Abstract
Spontaneous gene repair, also called revertant mosaicism, has been documented in several genetic disorders involving organs that undergo self-regeneration, including the skin. Genetic reversion may occur through different mechanisms, and in a single individual, the mutation can be repaired in various ways. Here we describe a disseminated pattern of revertant mosaicism observed in 6 patients with Kindler syndrome (KS), a genodermatosis caused by loss of kindlin-1 (encoded by FERMT1) and clinically characterized by patchy skin pigmentation and atrophy. All patients presented duplication mutations (c.456dupA and c.676dupC) in FERMT1, and slipped mispairing in direct nucleotide repeats was identified as the reversion mechanism in all investigated revertant skin spots. The sequence around the mutations demonstrated high propensity to mutations, favoring both microinsertions and microdeletions. Additionally, in some revertant patches, mitotic recombination generated areas with homozygous normal keratinocytes. Restoration of kindlin-1 expression led to clinically and structurally normal skin. Since loss of kindlin-1 severely impairs keratinocyte proliferation, we predict that revertant cells have a selective advantage that allows their clonal expansion and, consequently, the improvement of the skin condition.
Introduction
Spontaneous gene repair, also called revertant mosaicism, has been observed in genetic disorders primarily involving organs that undergo self-regeneration, such as bone marrow, liver, and skin (1). Depending on the frequency of the reversion and the clonal expansion of the reverted cells, the phenotype may improve in a sense of natural healing (2). Genetic reversion can occur through recombination, back, or second-site mutations, and in a single individual, the disease-causing mutation can be mended in various ways (3). The accessibility of the skin facilitates the recognition of revertant mosaicism, which has previously been described in epidermolysis bullosa (4, 5) and ichthyosis (6).
Kindler syndrome (KS; OMIM 173650) is an autosomal-recessive skin fragility disorder that results from loss-of-function mutations in FERMT1, which encodes kindlin-1, a key regulator of integrin activation. KS is characterized by congenital skin blistering and photosensitivity, evolving to poikiloderma with pronounced skin atrophy (7). Poikiloderma refers to a combination of hypo- and hyperpigmentation, atrophy, and telangiectasias and is morphologically characterized by thin epidermis without rete ridges, focal vacuolization of basal keratinocytes, and pigment incontinence. Typically, KS skin shows multiple planes of cleavage at the dermal-epidermal junction (DEJ) and reduplication and interruption of the epidermal basement membrane. Consistent with the epidermal atrophy, basal keratinocytes exhibit minimal proliferation (8, 9).
Here, we describe 6 patients with KS caused by inherited FERMT1 duplicating insertions and a disseminated pattern of revertant skin patches. The reversion mechanism in each investigated skin patch was back mutation by slipped mispairing in direct nucleotide repeats. In addition, mitotic recombination was documented in some reverted areas.
Results and Discussion
Patient 1 (P1), who exhibited the hallmark feature of KS (poikiloderma with pronounced skin atrophy), was the index case, with innumerable normal-appearing skin patches. Subsequently, we observed a similar pattern in P2. Notably, both patients had FERMT1 duplication insertion mutations, which led to frame shifts, premature termination codons, and loss of full-length kindlin-1, as demonstrated by immunoblotting of keratinocyte lysates (10). A systematic review of 24 additional KS patients revealed that all individuals harboring duplicating insertions (P3–P6; Table 1) exhibited the same skin pattern (Supplemental Figures 1 and 2; supplemental material available online with this article; doi: 10.1172/JCI61976DS1). P1 and P2 were available for systematic investigation.
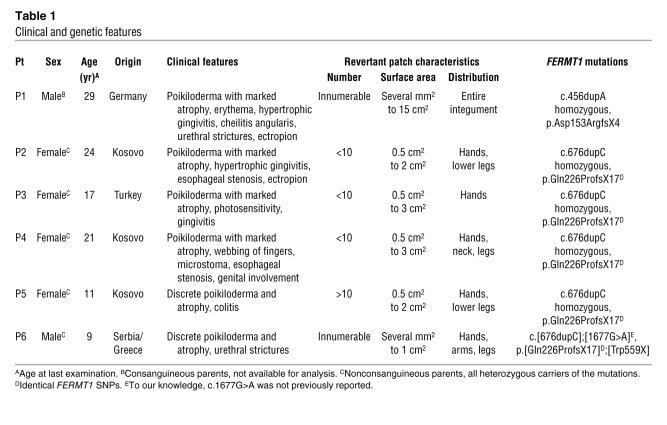
Table 1 .
