Abstract
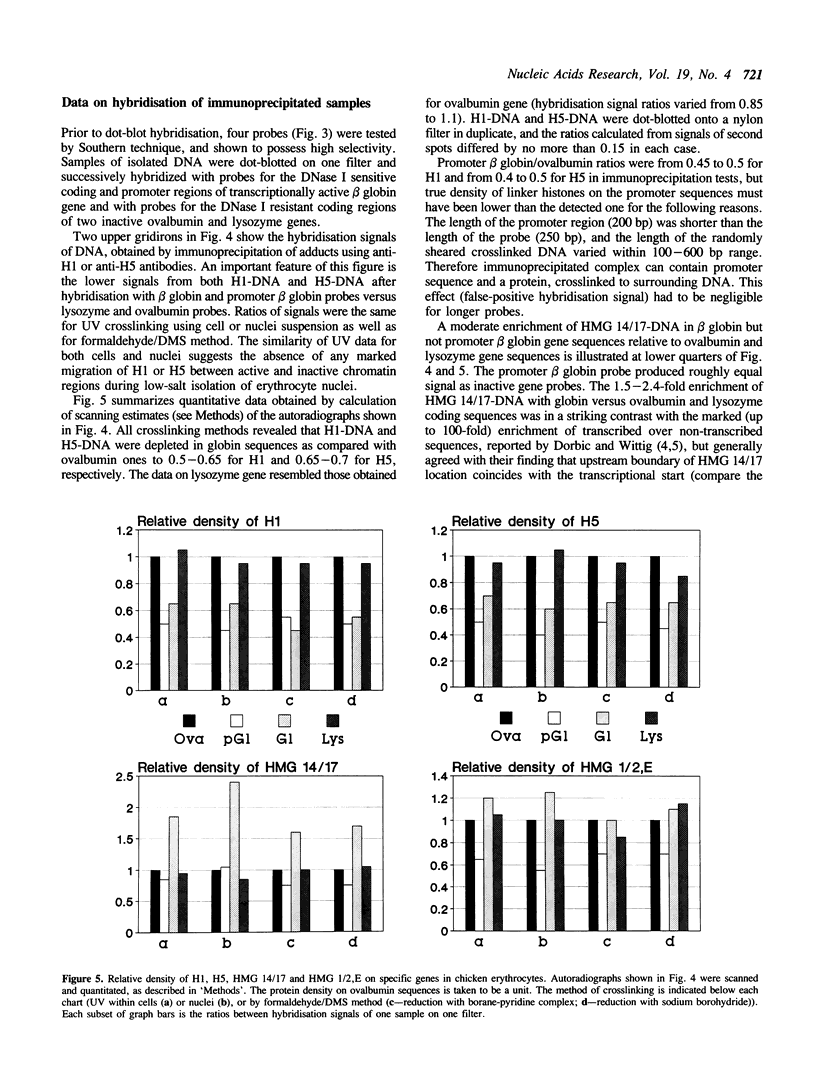
Quantitative analysis of distribution of chromosomal proteins on single copy DNA sequences has been further developed. Our approach consists of DNA-protein crosslinking within whole cells or isolated nuclei, specific immunoaffinity isolation of crosslinked complexes via protein and identification of crosslinked DNA by hybridisation with single-stranded DNA probes. The present study shows that transcribed chromatin of chicken embryonic erythrocyte beta globin gene is characterized by about 1.5-2.5-fold higher density of HMG 14/17 and 2-fold lower density of H1 and H5 as compared with non-transcribed chromatin of ovalbumin and lysozyme genes, whereas HMG 1/2, E proteins were equally distributed between DNA of both transcribed and non-transcribed genes. The depletion of H1/H5 in beta globin sequences was verified by the 'protein image' hybridisation technique (1). The DNase I hypersensitive site located 5' upstream from beta globin gene is deficient in all the proteins assayed, what implies a drastic disruption in the nucleosomal array. Minor quantitative changes of protein pattern suggest transient local perturbation of the chromatin on transcription.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Belyavsky A. V., Bavykin S. G., Goguadze E. G., Mirzabekov A. D. Primary organization of nucleosomes containing all five histones and DNA 175 and 165 base-pairs long. J Mol Biol. 1980 May 25;139(3):519–536. doi: 10.1016/0022-2836(80)90144-8. [DOI] [PubMed] [Google Scholar]
- Bianchi M. E., Beltrame M., Paonessa G. Specific recognition of cruciform DNA by nuclear protein HMG1. Science. 1989 Feb 24;243(4894 Pt 1):1056–1059. doi: 10.1126/science.2922595. [DOI] [PubMed] [Google Scholar]
- Blanco J., States J. C., Dixon G. H. General method for isolation of DNA sequences that interact with specific nuclear proteins in chromosomes: binding of the high mobility group protein HMG-T to a subset of the protamine gene family. Biochemistry. 1985 Dec 31;24(27):8021–8028. doi: 10.1021/bi00348a028. [DOI] [PubMed] [Google Scholar]
- Bolton A. E., Hunter W. M. The labelling of proteins to high specific radioactivities by conjugation to a 125I-containing acylating agent. Biochem J. 1973 Jul;133(3):529–539. doi: 10.1042/bj1330529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brotherton T. W., Ginder G. D. Preferential in vitro binding of high mobility group proteins 14 and 17 to nucleosomes containing active and DNase I sensitive single-copy genes. Biochemistry. 1986 Jun 3;25(11):3447–3454. doi: 10.1021/bi00359a053. [DOI] [PubMed] [Google Scholar]
- Caplan A., Kimura T., Gould H., Allan J. Perturbation of chromatin structure in the region of the adult beta-globin gene in chicken erythrocyte chromatin. J Mol Biol. 1987 Jan 5;193(1):57–70. doi: 10.1016/0022-2836(87)90626-7. [DOI] [PubMed] [Google Scholar]
- Caron F., Thomas J. O. Exchange of histone H1 between segments of chromatin. J Mol Biol. 1981 Mar 15;146(4):513–537. doi: 10.1016/0022-2836(81)90045-0. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras M. A., Bale W. F., Spar I. L. Iodine monochloride (IC1) iodination techniques. Methods Enzymol. 1983;92:277–292. [PubMed] [Google Scholar]
- Dorbic T., Wittig B. Chromatin from transcribed genes contains HMG17 only downstream from the starting point of transcription. EMBO J. 1987 Aug;6(8):2393–2399. doi: 10.1002/j.1460-2075.1987.tb02517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorbic T., Wittig B. Isolation of oligonucleosomes from active chromatin using HMG17-specific monoclonal antibodies. Nucleic Acids Res. 1986 Apr 25;14(8):3363–3376. doi: 10.1093/nar/14.8.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douillard J. Y., Hoffman T. Enzyme-linked immunosorbent assay for screening monoclonal antibody production using enzyme-labeled second antibody. Methods Enzymol. 1983;92:168–174. doi: 10.1016/0076-6879(83)92016-5. [DOI] [PubMed] [Google Scholar]
