Abstract
SIVs infecting wild-living apes in west central Africa have crossed the species barrier to humans on at least four different occasions, one of which spawned the AIDS pandemic. Although the chimpanzee precursor of pandemic HIV-1 strains must have been able to infect humans, the capacity of SIVcpz strains to replicate in human lymphoid tissues (HLTs) is not known. Here, we show that SIVcpz strains from two chimpanzee subspecies are capable of replicating in human tonsillary explant cultures, albeit only at low titers. However, SIVcpz replication in HLT was significantly improved after introduction of a previously identified human-specific adaptation at position 30 in the viral Gag matrix protein. An Arg or Lys at this position significantly increased SIVcpz replication in HLT, while the same mutation reduced viral replication in chimpanzee-derived CD4+ T cells. Thus, naturally occurring SIVcpz strains are capable of infecting HLTs, the major site of HIV-1 replication in vivo. However, efficient replication requires the acquisition of a host-specific adaptation in the viral matrix protein. These results identify Gag matrix as a major determinant of SIVcpz replication fitness in humans and suggest a critical role in the emergence of HIV/AIDS.
Introduction
AIDS, one of the most devastating infectious diseases that have emerged recently, is the result of zoonotic transmissions of viruses naturally infecting primates in Africa. It is now well established that HIV-1, the main cause of AIDS, emerged as a consequence of at least 4 cross-species transmissions of SIVs infecting wild-living apes (1, 2). Both SIVcpz-infected chimpanzees (Pan troglodytes troglodytes) and SIVgor-infected gorillas (Gorilla gorilla gorilla) transmitted their viruses to humans, generating HIV-1 groups M, N, O, and P (1–4). Of these, groups, M and N have been traced to geographically isolated chimpanzee communities in southeastern Cameroon (3). Group P appears to be of gorilla origin (4, 5), while the ape source of group O has not yet been defined (6). Although members of all 4 HIV-1 lineages can cause CD4+ T cell decline and AIDS, the extent of their spread in the human population has been very different. Group M is by far the most prevalent and has infected over 60 million individuals worldwide. Group O accounts for less than 1% of AIDS cases and is largely restricted to Cameroon, Gabon, and neighboring countries (7, 8). Groups N and P are much rarer still and have been found in only 17 and 2 individuals, all but 1 from Cameroon, respectively (4, 5, 9, 10). The reasons underlying this extreme variation in distribution of the 4 groups are poorly understood, but most likely involve differences in the degree of adaptation to the new human host (11).
Like other viruses, primate lentiviruses must utilize numerous host proteins and counteract a variety of innate restriction factors in order to replicate efficiently in their respective hosts (11, 12). Many cellular factors vary among different species (13), and it is thus not surprising that viruses tend to cross more readily between more closely related host species (14, 15). However, even the genomes of very closely related species, such as humans and chimpanzees, exhibit multiple differences (16, 17). To define how these differences may have influenced the zoonotic potential of ape lentiviruses, we have begun to look for host-specific adaptations. In a previous study, we searched SIVcpz proteomes for sites that were highly conserved in the ape precursors of HIV-1, but changed — in the same way — each time these viruses crossed the species barrier to humans. This analysis identified one site in the viral matrix protein (Gag30) that encoded a Met or Leu in all known strains of SIVcpz and SIVgor, but switched to an Arg in the inferred ancestors of HIV-1 groups M, N, and O and to a Lys in many current pandemic HIV-1 strains (18). We also noted that an HIV-1 strain that had been extensively passaged in chimpanzees underwent a reversion of this host-specific signature (19). When molecular clones of this virus were tested for replication in chimpanzee CD4+ T cells, only those that encoded a Met at Gag30 replicated efficiently, whereas isogenic controls that encoded a Lys at Gag30 did not, while the opposite was observed in human CD4+ T cells (18). These results suggested that the amino acid at position 30 in the viral matrix protein has been under strong host-specific selection pressure. However, the impact of Gag30 on the ability of SIVcpz strains to replicate in HLTs, the major site of viral replication in infected humans, was not assessed.
To determine more directly the extent to which the amino acid residue at Gag30 influences the zoonotic potential of ape SIVs, we used noninvasive methods to generate a panel of infectious SIVcpz clones and examined their ability to replicate in human lymphoid tissues (HLTs) in the presence and absence of the species-specific signature. In addition to lymphocyte cultures, we selected human tonsil tissue to examine viral replication fitness, since these cultures do not require exogenous activation for viral infection (20, 21). Moreover, tonsil cultures maintain their cell complexity and cytoarchitecture and are thus more likely to recapitulate conditions of viral growth and transmission in vivo (20, 21). Analyzing 4 genetically diverse SIVcpz strains, including the closest relatives of HIV-1 groups M and N, we found that all chimpanzee viruses were capable of infecting and replicating in HLTs, albeit at lower titers than HIV-1 strains. However, introduction of the human-specific Gag30 substitution significantly increased their growth potential in HLT cultures (P < 0.0001). Thus, naturally occurring SIVcpz strains were capable of infecting HLT, but host-specific adaptations were required to enhance viral replication fitness and ensure successful colonization of humans.
