of CD4+ or CD8+ T cell responses to the fungus in NADPH deficiency. The activation of either T cell subset was contingent upon the nature of the fungal vaccine, the involvement of distinct innate receptor signaling pathways, and the mode of antigen routing and presentation in DCs. MHC class I–restricted CD8+ memory T cells failed to be generated in NADPH deficiency, due to defective DC endosomal alkalinization and autophagy. However, long-lasting antifungal protection could be successfully achieved upon vaccination with purified fungal antigens activating CD4+ T cells through the endosome/lysosome pathway. Thus, distinct intracellular pathways are exploited for CD4+ or CD8+ T cell priming to the fungus such that CD4+ T cell vaccination may overcome defective CD8+ T cell memory to A. fumigatus in CGD.
Results
CD8+ T cell vaccination against A.
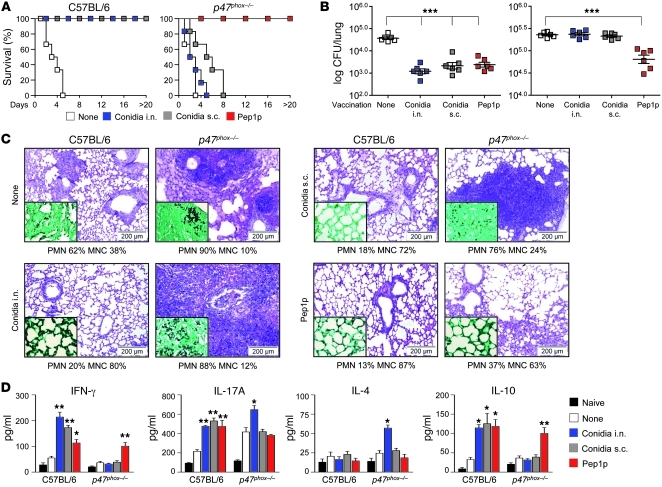
fumigatus is defective in p47phox–/– mice. We used well-established experimental models of vaccine-induced resistance against aspergillosis (22) to assess the ability of p47phox–/– mice (a model of CGD) to develop memory responses to A. fumigatus. Mice were intranasally preexposed to sublethal live A. fumigatus conidia or the protective (22) recombinant fungal aspartic protease (Pep1p) with CpG as adjuvant, and assessed for resistance to subsequent infection in terms of survival, fungal growth, and lung histopathology, as well as pattern of cytokine production. In contrast to C57BL/6 mice, p47phox–/– mice failed to develop vaccine-induced resistance to conidia, as revealed by the inability to survive the infection (Figure 1A), restrict fungal growth (Figure 1, B and C), control lung inflammation (Figure 1C), and activate antigen-specific T cells for IFN-γ and IL-10 production (Figure 1D). Failure to acquire vaccine-induced resistance was also observed upon subcutaneous exposure to viable conidia (Figure 1, A–D) or intranasal exposure to inactivated conidia (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI60862DS1); these findings point to a generalized T cell defect in response to conidia in p47phox–/– mice that was not secondary to the failure to clear the primary infection in the lung. Surprisingly, and similar to C57BL/6 mice, they developed full resistance upon Pep1p vaccination, which was associated with IFN-γ/IL-10 production by antigen-specific T cells (Figures 1, A–D). Pep1p vaccination was also observed with Pep1p coated on microparticles of the approximate size of conidia (Supplemental Figure 2), a finding that discloses the relative contribution of CpG to vaccination. Indeed, despite the ability of CpG alone to slightly increase resistance to reinfection in p47phox–/– mice (Supplemental Figure 3, A and B), this increased resistance was not as long-lasting as that induced by Pep1p, and was no longer apparent at 60 days after vaccination (Supplemental Figure 3, C and D). Similar to C57BL/6 mice (22), p47phox–/– mice also developed full resistance against two other protective Aspergillus recombinant antigens, the cell wall–associated transglycosidases (Gel1p and Crf1p), and not to the non-protective metalloprotease (Mep1p) (Supplemental Figure 4). Thus, the ability to respond to purified fungal antigens is fully retained in CGD mice, irrespective of the protease activity of the fungal antigen.
Figure 1. p47phox–/– mice fail to develop vaccine-induced resistance to A. fumigatus conidia.
Mice (6/group) received A. fumigatus conidia i.n. or s.c 14 days before reinfection with Aspergillus conidia i.n. Pep1p was given i.n. with CpG 14, 7, and 3 days before reinfection. Mice were given cyclophosphamide a day before reinfection. Vaccine-induced resistance was assessed in terms of (A) survival (%); (B) fungal growth (log10 CFU ± SEM); (C) lung histology (PAS staining and Gomori staining in the insets, visualized, respectively, with an original magnification of ×200 and ×1,000); and (D) cytokine production (ELISA) by purified lung T cells cultured in vitro with conidia- or Pep1p-pulsed DCs from the corresponding naive mice. In C, values represent percentages of polymorphonuclear (PMN) or mononuclear (MNC) cells in the BAL. Assays were done at 3 days after infection. Data are pooled or representative (histology) of 3 independent experiments.*P < 0.05, **P < 0.01, ***P < 0.001, vaccinated versus unvaccinated (None) mice. Scale bars: 200 μm.
Murine studies have shown that both CD8+ and CD4+ T cells mediate memory responses to fungi via the production of effector cytokines, such as IFN-γ, and, in the case of CD8+ T cells, through cytotoxic activity against fungus-laden cells (7–11). The requirement for IFN-γ in vaccine-mediated resistance was confirmed by vaccination of Ifng–/– or Il17ra–/– mice with Aspergillus conidia or Pep1p. Both vaccines retained the vaccinating potential in Il17ra–/– mice, albeit to a lesser degree for conidia, as compared with wild-type mice, but both failed to induce resistance in Ifng–/– mice (Supplemental Figure 5, A–C). Increased conidiocidal activity (Supplemental Figure 5D), more than induction of protective defensins (Supplemental Figure 5E), was associated with vaccine-induced resistance. Thus, as in humans (23), Th1, and to a lesser extent Th17, cells contribute to vaccine-induced resistance in murine aspergillosis.
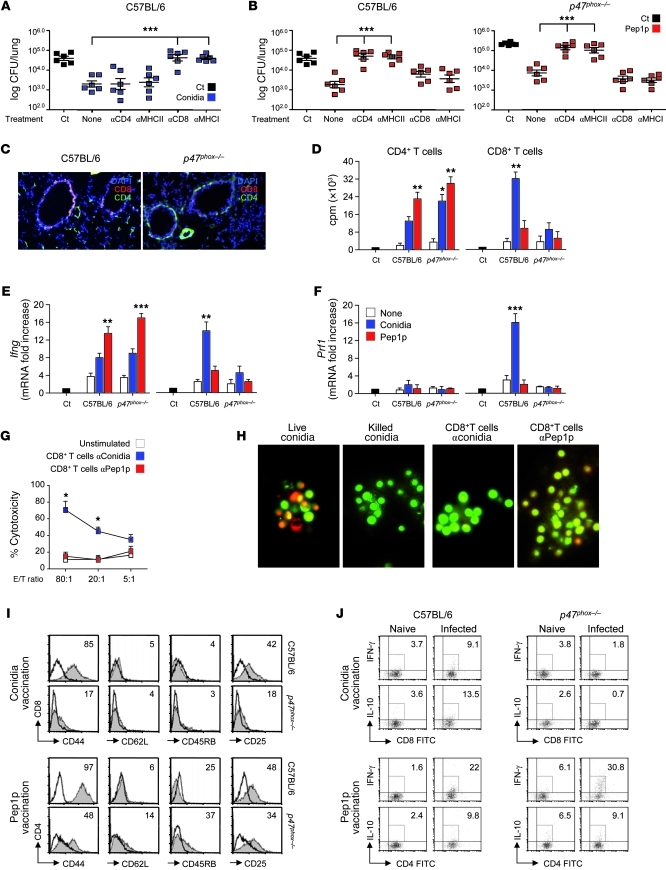
In terms of CD8+ or CD4+ T cell activation, conidia-induced resistance was abrogated in C57BL/6 mice upon blocking of class I antigens or CD8 (Figure 2A) and in CD8-deficient mice (Supplemental Figure 6), whereas Pep1p-induced resistance was abrogated in both types of mice upon blocking of class II antigens or CD4 (Figure 2B) and in CD4-deficient mice (Supplemental Figure 6). Actually, CD4+, more than CD8+, T cells were detected in the lungs of CGD mice in infection (Figure 2C), and CD4+ T cells from these mice responded to Pep1p stimulation in vitro in terms of proliferation (Figure 2D) and Ifng gene expression (Figure 2E) to the same or even a greater extent than C57BL/6 mice. In contrast, CD8+ T cells were activated in response to conidia in C57BL/6 mice only, as indicated by proliferation, Ifng and Prf1 expression (Figure 2, D–F), and cytotoxic activity against fungus-pulsed DCs (Figure 2G) or the fungus itself (Figure 2H). Consistent with the ability of antigens delivered in particulate form to induce robust CD8+ T cell activation (24, 25), CD8+ T cells were also induced by vaccination with Pep1p coated on microparticles (Supplemental Figure 2). Thus, despite the ability of CpG to prime CD8+ T cell immunity to “split” vaccines (26), soluble fungal antigens did not elicit class I MHC–restricted CD8+ T cell responses, unless delivered in particulate form. Corroborating these findings, phenotypic and functional analysis of ex vivo cells revealed that CD4+ T cells from Pep1p-vaccinated mice, both wild-type and CGD, and CD8+ T cells from conidia-vaccinated wild-type mice were CD44hiCD62LloCD45RBloCD25lo, consistent with an effector memory phenotype (Figure 2I), and stained positive for IFN-γ and IL-10 (Figure 2J). Because p47phox–/– mice did not overtly demonstrate an increased susceptibility to MCMV infection (Supplemental Figure 7), MHC class I–restricted CD8+ T cell activation in response to conidia was selectively defective, whereas MHC class II–restricted CD4+ T cell responses to soluble fungal antigens was apparently facilitated in these mice. In fact, consistent with published findings (27), a robust Aspergillus-specific humoral response was observed in Pep1p-vaccinated p47phox–/– mice (data not shown).
Figure 2. p47phox–/– mice fail to develop MHC class I–restricted CD8+ T cell responses to Aspergillus conidia.
