Abstract
Gamma-retroviral/lentiviral vectors (γRV/LV) with self-inactivating (SIN) long terminal repeats (LTRs) and internal moderate cellular promoters pose a reduced risk of insertional mutagenesis when compared with vectors with active LTRs. Yet, in a recent LV-based clinical trial for β-thalassemia, vector integration within the HMGA2 gene induced the formation of an aberrantly spliced mRNA form that appeared to cause clonal dominance. Using a method that we developed, cDNA linear amplification-mediated PCR, in combination with high-throughput sequencing, we conducted a whole transcriptome analysis of chimeric LV-cellular fusion transcripts in transduced human lymphoblastoid cells and primary hematopoietic stem/progenitor cells. We observed a surprising abundance of read-through transcription originating outside and inside the provirus and identified the vector sequences contributing to the aberrant splicing process. We found that SIN LV has a sharply reduced propensity to engage in aberrant splicing compared with that of vectors carrying active LTRs. Moreover, by recoding the identified vector splice sites, we reduced residual read-through transcription and demonstrated an effective strategy for improving vectors. Characterization of the mechanisms and genetic features underlying vector-induced aberrant splicing will enable the generation of safer vectors, with low impact on the cellular transcriptome.
Introduction
Retroviruses, transposons, and gene therapy vectors integrate into the genome of host cells and are able to trigger oncogenesis by a process known as insertional mutagenesis, which consists of the deregulation of proto-oncogenes found at or nearby the insertion site via different molecular mechanisms (1, 2). As demonstrated in several different mouse models of oncogene tagging and in gene therapy clinical trials (3–8), enhancer-mediated activation is the most prominent mechanism involved in oncogene activation. Such enhancer-mediated activation involves short- and long-range interaction of viral enhancer sequences with cellular promoters to increase the mRNA levels of a proto-oncogene (9).
However, additional mechanisms of proto-oncogene activation may involve the generation of chimeric transcripts originating from the interaction of promoter elements or splice sites contained in the genome of the insertional mutagen with the cellular transcriptional unit targeted by integration (10–13). Chimeric fusion transcripts comprising vector sequences and cellular mRNAs can be generated either by read-through transcription starting from vector sequences and proceeding into the flanking cellular genes or vice versa (6, 10, 12, 14). In vitro genotoxicity assays and mouse studies show that when retroviruses, transposons, and gamma-retroviral/lentiviral vectors (γRV/LVs) with active long terminal repeats (LTRs) integrate downstream of the promoters of cellular genes in the same transcriptional orientation, gene transcription is put under the control of the viral promoter present in the 5′ or 3′ LTR (4, 11). In our previous study, using a tumor-prone mouse model for LV genotoxicity testing, we found a tumor harboring an integration of an LV with active LTRs within the Braf transcription unit (5). This integration led to the formation of an aberrant transcript encoding for a truncated Braf protein lacking the regulatory domain and endowed with oncogenic activity (15, 16). Specifically, the canonical LV splice donor sequence placed downstream the active LTR proficiently interacted with the splice acceptor of the thirteenth exon of Braf to form this aberrant transcript (5). The same mechanism of vector LTR-driven read-through and splicing capture was also responsible for several independent gene activation events in an in vitro genotoxicity assay (11, 12). Aberrant transcript formation can even be caused by vectors with self-inactivating (SIN) LTRs, which are devoid of strong enhancer-promoter sequences. Indeed, the R region in the LTR sequence contains both the canonical viral polyA signal in the same orientation and a cryptic polyA signal in the opposite orientation (17). Therefore, proviral intragenic integrations in both orientations can potentially elicit the premature termination of gene transcription. These polyadenylation sites, however, are used only in a context-dependent manner and when specific requirements are met (18, 19). Moreover, viral-cellular fusion transcripts terminating at cryptic polyadenylation sites located in the host cellular genome were found in keratinocytes transduced with SIN LVs (20). Thus, a complex interplay among the presence, relative strength, position, and distance of promoters; splice site consensus sequences; and mRNA polyadenylation signals is ultimately responsible for the production of specific aberrant mRNAs (17, 21). These mechanisms also appear to have clinical relevance: in a recent LV-based gene therapy trial for the treatment of β-thalassemia, a transplanted patient displayed a dominant myeloid cell clone harboring an integrated vector copy within HMGA2. Vector integration triggered the fusion of the splice donor sequence of the third exon of HMGA2 with a cryptic splice acceptor sequence present within an insulator element inserted in the vector LTR. Interestingly, this new splicing event caused activation of the viral polyadenylation signal in the LV LTR and thus induced premature HMGA2 transcript termination. This aberrant mRNA, lacking let7 miRNA binding sites, displayed a higher stability that in turn lead to increased protein levels. Although not fully proven, the activation of HMGA2 has been suggested to be causative of the clonal dominance (14). Thus, there is emerging evidence that the potential of inducing aberrant transcripts might constitute a previously unappreciated genotoxicity factor for gene therapy vectors. How to reduce these splicing capture events and aberrant transcript formation triggered by vector integration is still unclear. In order to reduce splicing capture events and aberrant transcript formation it will be necessary to better understand this phenomenon by (a) identifying the genes that are mostly affected by aberrant splicing; (b) quantifying the overall read-through transcription of LV sequences; and (c) identifying splice site consensus sequences within the vector that mostly interfere with cellular splicing at the genome-wide level. From this perspective, it is conceivable that safety studies that so far have been mainly centered on a genomic integration DNA world should be expanded into the RNA world to explore the impact of vector integration on the mRNA structure of cellular genes at the whole transcriptome level.
This unmet need prompted us to devise a method to retrieve aberrant fusion events between specific portions of the LV genome and cellular mRNA sequences. With this aim, we developed a PCR method, referred to as cDNA linear amplification-mediated PCR (cLAM-PCR), capable of retrieving chimeric LV-cellular transcripts in a high-throughput fashion from the whole transcriptome of LV-transduced cells and characterized the type of aberrant splicing events in primary human CD34+ hematopoietic stem progenitor cells (HSPCs) as well as a human cell line. Our findings highlight a surprising abundance of read-through transcription originating outside and inside the LV provirus, identify the major vector sequences contributing to the aberrant splicing process, establish a previously unnoticed advantage of SIN LV over vectors carrying active LTRs in terms of propensity to engage in aberrant splicing, and dictate a strategy to further reduce residual read-through transcription.