Results
To examine the growth kinetics of chimpanzee lentiviruses in human CD4+ T cells and lymphatic tissues, we generated a panel of replication-competent molecular clones of SIVcpz from wild-living chimpanzees. This was accomplished as previously described by synthesizing viral consensus sequences amplified from fecal samples of naturally infected apes (3, 6, 22, 23). Three of these viruses were derived from members of the central chimpanzee subspecies (P.t. troglodytes) sampled in Cameroon (MT145, MB897, and EK505), while the fourth strain was obtained from an eastern chimpanzee (Pan troglodytes schweinfurthii) sampled in Tanzania (TAN2) (Table 1). Importantly, SIVcpzPtt strains MB897 and EK505 represent the closest genetic relatives of HIV-1 groups M and N, respectively, while SIVcpzPts strain TAN2 is highly divergent from all HIV-1 groups and represents a viral lineage that has never been found in humans (Figure 1). For control, we also examined reference strains of HIV-1 (NL4-3, JRCSF, SG3, YU2) as well as 2 HIV-1 clones (JC16 and NC7) that were obtained after prolonged in vivo passage in chimpanzees (refs. 19, 24–29, and Table 1). To account for differences in coreceptor usage, we also included a CCR5-tropic version of NL4-3 (25). Finally, we obtained site-directed mutants of all of these viruses by introducing species-specific signatures at position 30 of the p17 matrix protein (18). In all SIVcpz constructs and in the chimpanzee-adapted JC16 and NC7 clones, Gag30 was changed from a hydrophobic Met or Leu residue to a basic Arg or Lys residue, while reciprocal changes were introduced in all reference strains of HIV-1 (for NL4-3, both Met and Leu containing mutants were generated).
Table 1 .
HIV-1 and SIVcpz infectious molecular clones analyzed
Figure 1. Evolutionary relationships of HIV-1 (including groups M, N, O, and P), SIVcpzPtt (upper clade) and SIVcpzPts (lower clade), and SIVgor Gag protein sequences, shown in red, blue, and green, respectively.
The 4 molecular clones of SIVcpz analyzed in this study are highlighted in yellow. The tree was constructed using maximum likelihood methods *Bootstrap support of greater than 80%. Scale bar: 0.1 amino acid replacements per site.
To examine early infection events, viral stocks were generated by transfection of 293T cells and used to infect TZM-bl cells, which express high levels of the HIV/SIV receptor (CD4) and coreceptors (CCR5 and CXCR4). TZM-bl cells also contain LTR-driven β-galactosidase and luciferase reporter cassettes that are activated by Tat expression (30, 31). Using viral stocks that were equilibrated by p24 content, we found that all viruses were capable of infecting TZM-bl cells, albeit to varying degrees (Figure 2A). Overall, HIV-1 strains resulted in significantly higher (Tat-driven) β-galactosidase activity than SIVcpz strains (Figure 2B), indicating that they were able to enter TZM-bl cells and integrate their proviral genomes more efficiently. However, changing a Met or Leu at Gag30 to an Arg increased the infectiousness of SIVcpz by 2.5-fold, while the reciprocal mutations (K/R30M/L) in HIV-1 strains reduced their infectivity (Figure 2B). The effect of mutations at Gag30 in WT HIV-1 and SIVcpz strains was highly significant (Figure 2C), although the magnitude was strain dependent. For example, a K30M change impaired the infectiousness of the HIV-1 strains JRCSF and YU2C, but had little effect on SG3. Similarly, the reciprocal M30R change enhanced the infectiousness of the SIVcpz strains EK505 and MB897 more significantly than that of MT145 (Figure 2A).
Figure 2. Infectivity of WT and Gag30 mutant HIV-1 and SIVcpz strains in TZM-bl cells.
(A) TZM-bl indicator cells were infected with WT and Gag30 mutant SIVcpz and HIV-1 constructs as indicated. Infections were performed in triplicate with virus stocks containing 1 ng of p24 antigen (average values ± SDs are shown). CPZa, chimpanzee adapted. (B) Relative infectivity of WT and Gag30 mutant HIV-1 and SIVcpz constructs. Shown are minimum and maximum values, 25% and 75% percentiles, and median values. (C) Modulation of viral infectivity by host-specific adaptation at Gag30. Changing a Lys or Arg to a Met or Leu reduced the infectivity of HIV-1 by 50%, while the reciprocal mutations more than doubled the infectivity of SIVcpz. Values are shown in relation to those of the corresponding parental HIV-1 or SIVcpz constructs set to 100%.
We previously reported that chimpanzee-adapted HIV-1 strains encoding Met instead of Lys or Arg at Gag30 had a growth advantage in chimpanzee, but not human, CD4+ T cells (18). To examine the effect of Gag30 on viral replication in cells from different hosts, we compared WT and mutant SIVcpz and HIV-1 constructs in human and chimpanzee CD4+ T lymphocytes. Interestingly, we found that K/R30M/L mutations had little, if any, effect on the replication fitness of HIV-1 in human CD4+ T cells (Figure 3A). This was also true for SIVcpz strains, which replicated to similar titers regardless of whether they encoded a WT Gag30 residue or the human K/R adaptation (Figure 3A). Direct competition experiments confirmed this, revealing roughly equal ratios of WT and mutant viruses in most CD4+ T cell cultures 7 days after infection, although in some cultures, viruses containing the human-specific Arg or Lys residue became predominant (examples shown in Figure 3B). In contrast, a chimpanzee-specific Met or Leu was usually required for efficient viral growth in chimpanzee-derived CD4+ T cells. This was particularly true for SIVcpz strains MB897 and EK505, where a human-specific Arg residue caused a severe fitness loss in chimpanzee CD4+ T cells (Figure 3A). An Arg-to-Met substitution was also essential for replication of the HIV-1 strain JRCSF in chimpanzee T cells, but had only marginal effects on the growth of SG3 and YU2C (Figure 3C). On average, the WT (R/K) HIV-1 constructs replicated significantly (P = 0.0006) more efficiently in human-derived than in chimpanzee-derived T cells, whereas WT (M/L) SIVcpz constructs replicated with similar efficacy in cells from both species (Figure 3D). However, an Arg at Gag30 caused a significant reduction in SIVcpz replication in chimpanzee cells (50.2 ± 10.0%; P = 0.0009; n = 24), while a modest enhancement was observed in human CD4+ T cells (141.1 ± 23.5%; P = 0.0094; n = 16) (Figure 3E). Interestingly, the HIV-1 group M and N relatives MB897 and EK505 replicated substantially more efficiently than MT145 and TAN2 in human, but not in chimpanzee, cells (Figure 3C).