Clinical and genetic features
P1, a 29-year-old male born to consanguineous parents, was homozygous for c.456dupA (11). During a 3-year follow-up, we noted normal-appearing patches with preserved texture, measuring between several mm2 and 15 cm2, disseminated over the entire integument, that contrasted the atrophic, dry, hyperpigmented, and often erythematous skin (Figure 1, A–C). Immunofluorescence staining of a skin sample obtained from an atrophic area (P1-1) revealed absence of kindlin-1 at the DEJ (Figure 2B). In contrast, 3 normal-appearing patches (P1-2, P1-3, and P1-4) showed positive kindlin-1 signals over the entire length of the biopsy, similar to normal skin (Supplemental Table 1).
Figure 1. Clinical features indicative of revertant mosaicism in KS.
(A–C) P1 exhibited numerous normal-appearing skin patches of several mm2 to 15 cm2 on the entire integument, which remained constant over the 3-year observation period. Shown are the right axilla, upper thorax, and right arm (A); the left arm (B); and the left leg (C). (D and E) P2 had normal-appearing skin patches on the right hand (D) and the right lower leg (E). Revertant areas investigated here are outlined in black; biopsy sites are circled in blue or directly visible.
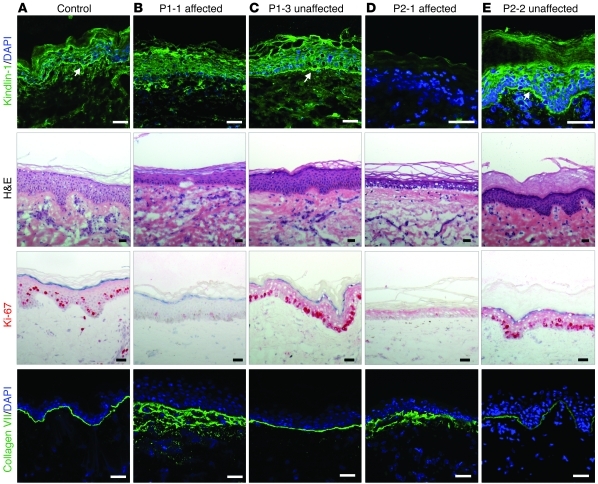
Figure 2. Kindlin-1 expression and skin morphology correlate with clinical phenotype.
Skin of a control individual (A) and affected (B and D) and unaffected (C and E) skin of P1 and P2 were stained with H&E and antibodies to kindlin-1, Ki-67, and collagen VII. In the affected skin P1-1 and P2-1, loss of kindlin-1 expression was associated with thin epidermis, microblisters at the DEJ, strong reduction of Ki-67–positive keratinocytes, and irregular collagen VII staining at the DEJ and in the upper dermis. In contrast, in the normal-appearing patches P1-3 and P2-2 with restored kindlin-1 expression (arrows), skin morphology, Ki-67 staining, and collagen VII distribution were comparable to those of the control. Scale bars: 50 μm.
P2, a 24-year-old female homozygous for c.676dupC, exhibited multiple normal-appearing patches, measuring 0.5–2 cm2, that were clearly distinguishable from the atrophic integument (Figure 1, D and E). Kindlin-1 immunofluorescence was negative in the affected skin, but positive (comparable to control skin) in 2 normal-appearing patches (Figure 2, D and E, and Supplemental Table 1).
In both P1 and P2, restored kindlin-1 expression visibly improved skin morphology (i.e., preservation of rete ridges, epidermal thickness, and keratinocyte proliferation; Figure 2, C and E). Collagen VII staining was used to evaluate the integrity of the DEJ. In normal-appearing areas with positive kindlin-1 immunostaining, a linear collagen VII signal was seen that contrasted the irregular pattern in affected areas (Figure 2, B–E), which indicated that restoration of kindlin-1 normalized the DEJ.