Mice (6/group) received conidia or Pep1p plus CpG as described for Figure 1 and were concomitantly treated with the indicated antibodies or an isotype-matched control antibody (None). Fungal growth (log10 CFU ± SEM) in conidia-vaccinated (A) or Pep1p-vaccinated (B) mice at 3 days after reinfection. Control (Ct), infected, unvaccinated mice. Pooled data from 3 experiments are shown. (C) Lung immunohistochemistry 3 days after reinfection. Cell surface markers were Alexa Fluor 488–anti-CD4 and Alexa Fluor 647–anti-CD8 antibody. Cell nuclei were stained with DAPI (blue). Representative pictures (of 3 experiments) were taken with an original magnification of ×200. (D) Proliferation of CD4+ and CD8+ T cells purified from lungs 1 week after a primary i.n. infection. DNA synthesis was measured by 3H-thymidine uptake after 72 hours coculture with conidia- or Pep1p-pulsed DCs from the corresponding naive mice. Ct, T cells alone. Relative expression of (E) Ifng and (F) Prf1 by RT-PCR in CD4+ and CD8+ T cells exposed to conidia- or Pep1p-pulsed DCs for 24 hours. (G) Cytolytic activity of CD8+ T cells, obtained as in D, against conidia-pulsed DCs at different E/T ratios. Shown is the percentage of specific cytotoxic activity determined by a standard 4-hour 51Cr-release assay. (H) Conidiocidal activity of culture supernatants from CD8+ T cells exposed as in D (visualized with an original magnification of ×400). Activation/memory marker expression (I) and intracellular cytokine staining (J) by lung CD8+ and CD4+ T cells purified from conidia- or Pep1p-vaccinated mice, respectively, 3 days after reinfection. Histograms were generated from pooled samples of 6 mice/group. Representative histograms from a single experiment are shown. Values are the percentages of positive cells. *P < 0.05, **P < 0.01, ***P < 0.001 treated versus untreated mice (A and B) and pulsed DC–stimulated versus unstimulated cells (None) (D–G).
CD4+ or CD8+ T cell vaccination requires distinct TLR signaling.
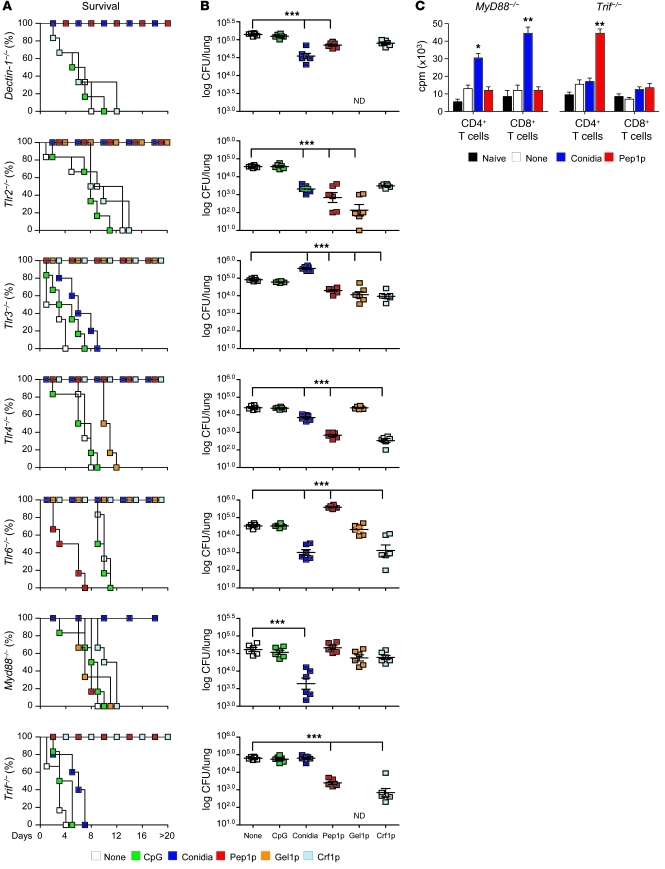
TLR signaling triggers a variety of responses that direct and enhance antigen presentation (28, 29). Lower levels of MHC class II antigen presentation have indeed been observed in the absence of myeloid differentiation primary response gene 88 (MyD88) (28) and reduced cross-presentation in the absence of TIR domain–containing adapter-inducing interferon–β (TRIF) (30). To assess the contribution of the MyD88 and TRIF pathways and upstream TLRs in the generation of vaccine-induced resistance, we assessed the vaccinating potential of conidia or Pep1p in Myd88–/–, Trif–/–, selected TLR-deficient, or Dectin-1–deficient mice. We found that the vaccinating capacity of live conidia was lost in Tlr3–/– and Trif–/– mice, whereas the vaccinating capacity of Pep1p was lost in Tlr6–/– and Myd88–/– mice (Figure 3, A and B), and this was paralleled by a failure to induce the proliferation of CD8+ T cells by conidia in Trif–/– mice and of CD4+ T cells by Pep1p in Myd88–/– mice (Figure 3C). The vaccinating potential of the two other protective fungal antigens (Crf1p and Gel1p) was also dependent on MyD88 and Dectin-1/TLR2 (Crf1p) or TLR4/TLR6 (Gel1p) (Figure 3, A and B). We concluded that the TLR3/TRIF pathway is required for CD8-mediated resistance to live conidia, whereas the MyD88 pathway essentially contributes to CD4-mediated resistance to purified fungal antigens with the participation of different TLRs. As both TLR3 expression (Supplemental Figure 8A) and responsiveness to TLR3 (Supplemental Figure 8B) were preserved in CGD mice, mechanisms other than TLR expression are apparently responsible for defective CD8+ T cell memory in CGD.
Figure 3. CD4+ or CD8+ T cell vaccination to A. fumigatus requires distinct TLR signaling.
Dectin-1, TLR-, MyD88-, or TRIF-deficient mice were administered A. fumigatus conidia or the protective recombinant fungal antigens (Pep1p, Gel1p, or Crf1p) plus CpG and assessed for resistance to reinfection in terms of (A) survival (%), (B) fungal growth (log10 CFU ± SEM), and (C) proliferation of CD4+ or CD8+ T cells, purified from Pep1p- or conidia-vaccinated mice, respectively, at 3 days after infection, and cultured in vitro with conidia- or Pep1p-pulsed DCs from the corresponding naive mice. DNA synthesis was measured by 3H-thymidine uptake after 72 hours coculture. *P < 0.05, **P < 0.01, ***P < 0.001, vaccinated versus unvaccinated (None) mice. ND, not done. Data are representative of 3 experiments.
CD4+ and CD8+ T cells are activated through distinct intracellular antigen presentation pathways.
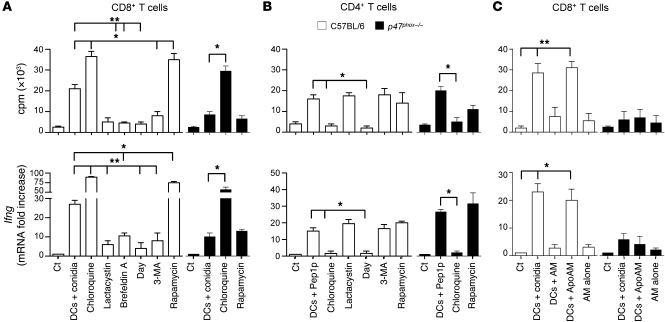
In the classical antigen presentation pathways, endogenous antigens are processed by the proteasome and loaded on MHC class I molecules in the ER to activate cytotoxic CD8+ T cells. In contrast, exogenous antigens taken up by endocytosis or phagocytosis are degraded and loaded on MHC class II in lysosome-derived organelles for CD4+ T cell activation (31, 32). However, evidence for alternative cross-presentation of exogenous antigens on MHC class I for CD8+ T cells has also emerged, such as alternate vacuolar MHC class I processing (31), a pathway for particulate antigens with no known mechanism for cytosolic penetration (24). To define which of the above pathways were activated in CD4+ or CD8+ T cell vaccination, we assessed CD8+ or CD4+ T cell priming by lung DCs pulsed with conidia or Pep1p, respectively, in conditions of efficient (chloroquine) or inhibited (diphenyleneiodonium [DPI]) endosomal alkalinization, as alkalinization of the phagosomal and endosomal pH in DCs prevented the activation of lysosomal proteases, thus rescuing internalized particulate antigens from rapid degradation and allowing their loading onto MHC class I (20, 21, 32). In addition, we used brefeldin A (33) or lactacystin (34) to inhibit the alternate vacuolar or the classical cytosolic MHC class I pathway, respectively, and 3-methyladenine (3-MA) or rapamycin to inhibit or promote, respectively, (35, 36), autophagy, a highly conserved cellular process of self-degradation, involved in MHC class I and class II antigen presentation as well as in cross-presentation (37–39). We found that CD8+ T cell priming (proliferation and Ifng gene expression) was inhibited by DPI, lactacystin, and 3-MA; was dependent on brefeldin A; and was promoted by chloroquine and rapamycin (Figure 4A). Experiments with p47phox–/– DCs showed that treatment with chloroquine, but not rapamycin, restored CD8+ T cell activation (Figure 4A). These findings indicate that handling of conidia by DCs for efficient CD8+ T cell activation occurs through the cytosolic and vacuolar MHC class I pathways and is facilitated by endosomal alkalinization and autophagy. In contrast, chloroquine but not 3-MA or rapamycin inhibited Pep1p-induced CD4+ T cell activation, indicating a requirement for endosomal acidification but not autophagy in class II–restricted CD4+ T cell activation (Figure 4B). Thus, CD4+ and CD8+ T cells appeared to be activated through distinct intracellular antigen presentation pathways. While the MHC class II pathway leading to CD4+ T cell activation operated normally in CGD, the MHC class I pathway to phagocytosed conidia was instead impaired, demonstrating that our understanding of antigen handling based on model antigens extends to pathogens (21).
Figure 4. CD4+ and CD8+ T cells are activated through distinct intracellular antigen presentation pathways.
CD8+ or CD4+ T cells were purified from lungs of C57BL/6 or p47phox–/– mice a week after the intranasal infection and exposed to conidia-pulsed (A) or Pep1p-pulsed (B) DCs purified from lungs of the corresponding naive mice. Prior to the 2-hour pulsing with conidia or Pep1p, DCs were exposed to the indicated antigen presentation pathway inhibitors for 120 minutes. (C) CD8+ T cells, purified as in A, were exposed to DCs and/or intact or apoptotic (Apo) alveolar macrophages (AM), purified from lungs of the corresponding naive mice. AMs were pulsed to live conidia before the induction of apoptosis with LPS plus ATP. Cells were assessed for proliferation and Ifng expression by RT-PCR, 72 hours after the coculture. DNA synthesis was measured by 3H-thymidine uptake. Data are from 4 independent experiments.*P < 0.05, **P < 0.01, inhibitors versus no inhibitors; and DC-exposed versus T cells alone (Ct).