Figure 3. Replication potential of WT and Gag30 mutant HIV-1 and SIVcpz in human and chimpanzee CD4+ T lymphocytes.
(A) Replication kinetics are shown for isogenic pairs of WT and Gag30 mutant HIV-1 (red) and SIVcpz (green) clones in human (HU) and chimpanzee (CPZ) CD4+ T lymphocytes. Each virus pair was tested in cells from 6 chimpanzee and 4 human donors by measuring RT activity in culture supernatants (one representative curve is shown). (B) Competition of WT and Gag30 mutant HIV-1 and SIVcpz constructs. Human CD4+ T cells were infected with virus stocks that were normalized for p24 content and mixed at a 1:1 ratio. Sequence chromatograms of RT-PCR amplification products obtained from the input virus and cell culture supernatants collected at 7 days after infection are shown. (C) Cumulative virus production over 13 days of culture in CD4+ T lymphocytes from 4 human (left) and 6 chimpanzee (right) donors infected with the indicated HIV-1 or SIVcpz variants. Data are shown as mean ± SEM. (D) Mean production of HIV-1 or SIVcpz constructs containing an R/K or M/L at Gag30 in human- or chimpanzee-derived CD4+ T cells. Data are shown as mean ± SEM obtained for all 3 HIV-1 and 4 SIVcpz strains analyzed. (E) Replication of SIVcpz M/L30R mutants in human and chimpanzee cells. Cumulative RT production levels of the Gag30 mutants are shown in relation to those of their parental constructs set to 100%. WT JRCSF and YU2C strains did not replicate at detectable levels in most chimpanzee cells, thus precluding a direct comparison.
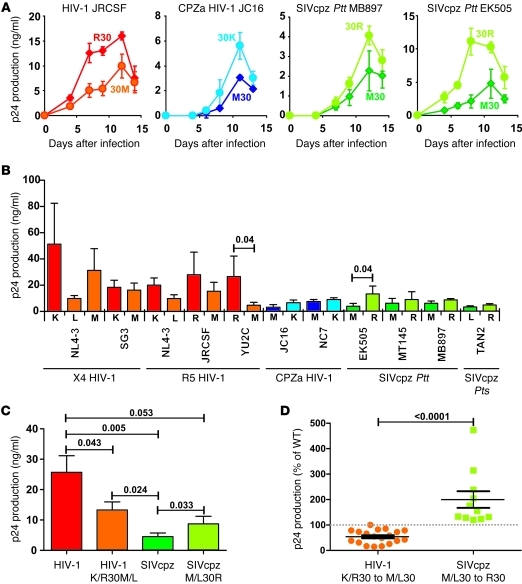
The fact that a basic residue at Gag30 emerged independently in all groups of HIV-1 (18), including 1 of the 2 known group P viruses (4, 5), suggested that it conferred a significant fitness advantage in human cells. It was thus surprising to find that introduction of this adaptive change into 4 SIVcpz genomes had little effect on their replication potential in human-derived CD4+ T cells (Figure 3). Lymphocyte cultures are maximally stimulated and thus do not reflect conditions of viral growth in vivo. Human tonsil explant cultures support HIV-1 replication without exogenous stimulation and largely maintain the complexity of cell populations and tissue architecture (20, 21). We thus reasoned that this culture system might be more appropriate for examining the replicative capacity of SIVcpz strains in HLTs. Human tonsils were dissected, set up in culture, and infected as described previously (20, 21). The results showed that M30K or M30R changes increased the replication potential of SIVcpz and chimpanzee-adapted HIV-1 strains in this culture system, while R30M or K30L/M changes decreased the replication potential of HIV-1 strains (Figure 4, A and B). Again, the effect of the Gag30 changes on viral replication was strain dependent and most pronounced in SIVcpz strain EK505, where a Met to Arg change at position 30 increased the ability of this virus to replicate in human tissue about 4-fold. Notably, changing Lys to Leu, which is the predominant amino acid among known SIVcpzPts strains, reduced the replicative potential of HIV-1 NL4-3 more severely than a substitution to Met, found in most SIVcpzPtt strains (Figure 4B). On average, WT HIV-1 strains produced 5-fold higher cumulative levels of p24 antigen than WT SIVcpz strains in HLTs (Figure 4C). Introduction of a chimpanzee-specific Met or Leu into HIV-1 constructs reduced p24 production in HLT to 54% ± 6% (n = 19) of WT levels (Figure 4C). In contrast, introduction of a human-specific Arg into SIVcpz constructs increased virus production to 200% ± 33% (n = 11) of WT levels (Figure 4D). As expected, the negative effect of a Met or Leu at Gag30 on the replication potential of both HIV-1 and SIVcpz was more pronounced in ex vivo–infected HLT than in human CD4+ T cell cultures (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI61429DS1), although there was a significant correlation between the magnitude of the disruptive effect in the 2 culture systems for each strain (Supplemental Figure 1B). In summary, the results showed that SIVcpz constructs containing human Gag30 signatures replicated considerably more efficiently in ex vivo HLT than WT SIVcpz, suggesting a key role in human adaptation. It should be noted, however, that this change was not sufficient to raise SIVcpz replication levels to those of contemporary HIV-1 strains, since HIV-1 WT constructs still produced 3-fold higher amounts of p24 than SIVcpz containing the Arg Gag30 substitution (Figure 4C).
Figure 4. Replication kinetics of WT and Gag30 mutant HIV-1 and SIVcpz in human tonsil explant cultures.