Results
cLAM-PCR technique to study LV-induced aberrant splicing in primary human stem progenitor cells.
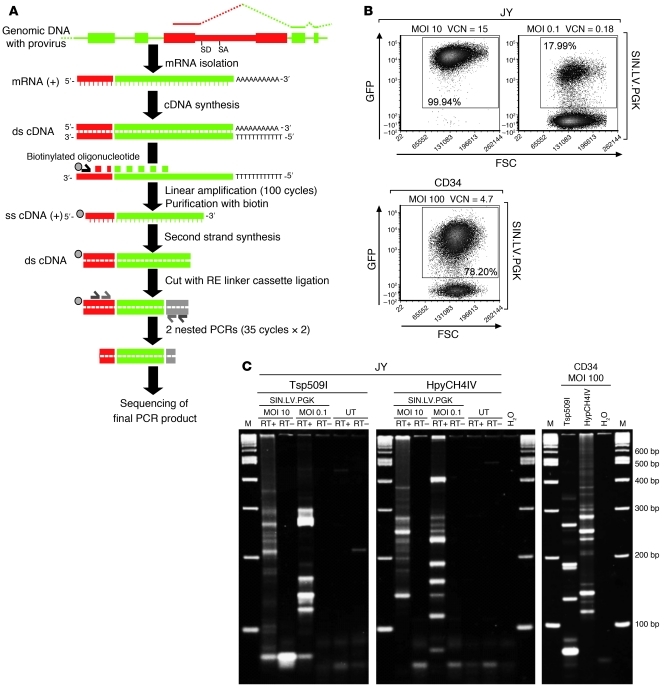
cLAM-PCR is aimed at retrieving in a high-throughput fashion aberrantly spliced mRNAs that contain LV sequences fused with cellular transcripts from the whole transcriptome of LV-transduced cells (schematics in Figure 1A). Similar to the previously published linear amplification-mediated PCR (LAM-PCR) technique (22), a biotinylated oligonucleotide was designed on a sequence complementary to the HIV backbone and used for linear amplification on single- or double-stranded cDNA from LV-transduced cells. The resulting single-stranded DNA molecules will contain expressed portions of the HIV backbone and, in chimeric transcripts, may also contain unknown cellular sequences. The linear amplification products were then purified with streptavidin-coupled paramagnetic beads and subsequently subjected to double-strand synthesis and digested with a restriction enzyme to ligate a linker cassette. The restriction enzymes used in this study were Tsp509I (AATT) and HpyCH4IV (AGCT), as their efficacy in LAM-PCR protocols has been previously confirmed (10, 22). The resulting products were then amplified by exponential PCR using nested oligonucleotides complementary to the HIV backbone and the linker cassette. The final cLAM-PCR products were sequenced by 454 pyrosequencing and analyzed by dedicated high-throughput computational pipeline. This computational pipeline has been developed to recognize and annotate chimeric LV-genome transcripts that contain LV sequences fused to host cell sequences. LV sequences were recognized, and the nucleotide position at the fusion point was identified (splice site) on the LV genome. The remaining sequence portion, after removal of the LV and linker cassette sequences, was mapped on the appropriate genome by BLAST. With this technique, by designing the proper oligonucleotide sets in different portions of the LV backbone, it is possible to interrogate different LV sequences for their ability to generate aberrant splicing events. The most obvious choice was to design an oligonucleotide set upstream of the canonical LV splice donor site and in forward orientation with respect to the HIV genome (oligonucleotide set: UPLVSD). The second cLAM oligonucleotide set was designed downstream of the canonical splice acceptor site sequence, in reverse orientation with respect to HIV transcription (oligonucleotide set: DWLVSA). These 2 cLAM oligonucleotide sets encompass the 1,165-bp HIV intron, which, based on our previous results of LV.SF.LTR-induced Braf activation in Cdkn2a–/– tumors, likely plays a relevant role in the splicing capture process (5).
Figure 1. cLAM-PCR procedure for the retrieval of LV cellular fusion transcripts.
(A) Scheme of the experimental procedure for cLAM-PCR. Total mRNA is retrotranscribed into double-stranded cDNA (ds cDNA) using oligo-dT primers. Linear PCR uses a biotinylated primer located upstream/downstream of a known LV splice site, allowing extension into vector or an unknown cellular portion of a chimeric transcript. Single-stranded product is purified by streptavidin-coupled magnetic beads, double stranded using Klenow enzyme, and cut using restriction enzymes (REs). A linker cassette compatible with the restriction enzyme cut is ligated, and 2 sequential nested PCRs are performed. The final PCR products are then sequenced. ss cDNA, single-stranded cDNA. (B) FACS plots showing the percentage of GFP+ in JY cells and CD34+ HSPCs after SIN.LV.PGK transduction. The VCN and the MOI are indicated. Numbers in the graph indicate the percentage of GFP+ cells. (C) Representative band pattern of cLAM-PCR performed on mRNA from SIN.LV.PGK-transduced cells. Retrotranscribed mRNA (RT+) and negative controls (RT–) were used. By sequencing, bands in retrotranscribed mRNA samples corresponded to aberrant transcripts or unspliced internal control sequences. Rare faint bands in negative controls corresponded to oligonucleotide dimers or concatemers. H2O, negative PCR control from the linear amplification reaction to the second exponential phase. M, marker.
We investigated the aberrant splicing induced by an LV with SIN LTRs, containing the human phosphoglycerate kinase (hPGK) promoter in internal position driving the expression of the GFP (SINLV.PGK). This vector was used to transduce a human B-lymphoblastoid cell line (JY cells) and the clinically relevant human primary cord blood–derived CD34+ HSPCs. JY cells were transduced at different MOI, 0.1 or 10, obtaining an average vector copy number (VCN) of 0.18 and 15 and a percentage of vector-marked cells of 18% and 100%, respectively. Human CD34+ HSPCs were transduced at MOI 100, using an established clinical protocol (23), obtaining an average VCN of 4.7 and a percentage of vector-marked cells of 78% (Figure 1B).
Spreadex gel electrophoresis of cLAM-PCR products obtained from transduced JY and CD34+HSPCs showed several bands of variable molecular size, ranging from 100 to 600 bp. On the other hand, non-retrotranscribed RNA controls (Figure 1C, shown as RT-) yielded rare and faint bands corresponding to primer dimers or concatemers. The complexity of band patterns correlated with the marking levels of the samples tested. Cells transduced with high vector loads produced many bands of different molecular size, while samples from low MOI showed smaller numbers of bands (Figure 1C).
