Abstract
In contrast to the well-studied classic MAPKs, such as ERK1/2, little is known concerning the regulation and substrates of the atypical MAPK ERK3 signaling cascade and its function in cancer progression. Here, we report that ERK3 interacted with and phosphorylated steroid receptor coactivator 3 (SRC-3), an oncogenic protein overexpressed in multiple human cancers at serine 857 (S857). This ERK3-mediated phosphorylation at S857 was essential for interaction of SRC-3 with the ETS transcription factor PEA3, which promotes upregulation of MMP gene expression and proinvasive activity in lung cancer cells. Importantly, knockdown of ERK3 or SRC-3 inhibited the ability of lung cancer cells to invade and form tumors in the lung in a xenograft mouse model. In addition, ERK3 was found to be highly upregulated in human lung carcinomas. Our study identifies a previously unknown role for ERK3 in promoting lung cancer cell invasiveness by phosphorylating SRC-3 and regulating SRC-3 proinvasive activity by site-specific phosphorylation. As such, ERK3 protein kinase may be an attractive target for therapeutic treatment of invasive lung cancer.
Introduction
ERK3, also known as MAPK6, is a member of the atypical MAPK subfamily (1). Other atypical MAPKs include ERK4 (MAPK4) and Nemo-like kinase (NLK). ERK3 and ERK4 possess a single Ser-Glu-Gly phospho-acceptor motif in their activation loop, instead of the Thr-Xaa-Tyr motif conserved in the classic MAPKs, such as ERK1/2 (2). Unlike ERK1 and ERK2, which have been studied extensively, little is known about the upstream stimuli and activators of ERK3 or about its downstream targets. As opposed to most other ERK kinases, phosphorylation of the activation loop of ERK3 is not affected by classic mitogenic stimuli, including serum and phorbol esters, such as PMA (3). To date, the only known ERK3 substrate is another kinase, MAPK-activated protein kinase 5 (MK5) (4–6). Several lines of evidence suggest that ERK3 is involved in cell differentiation and cell cycle regulation (7–9). A physiological role for ERK3 was revealed by ERK3 gene disruption in mice (10); ERK3-deficient mice display intrauterine growth restriction and neonatal lethality, mainly due to pulmonary immaturity. Interestingly, we noted that ERK3 expression was upregulated in several human cancer microarray databases (11–14), but the function of ERK3 in general or lung cancer development and progression is virtually unknown.
Steroid receptor coactivator 3 (SRC-3) is overexpressed in multiple cancers, including breast, prostate, lung, pancreatic, and intestinal cancers, and has been defined as a common bona fide oncogene (15–17). SRC-3 acts as a coactivator of nuclear receptors and other transcription factors. Interestingly, the SRC-3 gene promotes not only cell proliferation and transformation but also cancer cell migration and invasion (17, 18). In cultured cells, SRC-3 promotes cancer cell invasion by coactivating PEA3- and AP-1–regulated MMP expression (19–21), but the invasive signals to SRC-3 are completely unknown. SRC-3 is phosphorylated at multiple residues upon the stimulation of growth factors or hormones (22–24). These posttranslational modifications (PTMs) regulate SRC-3 protein stability and its activities in gene transcription. However, whether SRC-3 PTMs are involved in cancer cell invasion and tumor metastasis have not been elucidated. Also, nothing is known concerning responsible kinases creating PTMs that promote invasion.
In this study, we found that ERK3 regulates invasion by interacting with and phosphorylating SRC-3 specifically at serine 857 (S857); phosphorylation at S857 by ERK3 is essential for the interaction of SRC-3 with the ETS transcription factor PEA3, its regulation of MMP gene expression, and its proinvasive activity in lung cancer cells. Importantly, depletion of ERK3 and SRC-3 almost abolishes the ability of lung cancer cells to invade and form tumors in the lungs in a xenograft mouse model. In addition, we found that ERK3 expression is highly upregulated in human lung carcinomas (7.363 fold; P = 3.444 × 10–7). Our study identifies what we believe to be a novel function of ERK3 in promoting lung cancer cell invasion through phosphorylation of SRC-3 and its regulation of SRC-3 proinvasive activity by site-specific phosphorylation. Since invasion and metastasis are criteria critical for survival from cancer, our observations have obvious clinical importance.
Results
ERK3 interacts with and phosphorylates SRC-3 at S857.
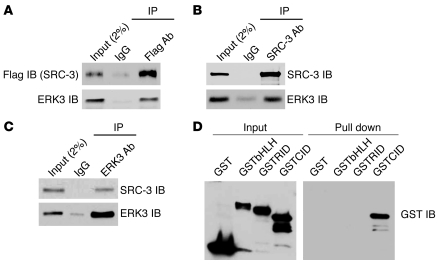
Although ERK3 was identified 20 years ago (2), little is known about its upstream activators and its downstream substrates. As a continuation of our coregulator immunoprecipitation–mass spectrometry analyses in the NURSA project (25), we identified ERK3 as a coimmunoprecipitating protein with SRC-3. The interaction between these 2 proteins was confirmed by immunoprecipitation–Western blotting analysis in H1299 lung cancer cells. Immunoprecipitation of Flag-tagged SRC-3 from transfected H1299 cells using a Flag Ab (Figure 1A) or endogenous SRC-3 from parental H1299 cells using a SRC-3–specific Ab (Figure 1B) substantiated coimmunoprecipitation of ERK3 in both cases. Similarly, immunoprecipitation of endogenous ERK3 from H1299 cells using an ERK3-specific Ab (Figure 1C) also pulled down SRC-3 protein. To identify which region in SRC-3 is important for its interaction with ERK3, an in vitro pull down assay was performed by incubating purified ERK3 protein immobilized on beads (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI61492DS1) with GST-SRC-3 fragment fusion proteins. As shown in Figure 1D, the CBP-interacting domain-containing (CID) region of SRC-3 interacts with ERK3. Taken together, these results demonstrate that SRC-3 directly interacts with the ERK3 through the CID region.
Figure 1. ERK3 interacts with SRC-3.
(A) H1299 cells were transfected with pSG5SRC-3Flag. The interaction between endogenous ERK3 and exogenously expressed SRC-3 was analyzed by coimmunoprecipitation using a Flag Ab or a corresponding control IgG, followed by Western blotting. (B and C) The interaction between endogenous ERK3 and SRC-3 in H1299 cells was analyzed by coimmunoprecipitation using (B) a SRC-3 Ab or (C) an ERK3 Ab. (D) ERK3 interacts with SRC-3 at the CID region in vitro. The N-terminal basic helix-loop-helix domain-containing region of SRC-3 (bHLH), receptor interaction domain-containing region (RID), and CID region were expressed as GST-fusion proteins in E. coli. A protein-protein pull down assay was performed by incubating each purified GST-SRC-3 fragment fusion protein with purified Myc-ERK3 immobilized on agarose beads. Western blots were probed with a GST Ab.
