Abstract
Light is a fundamental signal that regulates important physiological processes such as development and circadian rhythm in living organisms. Phytochromes form a major family of photoreceptors responsible for red light perception in plants, fungi and bacteria1. They undergo reversible photoconversion between red-absorbing (Pr) and far-red-absorbing (Pfr) states, thereby ultimately converting a light signal into a distinct biological signal that mediates subsequent cellular responses2. Several structures of microbial phytochromes have been determined in their dark-adapted Pr or Pfr states3-7. However, the structural nature of initial photochemical events has not been characterized by crystallography. Here we report the crystal structures of three intermediates in the photoreaction of Pseudomonas aeruginosa bacteriophytochrome (PaBphP). We employed cryo-trapping crystallography to capture intermediates, and followed structural changes by scanning the temperature at which the photoreaction proceeds. Light-induced conformational changes in PaBphP originate in ring D of the bilin chromophore, and E to Z isomerization about the C15=C16 double bond between rings C and D is the initial photochemical event. As the chromophore relaxes, the twist of the C15 methine bridge about its two dihedral angles is reversed. Structural changes extend further to rings B and A, and to the surrounding protein regions. These data suggest that absorption of a photon by the Pfr state of PaBphP converts a light signal into a structural signal via twisting and untwisting of the methine bridges in the linear tetrapyrrole within the confined protein cavity.
Cryo-trapping and time-resolved room-temperature experiments are two main experimental strategies to study the structures of intrinsically short-lived reaction intermediates8. To establish the molecular mechanism of Pfr/Pr photoconversion, we generated and cryotrapped intermediates between the reactant Pfr state and product state(s) in fully photoactive crystals of the photosensory core module (PCM)5 of PaBphP. We followed progress of the reaction by applying a “trap-pump-trap-probe” strategy at variable pump temperatures (Fig. 1, Fig. S1, Methods Summary). Here, temperature mimics time: the higher the pump temperature, the greater the structural relaxation and the further a reaction proceeds along its pathway.
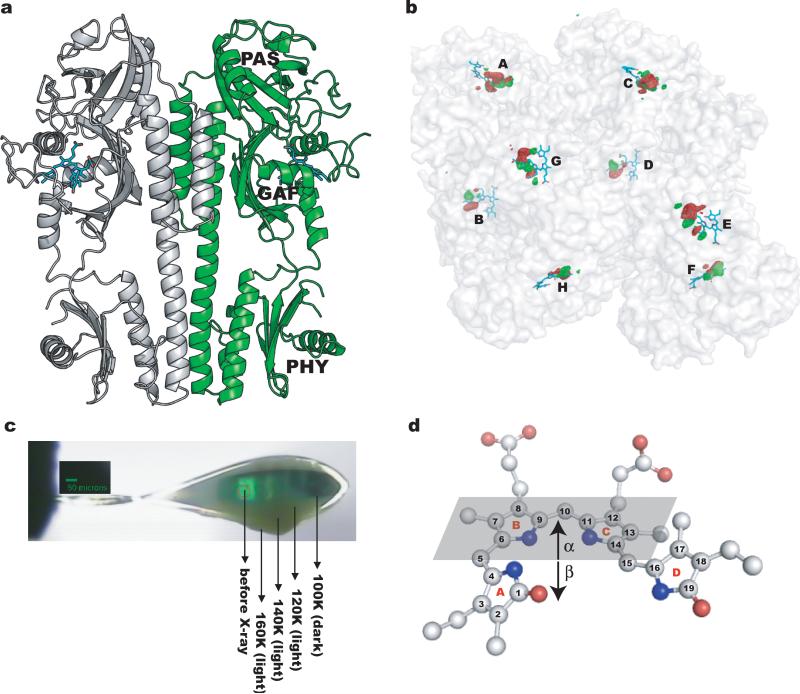
Fig. 1.