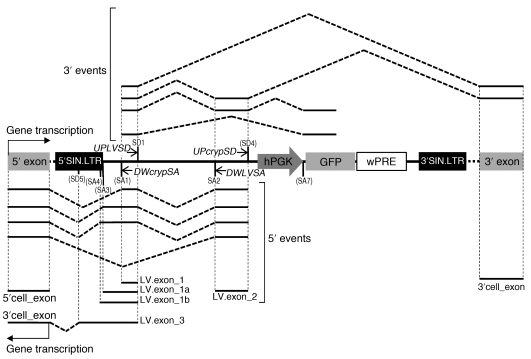
cLAM-PCR products were shotgun cloned into plasmids and sequenced by the Sanger method or tagged by PCR with adapter primers designed to include a sequence bar code tag (DNA bar coding) and subjected to 454 pyrosequencing. The information contained in the DNA bar codes allows the simultaneous sequencing of pooled amplicons from different samples (3). By this approach, we identified a total of 8 splice sites within the LV backbone that participate in the aberrant splicing process with variable efficiency. Among the splice sites identified by cLAM-PCR, 2 were already known to play an important role in HIV life cycle (canonical splice donor SD1 and acceptor SA2 sites) (9) and, to our knowledge, 6 are novel (SA1, SA3, SA4, SD4, SD5, and SA7) (Figure 2). Based on these preliminary data, 2 additional cLAM primer sets were designed to interrogate the activity of SA1 and SD4 (DWcrypSA and UPcrypSD sets, respectively) (Figure 2).
Figure 2. cLAM-PCR procedure for the retrieval of LV cellular fusion transcripts.
Cryptic splice sites identified by cLAM in the LV backbone are shown in parentheses: SA1, SD4, SA7, SA3, SA4, and SD5. cLAM-PCR primer sets, UPLVSD and DWLVSA as well as UPcrypSD and DWcrypSA, are indicated. LV.exon_1, LV.exon_1a, LV.exon_1b, LV.exon_2, and LV.exon_3, as defined by their boundary splice sites, are indicated.
Sequence analysis of aberrantly spliced transcripts and LV sequences participating in the aberrant splicing process.
Overall, using the 4 cLAM-PCR primer sets, we obtained 39,430 sequencing reads from SINLV.PGK-transduced JY cells and CD34+ HSCPs. A dedicated bioinformatics pipeline was used to eliminate the LV sequence complementary to the oligonucleotide used and the linker cassette. The remaining sequence was mapped on the LV and the human genomes to precisely identify the sequences involved in the fusion process. The majority of the sequencing reads (n = 28,216; 71.5%) were too short to be univocally mapped on the LV or human genome (less than 20 nucleotides) or lack the LV sequences required to validate the PCR products as genuine LV-originated transcripts. Although the process may appear to be relatively inefficient, we were still able to validate 11,214 sequence reads, sequencing reads (28.5%) as genuine transcripts containing LV backbone sequences. After exclusion of sequencing reads containing only LV genome (n = 8,720) and pooling all the redundant sequencing reads (n = 2,494), we identified 317 unique LV fusion transcripts with cellular gene exons or genomic sequences. The fusion transcripts were generated using the LV canonical splice acceptor or donor sites (17%) or the other splice sites within the LV backbone (SA1, 14.8%; SA3, 10.7%; SA4, 37.2%; SD4, 3.5%; SD5, 16.7%). Overall, the retrieved transcripts were fusions between LV splice sites and (a) known gene exons (88.6%); (b) cryptic splice sites located in known gene introns (6.6%); (c) 3′ UTRs (0.6%); and (d) cryptic splice sites located in intergenic regions (4.1%) (Figure 3 and Supplemental Table 1; supplemental material available online with this article; doi: 10.1172/JCI62189DS1). The latter cases are quite peculiar, as it appears that LV genomic integrations are able to tag unknown human transcripts or induce the formation of novel transcripts. All the splice acceptor sites identified within the LV backbone have the typical AG dinucleotide (Supplemental Table 1). Two out of three LV splice donors have the GT dinucleotide, while splice donor SD5 has a GC dinucleotide. We observed that 237 fusion transcripts (75%) show the expected GT/AG junction, a frequency lower than the 98%–99% reported for the genomic splice junctions (24). On the other hand, 53 fusion transcripts (16.7%) were generated by using the LV splice donor SD5, thus generating GC/AG sequences at the putative splice junctions. The remaining 27 fusion transcripts (7.3%) contained noncanonical splice junctions (for example, GC/AG, TC/AG and AC/AG). Interestingly, the latter class of transcripts was mainly (25 out of 27) the fusion among LV sequences and cryptic splice sites located in gene introns (n = 6), cryptic splice sites located in intergenic regions (n = 4), or within exonic sequences (n = 15).
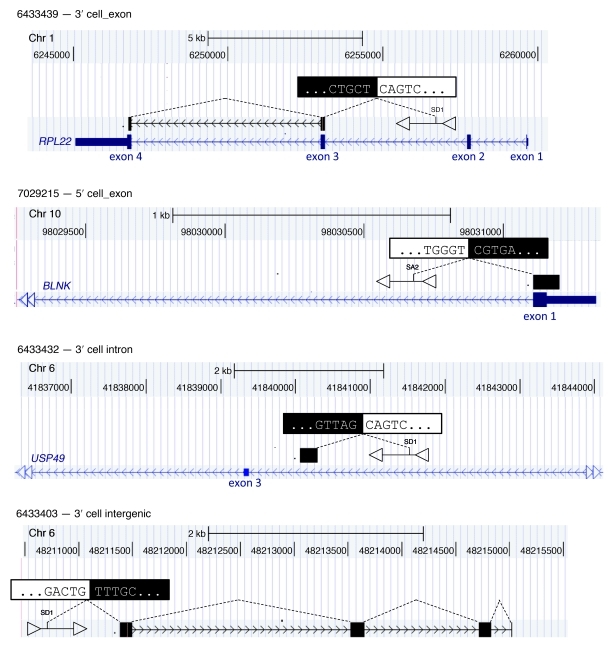
Figure 3. Examples of chimeric LV/cellular gene/genome transcripts.