Given the ability of macrophages to cross-present phagocytosed antigens (40) and to promote cross-presentation in DCs via transfer of fungal antigens upon apoptosis (41), we assessed both functions in lung macrophages from wild-type and CGD mice. While neither CD8+ T cell proliferation nor Ifng gene expression was observed with macrophages from either type of mice (Figure 4C), DCs efficiently activated CD8+ T cells upon coculture with apoptotic macrophages that had phagocytosed live conidia but failed to do so with non-apoptotic macrophages (Figure 4C). Because A. fumigatus conidia inhibit apoptosis of alveolar macrophages via caspase-3 inhibition (42) and we failed to detect macrophage apoptosis after phagocytosis of conidia (data not shown), it is unlikely that DC ingestion of apoptotic macrophages actually contributes to cross-presentation in vivo.
Distinct intracellular routing of conidia and soluble fungal antigens in DCs.
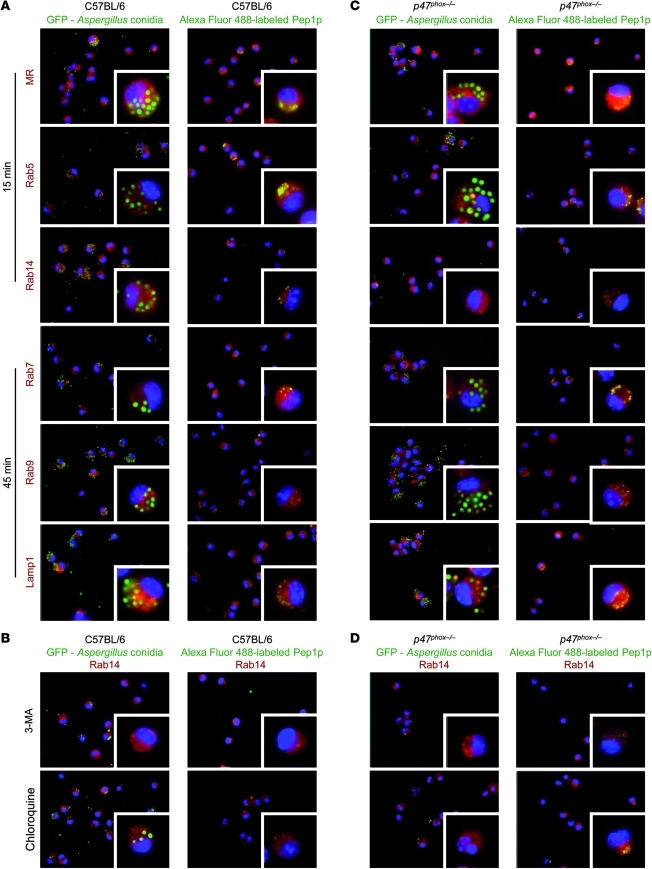
To find out whether distinct intracellular routings for live conidia and soluble antigens in DCs could explain the activation of either T cell type, we followed the intracellular location of GFP-conidia or Alexa Fluor 488–labeled Pep1p by determining the co-localization and quantifying the degree of overlap (Supplemental Table 1) with members of the Rab small GTPase family, known to be specifically localized to different intracellular organelles and to regulate endocytic traffic (43). We used Rab5 for its association with the clathrin-dependent endocytic pathway and early endosomes (44); Rab7 to detect early-to-late endosomal maturation (45); Rab9 because it stimulates late endosome to trans-Golgi network (TGN), a major secretory pathway (46); and Rab14 because it reduces routing of antigens from early endosomes to the acidic lysosomal environment, thus limiting antigen degradation and favoring cross-presentation in DCs (47). We also assessed co-localization with the mannose receptor (MR) that targets the mildly acidic stable early endosomal compartment where presentation on MHC I molecules and Th1 polarization occur (32, 48); and with lysosomal-associated membrane protein 1 (Lamp1), a transmembrane glycoprotein that is localized primarily in late endosomes and lysosomes (43). We found that Aspergillus conidia colocalized with MR, Rab5, and Rab14 at 15 minutes after phagocytosis (Figure 5A), being evident as early as after 5 minutes (data not shown). Conidia were also found to be associated with Rab7, Rab9, and Lamp1 at 45 minutes (Figure 5A) and, in the case of Lamp1, also at 120 minutes (data not shown). Thus, phagocytosed conidia are routed to both the late endosome/lysosome and the Rab14+ compartments. Of great interest, the routing of conidia to the Rab14+ compartment (Figure 5B) but not to Lamp1 (data not shown) was unaffected by chloroquine but was greatly impaired upon blocking of autophagy with 3-MA (Figure 5B), a finding suggesting the participation of autophagy in the escape of conidia from the endosome/lysosome compartment to the Rab14+ compartment. At variance with conidia, the Alexa Fluor 488–labeled Pep1p colocalized sequentially with MR, Rab5, Rab7, Rab9, and Lamp1, with minimal or no co-localization with Rab14 (Figure 5A). Thus, the routing to the late endosome/lysosome compartment is apparently required for adequate processing and presentation of soluble fungal antigens for CD4+ T cell activation. This requirement was further confirmed by analyzing the co-localization of the non-protective fungal antigen Mep1p. Similar to Pep1p, Mep1p colocalized with MR and Rab5 but not Rab14 (Supplemental Figure 9). At variance with Pep1p, it rapidly (at 15 minutes) and maximally (at 45 minutes, data not shown) colocalized with Rab9 and weakly with Lamp1 (Supplemental Figure 9), a finding suggesting that transporting antigens from late endosomes to the TGN prevents adequate processing and presentation of antigens for CD4+ T cell activation. These results indicate a different routing for live conidia and soluble fungal antigens in DCs. Like soluble fungal antigens, phagocytosed conidia are routed to the endosome/lysosome compartment, from where conidia are diverted to the Rab14+ compartment through autophagy.
Figure 5. Distinct intracellular routing of Aspergillus conidia and soluble fungal antigens in DCs and its subversion in the absence of NADPH.
Immunofluorescence imaging of purified DCs from lungs of C57BL/6 (A) or p47phox–/– (C) mice after in vitro exposure to GFP-conidia or Alexa Fluor 488–Pep1p at 37°C for 2 hours and chasing for 15 and 45 minutes. (B and D) DCs were preincubated with 5 mM 3-MA or 5 μg/ml chloroquine for 30 minutes prior to exposure to GFP-conidia or to Alexa Fluor 488–Pep1p. Formaldehyde-fixed DCs were incubated with primary antibodies against MR, Rab5, Rab7, Rab9, Rab14, and Lamp1, followed by secondary anti-rabbit IgG–TRITC antibody. Nuclei were counterstained with DAPI. Images were acquired using a fluorescence microscope (BX51) with an original magnification of ×1,000 and analySIS image processing software. Shown are merged images of DCs (a single cell is magnified in the inset visualized with an original magnification of ×1,000) pulsed with GFP-conidia or fungal antigens (green) and red-stained for each endosomal compartment. Shown are representative data from 4 independent experiments. For quantitative analysis of co-localization, see Supplemental Table 1.
The intracellular routing of conidia in DCs is altered in NOX2 deficiency.
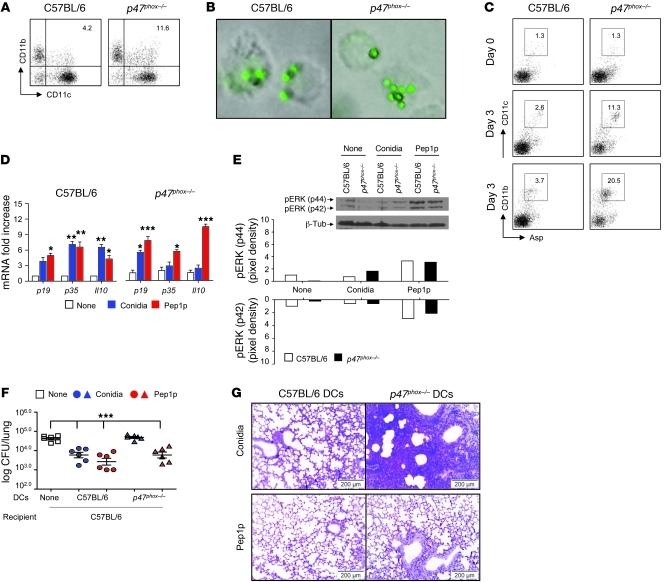
We next assessed the routing (co-localization and its quantification, Supplemental Table 1) of GFP-conidia or Alexa Fluor 488–labeled Pep1p in p47phox–/– DCs. Similar to wild-type DCs, Aspergillus conidia colocalized with MR and Rab5 at 15 minutes after phagocytosis (Figure 5C) and with Rab7/Rab9/Lamp1at 45 minutes (Figure 5C). In contrast, co-localization with Rab14 was observed neither at 15 minutes (Figure 5C) nor at later time points (data not shown). Thus, the rerouting of conidia away from the lysosomal compartment is defective in the absence of NOX2. Similar to what was observed in wild-type DCs, Pep1p colocalized sequentially with MR, Rab5, Rab7, Rab9, and Lamp1, with minimal or no co-localization with Rab14 (Figure 5C). However, chloroquine but not 3-MA greatly promoted the co-localization of Pep1p with Rab14 (Figure 5D), a finding suggesting that endosomal alkalinization, while rescuing peptides from degradation, may favor rerouting of soluble antigens to the Rab14+ compartment. These results indicate that live conidia failed to be sorted in the Rab14+ compartment in the absence of NOX2-dependent autophagy, while endosomal alkalinization apparently favors the Rab14+ sorting of soluble fungal antigens. Further studies confirmed that CD11bloCD103+ DCs or CD8α+ DCs, known to mediate CD8+ T cell cross-priming (49, 50), were not defective in the lungs of CGD mice (Supplemental Figure 10), whereas a population of CD11chiCD11bhi myeloid DCs, known to prime CD4+ T cell responses to A. fumigatus (51), was expanded in these mice (Figure 6A), as already described (52). p47phox–/– DCs phagocytosed conidia in vitro (Figure 6B) and in vivo (Figure 6C) and responded to conidia in terms of Il23p19 expression (Figure 6D) and ERK phosphorylation (Figure 6E) but not in terms of Il12p35 and Il10 expression (Figure 6D), a finding indicating that handling of conidia is somehow altered in NADPH deficiency. Upon Pep1p stimulation, in contrast to conidia stimulation, p47phox–/– DCs responded to the same or a greater extent as C57BL/6 DCs (Figure 6, D and E) and, pulsed with Pep1p, were fully able to confer vaccine-induced resistance in vivo upon adoptive transfer into C57BL/6 mice with aspergillosis (Figure 6, F and G). Thus, the intracellular routing for live conidia and soluble antigens in DCs is apparently subverted in conditions of NADPH deficiency.