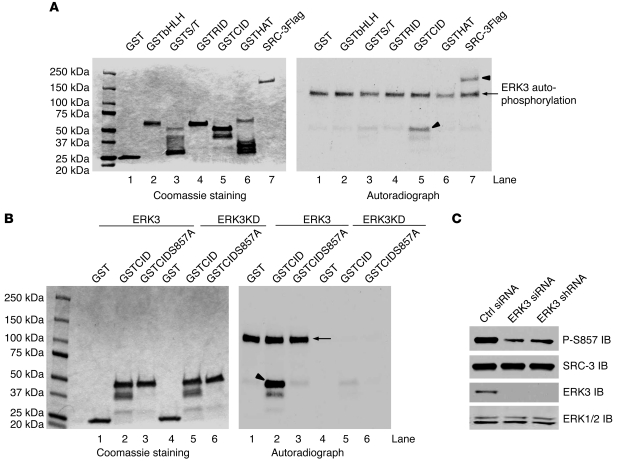
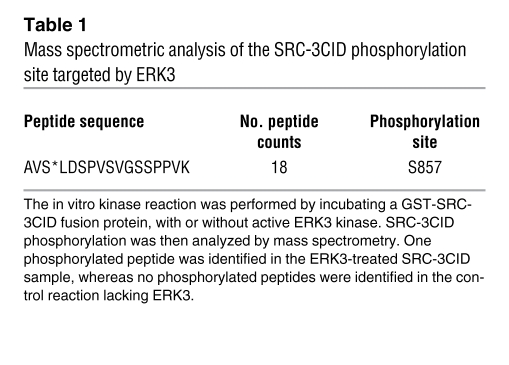
To determine the functional interplay between these 2 proteins, we first tested whether ERK3 phosphorylates SRC-3. By performing an in vitro kinase assay using purified ERK3 kinase (the eluted ERK3; Supplemental Figure 1), we found that ERK3 phosphorylated full-length SRC-3Flag (indicated by arrowhead in lane 7, right panel, Figure 2A). To determine which region of SRC-3 was phosphorylated by ERK3, a series of GST-SRC-3 fragment fusion proteins (Figure 2A) were added as substrates to the in vitro kinase reaction. As shown in Figure 2A, ERK3 predominantly phosphorylated the CID-containing region of SRC-3 (indicated by arrowhead in lane 5, right panel, Figure 2A). To further identify the specific phosphorylation site(s) of SRC-3 by ERK3, GST-CID fusion protein was phosphorylated by ERK3 in vitro and then analyzed by mass spectrometry. Only S857 within the CID region of SRC-3 was phosphorylated (Table 1). Consistent with this identification, mutation of S857 to alanine abolished phosphorylation of the CID by ERK3 (compare lane 3 to lane 2, right panel, Figure 2B). We developed a phospho-specific Ab against this S857 residue (22). To demonstrate the importance of ERK3 in SRC-3 phosphorylation at S857 in cultured cells, endogenous ERK3 in H1299 cells was knocked down by RNAi targeting of ERK3 (either by siRNA or shRNA), and SRC-3 phosphorylation at S857 was assayed by Western blotting using the phospho-S857–specific Ab. The efficacy and specificity ERK3 siRNA or shRNA were demonstrated by the depletion of ERK3 protein and lack of change on ERK1/2 expression, respectively. Importantly, knockdown of ERK3 inhibited SRC-3 phosphorylation at S857.
Figure 2. ERK3 phosphorylates SRC-3 at S857.
(A) ERK3 phosphorylates SRC-3 at the CID region in vitro. SRC-3Flag protein was expressed in Sf9 cells and purified using anti-Flag beads. The N-terminal basic helix-loop-helix domain-containing region of SRC-3 (aa 1–320), serine/threonine-rich region (S/T; aa 321–580), receptor interaction domain-containing region (aa 581–840; lane 5), CID region (aa 841–1080), and histone acetyltransferase domain-containing region (HAT; aa 1081–1424) were expressed as GST-fusion proteins in E. coli. In vitro kinase assay was performed by incubating purified SRC-3Flag (lane 7) or GSTSRC-3 fragment proteins (see Coomassie staining) with purified ERK3 kinase. ERK3 autophosphorylation (arrow, lane 7) and phosphorylation of the substrates (arrowheads, lane 5 and 7) are shown in the autoradiograph in the right panel. (B) ERK3 phosphorylates SRC-3 at S857 within the CID region in vitro. GST, GSTCID, and GSTCIDS857A proteins (see Coomassie staining) were incubated with either purified active ERK3 protein or ERK3KD protein. The arrow and arrowhead indicate ERK3 autophosphorylation and GSTCID phosphorylation, respectively. (C) Knockdown of ERK3 decreases SRC-3 phosphorylation at S857 in H1299 cells. ERK3 was knocked down by transfecting cells with ERK3 siRNA or by transducing cells with lentiviruses expressing ERK3 shRNA. SRC-3 phosphorylation at S857 was analyzed by Western blotting using an Ab that specifically recognizes phosphorylated S857.
Table 1 .
Mass spectrometric analysis of the SRC-3CID phosphorylation site targeted by ERK3
ERK3 and SRC-3 signaling axis regulates lung cancer cell migration and invasion.
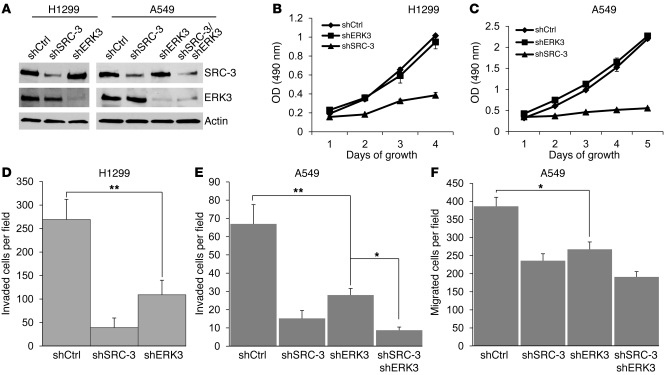
Genetic disruption of the murine Erk3 gene resulted in a lethal lung developmental defect (10), but the function of ERK3 in lung cancer is unclear. SRC-3 gene amplification and protein overexpression have been observed in lung cancer, and depletion of SRC-3 greatly inhibited growth of lung cancer cells (26, 27). The finding that SRC-3 is a novel target of ERK3 to our knowledge suggests that ERK3 may play an important role in lung cancer. As such, we determined the functions of ERK3 in lung cancer cell growth, migration, and invasion. H1299 and A549 cell pools with stable knockdown of ERK3, SRC-3, or both were generated by lentiviral expression of shRNAs targeting ERK3, SRC-3, or both, respectively (Figure 3A). Consistent with the published effects of SRC-3 in cellular proliferation, knockdown of SRC-3 inhibited the proliferation of H1299 (Figure 3B) and A549 cells (Figure 3C), whereas depletion of ERK3 had no significant effect. In contrast, knockdown of either ERK3 or SRC-3 greatly decreased H1299 (Figure 3D) and A549 (Figure 3E) cell invasion and inhibited A549 cell migration (Figure 3F). The inhibition of lung cancer cell invasion by knockdown of ERK3 and SRC-3 was confirmed in 2 other lung cancer cell lines, H441 and H520 (Supplemental Figure 2, A and B). Interestingly, exogenous overexpression of Myc-tagged ERK3 significantly increased invasion of A549 (lane 2, Figure 4A) and H520 cells (Supplemental Figure 3A). However, ERK3 lost the capability to promote invasion of A549 (lane 4, Figure 4A) and H520 cells (Supplemental Figure 3A) when SRC-3 was depleted, suggesting that ERK3 promotes A549 cell invasion in a SRC-3–dependent manner. Cell migration and invasion are regulated by cytoskeletal organization, including actin polymerization (28). It is noteworthy that, despite their important role in promoting cell invasion, knockdown of either ERK3 or SRC-3 did not significantly alter actin polymerization (Supplemental Figure 4) in H1299 cells.
Figure 3. Knockdown of SRC-3 and ERK3 inhibits lung cancer cell invasion and migration.
(A) H1299 and A549 cells were stably transduced with lentiviral shSRC-3, shERK3, or the nontargeting control shRNA (shCtrl). Knockdown of SRC-3 and ERK3 was verified by Western blotting analysis using a SRC-3 Ab and an ERK3 Ab, respectively. (B and C) MTS cell proliferation assay of (B) H1299 and (C) A549 cell pools stably expressing shSRC-3, shERK3, or shCtrl. Depletion of SRC-3 inhibits the proliferation of lung cancer (B) H1299 and (C) A549 cells, whereas depletion of ERK3 has no significant effect. (D) Transwell Matrigel cell invasion assay of H1299 cell pools stably expressing shSRC-3, shERK3, or shCtrl. Depletion of SRC-3 or ERK3 inhibits H1299 cell invasion. (E) Transwell Matrigel cell invasion assay of A549 cell pools stably expressing shCtrl, shSRC-3, or shERK3 or coexpressing shSRC-3 and shERK3. Depletion of SRC-3 or ERK3 inhibits A549 cell invasion. (F) Transwell cell migration assay of A549 stable cell pools. Depletion of SRC-3 or ERK3 inhibits A549 cell migration. Values are mean ± SEM of 4 separate experiments. *P < 0.05, **P < 0.005 (Student’s t test).
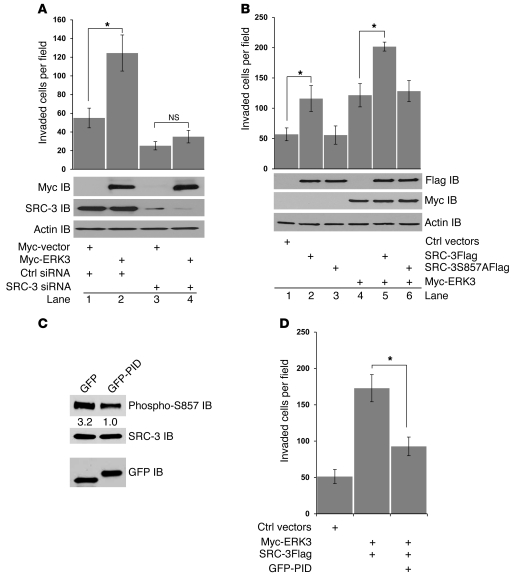
Figure 4. ERK3 promotes A549 cell invasion in a SRC-3–dependent manner, and S857 phosphorylation of SRC-3 is critical in promoting cell invasion.