(A) Representative growth curves of WT and Gag30 mutant HIV-1, CPZ-adapted HIV-1 (CPZa), and SIVcpz constructs. Infections were performed in triplicate. Data shown are average values ± SD. (B) Virus production by tonsillary explant cultures infected with SIVcpz and HIV-1 constructs. Cumulative virus production is shown over 14 days. Matched tissues from 7 to 12 donors were inoculated with the WT HIV-1 (left), chimpanzee-adapted HIV-1 (middle). and SIVcpz (right) constructs or with their respective Gag30 mutants, and for each HLT culture, virus production was measured over 14 days. Data are shown as mean ± SEM. (C) Average cumulative p24 antigen levels in HLT cultures from 7 to 12 donors. Data are shown as mean ± SEM. (D) Host-specific adaptation increases replication fitness. Mutation of K/R30M/L reduces HIV-1 replication, and substitution of M/L30R increases SIVcpz replication in HLTs. Cumulative p24 antigen production levels of the Gag30 mutants are shown in relation to those of their respective parental constructs set to 100%.
To quantify the effect of an Arg residue at Gag30 on viral replication fitness in human cells, we performed a series of competition experiments in ex vivo HLTs. Stocks of WT and mutant viruses were mixed in equal ratios and used to infect the tonsil explant cultures. Virus production was then monitored by RT-PCR followed by direct sequencing of the p17 coding region that encompasses Gag30. Since WT and mutant HIV-1 and SIVcpz clones differ by only 1 or 2 nucleotides within the Gag30 codon, PCR amplifications were unbiased with respect to these mutations. Direct sequence analysis of amplification products indicated that the fluorescence levels of WT and mutant nucleotides reflected their input ratios and thus allowed the generation of standard curves (Figure 5A). We then analyzed equal mixtures of isogenic WT and Gag30 mutant before and after culture in tonsil explant cultures. The results showed that WT and mutant viruses were present at similar ratios (46% to 54%) in the infection stock (Figure 5B). However, after 2 weeks of HLT culture, viruses containing an Arg or Lys residue at Gag30 outcompeted those that encoded a Met or Leu residue (Figure 5B and Table 2). This selection bias for a basic residue at Gag30 resulted in a significantly higher percentage of viral sequences encoding Arg or Lys than Met or Leu (Supplemental Figure 2A). Notably, the proportion of HIV-1 and SIVcpz constructs containing M/L at Gag30 at the end of the competition experiments correlated with their relative replication fitness in ex vivo–infected HLTs (Supplemental Figure 2B). Thus, a basic residue at position 30 of the p17 matrix protein can increase SIVcpz replication fitness in human lymphoid cultures up to 5-fold, although the magnitude of this effect is clearly strain specific.
Figure 5. Host-specific adaptation at Gag30 enhances SIVcpz replication fitness in ex vivo HLTs.
(A) Detection of WT and mutant sequences in viral mixtures. Left: WT and K30M mutant HIV-1 NL4-3 stocks were normalized for p24 content, mixed at different ratios as indicated, and sequenced directly. Right: example for a standard curve. The peak fluorescence of the 2 WT A residues is expressed as a fraction of the total fluorescence. (B) Sequence chromatograms of mixed viral cultures at input and 14 days after infection. Percentages of WT and mutant sequences provided in B were estimated from standard curves. Values of less than 10% may represent nonspecific background.
Table 2 .
Selective advantage of HIV-1 and SIVcpz constructs encoding R/K at Gag30
Discussion
SIV infection is endemic in wild-living apes throughout equatorial Africa, yet only certain lineages of SIVcpz and SIVgor have been found to infect and cause disease in humans (1–3). This has raised the question of whether certain strains of SIVcpz/SIVgor are more prone than others to cross from apes to humans. Although all SIVcpz and SIVgor strains identified to date grow efficiently in human CD4+ T cells (6, 22), these maximally activated cultures are unlikely to represent a reliable surrogate of zoonotic potential. In the present study, we show that 4 genetically diverse SIVcpz strains from both central and eastern chimpanzees are capable of establishing a productive infection in HLT, the major site of HIV-1 replication in vivo. This is remarkable, given the number of different host proteins and innate restriction factors that primate lentiviruses must utilize or counteract (11, 32–34) and suggests that most (if not all) SIVcpz strains have the biological properties required for cross-species infection of humans. However, replication fitness of SIVcpz doubled upon the introduction of a previously identified human signature at position 30 of the viral matrix protein. Thus, even chimpanzee-to-human transmission seems to impose substantive adaptive hurdles to ape lentiviruses.
Although replacement of Met/Leu with Arg at Gag30 enhanced the replication potential of all 4 SIVcpz strains in HLTs, their overall growth rates still remained 3-fold lower than those of the HIV-1 strains, indicating additional adaptations. One of these likely includes effective antitetherin activity. Tetherin (also termed Bst-2) inhibits the release of virions from infected cells by “tethering” them at the plasma membrane (35, 36). Like most primate lentiviruses, SIVcpz uses its Nef protein to antagonize tetherin (37–39). However, the SIVcpz Nef protein is inactive against human tetherin due to a 5–amino acid deletion in the cytoplasmic domain of the human protein that confers resistance to Nef binding (37–39). This lack of antitetherin activity must explain, at least in part, why SIVcpz constructs containing the human Gag30 adaptation still replicate less efficiently than HIV-1 strains in HLT cultures. In fact, previous studies have shown that a defective vpu gene decreased replication of HIV-1 in ex vivo–infected human tonsillary tissue by about 3-fold, although it was not determined whether this reduction was due to lack of antitetherin activity or other Vpu functions, such as degradation of CD4 (40). Nonetheless, the results suggest that effective tetherin antagonism may be required for efficient replication of SIVcpz in HLTs.
