Abstract
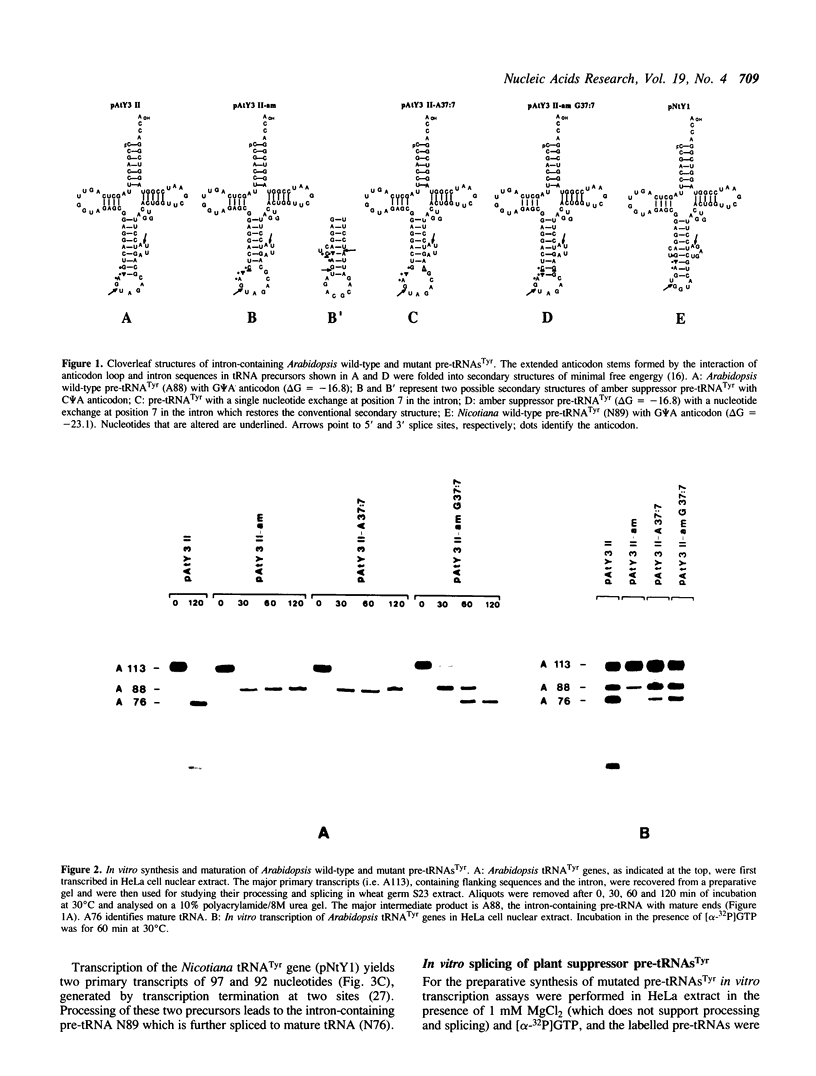
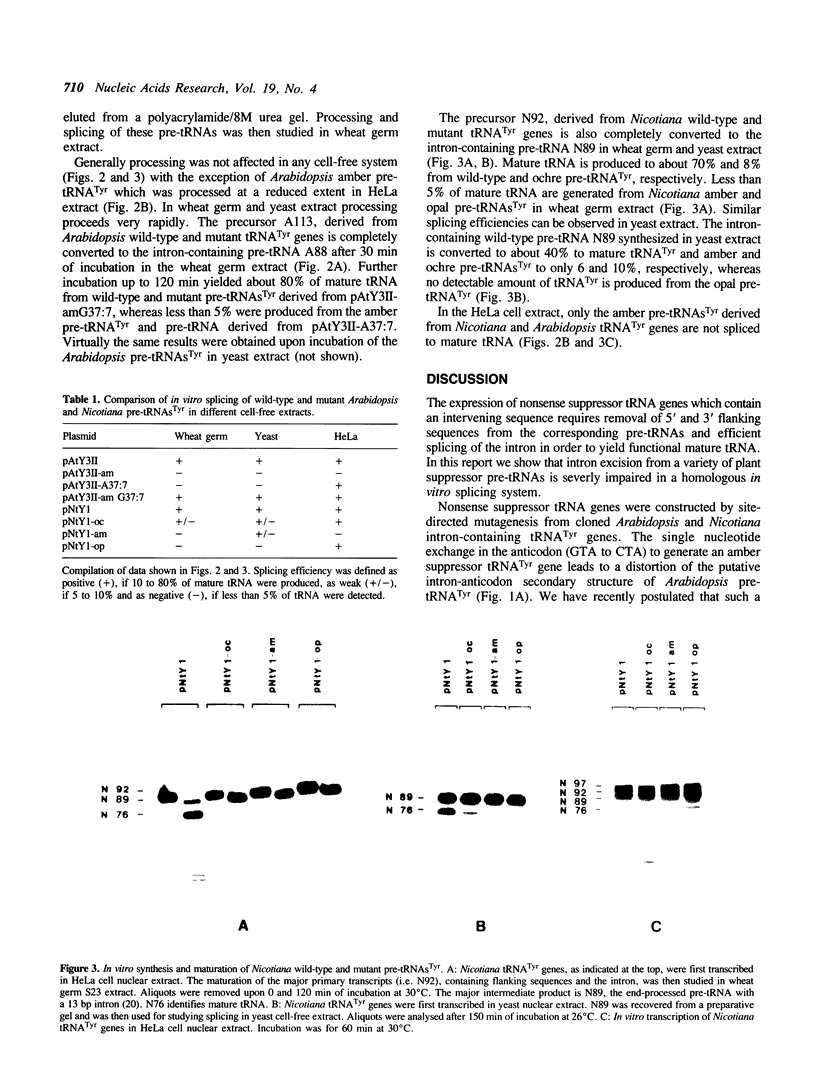
Oligonucleotide-directed mutagenesis was used to generate amber, ochre and opal suppressors from cloned Arabidopsis and Nicotiana tRNA(Tyr) genes. The nonsense suppressor tRNA(Tyr) genes were efficiently transcribed in HeLa and yeast nuclear extracts, however, intron excision from all mutant pre-tRNAs(Tyr) was severely impaired in the homologous wheat germ extract as well as in the yeast in vitro splicing system. The change of one nucleotide in the anticodon of suppressor pre-tRNAs leads to a distortion of the potential intron-anticodon interaction. In order to demonstrate that this caused the reduced splicing efficiency, we created a point mutation in the intron of Arabidopsis tRNA(Tyr) which affected the interaction with the wild-type anticodon. As expected, the resulting pre-tRNA was also inefficiently spliced. Another mutation in the intron, which restored the base-pairing between the amber anticodon and the intron of pre-tRNA(Tyr), resulted in an excellent substrate for wheat germ splicing endonuclease. This type of amber suppressor tRNA(Tyr) gene which yields high levels of mature tRNA(Tyr) should be useful for studying suppression in higher plants.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnold G. J., Schmutzler C., Thomann U., van Tol H., Gross H. J. The human tRNAVal gene family: organization, nucleotide sequences and homologous transcription of three single-copy genes. Gene. 1986;44(2-3):287–297. doi: 10.1016/0378-1119(86)90193-9. [DOI] [PubMed] [Google Scholar]
- Atkin A. L., Roy K. L., Bell J. B. Construction of an opal suppressor by oligonucleotide-directed mutagenesis of a Saccharomyces cerevisiae tRNA(Trp) gene. Mol Cell Biol. 1990 Aug;10(8):4379–4383. doi: 10.1128/mcb.10.8.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capone J. P., Sharp P. A., RajBhandary U. L. Amber, ochre and opal suppressor tRNA genes derived from a human serine tRNA gene. EMBO J. 1985 Jan;4(1):213–221. doi: 10.1002/j.1460-2075.1985.tb02338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingermann T., Nerke K., Blöcker H., Frank R. Structural requirements for the synthesis of tRNATrp from Dictyostelium discoideum in yeast. Biochimie. 1988 Jun;70(6):711–719. doi: 10.1016/0300-9084(88)90099-5. [DOI] [PubMed] [Google Scholar]
- Doerig R. E., Suter B., Gray M., Kubli E. Identification of an amber nonsense mutation in the rosy516 gene by germline transformation of an amber suppressor tRNA gene. EMBO J. 1988 Aug;7(8):2579–2584. doi: 10.1002/j.1460-2075.1988.tb03107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiduschek E. P., Tocchini-Valentini G. P. Transcription by RNA polymerase III. Annu Rev Biochem. 1988;57:873–914. doi: 10.1146/annurev.bi.57.070188.004301. [DOI] [PubMed] [Google Scholar]