(A) Transwell Matrigel cell invasion assay of A549 cells transiently transfected with Myc-ERK3 or the empty vector or together with either SRC-3 siRNA or nontargeting control siRNA. ERK3 overexpression and SRC-3 knockdown were demonstrated by Western blotting using a Myc Ab and a SRC-3 Ab, respectively. ERK3 lost the capability of promoting A549 cell invasion when SRC-3 was depleted. *P < 0.05. (B) Transwell Matrigel cell invasion assay of A549 cells transiently transfected with SRC-3Flag, SRC-3S857AFlag, or Myc-ERK3 or cotransfected with 2 of the constructs as indicated. Exogenous expression of SRC-3 and/or ERK3 was analyzed by Western blotting using a Flag Ab and a Myc Ab, respectively. Intact S857 is required for SRC-3 to promote cell invasion. (C and D) Inhibition of PAKs by expression of the PID greatly decreased the phosphorylation of SRC-3 at S857 and the effect of ERK3 and SRC-3 expression in cell invasion. GFP or GFP-PID fusion protein was overexpressed in A549 cells. (C) The effect of GFP-PID overexpression on the phosphorylation of SRC-3 at S857 was determined by Western blotting using an Ab that specifically recognizes phosphorylated S857. Numbers represent the relative intensity of the protein bands. The band intensity in the right lane is set as “1.0.” (D) The effect of GFP-PID overexpression on A549 cell invasion that was stimulated by ERK3 and SRC-3 was analyzed by transwell Matrigel cell invasion assay. *P < 0.01.
Given that ERK3 phosphorylates SRC-3 at S857, we postulated that S857 phosphorylation could play an important role in SRC-3–mediated cell invasion. Indeed, mutation of S857 to alanine (S857A) abolished the function of SRC-3 in promoting cell invasion (comparing lane 3 with lane 2, Figure 4B and Supplemental Figure 3B). In addition, exogenous co-overexpression of ERK3 and SRC-3 further significantly enhanced lung cancer cell invasion in comparison with overexpression of either ERK3 or SRC-3 alone (Figure 4B and Supplemental Figure 3B). In contrast, the SRC-3S857A mutant was not able to cooperatively work with ERK3 to promote cell invasion (comparing ERK3 alone with coexpression of ERK3 and SRC-3S857A). Taken together, these results demonstrate that ERK3 and SRC-3 cooperatively promote lung cancer cell invasion, which is mediated by the phosphorylation site S857 in SRC-3.
ERK3 was recently shown to be activated by group I p21-activated kinases (PAKs) that include PAK1, PAK2, and PAK3 (29, 30). Group I PAKs are important regulators for cell migration and invasion. The activities of PAKs are autoinhibited due to the interaction of the PAK autoinhibitory domain (PID) with the kinase domain, whereas the binding of Rac-1 or cdc42 to PAKs will release the autoinhibition and activates PAKs (31). On the basis of this molecular mechanism, exogenous overexpression of GFP-PID was shown to effectively inhibit the activities of group I PAKs in the cells (32). To determine the role of PAKs in regulating ERK3/SRC-3–mediated cell invasion, GFP-PID was overexpressed in A549 cells (Figure 4C). Interestingly, overexpression of GFP-PID greatly reduced phosphorylation of SRC-3 at S857 (Figure 4C) and significantly decreased A549 cell invasion induced by ERK3/SRC-3 (Figure 4D). This result indicates an important role of PAK/ERK3/SRC-3 in this signaling pathway for promoting lung cancer cell invasion.
ERK3 and SRC-3 signaling axis regulates MMP gene expression.
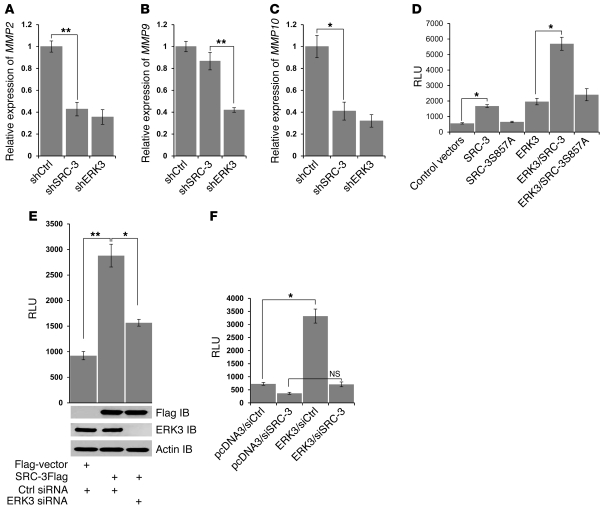
SRC-3 coactivates the ETS transcription factor PEA3 in upregulating the expressions of certain MMP genes, which are critical for their functions in promoting prostate and breast cancer cell invasion (19, 20). Thus, we tested whether ERK3 regulates MMP gene expression and whether ERK3-mediated phosphorylation of SRC-3 at S857 is important in this process. To determine the importance of ERK3 and SRC-3 signaling in regulating MMP gene expression, we investigated the effect of ERK3 and SRC-3 knockdown on endogenous MMP genes in lung cancer cells by real-time RT-qPCR analysis. We screened MMP2, MMP9, and MMP10, as these genes are PEA3/SRC-3 direct target genes in breast and prostate cancer cells (19, 20). Depletion of either ERK3 or SRC-3 significantly inhibited the expression of MMP2 and MMP10 in H1299 (Figure 5, A and C), A549 (Supplemental Figure 5, A and B), and H441 cells (Supplemental Figure 5, C and D), whereas MMP9 was only affected by the loss of ERK3 function (Figure 5B).
Figure 5. ERK3 and SRC-3 cooperatively regulate MMP gene expression.
(A–C) Depletion of SRC-3 or ERK3 significantly decreased (A) MMP2, (B) MMP9, and (C) MMP10 gene expression. MMP gene expression in H1299 cell pools stably expressing shSRC-3, shERK3, or shCtrl was determined by RT-qPCR analysis. *P < 0.01, **P < 0.005 (Student’s t test). (D) SRC-3 and ERK3 cooperatively promote MMP2 promoter-driven luciferase activity, in which S857 of SRC-3 plays an important role. A549 cells were singly transfected or cotransfected with the constructs as indicated. Luciferase activity is represented as relative luciferase units (RLUs) on the y axis. *P < 0.01 (Student’s t test). (E) Knockdown of ERK3 significantly decreased SRC-3–induced MMP2 promoter-driven luciferase activity. A549 cells were transfected with SRC-3Flag or the empty vector or together with either ERK3 siRNA or nontargeting control siRNA (Ctrl siRNA). *P < 0.05, **P < 0.001 (Student’s t test). (F) Knockdown of SRC-3 significantly decreased ERK3-induced MMP2 promoter-driven luciferase activity. A549 cells were transfected with Myc-ERK3 or the empty vector or together with either SRC-3 siRNA (siSRC-3) or nontargeting control siRNA (siCtrl). Values represent the mean ± SEM of 3 separate experiments. *P < 0.01 (Student’s t test).
To further test whether ERK3-mediated phosphorylation of SRC-3 at S857 is important for MMP gene expression, we transfected A549 cells with an MMP2 promoter-driven luciferase reporter and SRC-3 or SRC-3S857A in the presence or the absence of ERK3. As expected, overexpression of SRC-3 enhanced MMP2 promoter activity (Figure 5D), and, importantly, either mutation of S857 to alanine (Figure 5D) or knockdown of ERK3 (Figure 5E) remarkably inhibited its effect. Expression of ERK3 significantly increased MMP2 promoter-driven luciferase activity (Figure 5F). However, ERK3 was not able to significantly enhance MMP2 promoter-driven luciferase activity when SRC-3 was depleted (Figure 5F), suggesting that ERK3 promotes MMP2 gene transcriptional activity in a SRC-3–dependent manner. In addition, co-overexpression of ERK3 and SRC-3 further enhanced MMP2 promoter-driven luciferase activity in comparison with overexpression of either ERK3 or SRC-3 alone (Figure 5D). In contrast, ERK3 was not able to functionally cooperate with the SRC-3S857A mutant in this regard (comparing expression of ERK3 alone with coexpression of ERK3 and SRC-3S857A, Figure 5D). Similar effects of ERK3 and SRC-3 in regulating MMP2 promoter-driven luciferase activity were observed in H1299 cells (Supplemental Figure 5, E and F). Taken together, these results suggest that ERK3 and SRC-3 cooperatively regulate MMP2 gene promoter activity via the phosphorylation site S857 of SRC-3.