- Druckmann S., Mendelson E., Landsman D., Bustin M. Immunofractionation of DNA sequences associated with HMG-17 in chromatin. Exp Cell Res. 1986 Oct;166(2):486–496. doi: 10.1016/0014-4827(86)90493-3. [DOI] [PubMed] [Google Scholar]
- Earnshaw W. C. Anionic regions in nuclear proteins. J Cell Biol. 1987 Oct;105(4):1479–1482. doi: 10.1083/jcb.105.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour D. S., Lis J. T. Detecting protein-DNA interactions in vivo: distribution of RNA polymerase on specific bacterial genes. Proc Natl Acad Sci U S A. 1984 Jul;81(14):4275–4279. doi: 10.1073/pnas.81.14.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson V., Chalkley R. A new method for the isolation of replicative chromatin: selective deposition of histone on both new and old DNA. Cell. 1981 Jan;23(1):121–134. doi: 10.1016/0092-8674(81)90277-4. [DOI] [PubMed] [Google Scholar]
- Jackson V., Chalkley R. A new method for the isolation of replicative chromatin: selective deposition of histone on both new and old DNA. Cell. 1981 Jan;23(1):121–134. doi: 10.1016/0092-8674(81)90277-4. [DOI] [PubMed] [Google Scholar]
- Jackson V. Deposition of newly synthesized histones: new histones H2A and H2B do not deposit in the same nucleosome with new histones H3 and H4. Biochemistry. 1987 Apr 21;26(8):2315–2325. doi: 10.1021/bi00382a037. [DOI] [PubMed] [Google Scholar]
- Jantzen K., Fritton H. P., Igo-Kemenes T. The DNase I sensitive domain of the chicken lysozyme gene spans 24 kb. Nucleic Acids Res. 1986 Aug 11;14(15):6085–6099. doi: 10.1093/nar/14.15.6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y. J., Cole R. D. Exchange of H1 histone depends on aggregation of chromatin, not simply on ionic strength. J Biol Chem. 1986 Nov 25;261(33):15805–15812. [PubMed] [Google Scholar]
- Karpov V. L., Preobrazhenskaya O. V., Mirzabekov A. D. Chromatin structure of hsp 70 genes, activated by heat shock: selective removal of histones from the coding region and their absence from the 5' region. Cell. 1984 Feb;36(2):423–431. doi: 10.1016/0092-8674(84)90235-6. [DOI] [PubMed] [Google Scholar]
- Landsman D., Mendelson E., Druckmann S., Bustin M. Exchange of proteins during immunofractionation of chromatin. Exp Cell Res. 1986 Mar;163(1):95–102. doi: 10.1016/0014-4827(86)90561-6. [DOI] [PubMed] [Google Scholar]
- Levina E. S., Bavykin S. G., Shick V. V., Mirzabekov A. D. The method of crosslinking histones to DNA partly depurinated at neutral pH. Anal Biochem. 1981 Jan 1;110(1):93–101. doi: 10.1016/0003-2697(81)90117-2. [DOI] [PubMed] [Google Scholar]
- Lorch Y., LaPointe J. W., Kornberg R. D. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987 Apr 24;49(2):203–210. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- Losa R., Brown D. D. A bacteriophage RNA polymerase transcribes in vitro through a nucleosome core without displacing it. Cell. 1987 Aug 28;50(5):801–808. doi: 10.1016/0092-8674(87)90338-2. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Wood W. I., Dolan M., Engel J. D., Felsenfeld G. A 200 base pair region at the 5' end of the chicken adult beta-globin gene is accessible to nuclease digestion. Cell. 1981 Nov;27(1 Pt 2):45–55. doi: 10.1016/0092-8674(81)90359-7. [DOI] [PubMed] [Google Scholar]