Chimeric sequences are aligned on the human genome sequence using BLAT and shown on the UCSC genome browser. Sequences aligned to exonic sequences (black boxes) of know transcripts (chromosomal coordinates and size interval are shown above each panel). Orientation of vector and genes with respect to genome is indicated by orientation of triangles and arrows, respectively. Vector position and size are arbitrary. The 10 bases surrounding the vector/genomic junction are indicated: black text on white background indicates vector sequence, white text on black background indicates genomic sequence. In the 3 top panels, LV integrations in the same gene transcriptional orientation involved the canonical vector splice donor site SD1 sequence fused downstream of the SA site of a gene exon (i.e., RPL22, top panel); the vector splice acceptor sequence SA1 fused to cellular exons upstream (i.e., BLNK, second panel); and, in some cases, junctions with a splice site in an unannotated exon within gene introns were found (i.e., USP49, third panel). In some cases, fusion transcripts aligned discontinuously to genomic portions without annotated transcripts were identified (bottom panel). Chr, chromosome.
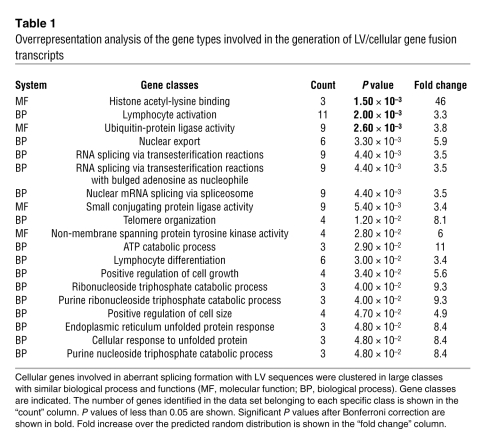
To understand whether the genes subjected to aberrant splicing were enriched for specific gene classes, we performed gene ontology analysis using the DAVID EASE online software ( http://david.abcc.ncifcrf.gov). From this analysis, we found that LV chimeric transcripts were significantly overrepresented for gene classes such as ubiquitin-protein ligase activity (P = 2.6 × 10–3, fold change = 3.8), nuclear export (P = 3.3 × 10–3, fold change = 5.9), lymphocyte activation (P = 2.0 × 10–3, fold change = 3.3), lymphocyte differentiation (P = 3.0 × 10–2, fold change = 3.4), positive regulation of growth (P = 3.4 × 10–2, fold change = 5.6), RNA splicing (P = 4.4 × 10–3, fold change = 3.5), nuclear mRNA splicing (P = 4.4 × 10–3, fold change = 3.5), and ATP catabolic process (P = 2.9 × 10–2, fold change = 11) (Table 1). This bias toward these gene classes overlapped only partially with the typical LV integration bias reported in hematopoietic cells (23, 25). To directly test the LV integration bias in our cells, we performed LAM-PCR on the genomic DNA of the same SINLV.PGK-transduced JY and CD34+ cell preparation used for cLAM-PCR. Overall, we mapped 1,630 unique LV integration sites and determined the integration bias into specific gene classes by gene ontology analysis. Similar to the reported LV integration bias reported in other hematopoietic cells, we observed the marked tendency to integrate into genes that are enriched for chromatin-remodeling functions (Supplemental Table 2).
Table 1 .
Overrepresentation analysis of the gene types involved in the generation of LV/cellular gene fusion transcripts
Interestingly, several gene classes significantly overrepresented in LV-mediated aberrant splicing formation, such as positive regulation of growth, RNA splicing, and lymphocyte activation and differentiation, are different from those found in our and other previously reported genomic integration profiles on hematopoietic cells (14, 23).
Impact of spice site recoding on vector infectivity and levels of read-through transcription on LV backbone.
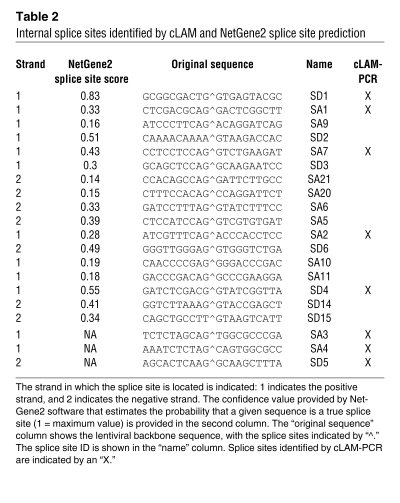
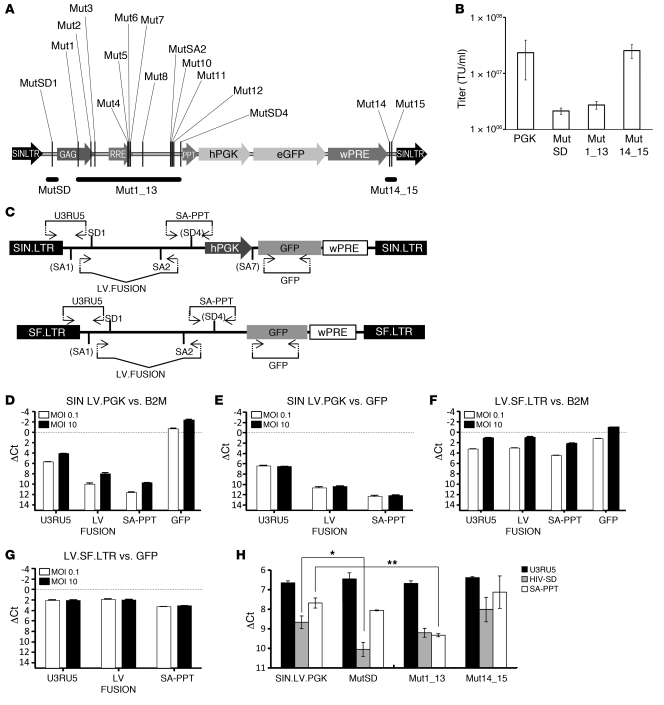
The identification of the 8 splice sites within the LV backbone by cLAM-PCR provides important information on how to recode the sequences to reduce the aberrant splicing potential events. Moreover, we used the NetGene2 server splice site prediction software ( http://www.cbs.dtu.dk/services/NetGene2/) to identify other potential splice sites within the LV backbone. The software identified 5 experimentally validated splice sites, with levels of confidence ranging from 0.28 to 0.83 (where 1 = a consensus splice site), and 15 additional putative splice sites (levels of confidence, >0.14) (Table 2). Since some splice sites are located in regions with a highly conserved secondary structure, 3 sets of mutations were distributed into the parental SINLV.PGK vector: a construct containing only the recoding of the canonical splice donor site SD1 (SINLV.MutSD.PGK.GFP.mwPREpre, referred to as MutSD); a construct containing 13 recoded splice sites comprising the region between SD1 and the cryptic SD4 (SINLV.Mut1_13.PGK.GFP.mwPREpre, referred to as Mut1_13); and a construct harboring 2 recoded splice sites near the 3′LTR (SINLV.Mut14_15.PGK.GFP.mwPRE, referred to as Mut14_15) (Figure 4A).