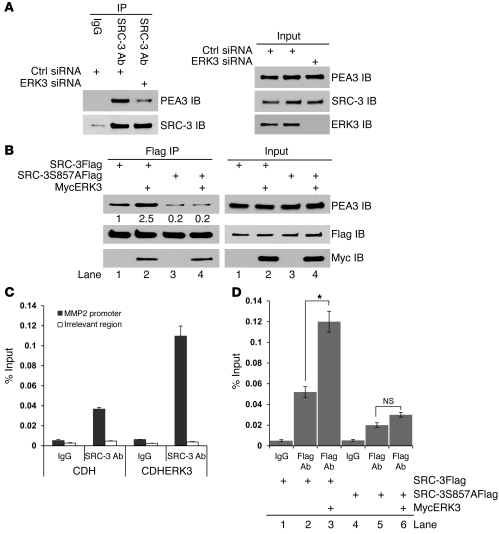
ERK3 promotes the interaction of SRC-3 with PEA3 and occupancy of SRC-3 on the MMP2 promoter, which is mediated by the phosphorylation site S857.
After revealing the role of ERK3 and SRC-3 signaling in upregulating MMP gene expression, we went further to elucidate the underlying molecular mechanisms. SRC-3 is known to interact with PEA3 and coactivate PEA3-regulated MMP gene expression (19, 20). Our evidence that ERK3 and SRC-3 cooperatively regulate MMP2 gene promoter activity through ERK3-mediated phosphorylation of SRC-3 at S857 indicates that ERK3 regulates the interaction of SRC-3 with PEA3 through S857 phosphorylation. Indeed, knockdown of ERK3 in H1299 cells greatly decreased the interaction of SRC-3 with PEA3 (Figure 6A), whereas overexpression of ERK3 increased the interaction between SRC-3 and PEA3 (compare lane 2 to lane 1, left panel, Figure 6B). Importantly, the SRC-3S857A mutant had much weaker ability to interact with PEA3 than did wild-type SRC-3 (compare lane 3 to lane 1, left panel, Figure 6B), although its expression levels were similar to those of wild-type (see input panel, Figure 6B). In addition, ERK3 was not able to promote the interaction of SRC-3S857A with PEA3 (compare lane 4 to lane 3, left panel, Figure 6B), suggesting that phosphorylation of S857 by ERK3 is critical for SRC-3 to interact with PEA3. Consistent with its effect on the interaction of SRC-3 with PEA3, exogenous expression of ERK3 promoted SRC-3 protein occupancy at the PEA3 binding site of the MMP2 gene promoter, no matter whether SRC-3 protein was endogenously (Figure 6C) or exogenously expressed (Figure 6D). In contrast, the SRC-3S857A mutant had greatly decreased occupancy at the MMP2 promoter as compared with that of wild-type SRC-3 (comparing lane 5 with lane 2, Figure 6D), and ERK3 was not able to promote its occupancy at this site (comparing lane 6 with lane 5, Figure 6D), indicating that phosphorylation of S857 by ERK3 is critical for SRC-3 to be recruited to the PEA3 binding site of the MMP2 gene promoter. Taken together, these results demonstrate that ERK3 upregulates MMP2 gene expression by phosphorylating SRC-3 at S857, thereby promoting the interaction of SRC-3 with PEA3 and its occupancy at the MMP2 promoter.
Figure 6. ERK3 promotes the interaction of SRC-3 with PEA3 and occupancy of SRC-3 on the PEA3 binding site of MMP2 gene promoter, which is mediated by the phosphorylation site S857.
(A) Knockdown of ERK3 decreases the interaction of SRC-3 with PEA3. H1299 cells were transfected with ERK3 siRNA or nontargeting control siRNA. Coimmunoprecipitation was performed using a SRC-3 Ab or a mouse IgG, followed by Western blotting. (B) ERK3 promotes the interaction of SRC-3 with PEA3, which is mediated by the phosphorylation site S857. H1299 cells were transfected with SRC-3Flag or SRC-3S857AFlag or together with Myc-tagged ERK3 (MycERK3). Coimmunoprecipitation was performed using a Flag Ab. Numbers below the PEA3 immunoblots in Flag immunoprecipitation represent the relative intensity of the protein bands. The band intensity in lane 1 is set as “1.0.” (C) Overexpression of ERK3 enhanced occupancy of SRC-3 on the MMP2 promoter. H1299 cells were stably transduced with either lentiviral vector CDH or lentiviral ERK3 (CDHERK3). ChIP assays were performed using either a SRC-3 Ab or goat IgG. SRC-3 protein occupancy on the PEA3 binding site of MMP2 gene promoter (700-bp upstream of transcription start site; ref. 19) was analyzed by quantitative real-time PCR and presented as the percentage of sheared chromatin input. (D) ERK3 promotes the occupancy of SRC-3, but not the SRC-3S857A mutant, on the MMP2 promoter. A549 cells were singly transduced or cotransduced stably with the lentiviral constructs as indicated. ChIP assays were performed using either a Flag Ab or mouse IgG. *P < 0.01.
ERK3 and SRC-3 signaling axis regulates MMP activities, which are critical for ERK3 and SRC-3 in promoting lung cancer cell invasion.
MMPs promote cell invasion by acting as secreted proteases and degrading cell matrix. To test whether ERK3 and SRC-3 signaling also regulates MMP activities, in addition to MMP gene expression, we measured the enzymatic activities of secreted MMP2 and MMP10 in the conditioned media from H1299 cells. Consistent with their effects in regulating MMP gene expression, depletion of either ERK3 or SRC-3 significantly reduced the activities of MMP2 and MMP10 in H1299 cells (Supplemental Figure 6, A and B). On the contrary, exogenous expression of either ERK3 or SRC-3 significantly increased the activity of MMP2 (Supplemental Figure 6C). Importantly, mutation of S857 to alanine (S857A) abolished the effect of SRC-3 in enhancing the activity of MMP2. In addition, co-overexpression of ERK3 and SRC-3 further enhanced the activity of MMP2 in comparison with overexpression of either ERK3 or SRC-3 alone (Supplemental Figure 6C). ERK3 was not able to functionally cooperate with the SRC-3S857A mutant in this regard (comparing expression of ERK3 alone with coexpression of ERK3 and SRC-3S857A, Supplemental Figure 6C).
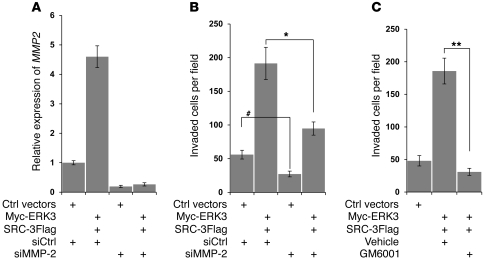
After determining the important roles of ERK3/SRC-3 signaling in regulating MMP gene expression and enzymatic activities, we went further to test whether MMPs are required for ERK3/SRC-3–mediated lung cell invasion. As shown in Figure 7, knockdown of MMP2 (Figure 7A), an important target of ERK3/SRC-3 signaling as defined in our study, significantly inhibited the function of ERK3 and SRC-3 in promoting A549 cell invasion (Figure 7B). In addition, treatment with GM6001 (a pan-MMP inhibitor) abolished the effect of ERK3 and SRC-3 expression in promoting cell invasion (Figure 7C). Taken together, these results suggest that MMPs are essential for ERK3 and SRC-3 signaling to promote lung cancer cell invasion.
Figure 7. Depletion of MMP2 expression or inhibition of MMP activity inhibits the effect of ERK3 and SRC-3 in promoting A549 cell invasion.