Figure 6. CD11b+ p47phox–/– DCs efficiently present fungal antigens.
(A and B) Flow cytometry of phagocytosis of live GFP-expressing conidia by lung DCs purified from uninfected mice. Values are percentages of positive cells on T and B cell–depleted lung cells. Phagocytosis was quantified via phase contrast and fluorescence microscopy at ×1,000 magnification (shown are representative microscopy images of 2 independent experiments). (C) Mice were infected i.n. with GFP-expressing A. fumigatus conidia, and the numbers of GFP+CD11c+ cells were assessed by flow cytometry at 3 days after infection. Values are percentages of positive cells on gated CD11c+ cells. DCs purified from lung of naive mice were unpulsed (None) or pulsed with live Aspergillus conidia or Pep1p for (D) 24 hours prior to being assessed for cytokine gene expression by RT-PCR and (E) 2 hours before the assessment of pERK44/42 phosphorylation by immunoblot analysis. Shown is a representative Western blot of 2 independent experiments and corresponding pixel density ratio normalized against β-tubulin (β-tub). (F and G) Adoptive transfer of C57BL/6 or p47phox–/– lung DCs pulsed with conidia or Pep1p into C57BL/6 mice. DCs were adoptively transferred by i.p. injection twice, a week apart, before the i.n. infection. Fungal growth in the lungs (F, log10 CFU ± SEM, representative of at least 3 independent experiments) and histology (G, PAS staining) were determined 3 days after infection. *P < 0.05, **P < 0.01, ***P < 0.001, pulsed versus unpulsed DCs (D) and mice receiving or not (None) DCs (F). Scale bars: 200 μm.
Autophagy restores defective cross-presentation of conidia in p47phox–/– mice.
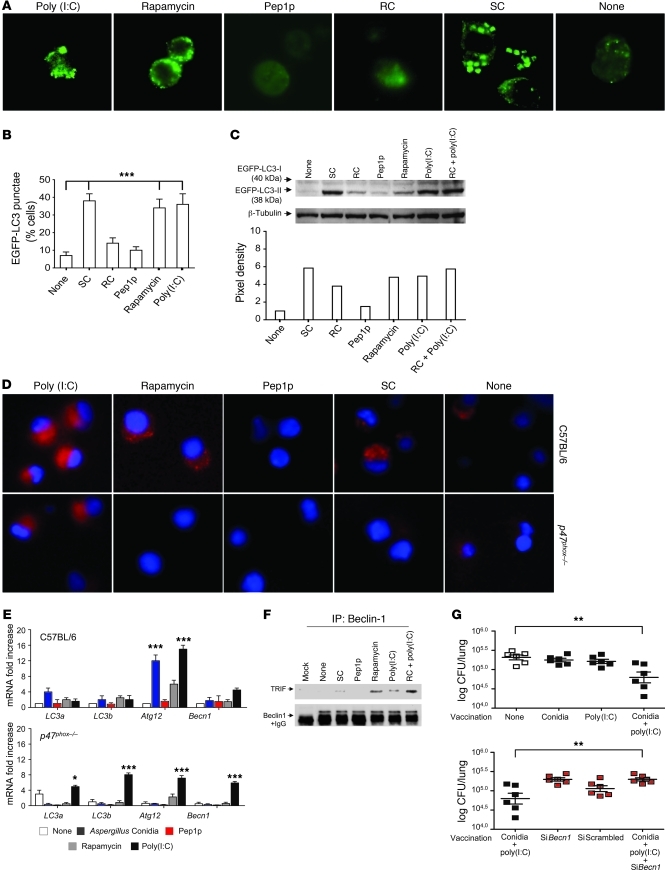
To assess whether restoring defective autophagy in CGD (53) could result in CD8+ T cell–mediated resistance in vivo, we first monitored the occurrence of autophagy in response to the fungus and/or fungal antigens. We transiently transfected RAW 264.7 cells with the EGFP-LC3 plasmid and exposed them to live resting or swollen conidia, to Pep1p, or to known activators of autophagy, such as rapamycin and poly(I:C). As with rapamycin and poly(I:C), autophagy was induced by swollen but not by resting conidia or Pep1p, as indicated by the increased number of cells with punctate dots containing EGFP-LC3 as opposed to the diffuse localization of the majority of EGFP-LC3 in untreated cells (Figure 7, A and B) and the increased ratio of LC3-II to LC3-I by immunoblotting (Figure 7C). Autophagy was promoted by resting conidia in the presence of poly(I:C) (Figure 7C). Similar results were obtained in purified lung DCs from C57BL/6 mice, as revealed by LC3 immunofluorescence (Figure 7D); mRNA expression levels of the autophagy genes LC3a, LC3b, Atg12, and Bcn1 (Figure 7E); and the association of Beclin-1, an essential autophagy effector, with TRIF (ref. 54 and Figure 7F). Autophagy was not observed in p47phox–/– DCs in response to the different stimuli, including rapamycin, with the exception of poly(I:C) (Figure 7, D and E), a finding suggesting that mTOR- and NADPH-independent autophagy may still be activated in CGD. The induction of autophagy to conidia via TLR3 would suggest that the exploitation of TLR3 signaling could restore CD8+ T cell memory in CGD mice. This proved to be the case, as the administration of conidia with poly(I:C) provided resistance to reinfection, a process partially antagonized by blocking autophagy (Figure 7G).
Figure 7. Autophagy restores defective cross-presentation of conidia in p47phox–/– mice.
(A) Fluorescence images of EGFP-LC3 transiently transfected RAW 264.7 cells exposed to A. fumigatus swollen (SC) or resting (RC) conidia, Pep1p, rapamycin, or poly(I:C) for 4 hours. Original magnification, ×1,000. (B) Number of RAW 264.7 cells with EGFP-LC3 punctae (mean ± SEM, determined by fluorescence microscopy) (n = 2). (C) Cell lysates of EGFP-LC3–transfected RAW 264.7 cells were probed with anti-GFP antibody followed by IgG-HRP secondary antibody. Normalization was performed on mouse β-tubulin. Quantification was obtained by densitometry image analysis using Image Lab 3.1.1 software. (D) Autophagy on purified lung DCs stimulated as above and incubated with anti-LC3 antibody followed by PE secondary antibody. Representative images (original magnification, ×400) are shown. DAPI was used to detect nuclei. (E) Autophagy gene expression by RT-PCR in lung DCs stimulated as above. None, unstimulated cells. Data are representative of 2 experiments. (F) Cell lysates from EGFP-LC3–transfected RAW 264.7 cells stimulated as in A were subjected to immunoprecipitation with antibody to Beclin-1. Immunoprecipitates were probed with polyclonal antibody to TRIF. Data are representative of 2 experiments. (G) p47phox–/– mice were vaccinated with Aspergillus conidia and poly(I:C) under the condition of Becn1 inhibition by siRNA administration. Western blotting of lung cells confirmed that Beclin-1 protein decreases upon siBecn1 administration. Fungal growth (log10 CFU ± SEM) in the lungs was assessed 3 days after infection. Data are representative of 2 independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, conidia + poly(I:C)–vaccinated versus unvaccinated (None) mice and with and without siBecn1.
Discussion
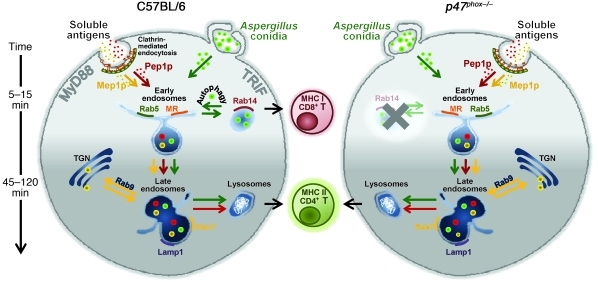
This study reveals that CD4+ and CD8+ T cell activation in vaccine-induced resistance to A. fumigatus is contingent upon the nature of the fungal vaccine, the involvement of distinct innate receptor signaling pathways, and the mode of antigen routing in DCs (Figure 8). Vaccine-induced resistance required IFN-γ, a cytokine with effector and immunoregulatory activity against fungi (15), and, partially, IL-17RA signaling, a finding pointing to a degree of similarity among respiratory fungal pathogens (55). MHC class II–restricted CD4+ Th1 cells provided long-lasting antifungal protection and disease control in CGD mice, in which MHC class I–restricted CD8+ T cells were not activated due to defective autophagy and endosomal alkalinization. Thus, understanding memory at basic levels may lead to a vaccination approach in CGD patients. As in CGD mice, CD4-dependent but not CD8-dependent memory responses were observed in BALB/c mice (data not shown), a finding indicating that the activation of either one of antifungal memory response is also contingent upon the host’s genetic background.
Figure 8. Distinct pathways of intracellular antigen routing in CD4+ and CD8+ T cell vaccination to Aspergillus fumigatus.
The activation of CD4+ and CD8+ T cells is contingent upon the nature of the fungal vaccine, the involvement of distinct innate receptor signaling pathways, and the mode of antigen routing and presentation in DCs. Specifically, both soluble fungal antigens and phagocytosed conidia are routed to the endosome/lysosome compartment that is apparently required for adequate processing and presentation of antigens for CD4+ T cell activation. At variance with soluble antigens, conidia are diverted from the early endosomes to the Rab14+ compartment, required for the alternative MHC class I presentation, through autophagy. MHC class I–restricted CD8+, but not MHC class II–restricted CD4+, memory T cells failed to be generated in mice with CGD due to defective DC endosomal alkalinization and autophagy, which impeded sorting to the Rab14+ compartment. Note that the non-protective fungal antigen Mep1p colocalized rapidly with Rab9 and weakly with Lamp1, a finding suggesting that transporting antigens from late endosome to the TGN prevents adequate processing and presentation of antigens for CD4+ T cell activation.