Table 2 .
Internal splice sites identified by cLAM and NetGene2 splice site prediction
Figure 4. Representation of aberrant splicing events within the LV backbone and quantification of transcription levels of LV backbone portions.
(A) Schematic representation of the position of the recoded splice sites within the LV backbone. The different mutations were distributed in 3 different vector constructs (indicated as MutSD, Mut1_13, and Mut14_15). RRE, rev-responsive element; wPRE, woodchuck hepatitis posttranscriptional regulatory element. (B) Titers of the 3 different recoded vectors. The titer is defined as number of transducing units per milliliter (TU/ml) of vector preparation. (C) Representation of the positions of the 4 TaqMan primer sets on SIN.LV.PGK and LV.SF.LTR vectors. U3RU5 recognizes the portion from the LV.LTR to the SD1, encompassing the cryptic splice acceptor SA1; LV.FUSION recognizes the internally spliced transcript (SD1 to SA2); SA-PPT recognizes the sequence downstream of the canonical splice acceptor SA2, encompassing the cryptic donor SD4; and GFP recognizes the GFP transgene sequence. (D–G) RT-qPCR results of transcription levels of different LV backbone portions performed on JY cells transduced with SIN.LV.PGK or LV.SF.LTR at MOI 0.1 (white bars) or MOI 10 (black bars). ΔCt values were obtained using β2 microglobulin (B2M) as normalizer to measure the relative expression levels with respect to the housekeeping cellular gene (vs. B2M). ΔCt values obtained using GFP as normalizer to measure the relative expression levels with respect to transgene expression (vs. GFP). (H) ΔCt values obtained using GFP as normalizer from JY cells transduced with SIN.LV.PGK. The recoded vectors are indicated. Probe sets used are indicated. Statistical evaluation was performed by 1-way ANOVA with Bonferroni’s correction (*P < 0.001; **P < 0.0001).
Each recoded LV construct was then tested by FACS to evaluate vector titer and by RT-qPCR to measure transgene expression and read-through transcription within the LV backbone as surrogate of aberrant splicing potential. The constructs harboring the mutated SD1 and the 13 mutations showed a substantial reduction in infectivity (10-fold reduction), while the vector with the 2 mutations near the 3′LTR (Mut14_15) had a comparable infectivity to that of the standard SINLV.PGK (Figure 4B and Supplemental Table 3). Mean fluorescence intensity (MFI) of GFP in single copy–transduced JY cells was similar among all the different vectors (MFI range 7,400–7,900), indicating that the recoding does not affect transgene expression in any case (Supplemental Table 3).
We set up RT-qPCR assays on cDNA from transduced JY cells to probe the transcription levels in different portions of the different LV backbones (Figure 4C). The oligonucleotides and probes used for the RT-qPCR were designed to amplify different portions of the LV backbone encompassing the splice sites identified in this study: the U3RU5 RT-PCR assay, encompassing the SA1; the LV.FUSION, encompassing the HIV1 intron and measuring only spliced LV mRNAs; SA.PPT, encompassing the SD4; the GFP assay, complementary to the GFP transgene sequence.
JY cells were initially transduced with 2 different LVs: the previously mentioned SINLV.PGK and an LV harboring the strong enhancer sequence of the spleen focus forming virus (SF) promoter within the LTR (LV.SF.LTR) and driving GFP expression (10). The latter vector was used as positive control of transcription within the LV backbone, as the SF promoter transcription starts upstream of the regions tested for expression, and the transcript is extended through the GFP transgene and terminates at the polyadenylation site in the 3′LTR.
The relative levels of read-through transcription within the LV backbone were normalized to the endogenous housekeeping β2 microglobulin (B2M) gene. The expression measured by the U3RU5 probe in SINLV.PGK-transduced cells at MOI 10 or 0.1 showed a ΔCt of 4.09 and 5.67, corresponding to 6.3% and 1.9% of the housekeeping gene expression level, respectively. Interestingly, the other probes (LV-FUSION and SA-PPT) showed a decrease in expression levels from 5′ to the 3′ of the LV backbone (Figure 4D). As the amounts of read-through transcription vary according to the number of vector integrations in expression-permissive genome, the relative expression level of each LV portion tested was also normalized to the GFP level, which depends directly on the integrated vector load. For both MOI, the U3RU5 probe showed a ΔCt of 6.4 ± 0.1 with respect to GFP, indicating that read-through transcription was more than 1.2% of the overall GFP transcripts produced by the internal expression cassette (Figure 4E). Also, with this normalization, both LV-FUSION and SA-PPT showed a progressive reduction of the expression levels from 5′ to the 3′ of the LV backbone. Using the same probe sets, we measured the transcription levels of the LV.SF.LTR backbone in transduced JY cells. The presence of the strong SF enhancer/promoter within the LTR resulted in a much higher level of expression compared with the backbone transcription measured for the SINLV.PGK, indicating a previously underappreciated advantage of the SIN LTR design (Figure 4, F and G). We then performed our RT-qPCR analysis on JY cells transduced with the recoded constructs (MutSD, Mut1_13, and Mut14_15). We used the U3RU5 and SA-PPT probe sets and an additional probe set encompassing the canonical HIV1 splice donor (HIV-SD). GFP normalized values show that splice site mutagenesis can further and significantly reduce the residual backbone transcription in MutSD and Mut1_13 vectors when compared with the expression of the parental vector (SINLV.PGK vs. MutSD with HIV-SD, P = 0.001 and SINLV.PGK vs. Mut1_13 with SA-PPT, P < 0.0001) (Figure 4H and Supplemental Figure 1).
Discussion
To address the concerns about LV-induced aberrant splicing in relevant cell types and in a high-throughput fashion, we adapted the sensitive LAM-PCR technique (22) to use cDNA from vector-transduced cells as template, rather than genomic DNA. In the 4 different LV regions interrogated by our probe sets, we identified several aberrantly spliced mRNAs containing LV sequences fused with cellular transcripts from a relevant cell type such as human primary HSPCs.