Trap-pump-trap-probe experiment. (a) Ribbon diagram of the PaBphP-PCM dimer. The BV chromophore is colored in cyan. (b) Experimental difference (Flight-Fdark) map at 130K (contoured at +/-5σ, where σ is the standard deviation of difference densities across the entire map). Strong positive (green) and negative (red) densities with peak signal greater than +/-12σ are clustered near the chromophores of the eight monomers (A-H) in the asymmetric unit. (c) Dark stripes of a mounted crystal correspond to segments from which X-ray datasets were collected. (d) A ball-and-stick representation of the chromophore in the Pfr state (PDB 3NHQ). The α-face of a pyrrole ring is defined when atom numbering follows a clockwise direction, with β defined as the opposite face. The α-face of an entire bilin chromophore is defined according to Rockwell et al.30.
We collected diffraction data from six crystals at 10 pump temperatures between 100 and 180K (Table S1), and calculated difference (Flight-Fdark) electron density maps from 14 light datasets and six reference dark datasets (Fig. S1a). In all maps, strong and highly significant difference electron densities are exclusively concentrated at the chromophore binding sites embedded in the GAF domain (Fig. 1b). Singular value decomposition (SVD) analysis9 of difference densities within a 5Å radius of the aligned chromophores revealed three major, independent, light-induced structures (Fig. S1c-d). Since difference densities are largely consistent among the eight monomers in the asymmetric unit and between different crystals illuminated at the same temperature, we averaged these densities by applying non-crystallographic symmetry (NCS), and focus here on the principal features common to the eight monomers.
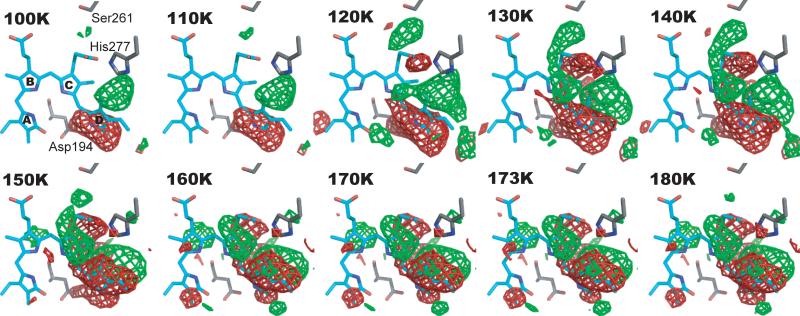
Difference densities vary markedly with pump temperature (Fig. 2). At the lowest temperatures they appear near ring D, which suggests that light-induced structural changes originate in ring D. As the temperature rises, difference densities expand to ring C and eventually to ring B and ring A, thus exhibiting a systematic structural progression as a function of temperature. Based on representative difference maps at 110, 130 and 173K, we modeled three light-induced chromophore structures, denoted L1, L2 and L3, respectively (Fig. 3a,b). We further refined the initial models of the L1, L2 and L3 structures jointly in real space against the NCS-averaged, SVD-filtered difference maps at all pump temperatures, and determined their relative concentrations at each temperature (Fig. 3g).
Fig. 2.
Difference maps (Flight-Fdark) at pump temperatures between 100 and 180K (contoured at +/- 4σ). The BV chromophore (cyan) is shown in the Pfr state. Positive (green) and negative (red) densities represent structural changes associated with formation of photoproduct state(s) and loss of the parent Pfr state, respectively. No significant difference densities with signal greater than +/-4σ are detected at longer range, beyond the 5Å radius around the chromophore.