Activation of CD4+ or CD8+ T cells occurred through distinct antigen uptake and presentation pathways. Purified fungal proteins were apparently routed to the endosome/lysosome-dependent MHC class II presentation pathway via MyD88 and the involvement of distinct upstream TLRs known to enhance the uptake of phagocytosed but also soluble antigens and thus potentiate antigen presentation (28). Further confirming the routing toward the MHC class II presentation pathway, treatment with nocodazole, which interferes with microtubule-dependent motility of early endosomes toward maturing endosomes, also blocked CD4+ T cell activation to fungal antigens (data not shown). Thus, similar to pinocytosed and scavenger receptor endocytosed antigens (14), fungal peptides are targeted toward a common pool of endosomes, which mature toward late endosomes and fuse with lysosomes, in which peptides for MHC class II–restricted presentation can be generated. Although primarily expressed on the plasma membrane, TLR2, TLR4, and TLR6 could also recycle through early endosomes; target and be activated by (56) the Rab family; and control the phagosomal pH (28). Moreover, intracellular TLRs, to signal efficiently, need to be proteolytically activated (29). Thus, it is likely that TLR trafficking to the diverse organelles may explain the involvement of distinct TLRs in the presentation of the different fungal antigens. However, precisely how this occurs is not known.
En route to lysosomes, purified fungal antigens also targeted to the mildly acidic stable early endosomal compartment, where the uptake by MR leads to presentation on MHC class I molecules and Th1 polarization (32, 48). Mannosylated fungal antigens (both N-linked and O-linked mannosylated) exhibited increased immunogenicity compared with proteins lacking mannosylation, were taken up by mannose-specific C-type lectin receptors, and colocalized in MHC class II+ compartments (57). We found that deglycosylated Pep1p no longer acted as a fungal vaccine capable of activating CD4-dependent memory to the fungus (data not shown), a finding confirming the importance of MR in the rapid internalization and concentration of a variety of glycosylated fungal antigens (57).
In contrast to soluble antigens, live conidia or soluble antigens delivered in a particulate form activated CD8-dependent memory through the TLR3/TRIF-dependent pathway. DC subsets are known to be capable of exporting internalized antigens from the endocytic compartment to the cytosol (58). In this regard, Aspergillus-derived antigens may be exported from endocytic compartment to cytosol for conventional MHC class I presentation, as CD8+ T cell activation was dependent on proteasomal degradation. However, the inhibition by brefeldin A, known to inhibit the alternate vacuolar MHC class I processing (33), suggests that Aspergillus conidia may exploit multiple class I presentation pathways for CD8+ T cell activation. Cross-priming itself involves a number of distinct mechanisms. How Aspergillus antigens are delivered to the cytosol or to the ER-resident, vacuolar-associated MHC class I molecules is presently unknown. We found that autophagy, while dispensable for the transit to the Lamp1 compartment, was instead required for diversion from the early endosome to the Rab14+ endosomal storage compartment, in which yeast interaction with MHC class I molecules has been described (47). Thus, like yeasts, conidia likely interact with MHC class I molecules in the Rab14+ compartment. Whether this actually occurs and the effects of conidia residency in this compartment versus the cytosolic compartment for CD8+ T cell responses are presently unknown but under investigation.
Primarily associated with MHC class II presentation, by delivering intracellular material for lysosomal degradation, macroautophagy has been recently shown to present tumor (59, 60) and viral antigens (39) on MHC class I, through the classical and vacuolar antigen presentation pathways (38). Indeed, by assisting TLRs in encountering their cognate ligands, autophagy contributed to the CD8+ T cell response to pathogens localized in autophagosomes containing components of the MHC class I presentation pathway (61). Irrespective of the mechanisms through which autophagy mediates Aspergillus sorting for MHC class I presentation, promoting autophagy in vivo provided CD8-dependent antifungal memory. Indeed, TLR3, known to activate DCs for cross-priming (62), activated both autophagy and CD8+ T cell memory in CGD mice. Although the net contribution of autophagy to the handling of the fungus and fungal control in infection will likely be more complex, our results are the first to our knowledge to show a role for autophagy in adaptive immunity to A. fumigatus and suggest that TLR3 adjuvants may optimize vaccination strategies in aspergillosis. Along the same line of reasoning, it will be of interest to assess the therapeutic potential of chloroquine, known to favor CD8+ T cell activation (63), as well as of inhibitors of cysteine and aspartic proteases to define whether excessive epitope degradation also contributes to defective cross-presentation of fungal antigens in CGD.
Overall, the present study shows that defective cross-presentation in the absence of NOX2 may contribute to the inability of CGD patients to successfully combat A. fumigatus. Because viral infections are not common in CGD (18), and CGD mice were not more susceptible to MMCV infection (the present study), it is likely that mechanisms of cross-presentation of viral versus fungal antigens are different, as has been speculated (31). From an immunological standpoint, our study highlights how understanding molecular and cellular mechanisms of memory at basic levels could be exploited to perfect vaccination strategies against inflammatory fungal diseases through the appropriate selection of fungal antigens and adjuvants and the targeting of DC pathways that enhance vaccine potency.
Methods
Mice.
Female C57BL/6 mice, 8–10 weeks old, were purchased from Charles River. Homozygous p47phox–/–, Dectin-1–/–, Tlr2–/–, Tlr3–/–, Tlr4–/–, Tlr6–/–, Trif–/–, Myd88–/–, Ifng–/–, and Il17ra–/– mice on a C57BL/6 background were bred under specific pathogen–free conditions at the Animal Facility of Perugia University. Cd4–/– and Cd8–/– mice were provided by Annette Oxenius, Institute of Microbiology, ETH Zürich, Zurich, Switzerland.
Fungal strains, infections, and treatments.
Viable conidia (>95%) from the A. fumigatus AF293 strain were obtained by growth on Sabouraud dextrose agar (Difco) at room temperature. Swollen conidia were obtained as described previously (22). A GFP-expressing strain of A. fumigatus (provided by M.M. Moore, Department of Biological Sciences, Simon Fraser University, Burnaby, British Columbia, Canada) was used to track the fate of viable fungi, as GFP expression is rapidly lost following cell death. For infection, mice were anesthetized by i.p. injection of 2.5% avertin (Sigma-Aldrich) before the intranasal instillation of a suspension of 2 × 107 conidia/20 μl saline. Mice were monitored for fungal growth (log10 CFU/organ, mean ± SEM), histopathology (PAS and Gomori staining of lung tissue sections), and lung immunofluorescence (see below). BAL fluid collection and morphometry were done as described previously (22). Histology sections and cytospin preparations were observed using a BX51 microscope (Olympus), and images were captured using a high-resolution DP71 camera (Olympus). Mice were treated with a total of 300 μg anti-CD4 (GK 1.5), anti-CD8 (YTS 169), anti–MHC class I-A (34-5-8S), or anti–MHC class II (M5/114) mAbs (Bioceros BV) at the time of vaccination. Control mice received isotype control rat IgG2a mAb (eBR2a) (eBioscience). Depletion of the corresponding T cell subsets with the anti-CD8 or anti-CD4 regimen was monitored in each experiment and was consistently between 95% and 98%, lasting for 3–5 days after treatment (data not shown and Supplemental Figure 11 for anti–MHC class I mAb). Treatment with anti-CD4 alone, but not anti-CD8, anti–MHC class I-A, or anti–MHC class II mAbs alone further increased the susceptibility to the infection in cyclophosphamide-treated mice as compared with control mice (data not shown).
Vaccination models.
Two vaccination models were used: one using live or inactivated (by autoclaving at 121°C for 30 minutes) A. fumigatus conidia or purified fungal antigens given with the immunoadjuvant murine CpG oligodeoxynucleotide 1862 or coupled to microparticles; the other using antigen-pulsed DCs (22). In the first model, mice received i.n. in 20 μl saline one of the following preparations: 2 × 107 live or heat-inactivated Aspergillus conidia, 14 days before the infection; 5 μg of the different purified fungal antigens plus 10 nM CpG-ODN; or 5 μg Pep1p plus 50 μg microparticles, administered 14, 7, and 3 days before the infection. Mice were immunosuppressed i.p. with 150 mg/kg of cyclophosphamide per day before infection. In the adoptive transfer model of antigen-pulsed DCs, purified lung CD11c+ DCs were pulsed with viable A. fumigatus conidia (1:1 cell/fungus ratio) for 2 hours before addition of 2.5 μg/ml amphotericin B (Sigma-Aldrich) to prevent A. fumigatus overgrowth or with 5 μg/ml of the recombinant fungal antigens Pep1p, Gel1p, and Crh1p produced in Pichia pastoris (22). Poly(I:C), 15 μg/20 μl (Sigma-Aldrich), was given with A. fumigatus conidia 14 days before the infection. For treatment with siRNA, each mouse was lightly anesthetized by inhaled diethyl ether and then given intranasal siRNA (10 mg/kg) or equivalent doses of nonspecific control siRNA duplex in a volume of 20 μl, in the TransITR-EE Starter Kit (Mirus). Intranasal siRNA was given twice together with conidia plus poly(I:C) vaccination. Pep1p coupling on 3-μm microparticles based on melamine resin (Fluka Analytical, Sigma-Aldrich) was done according to manufacturer’s instructions. Stable attachment of Pep1p was monitored by FACS analysis showing that more than 85% of microparticles were fluorescent 24 hours after coupling with 5 μg Alexa Fluor 488–conjugated Pep1p plus 50 μg microparticles. Microparticles coupled with FITC-Pep1p were used to monitor effective DC uptake in the lung in vivo, 24 hours after injection.
MCMV infection.
Infection with the Smith strain of MCMV, its titration in a standard plaque assay on murine embryonic fibroblast cells, and determination of cytolytic activity against 51Cr-labeled DCs and frequency of IFN-γ– or granzyme B–producing CD8+ T cells (ELISPOT assay) purified from spleens were done as described previously (64). For histology, sections of paraffin-embedded tissues were stained with H&E.
Cell preparation and cultures.