Fusions of LV sequences with exons of known cellular genes, new unannotated spliced transcripts aligning to discontinuous human genomic sequences, and spliced transcripts within the LV backbone were identified. The cellular genes involved in aberrant splicing formation were significantly enriched in molecular functions for chromatin modification and ubiquitin activity. The enrichment for these gene classes reflects the LV genomic integration bias directly confirmed in the same cells used in our experiments and that has been previously reported in hematopoietic cells from clinical trials and from humanized hematochimeric mice (23, 25). However, other gene classes, such as RNA splicing, lymphocyte activation and differentiation, and positive regulation of cell growth, appear to be specifically overrepresented in the aberrantly spliced products data set. Such differential enrichment in specific gene classes may reflect differences in gene expression levels among the differentiated HSC progeny, in which only a subset of vector integrations targeting the expressed genes of a specific lineage could produce detectable aberrant transcripts. Thus, integration sites and aberrant transcripts may not necessarily share exactly the same biases for gene classes. Interestingly, some transcripts mapped to exon-like discontinuous genomic regions without any corresponding annotated mRNA, suggesting the possibility that novel transcripts may be generated upon integration.
The biological impact of LV-mediated aberrant splicing on cells is unknown. Transcripts originating from the vector can use splice donor sequences to fuse with splice acceptors of cellular gene exons and generate truncated proteins. These events, frequently observed when using vectors with active LTRs (10), should be relatively rare using SIN LVs. Indeed, here we show a substantially reduced transcription of the vector backbone when comparing LV with SIN and active LTRs. Stable protein coding transcripts from the genes flanking the integration site may be truncated prematurely by LV splice acceptor and polyadenylation sites present in the LTRs or may acquire additional LV exonic sequences. These events can also occur with SIN LVs, as shown here. Conceivably, the mRNAs produced by read-through transcription within vector sequences undergo regulation by the nonsense mRNA decay (18, 19, 26). As shown in the work of Almarza et al. (20), the noncoding transcripts generated by read-through transcription from the internal promoter of an integrated SINLV are regulated by the nonsense mRNA decay mechanism. This would affect the abundance of specific transcripts with respect to all the potential aberrant transcripts that can be generated by all genomic integrations. Therefore, only a subset of expressed genes targeted by vector integration will produce stable mRNAs potentially encoding for aberrant proteins.
The cLAM-PCR provides qualitative information on the cellular genes and on the LV sequences that play a role in the aberrant splicing process. However, cLAM-PCR is not well suited to quantify how frequently this phenomenon occurs in LV-transduced cells. The use of restriction enzymes in the cLAM-PCR protocol, as it is true for the LAM-PCR, generates amplification products of heterogeneous size, inducing biases in the representation of the fusion products (10, 27). These biases prevent both a reliable quantification of aberrantly spliced forms produced by a gene targeted by vector integration and the relative usage of the different splice sites within the vector. Moreover, the amount of aberrant transcripts generated will probably vary depending on the vector design, the strength of the promoter of the targeted gene that drives the transcription of the chimeric mRNA, and/or the loss or gain of mRNA destabilizing/stabilizing signals. Thus, to quantify aberrant transcripts, it is necessary to isolate single cell–derived clones to map the integration site and design gene expression assays aimed to detect specific LV/gene fusion transcripts.
To overcome the need to study isolated cell clones, we devised an RT-qPCR approach to quantitatively measure the overall amount of transcription within different LV backbone portions in cells bearing widespread LV integrations throughout the genome. Thus, this approach does not interrogate gene-specific aberrant transcription. Our analysis shows an interesting transcription pattern of the SIN LV backbone, with measurable levels of read-through transcription in the first portion near the 5′ LTR (about 6% of the β2 microglobulin gene expression level). More internal portions from 5′ to the 3′ of the LV genome showed a progressive decrease in read-through expression levels. The mechanism underlining these differences is unknown, but we may hypothesize that read-through transcripts entering the LV genome will tend to use the first available splice sites to link to the next gene exon or to the next polyadenylation site and terminate transcription. A very different picture appeared when LV with active LTRs was used: high levels of transcription were observed along the vector backbone, without the 5′ to 3′ decrease observed in SIN LV-transduced cells. Our data thus indicate that the presence of transcriptional enhancer/promoter sequences within the LTR not only increases the overall transcription but also the usage of the splice sites surrounding the canonical HIV1 intron. Thus, the SIN design not only alleviates the risk of enhancer-mediated oncogene activation but also sharply decreases splice capture and transcriptional interference with read-through genomic transcripts. Considering the high levels of transduction that can be reached with SINLV.PGK in primary HSPCs (VCN of 2 to 5) and the tendency of LVs to integrate into actively transcribed genes (28), the observed 1.2% of vector read-through transcription relative to the PGK promoter-driven transgene expression can be considered low, especially when compared with vectors with active LTRs.
Yet, some aberrant transcripts are still produced from SIN LV integrations. Therefore, to further reduce the probability of LV-induced aberrant splicing, we extensively recoded the LV backbone to eliminate 17 splice sites identified by cLAM and bioinformatics prediction. Unfortunately, some splice sites are located in regions that are important for vector RNA packaging, such as the canonical splice donor of HIV1 and proximal sequences in the gag gene, and their elimination by recoding led to a substantial reduction in vector titers. Further work to dissect the critical sequences whose mutations are detrimental to vector titer is ongoing and will be instrumental to maximize the number of mutations that are neutral to vector titer.
We also investigated the effect of vector recoding on read-through transcription by our RT-qPCR strategy. Indeed, recoded vectors showed a concordant decrease in read-through transcription with respect to nonmutated vector. However, read-through transcription was not abrogated, suggesting that additional mutations are required to fully prevent aberrant splicing. Future vector designs could be generated to contain a minimal set of essential splice sites and incorporate safety features to reduce the abundance or stability of aberrantly spliced transcripts.
Overall, cLAM-PCR coupled to high-throughput sequencing technology and RT-qPCR analysis allowed us to gain insights into the occurrence and specific features of vector-mediated splicing alteration in cells that are relevant for gene therapy applications. A deeper characterization of the mechanisms and the genetic features that cause vector-induced aberrant splicing enables the generation of safer vectors with low impact on the cellular transcriptome.