(A and B) A549 cells were transiently cotransfected with both Myc-ERK3 and SRC-3Flag or the empty vectors or together with either MMP2 siRNA (siMMP2) or nontargeting control siRNA. (A) MMP2 mRNA expression was determined by RT-qPCR analysis. (B) Cell invasion was measured by Transwell Matrigel cell invasion assay. (C) A549 cells were transiently cotransfected with both Myc-ERK3 and SRC-3Flag or the empty vectors. Transwell Matrigel cell invasion assay was performed by adding GM6001 (10 μM), an MMP inhibitor, or the vehicle at the same concentration in the upper chamber of the transwell. Values represent the mean ± SEM of 3 separate experiments. *P < 0.01, **P < 0.001, #P < 0.05 (Student’s t test).
ERK3 and SRC-3 are important for lung cancer cell invasiveness in vivo.
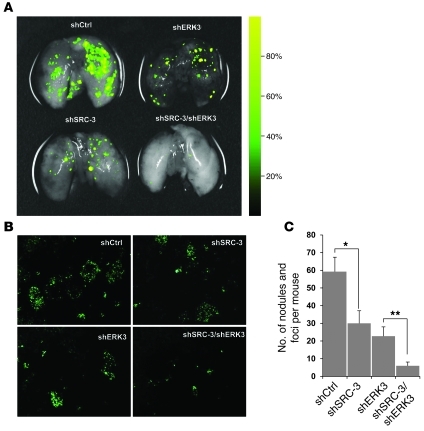
To determine whether ERK3 and SRC-3 are important for lung cancer cell invasiveness in vivo, we injected H1299 cells into mice via tail vein injection and analyzed cancer cell colonization and tumor formation in lungs 5 weeks after inoculation. As shown in Figure 8 and Supplemental Figure 7, knockdown of either ERK3 or SRC-3 greatly decreased the ability of H1299 cells to invade and form tumors in the lung. Most strikingly, knockdown of both ERK3 and SRC-3 almost abolished the ability of H1299 cells to form tumor nodules in the lung, indicating that ERK3/SRC-3 signaling plays a critical role in promoting lung cancer cell invasiveness in vivo.
Figure 8. Both SRC-3 and ERK3 are important for the invasiveness of H1299 lung cancer cells in vivo.
1 × 106 GFP-labeled H1299 cells with stable expression of the control shRNA (shCtrl), SRC-3 shRNA (shSRC-3), ERK3 shRNA (shERK3), or both shSRC3 and shERK3 were injected into SCID/Beige mice via tail vein. Five weeks after inoculation, formation of tumor nodules in the lungs was analyzed by GFP imaging of (A) the excised lungs or (B) frozen sections. Images shown in A are overlays of GFP-fluorescent images, with the corresponding white-light counterparts. The color scale bar at the right indicates GFP fluorescence intensity of the tumor nodules. Original magnification, ×50 (B). (C) The number of tumor nodules and foci in lungs was counted on the basis of GFP imaging. Values represent the mean ± SEM of 15 mice. *P < 0.05, **P < 0.001 (Student’s t test).
ERK3 expression is upregulated in human lung carcinomas.
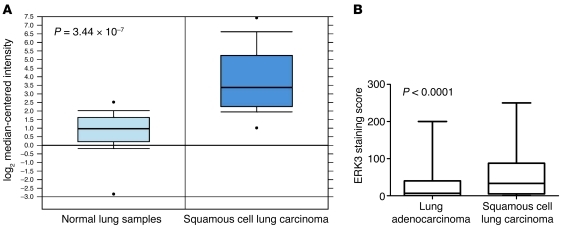
To investigate the clinical relevance of ERK3 expression in lung cancer, we first noted that, in an Oncomine mRNA microarray data set of non-small cell lung carcinomas (NSCLCs) (14), ERK3 mRNA expression in squamous cell lung carcinomas (a major type of NSCLCs), compared with that of normal lung tissues, was shown to have a 7.363-fold increase (P = 3.444 × 10–7; Figure 9A). To confirm the upregulation of ERK3 in lung cancer, we analyzed ERK3 protein expression by immunohistochemistry in NSCLC tissue microarrays (TMAs) that contain 302 lung adenocarcinomas and 154 squamous cell lung carcinomas. ERK3 immunostaining was mainly nuclear, and 65% (296 out of 456) of the tumors showed positive ERK3 staining (Supplemental Figure 8). Interestingly, ERK3 protein expression was significantly increased (P < 0.0001; Figure 9B) in squamous cell lung carcinomas in comparison with that in lung adenocarcinomas.
Figure 9. ERK3 is highly upregulated in human lung carcinomas.
(A) ERK3 gene expression is highly upregulated in squamous cell lung carcinomas compared with that in normal lung tissues. ERK3 gene expression in squamous cell lung carcinomas (n = 21) and normal lung samples (n = 17) was analyzed on Affimetrix U95A microarrays (14). The P value was calculated using unpaired 2-tailed t test. Horizontal bars indicate the median; boxes indicate 25th to 75th percentiles; and whiskers indicate 10th and 90th percentiles. (B) Immunohistological analysis of ERK3 protein expression in an NSCLC TMA that contains 302 lung adenocarcinomas and 154 squamous cell lung carcinomas. Statistical significance was determined by unpaired 2-tailed t test.
Discussion
In contrast with the extensive knowledge of the regulation of ERK1/2 signaling pathways and their phosphorylation substrates, little is known about the upstream activators and the downstream targets of ERK3. ERK3 phosphorylation within the activation loop (an indication of kinase activity) is not affected by classical MAPK stimuli, such as serum and phorbol esters. Although ERK3 was shown to be a negative regulator of cell cycle progression in untransformed cultured cells, such as mouse fibroblast NIH3T3 and myoblast C2C12 (7–9), we noted that ERK3 expression is upregulated in different databases for multiple types of cancers, including breast cancer (11), gastric cancer (12), and melanoma (13). Interestingly, ERK3 upregulation was shown to be associated with tumor invasion and metastasis in these studies, suggesting that ERK3 could play a role in promoting cancer invasiveness. Consistent with the above notion, this study identified the SRC-3 oncoprotein as a substrate of ERK3 and revealed an unknown function of ERK3 in promoting lung cancer cell invasion. MK5, another substrate of ERK3, was shown to promote HeLa cell migration by regulating actin cytoskeletal organization (33, 34). In our study, however, ERK3 knockdown did not cause significant changes in actin organization in lung cancer cells (Supplemental Figure 4). In contrast, ERK3 promoted strong cell invasion in a SRC-3–dependent manner (Figure 4A).
SRC-3 oncoprotein is phosphorylated upon a variety of extracellular stimuli, including growth factors, cytokines, and hormones (35). While phosphorylations of SRC-3 by PKCζ and ERK1/2 are important for its pro-proliferative activity (22–24), the functions of SRC-3 phosphorylation(s) and the responsible kinase(s) in cancer cell invasion and tumor metastasis have not been elucidated previously. Here, we identify S857 in SRC-3 as an ERK3 phosphorylation site that is essential for SRC-3 proinvasive activity. Interestingly, as an important step in understanding the upstream signaling events of the ERK3 kinase pathway, 2 recent studies (29, 30) reported that ERK3 was phosphorylated and activated by group I PAKs (PAK1, PAK2, and PAK3) that have been implicated in cell migration and invasion. Although the function of ERK3 in PAK-mediated cell migration and invasion was not investigated in these 2 studies, these findings, together with ours, imply an interesting PAK/ERK3/SRC-3 signaling pathway relationship for cancer cell migration and invasion. In support of this notion, inhibition of the activity of PAKs by overexpression of the PID greatly reduced SRC-3 phosphorylation at S857 and ERK3/SRC-3–mediated cell invasion of A549 lung cancer cells (Figure 4D).
Dysregulation of protein kinase activity is frequently implicated in cancer progression and metastasis, and therapeutic drugs have been developed by targeting these protein kinases (36). The clinical relevance of ERK3 signaling in lung cancer is substantiated by our finding that ERK3 is highly upregulated in NSCLCs. Advanced NSCLC is commonly present as a metastatic disease, which is currently incurable (37). The critical functions of ERK3 in promoting lung cancer cell invasiveness, together with its upregulation in advanced NSCLCs (Figure 9), strongly suggest that ERK3 should be considered for therapeutic targeting of this invasive disease. Another interesting finding from our study is that ERK3 expression is significantly higher in squamous cell lung carcinomas than in lung adenocarcinomas. EGFR kinase inhibitors, such as gefitinib and erlotinib, have been used in the clinic for treating patients with lung cancer (38). Compared with lung adenocarcinomas, squamous cell lung carcinomas have considerably less sensitivity to EGFR kinase inhibitors due to the less frequent overexpression of EGFR and the rare existence of EGFR mutations that increase drug sensitivity (38, 39). As such, the selective overexpression of ERK3 in squamous cell lung carcinoma suggests that ERK3 kinase could be an important therapeutic target for the treatment of this subtype of lung cancer.