CD4+ or CD8+ T cells were purified after incubation of non-adherent lung cells with FITC-labeled anti-CD4 or anti-CD8, followed by anti-FITC MicroBeads (Miltenyi Biotec) as described previously (22). T cells were purified by non-adherent lung cells with FITC-labeled anti-CD3, followed by anti-FITC MicroBeads. Macrophages were isolated from total lung cells after 2-hour plastic adherence at 37°C. DCs were isolated with MicroBeads (Miltenyi Biotec) conjugated to hamster anti–mouse CD11c mAbs (N418) before magnetic cell sorting (22). DCs or macrophages (106 cells/ml) were pulsed with live Aspergillus conidia (5:1 cell/fungus ratio), 5 μg/ml Pep1p, soluble or coated on microparticles, for 18 hours before RNA extraction and Western blotting or for 2 hours before additional coculture with purified CD4+ or CD8+ T cells (2 × 106 cells/ml) for 72 hours for cytokine production, cytokine gene expression, and lymphoproliferation. DNA synthesis was measured by 3H-thymidine uptake for 6 hours. DCs (2 × 105/ml) and CD8+ T cells (2 × 106/ml) were added to Aspergillus-pulsed macrophages (2 × 105/ml) for 72 hours. For apoptosis induction, macrophages were treated with LPS and ATP, as previously indicated (41), and cell death was confirmed by annexin V staining.
Flow cytometry and intracellular staining.
All staining reactions were performed at 4°C on cells first incubated with an Fc receptor mAb (2.4G2) to reduce nonspecific binding. Antibodies were as follows: anti-CD8α (clone 53-6.7), anti-CD4 (GK 1.5), anti-CD44 (Pgp1, Ly-24), anti-CD62L (LECAM-1 Ly-22), anti-CD45RB (C363-16A), anti-CD25 (3C7), anti CD11b (M1/70), anti-CD11c (N418), anti-CD103 (M290), anti-B220 (RA3-6B2) (all from BD Biosciences — Pharmingen), and anti–Siglec-H (HM1075F, HyCult Biotech). For intracellular staining, cells were stimulated with PMA/ionomycin, with addition of brefeldin, and then permeabilized with the CytoFix/CytoPerm kit (BD Biosciences) for intra-cytoplasmic detection of IFN-γ (Alexa Fluor 488, clone, XMG1.2, eBioscience), and IL-10 (phycoerythrin, clone JES5-16E3, BD Biosciences — Pharmingen). Cells were analyzed with a FACScan flow cytometer (BD) equipped with CELLQuest software.
Immunohistochemistry.
Snap-frozen OCT-embedded lungs were cryosectioned with a semiautomatic cryostat (MC4000, Histo-Line Laboratories) and stained with a combination of Alexa Fluor 488–anti–mouse CD4 (clone GK1.5, BioLegend) and Alexa Fluor 647–anti–mouse CD8 (clone 5H10-1, BioLegend) at room temperature for 1 hour. Pictures were taken with a fluorescence microscope (BX51, Olympus) using analySIS image processing software. DAPI (Molecular Probes, Invitrogen) was used to detect nuclei (original magnification, ×20).
Immunofluorescence.
Imaging was performed on DCs plated in complete medium into chambered coverglass (Lab-Tek/Nunc; Thermo Scientific) in a temperature-regulated environmental chamber. DCs were exposed to GFP-conidia (at a 1:4 cell/conidia ratio), Alexa Fluor 488–Pep1p, and Alexa Fluor 488–Mep1p peptides (1 μg/ml) in serum-free RPMI-1640 medium and incubated at 37°C for 160 minutes, and time chase experiments were done at 5, 15, 45, and 120 minutes. For inhibition experiments, cells were preincubated with 5 mM 3-MA or 5 μg/ml chloroquine for 30 minutes. After washout, cells were fixed in 2% formaldehyde for 40 minutes at room temperature and permeabilized in a blocking buffer containing 5% FBS, 3% BSA, and 0.5% Triton X-100 in PBS. The cells were then incubated at 4°C with primary antibodies produced in rabbit against mannose receptor (Abcam), Rab5, Rab7, Rab9, Rab14, and Lamp1 (Sigma-Aldrich) for 12 hours in a buffer containing 3% BSA and 0.1% Triton X-100 in PBS. After extensive washing with PBS, the cells were incubated at room temperature for 60 minutes with 1:400 secondary anti-rabbit IgG–TRITC antibody (Sigma-Aldrich). Nuclei were counterstained with DAPI. Images were acquired using a fluorescence microscope (BX51, Olympus) with a 100× objective and analySIS image processing software (Olympus). The co-localization program Fiji ( http://fiji.sc/wiki/index.php/Fiji) with the JACoP plug-in was used to quantify the degree of overlap by calculating the co-localization coefficients (Pearson’s correlation coefficient, overlap coefficient according to Manders, and the overlap coefficients as reported in Supplemental Table 1). Pep1p and Mep1p were fluorescently labeled using Alexa Fluor 488 Microscale Protein Labeling Kit (Molecular Probes, Invitrogen) following the manufacturer’s instructions. The dye peptide was maintained at 4°C with the addition of 2 mM sodium azide, protected from light until imaging experiments were performed.
Uptake tracking of GFP-expressing A. fumigatus in vivo.
To track DC uptake of GFP-expressing A. fumigatus in vivo, single-cell suspensions were prepared from lungs depleted of T and B cells, to avoid possible loss of GFP+ phagocytes. Cells were immunostained with the CD11c or the CD11b markers and analyzed by flow cytometry.
Phagocytosis and conidiocidal assay.
GFP-labeled conidia (1 × 106/ml) were added to DC monolayers (at a 1:1 ratio) previously adhered to glass coverslips in 24-well tissue culture plates, and phagocytosis was allowed to proceed for 2 hours at 37°C. The fluorescence of bound but uningested conidia was quenched with trypan blue (0.3 ml of 1 mg/ml in PBS for 15 minutes at 25°C), and the DCs were fixed in 1% paraformaldehyde. Phagocytosis was quantified via phase contrast and fluorescence microscopy at ×1,000 on a BX51 microscope (Olympus). For determination of fungicidal activity, total lung cells were incubated with unopsonized conidia at a 1:1 ratio at 37°C for 2 hours. Inhibition of colony forming units (expressed as mean percentage ± SEM), referred to as conidiocidal activity, was determined as described previously (22).
In vitro antigen presentation, cytotoxic, and conidiocidal assays.
To assess the intracellular mechanism of fungal antigen processing, a panel of antigen presentation pathway inhibitors (all from Sigma-Aldrich) at their optimum concentrations was added to DCs or macrophages for 120 minutes prior to the 2-hour pulsing with live conidia or Pep1p. We used 5 μg/ml chloroquine (an acidophilic weak base that inhibits endosomal acidification), 5 μg/ml brefeldin A (to inhibit the vacuolar pathway), 50 μM lactacystin (to inhibit proteasomal degradation), 5 mM 3-MA (to inhibit autophagy), 2 μg/ml DPI (to inhibit ROS generation), and 50 μM rapamycin (to induce autophagy). After pulsing, DCs were fixed in 0.8% paraformaldehyde (Sigma-Aldrich) for 5 minutes and extensively washed before use. Treated DCs (1 × 105) were incubated with CD4+ or CD8+ T cells (5 × 105), purified from lungs a week after the infection, in RPMI-1640 with 10% FCS, 2 mM l-glutamine, and antibiotics, in round-bottom, 96-well plates for lymphoproliferation (after 72 hours incubation) or in polypropylene tubes for collection of supernatants and for determination of gene expression and cytolytic activity of harvested cells (after 24 hours incubation). DNA synthesis was measured by 3H-thymidine labeling (Amersham Biosciences) for 6 hours. Cytolytic activity was tested using a standard 4-hour 51Cr release assay. Target cells (conidia-pulsed DCs) were labeled with 51Cr at 37°C for 45 minutes. A total of 104 51Cr-labeled targets/well were added to round-bottom, 96-well plates, to which effector cells were added in triplicate at the indicated E/T cell ratios to a total volume of 200 μl. The plates were incubated for 4 hours before 50 μl of supernatant was transferred into sample plates. A total of 150 μl/well scintillation liquid (OptiPhase SuperMix, PerkinElmer) was added, and the sample plates were read on a scintillation gamma counter. Spontaneous 51Cr-release was measured in control wells containing target cells with medium alone. Maximum release values were obtained by lysis of target cells with 2.5% Triton X-100 (Sigma-Aldrich). The percentage of specific release was calculated as follows: (experimental release – spontaneous release)/(maximal release – spontaneous release) × 100. A total of 106 live or heat-inactivated conidia of A. fumigatus were labeled in the dark with the fluorescent molecular stain FUN-1 (5 μM, Molecular Probes) with gentle shaking at 37°C for 30 minutes. After washing, FUN-1–stained live conidia were incubated overnight in fresh culture medium alone or with 500 μl of culture medium from naive CD8+ T cells or cells from infected mice either unexposed or exposed to conidia-pulsed DCs and then examined under fluorescence microscopy. Because lymphocytes take up FUN-1, cells could not be added directly in this assay. Metabolically active conidia accumulate orange fluorescence in vacuoles, while dormant and dead conidia stain green.
Cell line cultures, transfection, and autophagy.
RAW 264.7 cells (ATCC) were seeded in 100-mm petri dish (3.5 × 106) and transfected with the EGFP-LC3 plasmid (Addgene) using ExGen 500 in vitro Transfection Reagent (Fermentas) for 48 hours, according to the manufacturer’s instructions. Transiently transfected RAW 264.7 cells were exposed to A. fumigatus resting or swollen conidia (RC or SC, respectively) at a cell/fungi ratio of 1:1, 5 μg/ml Pep1p, 50 μM rapamycin (LC Laboratories), or 20 μg/ml poly(I:C) (Sigma-Aldrich) in the presence of 10 μg/ml pepstatin A and 10 μg/ml E64-D (both from Sigma-Aldrich) to inhibit autophagosomal degradation by lysosomal enzymes. Cells were incubated for 4 hours (as indicated by preliminary experiments performed at 2, 4, or 16 hours) at 37°C in 5% CO2. Cultures growing on coverslips were observed at ×100 magnification with the Olympus BX51 fluorescence microscope using an FITC filter. Results are expressed as number of cells with EGFP-LC3 punctae. An equal amount of cell lysate in 2× Laemmli buffer (Sigma-Aldrich) was probed with rabbit anti-GFP antibody (Abcam) and goat anti-rabbit IgG HRP-conjugated secondary antibody (Sigma-Aldrich) after separation in 12% Tris/glycine SDS gel and transfer to a nitrocellulose membrane. Normalization was performed by probing the membrane with mouse anti–β-tubulin antibody (Sigma-Aldrich) and goat anti-rabbit IgG HRP-conjugated secondary antibody (Sigma-Aldrich). Chemiluminescence detection was performed with LiteAblot Plus chemiluminescence substrate (EuroClone), using the ChemiDoc XRS+ imaging system (Bio-Rad), and quantification was obtained by densitometry image analysis using Image Lab 3.1.1 software (Bio-Rad). For autophagy on lung cells, 1 × 106 purified DCs were stimulated on glass slides in 24 multiwell plates as above and incubated for 2 hours at 37°C in 5% CO2. Cells were incubated with 1:200 diluted anti-LC3 antibody (Cell Signaling Technology) overnight at 4°C in PBS containing 3% normal bovine serum albumin, incubated with anti-rabbit–PE secondary antibody (Sigma-Aldrich), and fixed for 20 minutes in PBS containing 4% paraformaldehyde. Images were acquired using the Olympus BX51 fluorescence microscope with a ×40 objective and analySIS image processing software (Olympus). DAPI was used to detect nuclei.