Methods
Vector production and titration.
LV.SF.LTR and SINLV.PGK constructs were previously generated (10, 29). Concentrated LV stocks, pseudotyped with the vesicular stomatitis virus envelope, were produced by transient 4-plasmid cotransfection of 293T cells and titered on HeLa cells, as described previously (29). Recoded vectors were generated by DNA gene synthesis (GeneArt) and cloned in the SINLV.PGK transfer plasmid. A 728-bp DNA fragment harboring the mutation in the canonical HIV1 splice donor was cloned using NruI and NdeI restriction sites. A 1,331-bp DNA fragment harboring the mutations from 1 to 13 was cloned using NruI and XhoI restriction sites. A 751-bp DNA fragment harboring the mutations from 14 and 15 was cloned using SacII and AvrII restriction sites.
Isolation and transduction of human HSPCs and JY cells.
Cord blood–derived cells were harvested, cultured, and transduced as previously described (23). Human HSPCs were obtained by positive selection of CD34-expressing cells (CD34 Progenitor Cell Isolation Kit, MACS; Miltenyi Biotec) from cord blood from healthy donors. Soon after purification or thawing, cells were placed in culture at a concentration of 1 × 106 cells/ml to 1.5 × 106 cells/ml in the presence of cytokines (IL-3 [60 ng/μl], thrombopoietin [100 ng/μl], SCF [300 ng/μl], and Flt3 ligand [300 ng/μl]; PeproTech) for 24 to 48 hours of prestimulation. Cells were then transduced with the different LVs at a MOI as indicated for 12 hours. Cells were plated in Iscove’s modified Dulbecco’s medium (Euroclone) —10% fetal bovine serum (Euroclone) with cytokines (IL-3 [60 ng/μl]; IL-6 [60 ng/μl]; SCF [300 ng/μl]) — and cultured for a total of 14 days. Thereafter, cells were collected for molecular, biochemical, and flow cytometry studies. JY cells were grown in RPMI and 10% FBS supplemented with penicillin and streptomycin and transduced at a MOI of 10, 1, or 0.1 in a single round of infection.
Flow cytometry.
Before prestimulation and at the end of transduction, 5 × 104 cells were stained with 1 μl PE-conjugated anti-CD34 and FITC-conjugated anti-CD45 antibodies or IgG isotype controls (Dako). After 20 minutes on ice, cells were washed, resuspended in PBS with 2% FBS and 1% paraformaldehyde, and analyzed by flow cytometry (FACSCalibur; BD Biosciences — Immunocytometry Systems). The percentage of CD34+ cells was calculated on the gated CD45+ population.
At the end of the 14-day culture period, 1 × 105 GFP-transduced cells were collected, and GFP fluorescence (measured as the percentage of positive cells and MFI) was detected with detector channel FL1 calibrated to the FITC emission profile.
DNA and RNA isolation, cDNA synthesis, and quantitative PCR.
Genomic DNA was extracted from CD34+ liquid culture samples with the QIAamp DNA Blood Mini Kit (Qiagen) and from murine tissues with the Blood and Cells DNA Midi Kit (Qiagen) after overnight digestion with proteinase K (Roche).
Total RNA from JY or CD34+ cells was isolated with the RNeasy Mini Kit (Qiagen) following the manufacturer’s instructions. Double-stranded cDNA preparation was performed using the SuperScript Double-Stranded cDNA Synthesis Kit (Invitrogen). cDNA was used as the template for Custom Plus TaqMan Gene Expression Assays specific to each LV portion (Applied Biosystems). Amplification reactions were performed on a 7900HT Real-Time PCR Thermal Cycler (Applied Biosystems). The relative expression level of each gene was calculated by the ΔCt method and normalizing to β2 microglobulin (housekeeping gene) or GFP expression.
Gene expression assays used are listed in Supplemental Table 4.
cLAM-PCR amplification.
We used 500 ng double-stranded cDNA as template for cLAM-PCR. cLAM-PCR was initiated with a 100-cycle linear PCR using a biotinylated primer (UPLVSD_1, DWLVSA_1, UPcrypSD_1, or DWcrypSA_1), second-strand synthesis by the Klenow fragment and random hexamers, restriction digest using Tsp509I or HpyCH4IV, and ligation of a restriction site–complementary linker cassette. The biotinylated PCR product was captured via magnetic beads and reamplified by 2 nested PCRs using primers downstream to the first primer used (UPLVSD_2 and UPLVSD_3, or DWLVSA_2 and DWLVSA_3, or UPcrypSD_2 and UPcrypSD_3, or DWcrypSA_2) and primers complementary to the linker cassette (10, 22). Primer sequences for the 4 primer sets are as follows: (UPLVSD_1 GAAAGCGAAAGGGAAACCAGA, UPLVSD_2 GACGCAGGACTCGGCTTG, UPLVSD_3 ACGGCAAGAGGCGAGG; DWLVSA_1 TCGAGATCCGTTCACTAATCG, DWLVSA_2 ATGGATCTGTCTCTGTCTCTCTCT, DWLVSA_3 CCACCTTCTTCTTCTATTCCTTC; UPcrypSD_1 GAGGGGACCCGACAGG, UPcrypSD_2 CCGAAGGAATAGAAGAAGAAGG, UPcrypSD_3 CAGAGACAGATCCATTCGATTAGTG; DWcrypSA_1 CCTCGCCTCTTGCCGTGC, DWcrypSA_2 CTTCAGCAAGCCGAGTCC). Linker cassette primers were previously described (10, 22).
cLAM-PCR products were separated by Spreadex gel electrophoresis (Elchrom Scientific) to verify the presence and the number of bands. cLAM-PCR was shotgun cloned into the TOPO TA vector (Invitrogen) and sequenced by Sanger sequencing (GATC Biotech) or directly sequenced by 454 pyrosequencing after a PCR reamplification, with the use of oligonucleotides with specific 6-nucleotide sequence tags for sample identification. Sequences were validated and classified with specific scripts and aligned to the human genome (GRCh37/hg19) or with the use of the UCSC BLAT genome browser ( http://genome.ucsc.edu/cgi-bin/hgBlat).
Gene ontology analysis.
Analysis of overrepresentation of gene classes in integration data sets was performed with the DAVID EASE software ( http://david.abcc.ncifcrf.gov/home.jsp) using the stringency setting “high.”
Statistics.