Current data suggest that a fitness gain at Gag30 was a prerequisite for the emergence of HIV-1 groups M, N, and O. The fact that only 1 of the 2 recently discovered HIV-1 group P strains has switched from a Met to a Lys at Gag30 is interesting, since this could indicate that this group is still in the process of adaptation (41, 42). However, a single amino acid change at Gag30 seems easier to achieve than the acquisition of a new antitetherin function. Pandemic HIV-1 group M was able to master this by switching from Nef- to Vpu-mediated tetherin antagonism (38). However, this did not happen in HIV-1 groups O and P, whose Vpu proteins failed to gain this function (38, 39, 41, 42). The Vpu protein of HIV-1 group N gained some antitetherin activity, but lost its ability to degrade CD4. Thus, it is tempting to speculate that the adaptive change in Gag30 was a necessary first step to increase the replication potential of SIVcpz in HLTs such that additional mutations could subsequently accumulate to facilitate the gain of antitetherin function by its vpu gene.
Although SIVcpz is endemic in both central and eastern chimpanzees, only SIVcpzPtt strains have thus far been found in humans (1–3, 43). It is possible that humans are less frequently exposed to SIVcpz-infected eastern chimpanzees or that SIVcpzPts strains have been transmitted but gone unrecognized. However, it is also possible that SIVcpzPts strains are inherently less fit to infect humans. We found that the SIVcpzPts strain TAN2 produced about 2-fold lower viral titers in HLTs than the SIVcpzPtt strains MT145 and MB897 (Figure 4). Moreover, changing Gag30 from Lys to Leu, which is found in TAN2, impaired the replication of HIV-1 NL4-3 in HLT more severely than a change to Met found in all SIVcpzPtt strains (Figure 4). However, not all SIVcpzPtt strains replicated efficiently in HLT (Figure 4), and a recent molecular epidemiological study in the Democratic Republic of the Congo revealed that a number of SIVcpzPts strains encode Met at Gag30, as found in SIVcpzPtt (B.H. Hahn, unpublished observations). Thus, neither poor replication potential in HLTs nor coding differences at Gag30 can explain the absence of SIVcpzPts zoonotic infections. Further studies will need to define whether certain adaptations, such as the acquisition of antitetherin activity, may be more difficult to achieve for viruses endemic in eastern chimpanzees.
The mechanism or mechanisms of the Gag30-mediated effect on viral replication fitness remain to be determined. The MA protein is not only a structural component of the virion, but is also involved in viral entry/uncoating, cytoskeletal-mediated transport, nuclear import, virion assembly, and incorporation of the viral envelope glycoprotein into progeny virions (44, 45), interacting with as many as 20 different cellular proteins (34). These include 3 of the 4 clathrin adaptor protein complexes, AP-1, AP-2, and AP-3 (46–48), the microtubule-associated cellular motor protein KIF4 (49), calmodulin (50), 47-kDa tail interacting protein (TIP47) (51), elongation factor 1-α (EF1α) (52), the virion-associated nuclear shuttling protein (VAN) (53), and the barrier-to-autointegration factor (BAF) (54). Among these proteins, calmodulin, EF1α, BAF, AP-1 μ1A chain, and AP-2 μ2 chain are all predicted to be identical between chimpanzees and humans and thus unlikely to be responsible for the species-specific selection pressure. The chimpanzee and human AP-3 δ chain, TIP47, and VAN proteins differ by 3 or 4 amino acids, respectively, while the KIF4 motor protein is the most divergent, exhibiting 7.2% amino acid difference between human and chimpanzee homologs. Previous studies have shown that the AP-3 δ chain is involved in Gag trafficking, TIP47 mediates Env incorporation into particles, and VAN and KIF4 may play a role during postentry steps (48, 50, 52, 54). It is possible that Gag30, which is located in one of the N-terminal helices of MA, interacts with one or more of these proteins or with as-yet-unidentified host factors and may thus influence steps early in the viral life cycle. It should be noted, however, that the magnitude of the effect of Gag30 on viral fitness is strain dependent. Thus, as-yet-unidentified changes in the MA protein or elsewhere in the viral genome might minimize or even remove the need for a basic residue at Gag30.
The fact that we found a much less pronounced effect of Gag30 on the growth potential of SIVcpz in human CD4+ T cell cultures is not surprising. Fully activated CD4+ T cells represent optimal, albeit artificial, targets for viral replication. For example, they may express MA-interacting proteins at levels different from target cells that are not fully activated by exogenous stimuli. T cell activation is known to be associated with complex changes of the plasma membrane, the cytoplasm, the cytoskeleton and at the nucleus (55), all of which may minimize the dependency on MA protein functions. The mode of virus spread in the culture, i.e., cell-free versus cell-cell, may also have an impact on the magnitude of the Gag30 function. Perhaps most importantly, effective spread of HIV-1 and SIVcpz in vivo involves a complex interplay between different types of virally infected and noninfected cells, which are more accurately represented by lymphoid tissues than stimulated CD4+ T lymphocyte cultures. Thus, our finding that a basic residue significantly increases the replication fitness of SIVcpz strains in HLT strongly suggests that it is also relevant for viral replication and transmission in vivo.
In summary, our results demonstrate that nonadapted SIVcpz strains can replicate to some extent in HLTs. We also show that the acquisition of a basic residue at Gag30 in the viral matrix protein significantly increased the replication fitness of SIVcpz in the new human host, albeit not to the levels of current HIV-1 group M strains. Thus, additional changes, such as the development of effective tetherin antagonism, were required to achieve full replication fitness. Since exposures of humans to ape lentiviruses are likely to continue, further analyses of host-specific adaptation will be important to assess the future human zoonotic risk.
Methods
HIV-1 and SIVcpz constructs.
HIV-1 clones NL4-3, SG3, YU2, and JRCSF as well as SIVcpzPts clone TAN2 have been reported (refs. 21–28 and Table 1). To generate the SIVcpzPtt clones MT145, EK505, and MB897, subgenomic regions of the viral genome were amplified from fecal samples and sequenced directly; their consensus sequences were synthesized in 2 (MT145 and EK505) or 3 (MB897) fragments, which were then concatenated (Blue Heron Biotechnology). To enable directional cloning, unique restriction sites were added to the 5′ and 3′ termini of the proviruses (MluI and BamHI for MT145; MluI and NotI for EK505 and MB897, respectively). Full-length genomes were cloned into pCRXL (MT145 and EK505) or pBR-MCS (MB897) vectors and propagated in STBL2 cells (Life Technologies). The newly derived SIVcpzPtt clones have been submitted to the NIH Research and Reference Program, and their nucleotide sequence is available at GenBank (see Accession numbers). Site-directed mutagenesis of Gag30 was performed by splice overlap extension PCR, and all constructs were verified by sequence analysis.