Co-immunoprecipitation assays and Western blot analysis.
For co-immunoprecipitation assays, cells were lysed with the NP-40 lysis buffer and incubated with 1:100 diluted rabbit polyclonal antibody to Beclin-1 (Cell Signaling Technology) overnight at 4°C, with Protein A sepharose added (Sigma-Aldrich) for 1 hour at 4°C. Immunoprecipitates were subjected to SDS-PAGE gel electrophoresis and probed with polyclonal antibody to TRIF (Imgenex) and Beclin-1 followed by anti-rabbit IgG–peroxidase (Sigma-Aldrich) as secondary antibody. Control experiments included Western blotting on immunoprecipitates with an irrelevant antibody. For the phospho-ERK detection by Western blot, cells were lysed in Laemmli buffer, blotted, and incubated with rabbit polyclonal antibody recognizing phospho–p44/p42 ERK (Thr202/Tyr294), followed by HRP–anti-rabbit IgG, as per the manufacturer’s instruction (Cell Signaling Technology). Normalization against β-tubulin was performed by probing the membrane with mouse anti–β-tubulin antibody and goat anti-rabbit IgG HRP-conjugated secondary antibody (Sigma-Aldrich). Chemiluminescence detection was performed as above.
siRNA design.
The siRNA sequences specific for mouse Becn1 (sense, 5′-CUGAGAAUGAAUGUCAGAAdTdT-3′; antisense, 5′-UUCUGACAUUCAUUCUCAGdTdT-3′) were selected, synthesized, and annealed by the manufacturer and were used in combination with scrambled control siRNA (Sigma-Aldrich).
ELISA and real-time PCR.
The level of cytokines in culture supernatants was determined by ELISA Kit (R&D Systems). The detection limits of the assays were less than 10 pg/ml for IL-12p70, less than 3 pg/ml for IL-10, less than 30 pg/ml for IL-23, and less than 10 pg/ml for IL-17A. Real-time RT-PCR was performed using the iCycler iQ Detection System (Bio-Rad) and SYBR Green chemistry (Finnzymes Oy). Cells were lysed, and total RNA was extracted using RNeasy Mini Kit (QIAGEN) and was reverse transcribed with Sensiscript Reverse Transcriptase (QIAGEN) according to the manufacturer’s directions. PCR primers for cytokines were as described previously (22). The following primers were used: Atg12 sense, 5′-AATCATGCAGGGACACAGTT-3′, antisense, 5′-CACTCTTCCTGCTCAGGTGT-3′; Bcn1 sense, 5′-CAATCTCAAGTGGGGTCTT-3′, antisense, 5′-GGCAAACATCCCCTAAGG-3′; LC3a sense, 5′-AGCTTCGCCGACCGCTGTAAG-3′, antisense, 5′-CTTCTCCTGTTCATAGATGTCAGC-3′; LC3b sense, 5′-CGGAGCTTTGAACAAAGAGTG-3′, antisense, 5′-TCTCTCACTCTCGTACACTTC-3′. Amplification efficiencies were validated and normalized against Gapdh. The thermal profile for SYBR Green real-time PCR was 95°C for 3 minutes, followed by 40 cycles of denaturation for 30 seconds at 95°C and an annealing/extension step of 30 seconds at 60°C. Each data point was examined for integrity by analysis of the amplification plot. The mRNA-normalized data were expressed as relative cytokine mRNA in treated cells compared with that in unstimulated cells.
Statistics.
For multiple comparisons, P values were calculated by using 1-way ANOVA (Bonferroni post hoc test), as specified for individual P values described in figure legends. For single comparisons, P values were calculated by using 2-tailed Student’s t test. The data reported are either from one representative experiment of 3–5 independent experiments or were compiled from 3–5 experiments. At least 6 mice per group per experiment were used, as computed by power analysis to yield a power of at least 80% with an α value of 0.05.
Study approval.
Experiments were performed according to Italian Approved Animal Welfare Assurance A–3143–01, and protocols licensed by the Italian Ministry of Health. Infections were performed under avertin anesthesia, and all efforts were made to minimize suffering.
Supplementary Material
Acknowledgments
The studies were supported by the Specific Targeted Research Projects ‘‘ALLFUN’’ (FP7_HEALTH_2009_260338), the Italian Projects AIDS 2010 by ISS (Istituto Superiore di Sanità — contract number 40H40), and 2011.0124.021 RICERCA SCIENTIFICA E TECNOLOGICA by Fondazione Cassa di Risparmio di Perugia, all to to L. Romani.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(5):1816–1831. doi:10.1172/JCI60862.
See the related Commentary beginning on page 1608.
References
- 1.Aimanianda V, et al. Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature. 2009;460(7259):1117–1121. doi: 10.1038/nature08264. [DOI] [PubMed] [Google Scholar]
- 2.Felgentreff K, et al. Clinical and immunological manifestations of patients with atypical severe combined immunodeficiency. Clin Immunol. 2011;141(1):73–82. doi: 10.1016/j.clim.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 3.Chaudhary N, Staab JF, Marr KA. Healthy human T-Cell Responses to Aspergillus fumigatus antigens. . PLoS One. 2010;5(2):e9036. doi: 10.1371/journal.pone.0009036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hebart H, et al. Analysis of T-cell responses to Aspergillus fumigatus antigens in healthy individuals and patients with hematologic malignancies. . Blood. 2002;100(13):4521–4528. doi: 10.1182/blood-2002-01-0265. [DOI] [PubMed] [Google Scholar]
- 5.Perruccio K, et al. Transferring functional immune responses to pathogens after haploidentical hematopoietic transplantation. Blood. 2005;106(13):4397–4406. doi: 10.1182/blood-2005-05-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beck O, et al. Generation of highly purified and functionally active human TH1 cells against Aspergillus fumigatus. . Blood. 2006;107(6):2562–2569. doi: 10.1182/blood-2005-04-1660. [DOI] [PubMed] [Google Scholar]
- 7.Wuthrich M, et al. A TCR transgenic mouse reactive with multiple systemic dimorphic fungi. J Immunol. 2011;187(3):1421–1431. doi: 10.4049/jimmunol.1100921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wuthrich M, Filutowicz HI, Warner T, Deepe GS, Jr, Klein BS. Vaccine immunity to pathogenic fungi overcomes the requirement for CD4 help in exogenous antigen presentation to CD8+ T cells: implications for vaccine development in immune-deficient hosts. J Exp Med. 2003;197(11):1405–1416. doi: 10.1084/jem.20030109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fierer J, Waters C, Walls L. Both CD4+ and CD8+ T cells can mediate vaccine-induced protection against Coccidioides immitis infection in mice. . J Infect Dis. 2006;193(9):1323–1331. doi: 10.1086/502972. [DOI] [PubMed] [Google Scholar]
- 10.Huffnagle GB, Yates JL, Lipscomb MF. Immunity to a pulmonary Cryptococcus neoformans infection requires both CD4+ and CD8+ T cells. . J Exp Med. 1991;173(4):793–800. doi: 10.1084/jem.173.4.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carvalho A, et al. TLR3 essentially promotes protective class I–restricted memory CD8+ T–cell responses to Aspergillus fumigatus in hematopoietic transplanted patients. . Blood. 2012;119(4):967–977. doi: 10.1182/blood-2011-06-362582. [DOI] [PubMed] [Google Scholar]
- 12.Bevan MJ. Understand memory, design better vaccines. Nat Immunol. 2011;12(6):463–465. doi: 10.1038/ni.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dudziak D, et al. Differential antigen processing by dendritic cell subsets in vivo. Science. 2007;315(5808):107–111. doi: 10.1126/science.1136080. [DOI] [PubMed] [Google Scholar]
- 14.Burgdorf S, Kautz A, Bohnert V, Knolle PA, Kurts C. Distinct pathways of antigen uptake and intracellular routing in CD4 and CD8 T cell activation. Science. 2007;316(5824):612–616. doi: 10.1126/science.1137971. [DOI] [PubMed] [Google Scholar]
- 15.Romani L. Immunity to fungal infections. Nat Rev Immunol. 2011;11(4):275–288. doi: 10.1038/nri2939. [DOI] [PubMed] [Google Scholar]
- 16.Segal BH, Veys P, Malech H, Cowan MJ. Chronic granulomatous disease: lessons from a rare disorder. Biol Blood Marrow Transplant. 2011;17(1 suppl):S123–S131. doi: 10.1016/j.bbmt.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuhns DB, et al. Residual NADPH oxidase and survival in chronic granulomatous disease. N Engl J Med. 2010;363(27):2600–2610. doi: 10.1056/NEJMoa1007097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van den Berg JM, et al. Chronic granulomatous disease: the European experience. PLoS One. 2009;4(4):e5234. doi: 10.1371/journal.pone.0005234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Ravin SS, et al. Chronic granulomatous disease as a risk factor for autoimmune disease. J Allergy Clin Immunol. 2008;122(6):1097–1103. doi: 10.1016/j.jaci.2008.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mantegazza AR, et al. NADPH oxidase controls phagosomal pH and antigen cross-presentation in human dendritic cells. Blood. 2008;112(12):4712–4722. doi: 10.1182/blood-2008-01-134791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Savina A, et al. NOX2 controls phagosomal pH to regulate antigen processing during crosspresentation by dendritic cells. Cell. 2006;126(1):205–218. doi: 10.1016/j.cell.2006.05.035. [DOI] [PubMed] [Google Scholar]
- 22.Bozza S, et al. Immune sensing of Aspergillus fumigatus proteins, glycolipids, and polysaccharides and the impact on Th immunity and vaccination. . J Immunol. 2009;183(4):2407–2414. doi: 10.4049/jimmunol.0900961. [DOI] [PubMed] [Google Scholar]
- 23.Chai LY, et al. Anti-Aspergillus human host defence relies on type 1 T helper (Th1), rather than type 17 T helper (Th17), cellular immunity. . Immunology. 2010;130(1):46–54. doi: 10.1111/j.1365-2567.2009.03211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfeifer JD, Wick MJ, Roberts RL, Findlay K, Normark SJ, Harding CV. Phagocytic processing of bacterial antigens for class I MHC presentation to T cells. Nature. 1993;361(6410):359–362. doi: 10.1038/361359a0. [DOI] [PubMed] [Google Scholar]