- Mendelson E., Landsman D., Druckmann S., Bustin M. Immunofractionation of chromatin regions associated with histone H1o. Eur J Biochem. 1986 Oct 15;160(2):253–260. doi: 10.1111/j.1432-1033.1986.tb09964.x. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Mirzabekov A. D., Bavykin S. G., Belyavsky A. V., Karpov V. L., Preobrazhenskaya O. V., Shick V. V., Ebralidse K. K. Mapping DNA-protein interactions by cross-linking. Methods Enzymol. 1989;170:386–408. doi: 10.1016/0076-6879(89)70058-6. [DOI] [PubMed] [Google Scholar]
- Mirzabekov A. D., Shick V. V., Belyavsky A. V., Bavykin S. G. Primary organization of nucleosome core particle of chromatin: sequence of histone arrangement along DNA. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4184–4188. doi: 10.1073/pnas.75.9.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacheva G. A., Guschin D. Y., Preobrazhenskaya O. V., Karpov V. L., Ebralidse K. K., Mirzabekov A. D. Change in the pattern of histone binding to DNA upon transcriptional activation. Cell. 1989 Jul 14;58(1):27–36. doi: 10.1016/0092-8674(89)90399-1. [DOI] [PubMed] [Google Scholar]
- Nicolas R. H., Wright C. A., Cockerill P. N., Wyke J. A., Goodwin G. H. The nuclease sensitivity of active genes. Nucleic Acids Res. 1983 Feb 11;11(3):753–772. doi: 10.1093/nar/11.3.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridsdale J. A., Davie J. R. Selective solubilization of beta-globin oligonucleosomes at low ionic strength. Biochemistry. 1987 Jan 13;26(1):290–295. doi: 10.1021/bi00375a040. [DOI] [PubMed] [Google Scholar]
- Ridsdale J. A., Rattner J. B., Davie J. R. Erythroid-specific gene chromatin has an altered association with linker histones. Nucleic Acids Res. 1988 Jul 11;16(13):5915–5926. doi: 10.1093/nar/16.13.5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha E., Davie J. R., van Holde K. E., Weintraub H. Differential salt fractionation of active and inactive genomic domains in chicken erythrocyte. J Biol Chem. 1984 Jul 10;259(13):8558–8563. [PubMed] [Google Scholar]
- Rocha E., Davie J. R., van Holde K. E., Weintraub H. Differential salt fractionation of active and inactive genomic domains in chicken erythrocyte. J Biol Chem. 1984 Jul 10;259(13):8558–8563. [PubMed] [Google Scholar]
- Roche J., Gorka C., Goeltz P., Lawrence J. J. Association of histone H1(0) with a gene repressed during liver development. Nature. 1985 Mar 14;314(6007):197–198. doi: 10.1038/314197a0. [DOI] [PubMed] [Google Scholar]
- Rose S. M., Garrard W. T. Differentiation-dependent chromatin alterations precede and accompany transcription of immunoglobulin light chain genes. J Biol Chem. 1984 Jul 10;259(13):8534–8544. [PubMed] [Google Scholar]
- Sandeen G., Wood W. I., Felsenfeld G. The interaction of high mobility proteins HMG14 and 17 with nucleosomes. Nucleic Acids Res. 1980 Sep 11;8(17):3757–3778. doi: 10.1093/nar/8.17.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale R. L., Annunziato A. T., Smith R. D. High mobility group proteins: abundance, turnover, and relationship to transcriptionally active chromatin. Biochemistry. 1983 Oct 11;22(21):5008–5015. doi: 10.1021/bi00290a020. [DOI] [PubMed] [Google Scholar]
- Sheflin L. G., Spaulding S. W. High mobility group protein 1 preferentially conserves torsion in negatively supercoiled DNA. Biochemistry. 1989 Jun 27;28(13):5658–5664. doi: 10.1021/bi00439a048. [DOI] [PubMed] [Google Scholar]
- Shick V. V., Belyavsky A. V., Mirzabekov A. D. Primary organization of nucleosomes. Interaction of non-histone high mobility group proteins 14 and 17 with nucleosomes, as revealed by DNA-protein crosslinking and immunoaffinity isolation. J Mol Biol. 1985 Sep 20;185(2):329–339. doi: 10.1016/0022-2836(85)90407-3. [DOI] [PubMed] [Google Scholar]
- Singh J., Dixon G. H. High mobility group proteins 1 and 2 function as general class II transcription factors. Biochemistry. 1990 Jul 3;29(26):6295–6302. doi: 10.1021/bi00478a026. [DOI] [PubMed] [Google Scholar]