Methods
Expression plasmids.
The expression plasmid of SRC-3 with a C-terminal Flag tag (pSG5SRC-3Flag) was described previously (24). The constructs of GST-SRC-3bHLH (aa 1–320), GST-SRC-3S/T (aa 321–580), GST-SRC-3RID (aa 581–840), GST-SRC-3CID (aa 841–1080), and GST-SRC-3HAT (aa 1081–1421) were generated as described previously (22). pSG5SRC-3S857AFlag and pGST-SRC-3CIDS857A, both of which have a point mutation of serine 857 to alanine, were generated by site-directed mutagenesis using the QuikChange Kit (Stratagene), and the sequences of the resulting mutants were verified by sequencing. The expression constructs of ERK3 with 6 Myc tags at the N terminus (pcDNA3-Myc6-ERK3) and its kinase-dead form (pcDNA3-Myc6-ERK3KD) were provided by Sylvain Meloche at University of Montreal, Montreal, Quebec, Canada. pCDH-Myc6-ERK3 was generated by inserting the Myc6-ERK3 fragment released from pcDNA3-Myc6-ERK3 by Dra I/Stu I digestion into pCDH-CMV-MCS-EF1-Puro (System Biosciences), digested with Swa I. pCDHSRC-3Flag was generated by inserting the SRC-3Flag fragment released from pSG5SRC-3Flag by Apa I/NheI digestion (blunt-ending Apa I site with Klenow enzyme) into pCDH-CMV-MCS-EF1-Puro, digested with Xba I/Nhe I (blunt-ending Xba I site with Klenow enzyme). Similarly, pCDHSRC-3S857AFlag was generated by inserting the SRC-3S857AFlag fragment into pCDH-CMV-MCS-EF1-Puro.
Cell culture and transfection.
HeLa, HEK293T, and lung cancer cell lines, including H1299, A549, H441, and H520, were purchased from ATCC. H1299, A549, H441, and H520 cells were maintained in RPMI 1640 medium supplemented with 10% FBS. HeLa and HEK293T cells were cultured in Dulbecco’s modified Eagle medium supplemented with 10% FBS. All the culture media and supplements were purchased from Invitrogen. Plasmids and siRNAs were transfected into the cells using Fugene HD Reagent (Roche) and TransIT-TKO Transfection Reagent (Mirus Bio Corporation), respectively, following the manufacturer’s instructions. Unless specifically indicated, cells were harvested for various analyses 48 hours after transfection.
SRC-3 siRNA ON-TARGETplus SMARTpool and nontargeting siRNA pool were purchased from Dharmacon. The silencer select siRNAs targeting human ERK3 and human MMP2 and the Silencer Negative Control #1 were purchased from Ambion.
Immunoprecipitation and Western blotting.
Cells were lysed with EBC lysis buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 0.5% NP-40, 1 mM PMSF, 1 mM Complete protease inhibitors [Roche Diagnostics], 10 mM NaF, 1 mM sodium orthovanadate, and 1 mM Phosphatase Inhibitor Cocktail III [Sigma-Aldrich]). One mg of total protein lysate was used for each immunoprecipitation using either each specific Ab or the corresponding IgG control. The supernatant was precleared with 40 μl protein A/G agarose beads for 1 hour at 4°C with constant rotation. The samples were then incubated with the desired Ab for 2 hours, followed by the addition of 40 μl protein A/G agarose beads for additional 1 hour. The beads were washed 3 times (5 minutes per wash) with lysis buffer. Proteins were boiled off the beads in 2× Laemmli sample buffer and resolved on 4%–15% SDS-PAGE gels (Bio-Rad). 2% of the amount of protein supernatant for immunoprecipitation was loaded as the input control. Western blotting was performed by first blocking nitrocellulose membranes with 5% nonfat milk in PBS-T buffer for 30 minutes, followed by overnight incubation with the primary Ab at 4°C and 1-hour incubation with appropriate secondary Ab at room temperature. The Western blot was visualized by chemiluminescence (Amersham) and quantified by ImageJ software ( http://rsbweb.nih.gov/ij/). Abs used for immunoprecipitation were as follows: anti–SRC-3 (BD Biosciences), anti-Flag (Sigma-Aldrich), and anti-ERK3 (Epitomics). Primary Abs used in Western blotting were as follows: anti–SRC-3 (BD Biosciences), anti-Flag (Sigma-Aldrich), anti-ERK3 (Epitomics), anti-ERK1/2 (Cell Signaling Technology), anti-Myc (Millipore), anti-PEA3 (Santa Cruz Biotechnology Inc.), anti–β-actin (Chemicon International), and anti–phospho-SRC-3 (S857) (22).
In vitro phosphorylation assay.
Myc-tagged ERK3 kinase or its kinase-dead form (ERK3KD) were expressed in 293T cells and purified by immunoprecipitation using anti-Myc affinity agarose beads (Sigma-Aldrich), following the manufacturer’s instruction. pcDNA3-Myc6-ERK3 or pcDNA3-Myc6-ERK3KD were transiently overexpressed in 293T cells. Cells were lysed with lysis buffer (50 mM Tris [pH 7.5], 200 mM NaCl, 0.5% NP-40, 10% glycerol, 0.4 mM EDTA, 1 mM Complete protease inhibitors [Roche Diagnostics], and 1 mM Phosphatase Inhibitor Cocktail III [Sigma-Aldrich]). The protein lysate supernatant was precleared with protein A/G agarose beads for 1 hour at 4°C with constant rotation. The samples were then incubated with anti-Myc affinity agarose beads (Sigma-Aldrich) for 3 hours. The beads were washed 3 times (10 minutes per wash) with wash buffer (50 mM Tris [pH 7.5], 300 mM NaCl, 0.5% NP-40, 10% glycerol, 0.2 mM EDTA, 1 mM Complete protease inhibitors [Roche Diagnostics], 10 mM β-glycero-phosphate, and 1 mM Phosphatase Inhibitor Cocktail III [Sigma-Aldrich]). Purified proteins were eluted off in wash buffer containing 500 μg/ml Myc peptide and 200 μg/ml insulin.
SRC-3Flag protein was expressed in Sf9 cells and purified using anti-Flag beads, following the procedures as described previously (24). The N-terminal basic helix-loop-helix domain-containing region of SRC-3 (aa 1–320), serine/threonine-rich region (aa 321–580), receptor interaction domain-containing region (aa 581–840), CID (aa 841–1080), and histone acetyltransferase domain-containing region (aa 1081–1424) were expressed as GST-fusion proteins in E. coli and purified using a GST-fusion protein purification kit (Invitrogen), following the manufacturer’s protocol.
In vitro phosphorylation assay was carried out in 40 mM Tris HCl (pH 7.5), 10 mM MgCl2, 0.1 mM EGTA, 1 mM dithiothreitol, and 5 mM β-glycero-phosphate. Each reaction contained 30 ng of purified ERK3 kinase or ERK3KD, 1 μg of purified protein substrate, 5 μCi 32P-ATP (Perkin Elmer), and 25 μM cold ATP in a total volume of 30 μl. The reaction was carried out at 30°C for 30 minutes and then stopped by adding 10 μl of 4 X SDS sample buffer. Proteins were resolved by SDS-PAGE gel, stained with Coomassie Brilliant Blue (Bio-Rad), and visualized by autoradiography.
In vitro protein-protein pull-down assay.
Each purified GST-SRC-3 fragment fusion protein was incubated with purified Myc-ERK3 protein immobilized on agarose beads with constant rotation in cold room for 2 hours. Beads were spun down and washed 5 times (5 minutes per wash). Proteins were then eluted off the beads with 2× Laemmli sample buffer. The Western blot was probed with a GST Ab.
Generation of cell pools stably expressing shRNAs by lentiviral transduction.