- 25.Shen H, et al. Enhanced and prolonged cross-presentation following endosomal escape of exogenous antigens encapsulated in biodegradable nanoparticles. Immunology. 2006;117(1):78–88. doi: 10.1111/j.1365-2567.2005.02268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wagner H, Heit A, Schmitz F, Bauer S. Targeting split vaccines to the endosome improves vaccination. Curr Opin Biotechnol. 2004;15(6):538–542. doi: 10.1016/j.copbio.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 27.Vasilevsky S, Liu Q, Koontz SM, Kastenmayer R, Shea K, Jackson SH. Role of p47phox in antigen-presenting cell-mediated regulation of humoral immunity in mice. Am J Pathol. 2011;178(6):2774–2782. doi: 10.1016/j.ajpath.2011.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blander JM. Coupling Toll-like receptor signaling with phagocytosis: potentiation of antigen presentation. Trends Immunol. 2007;28(1):19–25. doi: 10.1016/j.it.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 29.Watts C, West MA, Zaru R. TLR signalling regulated antigen presentation in dendritic cells. Curr Opin Immunol. 2010;22(1):124–130. doi: 10.1016/j.coi.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 30.Burgdorf S, Scholz C, Kautz A, Tampe R, Kurts C. Spatial and mechanistic separation of cross-presentation and endogenous antigen presentation. Nat Immunol. 2008;9(5):558–566. doi: 10.1038/ni.1601. [DOI] [PubMed] [Google Scholar]
- 31.Amigorena S, Savina A. Intracellular mechanisms of antigen cross presentation in dendritic cells. Curr Opin Immunol. 2010;22(1):109–117. doi: 10.1016/j.coi.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 32.Burgdorf S, Kurts C. Endocytosis mechanisms and the cell biology of antigen presentation. Curr Opin Immunol. 2008;20(1):89–95. doi: 10.1016/j.coi.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 33.Chen L, Jondal M. Brefeldin A inhibits vesicular MHC class I processing in resting but not in CpG- and disruption-activated DC. Mol Immunol. 2008;46(1):158–165. doi: 10.1016/j.molimm.2008.07.018. [DOI] [PubMed] [Google Scholar]
- 34.Fenteany G, Standaert RF, Lane WS, Choi S, Corey EJ, Schreiber SL. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science. 1995;268(5211):726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 35.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584(7):1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seglen PO, Gordon PB. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci U S A. 1982;79(6):1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Munz C. Enhancing immunity through autophagy. Annu Rev Immunol. 2009;27:423–449. doi: 10.1146/annurev.immunol.021908.132537. [DOI] [PubMed] [Google Scholar]
- 38.Chemali M, Radtke K, Desjardins M, English L. Alternative pathways for MHC class I presentation: a new function for autophagy. Cell Mol Life Sci. 2011;68(9):1533–1541. doi: 10.1007/s00018-011-0660-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.English L, et al. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat Immunol. 2009;10(5):480–487. doi: 10.1038/ni.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pozzi LA, Maciaszek JW, Rock KL. Both dendritic cells and macrophages can stimulate naive CD8 T cells in vivo to proliferate, develop effector function, and differentiate into memory cells. J Immunol. 2005;175(4):2071–2081. doi: 10.4049/jimmunol.175.4.2071. [DOI] [PubMed] [Google Scholar]
- 41.Lin JS, Yang CW, Wang DW, Wu-Hsieh BA. Dendritic cells cross-present exogenous fungal antigens to stimulate a protective CD8 T cell response in infection by Histoplasma capsulatum. . J Immunol. 2005;174(10):6282–6291. doi: 10.4049/jimmunol.174.10.6282. [DOI] [PubMed] [Google Scholar]
- 42.Volling K, Brakhage AA, Saluz HP. Apoptosis inhibition of alveolar macrophages upon interaction with conidia of Aspergillus fumigatus. . FEMS Microbiol Lett. 2007;275(2):250–254. doi: 10.1111/j.1574-6968.2007.00883.x. [DOI] [PubMed] [Google Scholar]
- 43.Gruenberg J, van der Goot FG. Mechanisms of pathogen entry through the endosomal compartments. Nat Rev Mol Cell Biol. 2006;7(7):495–504. doi: 10.1038/nrm1959. [DOI] [PubMed] [Google Scholar]
- 44.Sato M, Sato K, Fonarev P, Huang CJ, Liou W, Grant BD. Caenorhabditis elegans RME-6 is a novel regulator of RAB-5 at the clathrin-coated pit. Nat Cell Biol. 2005;7(6):559–569. doi: 10.1038/ncb1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang T, Ming Z, Xiaochun W, Hong W. Rab7: role of its protein interaction cascades in endo-lysosomal traffic. Cell Signal. 2011;23(3):516–521. doi: 10.1016/j.cellsig.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 46.Lombardi D, Soldati T, Riederer MA, Goda Y, Zerial M, Pfeffer SR. Rab9 functions in transport between late endosomes and the trans Golgi network. Embo J. 1993;12(2):677–682. doi: 10.1002/j.1460-2075.1993.tb05701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saveanu L, et al. IRAP identifies an endosomal compartment required for MHC class I cross-presentation. Science. 2009;325(5937):213–217. doi: 10.1126/science.1172845. [DOI] [PubMed] [Google Scholar]
- 48.Singh SK, et al. Design of neo-glycoconjugates that target the mannose receptor and enhance TLR-independent cross-presentation and Th1 polarization. Eur J Immunol. 2011;41(4):916–925. doi: 10.1002/eji.201040762. [DOI] [PubMed] [Google Scholar]
- 49.del Rio ML, Rodriguez-Barbosa JI, Kremmer E, Forster R. CD103- and CD103+ bronchial lymph node dendritic cells are specialized in presenting and cross-presenting innocuous antigen to CD4+ and CD8+ T cells. J Immunol. 2007;178(11):6861–6866. doi: 10.4049/jimmunol.178.11.6861. [DOI] [PubMed] [Google Scholar]
- 50.den Haan JM, Lehar SM, Bevan MJ. CD8(+) but not CD8(–) dendritic cells cross-prime cytotoxic T cells in vivo. J Exp Med. 2000;192(12):1685–1696. doi: 10.1084/jem.192.12.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hohl TM, et al. Inflammatory monocytes facilitate adaptive CD4 T cell responses during respiratory fungal infection. Cell Host Microbe. 2009;6(5):470–481. doi: 10.1016/j.chom.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee K, Won HY, Bae MA, Hong JH, Hwang ES. Spontaneous and aging-dependent development of arthritis in NADPH oxidase 2 deficiency through altered differentiation of CD11b+ and Th/Treg cells. Proc Natl Acad Sci U S A. 2011;108(23):9548–9553. doi: 10.1073/pnas.1012645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang J, et al. Activation of antibacterial autophagy by NADPH oxidases. Proc Natl Acad Sci U S A. 2009;106(15):6226–6231. doi: 10.1073/pnas.0811045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi CS, Kehrl JH. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J Biol Chem. 2008;283(48):33175–33182. doi: 10.1074/jbc.M804478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wuthrich M, et al. Vaccine-induced protection against 3 systemic mycoses endemic to North America requires Th17 cells in mice. J Clin Invest. 2011;121(2):554–568. doi: 10.1172/JCI43984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Husebye H, et al. The Rab11a GTPase controls Toll-like receptor 4-induced activation of interferon regulatory factor-3 on phagosomes. Immunity. 2010;33(4):583–596. doi: 10.1016/j.immuni.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lam JS, Huang H, Levitz SM. Effect of differential N-linked and O-linked mannosylation on recognition of fungal antigens by dendritic cells. PLoS One. 2007;2(10):e1009. doi: 10.1371/journal.pone.0001009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rodriguez A, Regnault A, Kleijmeer M, Ricciardi-Castagnoli P, Amigorena S. Selective transport of internalized antigens to the cytosol for MHC class I presentation in dendritic cells. Nat Cell Biol. 1999;1(6):362–368. doi: 10.1038/14058. [DOI] [PubMed] [Google Scholar]
- 59.Li Y, Wang LX, Yang G, Hao F, Urba WJ, Hu HM. Efficient cross-presentation depends on autophagy in tumor cells. Cancer Res. 2008;68(17):6889–6895. doi: 10.1158/0008-5472.CAN-08-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Uhl M, Kepp O, Jusforgues-Saklani H, Vicencio JM, Kroemer G, Albert ML. Autophagy within the antigen donor cell facilitates efficient antigen cross-priming of virus-specific CD8+ T cells. Cell Death Differ. 2009;16(7):991–1005. doi: 10.1038/cdd.2009.8. [DOI] [PubMed] [Google Scholar]
- 61.Andrade RM, Wessendarp M, Gubbels MJ, Striepen B, Subauste CS. CD40 induces macrophage anti-Toxoplasma gondii activity by triggering autophagy-dependent fusion of pathogen-containing vacuoles and lysosomes. . J Clin Invest. 2006;116(9):2366–2377. doi: 10.1172/JCI28796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith TR, Tang X, Maricic I, Garcia Z, Fanchiang S, Kumar V. Dendritic cells use endocytic pathway for cross-priming class Ib MHC-restricted CD8alphaalpha+TCRalphabeta+ T cells with regulatory properties. J Immunol. 2009;182(11):6959–6968. doi: 10.4049/jimmunol.0900316. [DOI] [PubMed] [Google Scholar]
- 63.Accapezzato D, et al. Chloroquine enhances human CD8+ T cell responses against soluble antigens in vivo. J Exp Med. 2005;202(6):817–828. doi: 10.1084/jem.20051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bozza S, et al. Pentraxin 3 protects from MCMV infection and reactivation through TLR sensing pathways leading to IRF3 activation. Blood. 2006;108(10):3387–3396. doi: 10.1182/blood-2006-03-009266. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.