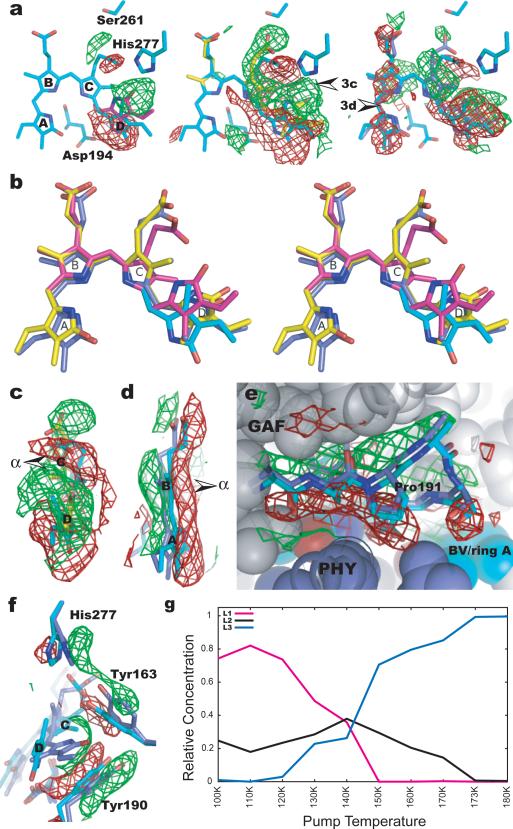
Fig. 3.
Light-induced structural changes. (a) Representative difference (Flight-Fdark) maps at 110, 130 and 173K. Arrows indicate viewpoints of Fig. 3c and 3d. (b) Stereoview of the superposition of the chromophore conformations in the Pfr (cyan), L1(magenta), L2(yellow) and L3 (blue) structures. (c) Sideview of difference map at 130K (contoured at +/-3σ) shows a twist in the C15 methine bridge. (d) Sideview of difference map at 173K (contoured at +/-2σ) indicates a β–facial shift of rings B/A. Arrows in Figs. 3c, 3d mark the α-face of the chromophore. (e) Difference densities at 173K (contoured at +/-2σ) near the PXSDIP motif (residues 191-196) at the GAF-PHY interface. (f) Differences densities associated with the side chains of His277, Tyr163 and Tyr190 (map contours: +/-3σ at 173K). (g) Relative concentrations of the L1, L2 and L3 structures as a function of pump temperature.
In the L1 structure represented at 110K, strong positive and negative densities are roughly aligned in the plane of ring D of the bilin chromophore (Fig. 3a). Negative densities span the pyrrole nitrogen of ring D and the side chain of a highly conserved Asp194 from the PXSDIP sequence motif, indicating the rupture of a key hydrogen bond that stabilizes ring D in the Pfr state5. The corresponding positive densities between His277 and ring D identify formation of the L1 structure in which the chromophore has isomerized about the C15=C16 double bond to adopt the 15Za configuration, and ring D is significantly shifted in its plane towards His277. As a result, L1 exhibits a smaller and strained C14-C15-C16 bond angle in the methine bridge between rings C and D compared to the “stretched” bond angle in the Pfr state (Fig. S3) and to those of the Pr state in D. radiodurans BphP and R. palustris BphP34, 10.
In the L2 structure represented at 130K, strong difference densities near ring C suggest that ring C moves towards its α-face, and that its propionate side chain breaks the hydrogen bonds with His277, Tyr163 and Ser275 present in the Pfr and L1 structures to form a new hydrogen bond with Ser261 (Fig. 3a, Fig. 4). Difference densities associated with rings C and D (Fig. 3c) suggest that counter-twist occurs across the C15 methine bridge in forming L2, in which ring D assumes a β-facial disposition relative to ring C. This partially relaxes strain in the C15 methine bridge while the C10 methine bridge between rings B and C is concomitantly twisted in the opposite direction (Fig. S5b).
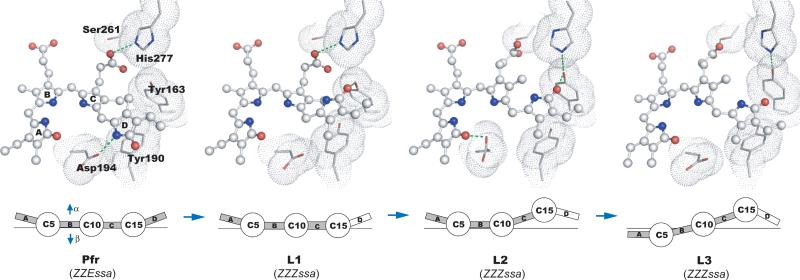
Fig. 4.