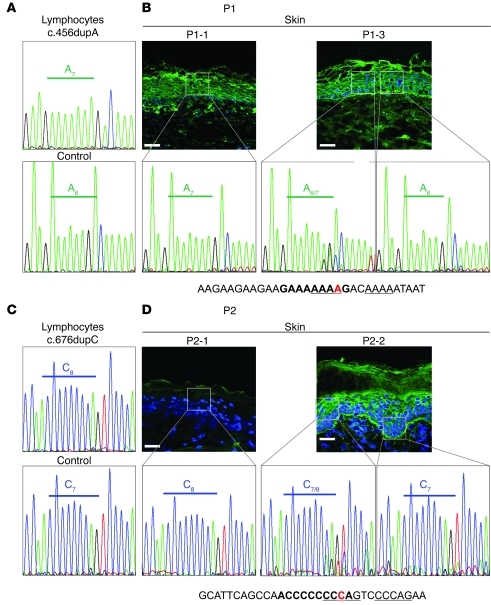
These results strongly supported the hypothesis that the normal-appearing kindlin-1–positive spots represent areas in which the inherited mutations had reverted. To test this, we used laser dissection microscopy (LDM) to collect keratinocytes from areas with altered or normal DEJ. DNA was extracted, and FERMT1 exons 4 and 5 were sequenced in P1 and P2, respectively. In both patients, the mutations were found in a heterozygous state in keratinocytes derived from all normal-appearing patches, whereas in diseased skin, the mutations were present in a homozygous state, similar to lymphocytes (Figure 3 and Supplemental Table 1). In the dermis below the reverted areas, the mutations were present in a homozygous state. Thus, the reversion had occurred only in keratinocytes, which are the sole kindlin-1–expressing cells in the skin (8, 9). The results were confirmed in P2 at the RNA level (data not shown), but no material was available from P1. Notably, in both patients, the same reversion mechanism (i.e., a single nucleotide deletion restoring the number of bases in the direct repeats) was found in all revertant patches, suggestive of a high propensity of the sequence to mutational events through slipped mispairing (12).
Figure 3. Identification of the reversion mechanism.
LDM was used to collect epidermal keratinocytes from areas with altered or normal DEJ morphology. (A) Partial sequence of FERMT1 exon 4 in lymphocytes of P1 showed the duplicating insertion in adenine repeats c.456dupA (A7). The control had the adenine repeat sequence A6. (B) In affected skin sample P1-1, the mutation was found in a homozygous state. Analysis of DNA from microdissected areas (insets) of P1-3 from unaffected skin revealed the mutation in a heterozygous state (A6/7) in the majority of the experiments, but the wild-type sequence (A6) was also found. The sequence in the vicinity of the revertant mutation is shown below; bold denotes the GA6-A6G motif, underline denotes a DNA-polymerase β frameshift hotspot; red denotes the mutation. (C) Partial sequence of FERMT1 exon 5 in lymphocytes of P2 and a control showing the duplicating insertion in a cytosine tract (C8). (D) The homozygous duplication c.676dupC was disclosed in the affected skin P2-1, whereas analysis of DNA from microdissected areas (insets) in unaffected skin P2-2 disclosed the mutation in a heterozygous state (C7/8), or the normal sequence (C7). The sequence in the vicinity of the revertant mutation is shown below; bold denotes the AC6-C6A motif, underline denotes the vertebrate topoisomerase II consensus cleavage site; red denotes the mutation. Scale bars: 50 μm.
Surprisingly, in both P1 and P2, the homozygous wild-type sequence was also disclosed in some areas of the normal-appearing skin samples (Figure 3), suggestive of a second mutational event in the revertant heterozygous keratinocytes — either a second back mutation, or mitotic recombination. To investigate this, we genotyped SNPs around FERMT1 in DNA extracted from lymphocytes of P1 and P2 and identified the borders of the homozygous regions, which spanned about 1.1 and 1.4 Mb, respectively. In the revertant patches with the homozygous wild-type FERMT1 sequences, recombination was confirmed by loss of heterozygosity of the SNP rs261360, located outside these homozygous regions (Supplemental Table 2).