- Hattori M., Sakaki Y. Dideoxy sequencing method using denatured plasmid templates. Anal Biochem. 1986 Feb 1;152(2):232–238. doi: 10.1016/0003-2697(86)90403-3. [DOI] [PubMed] [Google Scholar]
- Huibregtse J. M., Evans C. F., Engelke D. R. Comparison of tRNA gene transcription complexes formed in vitro and in nuclei. Mol Cell Biol. 1987 Sep;7(9):3212–3220. doi: 10.1128/mcb.7.9.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P. F., Abelson J. The yeast tRNATyr gene intron is essential for correct modification of its tRNA product. Nature. 1983 Apr 21;302(5910):681–687. doi: 10.1038/302681a0. [DOI] [PubMed] [Google Scholar]
- Kim D., Johnson J. Construction, expression, and function of a new yeast amber suppressor, tRNATrpA. J Biol Chem. 1988 May 25;263(15):7316–7321. [PubMed] [Google Scholar]
- Kim D., Raymond G. J., Clark S. D., Vranka J. A., Johnson J. D. Yeast tRNATrp genes with anticodons corresponding to UAA and UGA nonsense codons. Nucleic Acids Res. 1990 Jul 25;18(14):4215–4221. doi: 10.1093/nar/18.14.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laski F. A., Belagaje R., Hudziak R. M., Capecchi M. R., Norton G. P., Palese P., RajBhandary U. L., Sharp P. A. Synthesis of an ochre suppressor tRNA gene and expression in mammalian cells. EMBO J. 1984 Nov;3(11):2445–2452. doi: 10.1002/j.1460-2075.1984.tb02154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laski F. A., Belagaje R., RajBhandary U. L., Sharp P. A. An amber suppressor tRNA gene derived by site-specific mutagenesis: cloning and function in mammalian cells. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5813–5817. doi: 10.1073/pnas.79.19.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laski F. A., Ganguly S., Sharp P. A., RajBhandary U. L., Rubin G. M. Construction, stable transformation, and function of an amber suppressor tRNA gene in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6696–6698. doi: 10.1073/pnas.86.17.6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson J. M., Meuris P., Grunstein M., Abelson J., Miller J. H. Expression of a set of synthetic suppressor tRNA(Phe) genes in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6815–6819. doi: 10.1073/pnas.84.19.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappu S. S., Roy K. L., Bell J. B. Drosophila melanogaster tRNA(Ser) suppressor genes function with strict codon specificity when introduced into Saccharomyces cerevisiae. Gene. 1990 Jul 16;91(2):255–259. doi: 10.1016/0378-1119(90)90096-a. [DOI] [PubMed] [Google Scholar]
- Piper P. W., Wasserstein M., Engbaek F., Kaltoft K., Celis J. E., Zeuthen J., Liebman S., Sherman F. Nonsense suppressors of Saccharomyces cerevisiae can be generated by mutation of the tyrosine tRNA anticodon. Nature. 1976 Aug 26;262(5571):757–761. doi: 10.1038/262757a0. [DOI] [PubMed] [Google Scholar]
- Sharp S. J., Schaack J., Cooley L., Burke D. J., Söll D. Structure and transcription of eukaryotic tRNA genes. CRC Crit Rev Biochem. 1985;19(2):107–144. doi: 10.3109/10409238509082541. [DOI] [PubMed] [Google Scholar]