Light-induced molecular events in PaBphP. (Upper panel) The chromophores are in ball-and-stick representation, with their surrounding residues shown as van der Waals spheres. Green dotted lines indicate potential interactions with each cryo-trapped structure. (Lower panel) Schematic representation of changes in relative disposition of the four pyrrole rings of the BV chromophore, in which pyrrole rings A, B, C and D (boxes) are linearly connected by methine bridges (circles). The α- and β-faces of the chromophore are denoted by arrows.
In the L3 structure represented at 173K, structural changes extend to rings A and B (Fig. 3a). Difference densities sandwiching these rings (Fig. 3d) suggest that they move as a unit slightly towards the β-face of the chromophore while the C5 methine bridge retains its direction of twist. Motion is accompanied by small twists in the C5 and C10 methine bridges (Fig. S4b). Rings B and C are more coplanar in L3 than in L2, consistent with a more relaxed C10 methine bridge in L3.
These data demonstrate that E to Z isomerization about the C15=C16 double bond between rings C and D is the initial structural event in the Pfr to Pr photoreaction of PaBphP. This is consistent with findings in many phytochromes and BphPs2, 11, 12; but contrasts with a recent report based on room temperature NMR spectroscopy, in which rotation of ring A occurs between the Pr and Pfr states of an unusual, knotless phytochrome consisting of only the GAF domain13. In PaBphP, when ring D flips into the 15Za configuration within a protein cavity that is still optimized to accommodate the 15Ea configuration of the Pfr state, a highly distorted and strained C15 methine bridge is generated in L1. As L1 evolves to L2, counter-twist across the C15 methine bridge partially relieves the strain. Motion of ring C results in a C10 methine bridge twisted in the opposite direction in L2, further relaxation of which leads to the L3 structure. These data suggest a reaction trajectory that proceeds in the order Pfr →L1→ L2→L3 (Fig. 4).
To relate these cryo-trapped L1, L2 and L3 structures to spectroscopic intermediates in the Pfr to Pr photoreaction14-18, we measured visible absorption spectra on a crushed PaBphP-PCM crystal at temperatures between 100 and 180K using the X-ray experimental protocol (Fig. S5). Difference absorption spectra between illuminated and reference “dark” states show significant loss of the Pfr state, indicated by a negative peak at 768nm, and formation of blue-shifted photoproducts (Fig.S5b). SVD analysis revealed two significant basis difference absorption spectra, with blue-shifted peaks between 650 and 700nm. The basis spectrum with a blue-shifted peak at 684nm is largely populated between 100 and 150K, in agreement with the temperature dependence of the population of the L2 structure in X-ray data (Fig. S5c, Fig. 3g). Blue-shifted absorption peaks are consistent with the distorted tetrapyrrole conjugated system observed in L2 and L3.
The L1, L2 and L3 structures likely arise from early molecular events in the Pfr to Pr reaction of PaBphP. One or more may correspond to the Lumi-F spectroscopic intermediate detected on the femto- to pico-second time scale by room temperature time-resolved spectroscopy and characterized by blue-shifted absorption peaks18,14, 19. These structures are consistent with strong FTIR difference bands attributed to the carbonyl group of ring D resulting from 15Ea/15Za isomerization and the B-C methine stretching in cryotrapped Lumi-F of CphA18. L1 may be identified with early intermediates in Cph1, in which the C15-H out-of-plane (HOOP) mode was detected by ultrafast Raman spectroscopy20. The properties of L1 are also consistent with the NMR spectroscopic data on the cryotrapped Lumi-F of Cph1, which indicated that the C14-C15-C16 angle is distorted following 15Ea/15Za isomerization19. Furthermore, our temperature-scanning range (100-180K) coincides with the temperature range in which Lumi-F intermediates were trapped in several phytochrome systems 11, 18, 19. Formation of late Meta-F intermediates would require higher pump temperatures 19 to permit more extensive structural relaxation in the protein. However, at temperatures >180K the cryoprotectant solution undergoes a glass transition which causes severe deterioration in crystal diffraction 5,6.