Production of pseudotyped lentiviral particles and generation of stable cell pools by lentiviral transduction were performed, following the manufacturer’s instructions (Open Biosystems). Pseudotype lentiviruses were produced in TLA-293T cells by cotransfecting a pGIPZ lentiviral shRNAmir expression construct harboring a GFP marker and Trans-Lentiviral Packaging Plasmid Mix (Open Biosystems). Pseudoviral particles were harvested 48 hours after transfection and concentrated using PEG-it Virus Precipitation Solution (System Biosciences), following the manufacturer’s instructions. Cells were transduced with prepared virus with the addition of polybrene (5 μg/ml). Two days after transduction, cells were split and selected by puromycin (1 μg/ml) for 10 days. Knockdown of the expression of the targeted genes was verified by Western blotting analysis.
Generation of cell pools stably overexpressing cDNAs by lentiviral transduction.
Production of pseudotyped lentiviral particles and stable cell pools by lentiviral transduction were performed, following the manufacturer’s instructions (System Biosciences). Pseudotype lentiviruses were produced in 293TN cells by cotransfecting lentiviral pCDH-cDNA expressing construct and pPACK Packaging Plasmid Mix (System Biosciences). Pseudoviral particles were harvested 48 hours after transfection and concentrated using PEG-it Virus Precipitation Solution, following the manufacturer’s instructions. To generate cell pools stably overexpressing cDNAs by lentiviral transduction, cells were transduced with prepared virus with the addition of polybrene (5 μg/ml). Two days after transduction, cells were split and selected by puromycin (1 μg/ml) for 10 days.
Cell proliferation assay.
Cell proliferation was determined using the CellTiter 96 AQueous One Solution Cell Proliferation Assay Kit (Promega), following the manufacturer’s instructions.
Two-chamber transwell cell migration/invasion assay.
Cell migration and invasion were analyzed using a modified 2-chamber transwell system (BD Biosciences), following the manufacturer’s instructions. Cells were detached by trypsin-EDTA, washed once with 1× PBS, and then resuspended in serum-free medium. 0.5 ml of complete culture media was added to each bottom well. 1 × 105 cells were added in each transwell insert and allowed to migrate for 12 hours (for H1299 cells) or 24 hours (for A549 and HeLa cells) in a 37°C cell incubator. Cells in the upper surface of the transwell were removed using cotton swabs. Migrated cells attached on the undersurface were fixed with 4% paraformaldehyde for 10 minutes and stained with crystal violet solution (0.5% in water) for 10 minutes. Cells were counted under a microscope at ×100 magnification.
The cell invasion assay was performed by following the same procedures as those for the cell migration assay, except that transwell inserts were precoated with Growth Factor-Reduced Matrigel (BD Biosciences).
Luciferase reporter assay.
The luciferase reporter assay was performed as described previously (20). Luciferase activity was determined with the Promega Luciferase Assay Kit.
Actin polymerization assay.
Actin polymerization was determined using an Actin Polymerization Assay Kit (Cytoskeleton), following the manufacturer’s instructions. Baseline fluorescence was monitored for 5 minutes using a fluorescence microplate reader (Promega) with excitation at 355 nm and emission at 405 nm. For each reaction, 200 μg protein lysate from cells was added to the reaction mixture. Actin polymerization, as determined by an increase in pyrene fluorescence, was measured at different time points. Fluorescence intensity was normalized by protein content (relative fluorescence units/μg [RFU/μg]).
MMP activity assay.
20 ml of the conditioned media collected from cultured cells at 90% confluency were concentrated by centrifugation using the Amicon Ultra Centrifugal Filter Unit (Millipore). The activities of secreted MMP2 and MMP10 from H1299 cell pools were determined using the SensoLyte fluorimetric MMP2 and MMP10 activity assay kits (AnaSpec), following the manufacturer’s instructions. MMP activity is represented as relative fluorescence units on the y axes of graphs.
ChIP assay.
ChIP assay was performed using the EZ-Magna ChIP Kit (Upstate/Millipore), following the manufacturer’s protocol. The Abs and normal IgGs used for ChIP assay were as follows: anti–SRC-3 (C-20; Santa Cruz Biotechnology Inc.), anti-Flag (M2; Sigma-Aldrich), and normal goat and mouse IgGs (Santa Cruz Biotechnology Inc.). Protein occupancy on the PEA3-binding site of the MMP2 gene promoter (700-bp upstream of transcription start site; ref. 19) was analyzed by quantitative real-time PCR using SYBR Green technology (Roche Applied Science) and the following primers: the forward primer (5′-CCCCTGTTCAAGATGGAGTC-3′) and the reverse primer (5′-CCCAGGTTGCTTCCTTACCT-3′). An irrelevant region (1,800-bp downstream of transcription start site) served as a negative control. Results were presented as the percentage of sheared chromatin input.
In vivo cancer cell invasion and colonization in the lungs of mice.
GFP-labeled H1299 cells with stable expression of the control shRNA, SRC-3 shRNA, ERK3 shRNA, or both SRC-3 shRNA and ERK3 shRNA were trypsinized, washed once with 1× PBS, and resuspended in 1× PBS (5 × 106 cells/ml). 200 μl of cell solution was injected via tail vein into each SCID/Beige female mouse (Harlan Laboratories) at the age of 6 to 7 weeks. Fifteen mice were tested for each group. Mice were sacrificed with anesthesia 6 weeks after cell injection. Lungs were perfused with 1× PBS. Formation of tumor foci/nodules in the lungs was analyzed by GFP imaging of the excised lungs or frozen tissue sections. Part of the lung was fixed in 4% paraformaldehyde, embedded in paraffin, and sectioned at a thickness of 4 μM for histological examination by H&E staining.
TMA analysis of ERK3 protein expression.
To determine the expression of ERK3 in NSCLCs, we obtained 456 primary NSCLCs, including 302 adenocarcinomas and 154 squamous cell carcinomas, from surgically resected lung cancer specimens (stages I–IIIA) from the Lung Cancer Specialized Program of Research Excellence Tissue Bank at The University of Texas MD Anderson Cancer Center. We used archived, formalin-fixed, paraffin-embedded tumor tissue samples placed in TMA. The TMAs were constructed as previously described (26). ERK3 was immunostained on 5-μm-thick TMA histology sections with an ERK3 Ab (dilution 1:200; Cell Signaling Technology), following the procedures described previously (26). Nuclear ERK3 expression was quantified using a 4-value intensity score (0, 1+, 2+, or 3+) and the percentage (0%–100%) of the extent of reactivity. An immunohistochemical expression score was obtained by multiplying the intensity and reactivity extension values (range, 0–300), and these expression scores were used to determine expression levels.
Statistics.
Results are expressed as mean ± SEM. Statistical significance was determined by a 2-sided Student’s t test. A P value of less than 0.05 was considered statistically significant.
Study approval.
Animal work was done in accordance with a protocol approved by the Animal Care and Use Committee of Baylor College of Medicine.
Supplementary Material
Acknowledgments
The authors would like to thank Sylvain Meloche for providing pcDNA3-Myc6-ERK3 and pcDNA3-Myc6-ERK3KD constructs. We thank Luping Huang for help with the injection of lung cancer cells into mice via tail vein, Subhamoy Dasgupta for help with the analysis of tumor formation in lungs by GFP imaging, and other colleagues in the laboratory, including Ping Yi, Lei Wang, Qin Feng, Amber Johnson, and Bryan Nikolai, for providing reagents. This work was supported by NIH grants 5R01HD-8188, HD07857, 2U19DK062434, and P30 DK079638; Texas CPRIT; and the Welch Foundation to B.W. O’Malley.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(5):1869–1880. doi:10.1172/JCI61492.