- Stange N., Beier H. A cell-free plant extract for accurate pre-tRNA processing, splicing and modification. EMBO J. 1987 Sep;6(9):2811–2818. doi: 10.1002/j.1460-2075.1987.tb02577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stange N., Beier H. A gene for the major cytoplasmic tRNATyr from Nicotiana rustica contains a 13 nucleotides long intron. Nucleic Acids Res. 1986 Nov 11;14(21):8691–8691. doi: 10.1093/nar/14.21.8691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stange N., Gross H. J., Beier H. Wheat germ splicing endonuclease is highly specific for plant pre-tRNAs. EMBO J. 1988 Dec 1;7(12):3823–3828. doi: 10.1002/j.1460-2075.1988.tb03267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel M. C., Abelson J. Effect of intron mutations on processing and function of Saccharomyces cerevisiae SUP53 tRNA in vitro and in vivo. Mol Cell Biol. 1986 Jul;6(7):2663–2673. doi: 10.1128/mcb.6.7.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel M. C., Abelson J. Intron mutations affect splicing of Saccharomyces cerevisiae SUP53 precursor tRNA. Mol Cell Biol. 1986 Jul;6(7):2674–2683. doi: 10.1128/mcb.6.7.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekely E., Belford H. G., Greer C. L. Intron sequence and structure requirements for tRNA splicing in Saccharomyces cerevisiae. J Biol Chem. 1988 Sep 25;263(27):13839–13847. [PubMed] [Google Scholar]
- Szweykowska-Kulinska Z., Beier H. Nucleotide sequences of two nuclear tRNA(Tyr) genes from Triticum aestivum. Nucleic Acids Res. 1990 Apr 11;18(7):1894–1894. doi: 10.1093/nar/18.7.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple G. F., Dozy A. M., Roy K. L., Kan Y. W. Construction of a functional human suppressor tRNA gene: an approach to gene therapy for beta-thalassaemia. Nature. 1982 Apr 8;296(5857):537–540. doi: 10.1038/296537a0. [DOI] [PubMed] [Google Scholar]
- Wiederrecht G., Shuey D. J., Kibbe W. A., Parker C. S. The Saccharomyces and Drosophila heat shock transcription factors are identical in size and DNA binding properties. Cell. 1987 Feb 13;48(3):507–515. doi: 10.1016/0092-8674(87)90201-7. [DOI] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis: a simple method using two oligonucleotide primers and a single-stranded DNA template. DNA. 1984 Dec;3(6):479–488. doi: 10.1089/dna.1.1984.3.479. [DOI] [PubMed] [Google Scholar]
- van Tol H., Beier H. All human tRNATyr genes contain introns as a prerequisite for pseudouridine biosynthesis in the anticodon. Nucleic Acids Res. 1988 Mar 25;16(5):1951–1966. doi: 10.1093/nar/16.5.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tol H., Stange N., Gross H. J., Beier H. A human and a plant intron-containing tRNATyr gene are both transcribed in a HeLa cell extract but spliced along different pathways. EMBO J. 1987 Jan;6(1):35–41. doi: 10.1002/j.1460-2075.1987.tb04715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]