In both the L2 and L3 structures, the twist of the dihedral angles in the C15 methine bridge is quite distinct from those of the chromophore structures in the Pr and Pfr states (Figs. 3, 4 and S4b). Both the Pr and Pfr crystal structures of BphPs 3-5, 10 exhibit an α-facial disposition of ring D relative to ring C, which corresponds to a negative rotation of the red absorbance band in the circular dichroism (CD) spectra of BV-containing bacteriophytochromes such as PaBphP, DrBphP and Agp121, 22. Evidence for such counter-twist events in the methine bridges during photoconversion has been presented in Cph1 and Agp119, 22. We propose that the chromophore of PaBphP generates structural signals in response to light via subtle and local twist or counter-twist motions in the methine bridges within the confined protein cavity (Fig. 4), which alter specific interactions between the pyrrole rings and their immediate protein surroundings. It remains to be seen to what extent our findings in PaBphP apply to other members of the diverse and expanding phytochrome superfamily.
The protein moiety also plays an important role. First, following prompt E/Z isomerization, steric clashes between ring D and the side chain of Asp194 lead to initial strain in L1 that drives subsequent relaxation events. Second, the side chains of His277, Tyr163 and Tyr190 surrounding ring D also move slightly but concertedly to accommodate ring D as the chromophore evolves from Pfr to the L3 structure (Fig. 3f). Third, Ser261 appears to stabilize L2 and L3 via hydrogen bonds to the propionate group of ring C (Fig. 3a, 4). The single point mutant S261A inhibits formation of the Pr state upon illumination and significantly accelerates dark reversion23 (Fig. S6b). This provides further evidence that L2 and L3 are authentic photoproducts on the productive trajectory towards the product Pr state. Fourth, more extensive structural changes occur in L3 near the highly conserved PXSDIP sequence motif at the interface of the GAF and PHY domains. This segment and the side chain of Tyr190 are pushed away from the chromophore as rings B/A and ring D move towards the β-face of the chromophore (Fig. 3e). These movements expand the chromophore cavity to allow further relaxation, and may trigger further structural perturbations between the GAF domain and the arm of the PHY domain. As the reaction proceeds further, Tyr163 and Tyr190 may adjust their side chain rotamers to reshape the ring D pocket to accommodate the product Pr state23.
To examine light dependence of the HK activity in PaBphP, we conducted HK assays on wild type and the S261A mutant of full-length PaBphP under light and dark conditions (Fig. S6) and found that autophosphorylation of PaBphP is light-dependent, in contrast to earlier observations24. As in RpBphP225 and Cph126, PaBphP exhibits higher levels of autophosphorylation in the Pr state (here, the light state) than in the Pfr state. S261A exhibits a photoconversion phenotype that greatly prefers the Pfr state, and shows significantly reduced HK activity compared to WT. We predict that small light-induced structural changes, exemplified by the cryo-trapped intermediate structures L1, L2 and L3, propagate from the chromophore binding pocket of the N-terminal photosensory domains to the C-terminal HK domain via further tertiary and/or quaternary rearrangements27, which ultimately affect the HK activity and convert a light signal into a biological signal.
Methods Summary
Structural heterogeneity is intrinsic to all dynamic processes, and often challenges accurate interpretation of both cryo-trapping experiments at a single temperature and time-resolved crystallographic data 28, 29. This work presents a temperature-scan and analytical strategy to resolve structural heterogeneity and determine distinct, homogeneous structural species.