Phylogenetic analyses.
Gag protein sequences were inferred from gag gene regions (HXB2 coordinates: 790-2280) of published nucleotide sequences (HIV-1 M/A: M62320; HIV-1 M/B: K03455; HIV-1 N: AJ006022 and DQ017382; HIV-1 O: L20571 and L20587; HIV-1 P: GU111555 and HQ179987; SIVcpzANT: U42720; SIVcpzCAM13: AY169968; SIVcpzCAM5: AJ271369; SIVcpzDP943: EF535993; SIVcpzEK505: DQ373065; SIVcpzMB897: EF535994; SIVcpzMT145: DQ373066; SIVcpzTAN1: AF447763; SIVcpzTAN2: DQ374657; SIVcpzTAN3: DQ374658; SIVcpzTAN5: JN091691; SIVcpzUG38: JN091690; SIVcpzUS: AF103818; SIVgorCP684: FJ424871) and aligned using ClustalW (56). A maximum likelihood phylogeny with bootstrap support was determined using PhyML (57) using an LG+I+G evolutionary model (58) as favored by ProtTest (59).
Viral infectivity and replication.
Virus stocks were generated by transient transfection of 293T cells, and infectivity assays were performed using TZM-bl indicator cells as described (60).
Human and chimpanzee CD4+ T lymphocyte cultures.
CD4+ T lymphocytes from human or chimpanzee donors were purified from total PBMCs by positive selection, activated using Staphylococcal enterotoxin B, and subsequently cocultured with autologous monocyte-derived macrophages to achieve optimal activation as described (6, 18, 61). After 5 to 6 days, 5 × 105 activated CD4+ T cells were infected at an MOI of 0.1 (as determined using TZM-bl cells) using transfection-derived viral stocks. The RT activity in culture supernatants was measured every 3 days to monitor viral replication.
Infection of HLT ex vivo.
Human tonsils removed during routine tonsillectomies were dissected into 2- to 3-mm3 blocks, cultured, and infected with viral stocks normalized based on p24 content as described previously (20, 21, 40). Productive HIV-1 and SIVcpz infection were evaluated by measuring the amount of p24 core antigen released into the medium using the InnoTest HIV Antigen mAb ELISA (Innotest HIV Antigen mAb; Innogenetics). This ELISA recognizes the p24 proteins of divergent SIVcpz and HIV-1 strains with equal efficiency, and all results on viral replication were verified by RT assay.
Competition assay in HLT.
The tonsil blocks were each coinfected with stocks of WT and mutant virus containing 0.5 ng of p24 antigen. Every 2 to 3 days, the medium of the histoculture was replaced. After 2 weeks, the culture medium was collected and RT-PCR (SuperScript III One-Step RT-PCR with Platinum Taq; Invitrogen) was performed to amplify viral genomic RNA using primers flanking the Gag30 coding region in the p17 matrix proteins. The PCR fragments were purified from agarose gels and examined by direct sequence analysis.
Competition assay in human T cells.
Pairwise competition experiments were performed using human T cell–derived virus stocks to optimize target cell infection. Briefly, 293T cell–derived transfection supernatants were used to infect Staphylococcal enterotoxin B–activated human CD4+ lymphocytes, supernatants were collected at the replication peak (6 to 8 days after infection), and the virus titer was determined using the Innotest HIV Antigen mAb ELISA. For virus competition in human T cell culture, 1 million activated CD4+ lymphocytes were coinfected for 2 hours in 500 μl medium containing 12.5 ng of p24 for the WT and Gag30 mutant T cell–derived virus stocks. Cells were then washed 3 times and plated in 2 ml of complete medium (RPMI + 10% FCS +30 IU IL-2). Supernatants were collected on days 3, 5, 7, and 9 after infection to monitor virus replication, and the cell culture supernatants collected at 7 days after infection were used to analyze the outcome of the competition by direct sequencing of gag PCR products as described above. Cytopathic effects of viral replication precluded the meaningful analysis of later time points.
Accession numbers.
GenBank accession numbers of newly generated SIVcpz clones are as follows: JN835460 (EK505clone2), JN835461 (MB897FLclone2), and JN835462 (MT145clone2).
Study approval.
Chimpanzee blood samples were collected from captive individuals housed at the Yerkes National Primate Research Center during their annual health survey, a procedure approved by the Emory Institutional Animal Care and Use Committee. Studies involving human material were reviewed and approved by the University of Ulm Institutional Review Board, and individuals provided informed consent prior to donating blood or tonsillary tissue.
Statistics.
Statistical calculations were performed using 2-tailed unpaired (for comparison of different groups) or paired (to assess the impact of mutations in Gag30) Student’s t tests using Graph Pad Prism Version 5.0. P < 0.05 was considered significant. Correlations were calculated with the linear regression module.
Supplementary Material
Acknowledgments
We thank Matthias H. Kraus, Rebecca S. Rudicell, Martha Mayer, and Birgit Ott for technical assistance. This work was supported in part by the NIH (R21 AI080364, R01 AI50529, R01 AI58715, P30 AI27767), the Yerkes Regional Primate Research Center (RR-000165), the Bristol Myers Freedom to Discover Program, and the Deutsche Forschungsgemeinschaft.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(5):1644–1652. doi:10.1172/JCI61429.