These results demonstrated restoration of the FERMT1 reading frame, kindlin-1 expression, and skin structure in the revertant patches. Revertant keratinocytes proliferated normally, having a selective advantage compared with kindlin-1–negative cells (Figure 2), allowing them to expand. Nevertheless, large reverted areas (~15 cm2) may represent confluence of multiple overlapping or neighboring revertant events.
The slipped mispairing model involves the misalignment of short direct repeats during DNA replication and represents a common mutational mechanism (13–15), which has previously been elucidated in microorganisms (16–18). The primer strand containing the newly synthesized repeat dissociates from the template strand and misaligns, leading to either insertion or deletion of 1 or more direct repeats in the continued DNA synthesis. The fidelity of DNA replication is dependent on the local sequence environment and the DNA polymerase (19). PolyA and polyC repeats, as observed in our patients, are known hotspots for microdeletions and microinsertions (20, 21). Prior meta-analysis of the sequence vicinity of a large number of microdeletions and microinsertions in different genes revealed that short oligonucleotide motifs significantly correlate with the occurrence of both mutational events (12). Such motifs — GA6 and A6G, or AC6 and C6A — were present close to the mutations in our patients (Figure 3). Additionally, the DNA polymerase β frameshift hotspot AAAA, and the topoisomerase II consensus cleavage site CCCAG, which are potentially involved in site-specific recombination and putative deletion/insertion hotspots, were close to the mutations. Based on the identical molecular event in all analyzed spots and the finding that of our cohort of 26, all 6 KS patients with duplicating insertions exhibited multifocal revertant mosaicism, we strongly favor this mechanism as the cause of reversion. A recent report of an 8-year-old boy homozygous for c.676dupC and having a normal-appearing skin patch on his hand (5) strengthens our findings. Recognition of multiple patches in young children is difficult (22), since small normal-appearing areas may only become visible with age, against the progressive poikiloderma (ref. 7 and Supplemental Figure 2).
The multifocal mosaic pattern in KS is similar to the rare disorder ichthyosis with confetti, which is caused by dominant KRT10 mutations. In this disorder, multiple normal-appearing patches represent revertant areas in which the inherited mutation is rescued by different mitotic recombination events (6). In our cases, recombination occurred as a secondary mutational event, after the back mutation had already reverted the recessive condition.
These findings may open therapeutic perspectives for KS and other genodermatoses. The advantages of a therapy using revertant keratinocytes are the natural correction of the mutation and the lack of immune response. Grafting revertant keratinocytes in junctional epidermolysis bullosa demonstrated that the procedure is easy and well-tolerated by the patient, but cultivation of the reverted cells must be improved before it becomes clinically applicable (23).
Methods
Samples.
EDTA blood was obtained from patients and parents. 4-mm punch skin biopsies were obtained from P1 and P2 (Supplemental Table 1 and Figure 1).
Immunomorphological analysis.
H&E, immunohistochemistry, and indirect immunofluorescence staining of the skin was performed as described previously (8), using primary antibodies to kindlin-1 (8), Ki-67 (MIB-1; Dako), and collagen VII (LH7.2; Millipore) and the AEC (3-amino-9-ethylcarbazole) system (Dako) or Alexa Fluor 488–conjugated goat anti-rabbit and -mouse IgGs (Invitrogen). Nuclei were visualized with hematoxylin or DAPI. Stained sections were observed with light microscopy (Nikon 80i) or confocal laser scanning microscopy (LSM510; Carl Zeiss).
Mutation detection.
Mutation detection on DNA extracted from EDTA blood (Qiagen kit) was performed as described previously (24). All 15 exons and adjacent junctions of FERMT1 were amplified and sequenced in an ABI 3130XL genetic analyzer using Big Dye Terminator Chemistry (Applied Biosystems). DNA sequences were compared with the NCBI reference (NC_000020.10) using Mutation Surveyor DNA (2.61 Softgenetics). Mutations were verified by sequencing in both directions from independent PCRs. Additionally, 34 SNPs covering 7 Mb around FERMT1 were genotyped by direct sequencing from the patients’ lymphocytes (Supplemental Table 2).