We apply a “trap-pump-trap-probe” strategy to photoactive crystals of PaBphPPCM. We first trap the dark-adapted Pfr state (trap dark) by freezing the crystal (grown at 293K in the dark) in liquid nitrogen under safety green/blue light, and collected a reference crystallographic “dark” dataset at 100K for each crystal. The same crystal was then uniformly illuminated under white light for 10-15 minutes at elevated temperatures to generate reaction intermediates (pump), then cryo-cooled to 100K (trap light) to collect a “light” dataset from a fresh crystal volume (probe) (Fig. 1b). We calculated 14 (Flight-Fdark) difference maps from six crystals that cover 10 pump temperatures between 100 and 180K. Since each map contains eight monomers in the asymmetric unit, we obtained 112 independent difference maps for a PaBphP monomer. We spatially aligned these maps on a reference monomer C, and subjected difference densities within a 5Å radius of the aligned chromophores to singular value decomposition (SVD)9 that yielded 112 noise-reduced difference maps (Fig. S1c-d). Atomic models for the L1, L2 and L3 structures were initially built based on representative difference densities at 110, 130 and 173K, respectively. These models were further refined jointly in real space (together with a fixed reference Pfr conformation) against SVD-filtered difference maps at all pump temperatures. Based on least squares fitting between the calculated and observed difference densities, we also obtained relative concentrations of L1, L2 and L3 at each temperature. Fig. S1a summarizes methods and software used in X-ray data reduction and analysis.
Supplementary Material
Acknowledgements
We thank Andreas Möglich for comments and reading of the manuscript, and Vukica Šrajer of BioCARS for assistance in microspectrometer experiments on crystals. We also thank the staff of LSCAT and BioCARS at the Advanced Photon Source, Argonne National Laboratory for beam line access. Supported by National Institutes of Health grant GM036452 to K.M.. BioCARS is supported by National Institutes of Health grant RR07707 to K.M..
Methods
Purification, mutagenesis and crystallization
PaBphP-PCM containing residues 1-497 of P. aeruginosa PA4117 was expressed, purified, crystallized and cryo-protected in the dark as described5. Site-directed mutagenesis was carried out using the QuikChange Site-directed Mutagenesis Kit (Stratagene). Both wild type and the S261A mutant of the full-length PaBphP were purified using the same protocol for PaBphP-PCM.
Data collection and processing
Diffraction data were collected at LSCAT 21-IDG beamline, BioCARS 14-BMC and 14-IDB beamlines of Advanced Photon Source. Temperature was controlled by a cryostream cooler (Oxford Cryosystems). Illumination was applied to a mounted crystal using two unfiltered fiber optic lights from different directions for 10-15min, while the crystal spinned around the spindle axis of the goniometer at a speed of 30°/s. All diffraction images were processed using HKL200031. Light and dark data sets from the same crystal were scaled using ScaleIt in CCP432 (Table S1). A reference “dark” structure in the Pfr state was refined at 2.55Å resolution using Phenix33 (Table S2), and was used to calculate phases in generating difference Fourier maps by FFT in CCP4. Difference maps were masked and aligned using NCSMASK and MAPROT of CCP4, and were further subjected to SVD9 analysis in DynamiX (Fig. S1). Real space refinement of the L1, L2 and L3 structures, including the chromophore and several adjacent residues (Gln188-Asp194, His277, Ser261, Tyr163 and the covalent chromophore anchor Cys12), was carried out against NCS-averaged, SVD-filtered difference maps using DynamiX (Table S3). All modeling building was carried out using Coot34. Structure figures were generated using PyMol (http://pymol.org)
UV-visible spectroscopy
Absorption spectra of proteins in solution were recorded at room temperature with Shimadzu UV-1650 PC spectrophotometer. Visible spectra of crystals were measured using a microspectrophotometer Xspectra (4DX-ray Systems) at BioCARS.