References
- 1.Sharp PM, Hahn BH. Origins of HIV and the AIDS pandemic. Cold Spring Harb Perspect Med. 2011;1(1):a006841. doi: 10.1101/cshperspect.a006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Heuverswyn F, et al. SIV infection in wild gorillas. Nature. 2006;444(7116):164. doi: 10.1038/444164a. [DOI] [PubMed] [Google Scholar]
- 3.Keele BF, et al. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 2006;313(5786):523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Plantier JC, et al. A new human immunodeficiency virus derived from gorillas. Nat Med. 2009;15(8):871–872. doi: 10.1038/nm.2016. [DOI] [PubMed] [Google Scholar]
- 5.Vallari A, et al. Confirmation of putative HIV-1 group P in Cameroon. J Virol. 2011;85(3):1403–1407. doi: 10.1128/JVI.02005-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takehisa J, et al. Origin and biology of simian immunodeficiency virus in wild-living western gorillas. J Virol. 2009;83(4):1635–1648. doi: 10.1128/JVI.02311-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mauclere P, et al. Serological and virological characterization of HIV-1 group O infection in Cameroon. AIDS. 1997;11(4):445–453. doi: 10.1097/00002030-199704000-00007. [DOI] [PubMed] [Google Scholar]
- 8.Peeters M, et al. Geographical distribution of HIV-1 group O viruses in Africa. AIDS. 1997;11(4):493–498. doi: 10.1097/00002030-199704000-00013. [DOI] [PubMed] [Google Scholar]
- 9.Vallari A, et al. Four new HIV-1 group N isolates from Cameroon: Prevalence continues to be low. AIDS Res Hum Retroviruses. 2010;26(1):109–115. doi: 10.1089/aid.2009.0178. [DOI] [PubMed] [Google Scholar]
- 10.Delaugerre C, De Oliveira F, Lascoux–Combe C, Plantier J–C, Simon F. HIV–1 group N: travelling beyond Cameroon. Lancet. 2011;378(9806):1894. doi: 10.1016/S0140-6736(11)61457-8. [DOI] [PubMed] [Google Scholar]
- 11.Kirchhoff F. Immune evasion and counteraction of restriction factors by HIV-1 and other primate lentiviruses. Cell Host Microbe. 2010;8(1):55–67. doi: 10.1016/j.chom.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 12.Bushman FD, et al. Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS Pathog. 2009;5(5):e1000437. doi: 10.1371/journal.ppat.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ortiz M, Bleiber G, Martinez R, Kaessmann H, Telenti A. Patterns of evolution of host proteins involved in retroviral pathogenesis. Retrovirology. 2006;3:11. doi: 10.1186/1742-4690-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Longdon B, Hadfield JD, Webster CL, Obbard DJ, Jiggins FM. Host phylogeny determines viral persistence and replication in novel hosts. PLoS Pathog. 2011;7(9):e1002260. doi: 10.1371/journal.ppat.1002260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Streicker DG, Turmelle AS, Vonhof MJ, Kuzmin IV, McCracken GF, Rupprecht CE. Host phylogeny constrains cross-species emergence and establishment of rabies virus in bats. Science. 2010;329(5992):676–679. doi: 10.1126/science.1188836. [DOI] [PubMed] [Google Scholar]
- 16.Nature. 2005;437(7055):69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 17.McLean CY, et al. Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature. 2011;471(7337):216–219. doi: 10.1038/nature09774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wain LV, et al. Adaptation of HIV-1 to its human host. Mol Biol Evol. 2007;24(8):1853–1860. doi: 10.1093/molbev/msm110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mwaengo DM, Novembre FJ. Molecular cloning and characterization of viruses isolated from chimpanzees with pathogenic human immunodeficiency virus type 1 infections. J Virol. 1998;72(11):8976–8987. doi: 10.1128/jvi.72.11.8976-8987.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glushakova S, Baibakov B, Margolis LB, Zimmerberg J. Infection of human tonsil histocultures: a model for HIV pathogenesis. Nat Med. 1995;1(12):1320–1322. doi: 10.1038/nm1295-1320. [DOI] [PubMed] [Google Scholar]
- 21.Glushakova S, et al. Nef enhances human immunodeficiency virus replication and responsiveness to interleukin-2 in human lymphoid tissue ex vivo. J Virol. 1999;73(5):3968–3974. doi: 10.1128/jvi.73.5.3968-3974.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takehisa J, et al. Generation of infectious molecular clones of simian immunodeficiency virus from fecal consensus sequences of wild chimpanzees. . J Virol. 2007;81(14):7463–7475. doi: 10.1128/JVI.00551-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Heuverswyn F, et al. Genetic diversity and phylogeographic clustering of SIVcpzPtt in wild chimpanzees in Cameroon. Virology. 2007;368(1):155–171. doi: 10.1016/j.virol.2007.06.018. [DOI] [PubMed] [Google Scholar]
- 24.Adachi A, et al. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59(2):284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papkalla A, Münch J, Otto C, Kirchhoff F. Nef enhances human immunodeficiency virus type 1 infectivity and replication independently of viral coreceptor tropism. J Virol. 2002;76(16):8455–8459. doi: 10.1128/JVI.76.16.8455-8459.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghosh SK, et al. A molecular clone of HIV-1 tropic and cytopathic for human and chimpanzee lymphocytes. Virology. 1993;194(2):858–864. doi: 10.1006/viro.1993.1331. [DOI] [PubMed] [Google Scholar]
- 27. Li Y, Kappes JC, Conway JA, Price RW, Shaw GM, Hahn BH. Molecular characterization of human immunodeficiency virus type 1 cloned directly from uncultured human brain tissue: identification of replication-competent and -defective viral genome s J Virol. 1991. 65 8 3973 3985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, et al. Complete nucleotide sequence, genome organization, and biological properties of human immunodeficiency virus type 1 in vivo: evidence for limited defectiveness and complementation. J Virol. 1992;66(11):6587–6600. doi: 10.1128/jvi.66.11.6587-6600.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koyanagi Y, Miles S, Mitsuyasu RT, Merrill JE, Vinters HV, Chen IS. Dual infection of the central nervous system by AIDS viruses with distinct cellular tropisms. Science. 1987;236(4803):819–822. doi: 10.1126/science.3646751. [DOI] [PubMed] [Google Scholar]