- Solomon M. J., Larsen P. L., Varshavsky A. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell. 1988 Jun 17;53(6):937–947. doi: 10.1016/s0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- Strätling W. H., Dölle A., Sippel A. E. Chromatin structure of the chicken lysozyme gene domain as determined by chromatin fractionation and micrococcal nuclease digestion. Biochemistry. 1986 Jan 28;25(2):495–502. doi: 10.1021/bi00350a033. [DOI] [PubMed] [Google Scholar]
- Sun Y. L., Xu Y. Z., Bellard M., Chambon P. Digestion of the chicken beta-globin gene chromatin with micrococcal nuclease reveals the presence of an altered nucleosomal array characterized by an atypical ladder of DNA fragments. EMBO J. 1986 Feb;5(2):293–300. doi: 10.1002/j.1460-2075.1986.tb04212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. O., Rees C., Pearson E. C. Histone H5 promotes the association of condensed chromatin fragments to give pseudo-higher-order structures. Eur J Biochem. 1985 Feb 15;147(1):143–151. doi: 10.1111/j.1432-1033.1985.tb08730.x. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Wilson C. M. Selective radiolabelling and identification of a strong nucleosome binding site on the globular domain of histone H5. EMBO J. 1986 Dec 20;5(13):3531–3537. doi: 10.1002/j.1460-2075.1986.tb04679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Martinez S., Ruiz-Carrillo A. Nucleosomes containing histones H1 or H5 are closely interspersed in chromatin. Nucleic Acids Res. 1982 Apr 10;10(7):2323–2335. doi: 10.1093/nar/10.7.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremethick D. J., Molloy P. L. High mobility group proteins 1 and 2 stimulate transcription in vitro by RNA polymerases II and III. J Biol Chem. 1986 May 25;261(15):6986–6992. [PubMed] [Google Scholar]
- Villeponteau B., Landes G. M., Pankratz M. J., Martinson H. G. The chicken beta globin gene region. Delineation of transcription units and developmental regulation of interspersed DNA repeats. J Biol Chem. 1982 Sep 25;257(18):11015–11023. [PubMed] [Google Scholar]
- Waga S., Mizuno S., Yoshida M. Nonhistone protein HMG1 removes the transcriptional block caused by left-handed Z-form segment in a supercoiled DNA. Biochem Biophys Res Commun. 1988 May 31;153(1):334–339. doi: 10.1016/s0006-291x(88)81227-0. [DOI] [PubMed] [Google Scholar]
- Watt F., Molloy P. L. High mobility group proteins 1 and 2 stimulate binding of a specific transcription factor to the adenovirus major late promoter. Nucleic Acids Res. 1988 Feb 25;16(4):1471–1486. doi: 10.1093/nar/16.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H. Histone-H1-dependent chromatin superstructures and the suppression of gene activity. Cell. 1984 Aug;38(1):17–27. doi: 10.1016/0092-8674(84)90522-1. [DOI] [PubMed] [Google Scholar]
- Weisbrod S., Groudine M., Weintraub H. Interaction of HMG 14 and 17 with actively transcribed genes. Cell. 1980 Jan;19(1):289–301. doi: 10.1016/0092-8674(80)90410-9. [DOI] [PubMed] [Google Scholar]
- Woo S. L., Dugaiczyk A., Tsai M. J., Lai E. C., Catterall J. F., O'Malley B. W. The ovalbumin gene: cloning of the natural gene. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3688–3692. doi: 10.1073/pnas.75.8.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright J. M., Dixon G. H. Induction by torsional stress of an altered DNA conformation 5' upstream of the gene for a high mobility group protein from trout and specific binding to flanking sequences by the gene product HMG-T. Biochemistry. 1988 Jan 26;27(2):576–581. doi: 10.1021/bi00402a012. [DOI] [PubMed] [Google Scholar]