Histidine kinase autophosphorylation assay
The HK assay was modified based on published protocols26, 35. Reactions under light (continuous illumination with unfiltered fiber optical light) or dark (samples were protected from light in a covered box during dark incubation and reaction; handled under dim room light) were started by adding 10μl reaction buffer (50mM Tris HCl, pH 8.0; 2mM MgCl2; 2mM MnCl2; 0.1M KCl; 10% ethylene glycol; 80μM ATP and 32P-γ-ATP at 0.3mCi/ml) to each 10μl protein sample at the concentration of 1mg/ml. Reactions were stopped using standard stop buffer containing 0.1M EDTA and 0.1M DTT.
Footnotes
Author contributions: X.Y. initiated and designed research, collected X-ray and microspectroscopic carried out mutagenesis and HK assays; X.Y. and Z.R. analyzed and interpreted structures; Z.R. data analysis methods and analyzed data; J.K. purified proteins and grew crystals; K.M. initiated photoreceptor projects; X.Y., Z.R. and K.M. wrote the manuscript.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature
References
- 1.Montgomery BL, Lagarias JC. Phytochrome ancestry: sensors of bilins and light. Trends Plant Sci. 2002;7:357–66. doi: 10.1016/s1360-1385(02)02304-x. [DOI] [PubMed] [Google Scholar]
- 2.Rockwell NC, Su YS, Lagarias JC. Phytochrome Structure and Signaling Mechanisms. Annu Rev Plant Biol. 2006;57:837–58. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wagner JR, Brunzelle JS, Forest KT, Vierstra RD. A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature. 2005;438:325–31. doi: 10.1038/nature04118. [DOI] [PubMed] [Google Scholar]
- 4.Yang X, Stojkovic EA, Kuk J, Moffat K. Crystal structure of the chromophore binding domain of an unusual bacteriophytochrome, RpBphP3, reveals residues that modulate photoconversion. Proc Natl Acad Sci U S A. 2007;104:12571–6. doi: 10.1073/pnas.0701737104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang X, Kuk J, Moffat K. Crystal structure of Pseudomonas aeruginosa bacteriophytochrome: photoconversion and signal transduction. Proc Natl Acad Sci U S A. 2008;105:14715–20. doi: 10.1073/pnas.0806718105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Essen LO, Mailliet J, Hughes J. The structure of a complete phytochrome sensory module in the Pr ground state. Proc Natl Acad Sci U S A. 2008;105:14709–14. doi: 10.1073/pnas.0806477105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cornilescu G, Ulijasz AT, Cornilescu CC, Markley JL, Vierstra RD. Solution structure of a cyanobacterial phytochrome GAF domain in the red-light-absorbing ground state. J Mol Biol. 2008;383:403–13. doi: 10.1016/j.jmb.2008.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moffat K, Henderson R. Freeze trapping of reaction intermediates. Curr Opin Struct Biol. 1995;5:656–63. doi: 10.1016/0959-440x(95)80059-x. [DOI] [PubMed] [Google Scholar]
- 9.Rajagopal S, Schmidt M, Anderson S, Ihee H, Moffat K. Analysis of experimental time-resolved crystallographic data by singular value decomposition. Acta Crystallogr D Biol Crystallogr. 2004;60:860–71. doi: 10.1107/S0907444904004160. [DOI] [PubMed] [Google Scholar]
- 10.Wagner JR, Zhang J, Brunzelle JS, Vierstra RD, Forest KT. High resolution structure of deinococcus bacteriophytochrome yields new insights into phytochrome architecture and evolution. J Biol Chem. 2007;282:12298–309. doi: 10.1074/jbc.M611824200. [DOI] [PubMed] [Google Scholar]