- 30.Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infection by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72(4):2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei X, et al. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother. 2002;46(6):1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schoggins JW, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472(7344):481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bushman FD, et al. Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS Pathog. 2009;5(5):e1000437. doi: 10.1371/journal.ppat.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jäger S, et al. Global landscape of HIV-human protein complexes. Nature. 2011;481(7381):365–370. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451(7177):425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 36.Van Damme N, et al. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe. 2008;3(4):245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jia B, et al. Species-specific activity of SIV Nef and HIV-1 Vpu in overcoming restriction by tetherin/BST2. PLoS Pathog. 2009;5(5):1000429. doi: 10.1371/journal.ppat.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sauter D, et al. Tetherin-driven evolution of Vpu and Nef function and the emergence of pandemic and non-pandemic HIV-1 strains. Cell Host Microbe. 2009;6(5):409–421. doi: 10.1016/j.chom.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang F, et al. Nef proteins from simian immunodeficiency viruses are tetherin antagonists. Cell Host Microbe. 2009;6(1):54–67. doi: 10.1016/j.chom.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rücker E, Grivel JC, Münch J, Kirchhoff F, Margolis L. Vpr and Vpu are important for efficient human immunodeficiency virus type 1 replication and CD4+ T-cell depletion in human lymphoid tissue ex vivo. J Virol. 2004;78(22):12689–12693. doi: 10.1128/JVI.78.22.12689-12693.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang SJ, Lopez LA, Exline CM, Haworth KG, Cannon PM. Lack of adaptation to human tetherin in HIV-1 group O and P. Retrovirology. 2011;8:78. doi: 10.1186/1742-4690-8-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sauter D, et al. HIV-1 Group P is unable to antagonize human tetherin by Vpu, Env or Nef. . Retrovirology. 2011;8:103. doi: 10.1186/1742-4690-8-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rudicell RS, et al. High prevalence of simian immunodeficiency virus infection in a community of savanna chimpanzees. J Virol. 2011;85(19):9918–9928. doi: 10.1128/JVI.05475-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hearps AC, Jans DA. Regulating the functions of the HIV-1 matrix protein. AIDS Res Hum Retroviruses. 2007;23(3):341–346. doi: 10.1089/aid.2006.0108. [DOI] [PubMed] [Google Scholar]
- 45.Bukrinskaya A. HIV-1 matrix protein: a mysterious regulator of the viral life cycle. Virus Res. 2007;124(1–2):1–11. doi: 10.1016/j.virusres.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 46.Batonick M, Favre M, Boge M, Spearman P, Honing S, Thali M. Interaction of HIV-1 Gag with the clathrin-associated adaptor AP-2. Virology. 2005;342(2):190–200. doi: 10.1016/j.virol.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 47.Camus G, et al. The clathrin adaptor complex AP-1 binds HIV-1 and MLV Gag and facilitates their budding. Mol Biol Cell. 2007;18(8):3193–3203. doi: 10.1091/mbc.E06-12-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dong X, et al. AP-3 directs the intracellular trafficking of HIV-1 Gag and plays a key role in particle assembly. Cell. 2005;120(5):663–174. doi: 10.1016/j.cell.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 49.Tang Y, et al. Cellular motor protein KIF-4 associates with retroviral Gag. J Virol. 1999;73(12):10508–10513. doi: 10.1128/jvi.73.12.10508-10513.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghanam RH, Fernandez TF, Fledderman EL, Saad JS. Binding of calmodulin to the HIV-1 matrix protein triggers myristate exposure. J Biol Chem. 2010;285(53):41911–41920. doi: 10.1074/jbc.M110.179093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lopez-Verges S, Camus G, Blot G, Beauvoir R, Benarous R, Berlioz-Torrent C. Tail-interacting protein TIP47 is a connector between Gag and Env and is required for Env incorporation into HIV-1 virions. Proc Natl Acad Sci U S A. 2006;103(40):14947–14952. doi: 10.1073/pnas.0602941103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cimarelli A, Luban J. Translation elongation factor 1-alpha interacts specifically with the human immunodeficiency virus type 1 Gag polyprotein. . J Virol. 1999;73(7):5388–5401. doi: 10.1128/jvi.73.7.5388-5401.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gupta K, Ott D, Hope TJ, Siliciano RF, Boeke JD. A human nuclear shuttling protein that interacts with human immunodeficiency virus type 1 matrix is packaged into virions. J Virol. 2000;74(24):11811–11824. doi: 10.1128/JVI.74.24.11811-11824.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mansharamani M, et al. Barrier-to-autointegration factor BAF binds p55 Gag and matrix and is a host component of human immunodeficiency virus type 1 virions. J Virol. 2003;77(24):13084–13092. doi: 10.1128/JVI.77.24.13084-13092.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Samelson LE. Signal transduction mediated by the T cell antigen receptor: the role of adapter proteins. Annu Rev Immunol. 2002;20:371–394. doi: 10.1146/annurev.immunol.20.092601.111357. [DOI] [PubMed] [Google Scholar]
- 56.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 57.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 58.Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25(7):1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 59.Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21(9):2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 60.Münch J, et al. Nef-mediated enhancement of virion infectivity and stimulation of viral replication are fundamental properties of primate lentiviruses. J Virol. 2007;81(24):13852–13864. doi: 10.1128/JVI.00904-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Decker JM, et al. Effective activation alleviates the replication block of CCR5-tropic HIV-1 in chimpanzee CD4+ lymphocytes. Virology. 2009;394(1):109–118. doi: 10.1016/j.virol.2009.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.