Nested PCR was used to detect mutations in DNA from microdissected tissue (Supplemental Table 3). All PCRs were repeated with templates from at least 3 separate DNA isolations obtained by LDM, and all products were sequenced in both directions.
DNA and RNA isolation from skin sections.
For DNA recovery by LDM, 5-μm skin cryosections were mounted on 1.0-mm PEN membrane–covered slides (Zeiss). Collagen VII staining was used to visualize the DEJ because the signal is strong enough not to be bleached under LDM. Approximately 200 cells from affected and reverted areas were dissected with the Laser Robot Microbeam System (P.A.L.M. Microlaser Technology AG) and separately collected in the caps of 0.5-ml thin-walled tubes (Zeiss). For proteinase K digestion, 30 μl of a mix consisting of PCR buffer (JumpStart RED Taq DNA Polymerase), water, and proteinase K (Qiagen) was added, and the tubes remained inverted for 60 minutes at 55°C. Inactivation was subsequently achieved by heating to 98°C for 15 minutes (2). For RNA isolation, 8 10-μm skin sections were extracted with the Qiagen FFPE RNA kit (Qiagen). Reverse transcription was performed using the Advantage RT-for-PCR Kit (Clontech) with 0.2 μg total RNA, using oligodT primers. Primers were designed with Primer3 ( http://frodo.wi.mit.edu/primer3; Supplemental Table 3).
Cloning.
The entire epidermis was dissected with LDM from biopsies P1-2 and P2-3. Subsequently, DNA was isolated and submitted to nested PCR, and the amplicons were subcloned into the TOPO TA-cloning vector (Invitrogen). For each reaction, 15–20 clones were sequenced using the M13-RV primer.
Study approval.
The study was approved by the Ethics Committee of the University of Freiburg and conducted according to the Declaration of Helsinki. Patients provided informed consent prior to their participation.
Supplementary Material
Acknowledgments
The authors thank Gabriele Grüninger, Vera Morand, and Kaethe Thoma for expert technical assistance and Ayla Gulekon for sharing clinical data. This work was supported by grant HA 5663/1-1 from the German Research Foundation (DFG), by the E-RARE grants Kindlernet and RevertantEB, by the Freiburg Institute for Advanced Studies (FRIAS), by School of Life Sciences — LifeNet, and by the Kriezis scholarship of the “Ethnikon and Kapodistriakon University” of Athens to D. Kiritsi.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(5):1742–1746. doi:10.1172/JCI61976.
References
- 1.May M. Mutations to the rescue. Nat Med. 2011;17(4):405–407. doi: 10.1038/nm0411-405. [DOI] [PubMed] [Google Scholar]
- 2.Jonkman MF, et al. Revertant mosaicism in epidermolysis bullosa caused by mitotic gene conversion. Cell. 1997;88(4):543–551. doi: 10.1016/S0092-8674(00)81894-2. [DOI] [PubMed] [Google Scholar]
- 3.Youssoufian H, Pyeritz RE. Mechanisms and consequences of somatic mosaicism in humans. Nat Rev Genet. 2002;3(10):748–758. doi: 10.1038/nrg906. [DOI] [PubMed] [Google Scholar]
- 4.Jonkman MF, Pasmooij AM. Revertant mosaicism--patchwork in the skin. N Engl J Med. 2009;360(16):1680–1682. doi: 10.1056/NEJMc0809896. [DOI] [PubMed] [Google Scholar]
- 5.Lai-Cheong JE, Moss C, Parsons M, Almaani N, McGrath JA. Revertant mosaicism in kindler syndrome. J Invest Dermatol. 2012;132(3):730–732. doi: 10.1038/jid.2011.352. [DOI] [PubMed] [Google Scholar]
- 6.Choate KA, et al. Mitotic recombination in patients with ichthyosis causes reversion of dominant mutations in KRT10. Science. 2010;330(6000):94–97. doi: 10.1126/science.1192280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Has C, et al. Kindler syndrome: Extension of FERMT1 mutational spectrum and natural history. Hum Mutat. 2011;32(11):1204–1212. doi: 10.1002/humu.21576. [DOI] [PubMed] [Google Scholar]