For gene ontology analyses, we considered significant only those classes represented by at least 3 genes, a fold increase of more than 3, and a P value of less than 0.05. The results were corrected for multiple testing errors within each data set/system combination with Bonferroni’s method. For gene expression analyses, 1-way ANOVA test with Bonferroni’s multiple comparison post-hoc test was used to assess statistical significance of differences among all samples (P < 0.05). In all graphs, the mean ± standard deviations are indicated.
Study approval.
Cord blood–derived human CD34+ cells were collected upon informed consent, in the context of the TIGET01 protocol, which was approved by San Raffaele Scientific Institute Ethical Committee.
Supplementary Material
Acknowledgments
This work was supported by the Association for International Cancer Research grant to E. Montini (AICR 09-0784), EU Clinigene NoE grant to E. Montini (LSHB-CT-2006-018933), Italian Telethon to L. Naldini and E. Montini (TIGET grants), EU grant HEALTH-2009-222878 to L. Naldini (PERSIST), and Bill and Melinda Gates Foundation Grand Challenges Explorations grant (Round 7) to E. Montini (OPP1045909). We are grateful to Andrea Calabria for bioinformatics support.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(5):1667–1676. doi:10.1172/JCI62189.
See the related Commentary beginning on page 1600.
References
- 1.Uren AG, Kool J, Berns A, van Lohuizen M. Retroviral insertional mutagenesis: past, present and future. Oncogene. 2005;24(52):7656–7672. doi: 10.1038/sj.onc.1209043. [DOI] [PubMed] [Google Scholar]
- 2.Baum C. Insertional mutagenesis in gene therapy and stem cell biology. Curr Opin Hematol. 2007;14(4):337–342. doi: 10.1097/MOH.0b013e3281900f01. [DOI] [PubMed] [Google Scholar]
- 3.Hacein-Bey-Abina S, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118(9):3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kool J, Berns A. High-throughput insertional mutagenesis screens in mice to identify oncogenic networks. Nat Rev Cancer. 2009;9(6):389–399. doi: 10.1038/nrc2647. [DOI] [PubMed] [Google Scholar]
- 5.Montini E, et al. The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy. J Clin Invest. 2009;119(4):964–975. doi: 10.1172/JCI37630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Montini E, et al. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nat Biotechnol. 2006;24(6):687–696. doi: 10.1038/nbt1216. [DOI] [PubMed] [Google Scholar]
- 7.Ott MG, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006;12(4):401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- 8.Stein S, et al. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat Med. 2010;16(2):198–204. doi: 10.1038/nm.2088. [DOI] [PubMed] [Google Scholar]
- 9. Coffin JM, Hughes SH, Varmus H.Retroviruses . Plainview, New York, USA: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 10.Gabriel R, et al. Comprehensive genomic access to vector integration in clinical gene therapy. Nat Med. 2009;15(12):1431–1436. doi: 10.1038/nm.2057. [DOI] [PubMed] [Google Scholar]
- 11.Bokhoven M, et al. Insertional gene activation by lentiviral and gammaretroviral vectors. J Virol. 2009;83(1):283–294. doi: 10.1128/JVI.01865-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knight S, Bokhoven M, Collins M, Takeuchi Y. Effect of the internal promoter on insertional gene activation by lentiviral vectors with an intact HIV long terminal repeat. J Virol. 2010;84(9):4856–4859. doi: 10.1128/JVI.02476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lombardo A, et al. Site-specific integration and tailoring of cassette design for sustainable gene transfer. Nat Methods. 2011;8(10):861–869. doi: 10.1038/nmeth.1674. [DOI] [PubMed] [Google Scholar]
- 14.Cavazzana-Calvo M, et al. Transfusion independence and HMGA2 activation after gene therapy of human beta-thalassaemia. Nature. 2010;467(7313):318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436(7048):272–276. doi: 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]
- 16.Palanisamy N, et al. Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nat Med. 2010;16(7):793–798. doi: 10.1038/nm.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Q, Lucas A, Son S, Chang LJ. Overlapping enhancer/promoter and transcriptional termination signals in the lentiviral long terminal repeat. Retrovirology. 2007;4:4. doi: 10.1186/1742-4690-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Proudfoot NJ. Ending the message: poly(A) signals then and now. Genes Dev. 2011;25(17):1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108(4):501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- 20.Almarza D, Bussadori G, Navarro M, Mavilio F, Larcher F, Murillas R. Risk assessment in skin gene therapy: viral-cellular fusion transcripts generated by proviral transcriptional read-through in keratinocytes transduced with self-inactivating lentiviral vectors. Gene Ther. 2011;18(7):674–681. doi: 10.1038/gt.2011.12. [DOI] [PubMed] [Google Scholar]
- 21.Zaiss AK, Son S, Chang LJ. RNA 3’ readthrough of oncoretrovirus and lentivirus: implications for vector safety and efficacy. J Virol. 2002;76(14):7209–7219. doi: 10.1128/JVI.76.14.7209-7219.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt M, et al. High-resolution insertion-site analysis by linear amplification-mediated PCR (LAM-PCR). Nat Methods. 2007;4(12):1051–1057. doi: 10.1038/nmeth1103. [DOI] [PubMed] [Google Scholar]
- 23.Biffi A, et al. Lentiviral vector common integration sites in preclinical models and a clinical trial reflect a benign integration bias and not oncogenic selection. Blood. 2011;117(20):5332–5339. doi: 10.1182/blood-2010-09-306761. [DOI] [PubMed] [Google Scholar]
- 24.Burset M, Seledtsov IA, Solovyev VV. SpliceDB: database of canonical and non-canonical mammalian splice sites. Nucleic Acids Res. 2001;29(1):255–259. doi: 10.1093/nar/29.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cartier N, et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326(5954):818–823. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- 26.Gardner LB. Nonsense-mediated RNA decay regulation by cellular stress: implications for tumorigenesis. Mol Cancer Res. 2010;8(3):295–308. doi: 10.1158/1541-7786.MCR-09-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang GP, et al. DNA bar coding and pyrosequencing to analyze adverse events in therapeutic gene transfer. Nucleic Acids Res. 2008;36(9):e49. doi: 10.1093/nar/gkn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schroder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110(4):521–529. doi: 10.1016/S0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 29.Follenzi A, Ailles LE, Bakovic S, Geuna M, Naldini L. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat Genet. 2000;25(2):217–222. doi: 10.1038/76095. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.