References
- 1.Coulombe P, Meloche S. Atypical mitogen-activated protein kinases: structure, regulation and functions. Biochim Biophys Acta. 2007;1773(8):1376–1387. doi: 10.1016/j.bbamcr.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Boulton TG, et al. ERKs: a family of protein-serine/threonine kinases that are activated and tyrosine phosphorylated in response to insulin and NGF. Cell. 1991;65(4):663–675. doi: 10.1016/0092-8674(91)90098-J. [DOI] [PubMed] [Google Scholar]
- 3.Deleris P, Rousseau J, Coulombe P, Rodier G, Tanguay PL, Meloche S. Activation loop phosphorylation of the atypical MAP kinases ERK3 and ERK4 is required for binding, activation and cytoplasmic relocalization of MK5. J Cell Physiol. 2008;217(3):778–788. doi: 10.1002/jcp.21560. [DOI] [PubMed] [Google Scholar]
- 4.Schumacher S, et al. Scaffolding by ERK3 regulates MK5 in development. Embo J. 2004;23(24):4770–4779. doi: 10.1038/sj.emboj.7600467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seternes OM, et al. Activation of MK5/PRAK by the atypical MAP kinase ERK3 defines a novel signal transduction pathway. Embo J. 2004;23(24):4780–4791. doi: 10.1038/sj.emboj.7600489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kant S, Schumacher S, Singh MK, Kispert A, Kotlyarov A, Gaestel M. Characterization of the atypical MAPK ERK4 and its activation of the MAPK-activated protein kinase MK5. J Biol Chem. 2006;281(46):35511–35519. doi: 10.1074/jbc.M606693200. [DOI] [PubMed] [Google Scholar]
- 7.Coulombe P, Rodier G, Pelletier S, Pellerin J, Meloche S. Rapid turnover of extracellular signal-regulated kinase 3 by the ubiquitin-proteasome pathway defines a novel paradigm of mitogen-activated protein kinase regulation during cellular differentiation. Mol Cell Biol. 2003;23(13):4542–4558. doi: 10.1128/MCB.23.13.4542-4558.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Julien C, Coulombe P, Meloche S. Nuclear export of ERK3 by a CRM1-dependent mechanism regulates its inhibitory action on cell cycle progression. J Biol Chem. 2003;278(43):42615–42624. doi: 10.1074/jbc.M302724200. [DOI] [PubMed] [Google Scholar]
- 9.Hansen CA, Bartek J, Jensen S. A functional link between the human cell cycle-regulatory phosphatase Cdc14A and the atypical mitogen-activated kinase Erk3. Cell Cycle. 2008;7(3):325–334. doi: 10.4161/cc.7.3.5354. [DOI] [PubMed] [Google Scholar]
- 10.Klinger S, Turgeon B, Levesque K, Wood GA, Aagaard-Tillery KM, Meloche S. Loss of Erk3 function in mice leads to intrauterine growth restriction, pulmonary immaturity, and neonatal lethality. Proc Natl Acad Sci U S A. 2009;106(39):16710–16715. doi: 10.1073/pnas.0900919106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evtimova V, et al. Identification of breast cancer metastasis-associated genes by chip technology. Anticancer Res. 2001;21(6A):3799–3806. [PubMed] [Google Scholar]
- 12.Liang B, Wang S, Zhu XG, Yu YX, Cui ZR, Yu YZ. Increased expression of mitogen-activated protein kinase and its upstream regulating signal in human gastric cancer. World J Gastroenterol. 2005;11(5):623–628. doi: 10.3748/wjg.v11.i5.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nambiar S, et al. Signaling networks in cutaneous melanoma metastasis identified by complementary DNA microarrays. Arch Dermatol. 2005;141(2):165–173. doi: 10.1001/archderm.141.2.165. [DOI] [PubMed] [Google Scholar]
- 14.Bhattacharjee A, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A. 2001;98(24):13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torres-Arzayus MI, et al. High tumor incidence and activation of the PI3K/AKT pathway in transgenic mice define AIB1 as an oncogene. Cancer Cell. 2004;6(3):263–274. doi: 10.1016/j.ccr.2004.06.027. [DOI] [PubMed] [Google Scholar]
- 16.Zhou HJ, et al. SRC-3 is required for prostate cancer cell proliferation and survival. Cancer Res. 2005;65(17):7976–7983. doi: 10.1158/0008-5472.CAN-04-4076. [DOI] [PubMed] [Google Scholar]
- 17.Xu J, Wu RC, O’Malley BW. Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nat Rev Cancer. 2009;9(9):615–630. doi: 10.1038/nrc2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long W, et al. SRC-3Delta4 mediates the interaction of EGFR with FAK to promote cell migration. Mol Cell. 2010;37(3):321–332. doi: 10.1016/j.molcel.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin L, et al. The AIB1 oncogene promotes breast cancer metastasis by activation of PEA3-mediated matrix metalloproteinase 2 (MMP2) and MMP9 expression. Mol Cell Biol. 2008;28(19):5937–5950. doi: 10.1128/MCB.00579-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan J, et al. Steroid receptor coactivator-3/AIB1 promotes cell migration and invasiveness through focal adhesion turnover and matrix metalloproteinase expression. Cancer Res. 2008;68(13):5460–5468. doi: 10.1158/0008-5472.CAN-08-0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li LB, Louie MC, Chen HW, Zou JX. Proto-oncogene ACTR/AIB1 promotes cancer cell invasion by up-regulating specific matrix metalloproteinase expression. Cancer Lett. 2008;261(1):64–73. doi: 10.1016/j.canlet.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 22.Wu RC, et al. Selective phosphorylations of the SRC-3/AIB1 coactivator integrate genomic reponses to multiple cellular signaling pathways. Mol Cell. 2004;15(6):937–949. doi: 10.1016/j.molcel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 23.Oh AS, et al. Tyrosine phosphorylation of the nuclear receptor coactivator AIB1/SRC-3 is enhanced by Abl kinase and is required for its activity in cancer cells. Mol Cell Biol. 2008;28(21):6580–6593. doi: 10.1128/MCB.00118-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yi P, et al. Atypical protein kinase C regulates dual pathways for degradation of the oncogenic coactivator SRC-3/AIB1. Mol Cell. 2008;29(4):465–476. doi: 10.1016/j.molcel.2007.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malovannaya A, et al. Analysis of the human endogenous coregulator complexome. Cell. 2011;145(5):787–799. doi: 10.1016/j.cell.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai D, et al. Steroid receptor coactivator-3 expression in lung cancer and its role in the regulation of cancer cell survival and proliferation. Cancer Res. 2010;70(16):6477–6485. doi: 10.1158/0008-5472.CAN-10-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H, et al. Overexpression and gender-specific differences of SRC-3 (SRC-3/AIB1) immunoreactivity in human non-small cell lung cancer: an in vivo study. J Histochem Cytochem. 2010;58(12):1121–1127. doi: 10.1369/jhc.2010.956979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olson MF, Sahai E. The actin cytoskeleton in cancer cell motility. Clin Exp Metastasis. 2009;26(4):273–287. doi: 10.1007/s10585-008-9174-2. [DOI] [PubMed] [Google Scholar]
- 29.Deleris P, et al. Activation loop phosphorylation of ERK3/ERK4 by group I p21-activated kinases (PAKs) defines a novel PAK-ERK3/4-MAPK-activated protein kinase 5 signaling pathway. J Biol Chem. 2011;286(8):6470–6478. doi: 10.1074/jbc.M110.181529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De la Mota-Peynado A, Chernoff J, Beeser A. Identification of the atypical MAPK Erk3 as a novel substrate for p21-activated kinase (Pak) activity. J Biol Chem. 2011;286(15):13603–13611. doi: 10.1074/jbc.M110.181743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao ZS, Manser E. PAK and other Rho-associated kinases--effectors with surprisingly diverse mechanisms of regulation. Biochem J. 2005;386(pt 2):201–214. doi: 10.1042/BJ20041638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beeser A, Jaffer ZM, Hofmann C, Chernoff J. Role of group A p21-activated kinases in activation of extracellular-regulated kinase by growth factors. J Biol Chem. 2005;280(44):36609–36615. doi: 10.1074/jbc.M502306200. [DOI] [PubMed] [Google Scholar]
- 33.Tak H, et al. 14-3-3epsilon inhibits MK5-mediated cell migration by disrupting F-actin polymerization. Cell Signal. 2007;19(11):2379–2387. doi: 10.1016/j.cellsig.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 34.Kostenko S, Johannessen M, Moens U. PKA-induced F-actin rearrangement requires phosphorylation of Hsp27 by the MAPKAP kinase MK5. Cell Signal. 2009;21(5):712–718. doi: 10.1016/j.cellsig.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 35.Han SJ, Lonard DM, O’Malley BW. Multi-modulation of nuclear receptor coactivators through posttranslational modifications. Trends Endocrinol Metab. 2009;20(1):8–15. doi: 10.1016/j.tem.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noble ME, Endicott JA, Johnson LN. Protein kinase inhibitors: insights into drug design from structure. Science. 2004;303(5665):1800–1805. doi: 10.1126/science.1095920. [DOI] [PubMed] [Google Scholar]
- 37.Fidias P, Novello S. Strategies for prolonged therapy in patients with advanced non-small-cell lung cancer. J Clin Oncol. 2010;28(34):5116–5123. doi: 10.1200/JCO.2010.30.7074. [DOI] [PubMed] [Google Scholar]
- 38.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7(3):169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 39.Shigematsu H, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97(5):339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.