- 8.Herz C, Aumailley M, Schulte C, Schlotzer-Schrehardt U, Bruckner-Tuderman L, Has C. Kindlin-1 is a phosphoprotein involved in regulation of polarity, proliferation, and motility of epidermal keratinocytes. J Biol Chem. 2006;281(47):36082–36090. doi: 10.1074/jbc.M606259200. [DOI] [PubMed] [Google Scholar]
- 9.Ussar S, et al. Loss of kindlin-1 causes skin atrophy and lethal neonatal intestinal epithelial dysfunction. PLoS Genet. 2008;4(12):e1000289. doi: 10.1371/journal.pgen.1000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kern JS, et al. Chronic colitis due to an epithelial barrier defect: the role of kindlin-1 isoforms. J Pathol. 2007;213(4):462–470. doi: 10.1002/path.2253. [DOI] [PubMed] [Google Scholar]
- 11.Has C, et al. Kindlin-1 is required for RhoGTPase-mediated lamellipodia formation in keratinocytes. Am J Pathol. 2009;175(4):1442–1452. doi: 10.2353/ajpath.2009.090203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ball EV, Stenson PD, Abeysinghe SS, Krawczak M, Cooper DN, Chuzhanova NA. Microdeletions and microinsertions causing human genetic disease: common mechanisms of mutagenesis and the role of local DNA sequence complexity. Hum Mutat. 2005;26(3):205–213. doi: 10.1002/humu.20212. [DOI] [PubMed] [Google Scholar]
- 13.Streisinger G, et al. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb Symp Quant Biol. 1966;31:77–84. doi: 10.1101/SQB.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- 14.Efstratiadis A, et al. The structure and evolution of the human beta-globin gene family. Cell. 1980;21(3):653–668. doi: 10.1016/0092-8674(80)90429-8. [DOI] [PubMed] [Google Scholar]
- 15.Levinson G, Gutman GA. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987;4(3):203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- 16.Viguera E, Canceill D, Ehrlich SD. In vitro replication slippage by DNA polymerases from thermophilic organisms. J Mol Biol. 2001;312(2):323–333. doi: 10.1006/jmbi.2001.4943. [DOI] [PubMed] [Google Scholar]
- 17.Viguera E, Canceill D, Ehrlich SD. Replication slippage involves DNA polymerase pausing and dissociation. EMBO J. 2001;20(10):2587–2595. doi: 10.1093/emboj/20.10.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacob KD, Eckert KA. Escherichia coli DNA polymerase IV contributes to spontaneous mutagenesis at coding sequences but not microsatellite alleles. Mutat Res. 2007;619(1–2):93–103. doi: 10.1016/j.mrfmmm.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kunkel TA. DNA replication fidelity. J Biol Chem. 2004;279(17):16895–16898. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- 20.Bacon AL, Dunlop MG, Farrington SM. Hypermutability at a poly(A/T) tract in the human germline. Nucleic Acids Res. 2001;29(21):4405–4413. doi: 10.1093/nar/29.21.4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakagawa H, et al. Nine novel germline mutations of STK11 in ten families with Peutz-Jeghers syndrome. Hum Genet. 1998;103(2):168–172. doi: 10.1007/s004390050801. [DOI] [PubMed] [Google Scholar]
- 22.Has C. Mosaicism in the skin: the importance of mild or minimal skin lesions. Arch Dermatol. 2011;147(9):1094–1096. doi: 10.1001/archdermatol.2011.233. [DOI] [PubMed] [Google Scholar]
- 23.Gostynski A, Deviaene FC, Pasmooij AM, Pas HH, Jonkman MF. Adhesive stripping to remove epidermis in junctional epidermolysis bullosa for revertant cell therapy. Br J Dermatol. 2009;161(2):444–447. doi: 10.1111/j.1365-2133.2009.09118.x. [DOI] [PubMed] [Google Scholar]
- 24.Has C, et al. Molecular basis of Kindler syndrome in Italy: novel and recurrent Alu/Alu recombination, splice site, nonsense, and frameshift mutations in the KIND1 gene. J Invest Dermatol. 2006;126(8):1776–1783. doi: 10.1038/sj.jid.5700339. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.