- 11.Foerstendorf H, Mummert E, Schafer E, Scheer H, Siebert F. Fourier-transform infrared spectroscopy of phytochrome: difference spectra of the intermediates of the photoreactions. Biochemistry. 1996;35:10793–9. doi: 10.1021/bi960960r. [DOI] [PubMed] [Google Scholar]
- 12.Song C, et al. Two ground state isoforms and a chromophore D-ring photoflip triggering extensive intramolecular changes in a canonical phytochrome. Proc Natl Acad Sci U S A. 2010;108:3842–7. doi: 10.1073/pnas.1013377108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ulijasz AT, et al. Structural basis for the photoconversion of a phytochrome to the activated Pfr form. Nature. 2010;463:250–4. doi: 10.1038/nature08671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schumann C, et al. Subpicosecond midinfrared spectroscopy of the Pfr reaction of phytochrome Agp1 from Agrobacterium tumefaciens. Biophys J. 2008;94:3189–97. doi: 10.1529/biophysj.107.119297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Thor JJ, Ronayne KL, Towrie M. Formation of the early photoproduct lumi-R of cyanobacterial phytochrome cph1 observed by ultrafast mid-infrared spectroscopy. J Am Chem Soc. 2007;129:126–32. doi: 10.1021/ja0660709. [DOI] [PubMed] [Google Scholar]
- 16.van Wilderen LJ, Clark IP, Towrie M, van Thor JJ. Mid-infrared picosecond pump-dump-probe and pump-repump-probe experiments to resolve a ground-state intermediate in cyanobacterial phytochrome Cph1. J Phys Chem B. 2009;113:16354–64. doi: 10.1021/jp9038539. [DOI] [PubMed] [Google Scholar]
- 17.Muller MG, Lindner I, Martin I, Gartner W, Holzwarth AR. Femtosecond kinetics of photoconversion of the higher plant photoreceptor phytochrome carrying native and modified chromophores. Biophys J. 2008;94:4370–82. doi: 10.1529/biophysj.106.091652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwinte P, et al. The photoreactions of recombinant phytochrome CphA from the cyanobacterium Calothrix PCC7601: a low-temperature UV-Vis and FTIR study. Photochem Photobiol. 2009;85:239–49. doi: 10.1111/j.1751-1097.2008.00426.x. [DOI] [PubMed] [Google Scholar]
- 19.Rohmer T, et al. Phytochrome as molecular machine: revealing chromophore action during the Pfr --> Pr photoconversion by magic-angle spinning NMR spectroscopy. J Am Chem Soc. 2010;132:4431–7. doi: 10.1021/ja9108616. [DOI] [PubMed] [Google Scholar]
- 20.Dasgupta J, Frontiera RR, Taylor KC, Lagarias JC, Mathies RA. Ultrafast excited-state isomerization in phytochrome revealed by femtosecond stimulated Raman spectroscopy. Proc Natl Acad Sci U S A. 2009;106:1784–9. doi: 10.1073/pnas.0812056106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rockwell NC, Shang L, Martin SS, Lagarias JC. Distinct classes of red/far-red photochemistry within the phytochrome superfamily. Proc Natl Acad Sci U S A. 2009;106:6123–7. doi: 10.1073/pnas.0902370106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seibeck S, et al. Locked 5Zs-biliverdin blocks the Meta-RA to Meta-RC transition in the functional cycle of bacteriophytochrome Agp1. FEBS Lett. 2007;581:5425–9. doi: 10.1016/j.febslet.2007.10.043. [DOI] [PubMed] [Google Scholar]
- 23.Yang X, Kuk J, Moffat K. Conformational differences between the Pfr and Pr states in Pseudomonas aeruginosa bacteriophytochrome. Proc Natl Acad Sci U S A. 2009;106:15639–44. doi: 10.1073/pnas.0902178106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tasler R, Moises T, Frankenberg-Dinkel N. Biochemical and spectroscopic characterization of the bacterial phytochrome of Pseudomonas aeruginosa. Febs J. 2005;272:1927–36. doi: 10.1111/j.1742-4658.2005.04623.x. [DOI] [PubMed] [Google Scholar]
- 25.Giraud E, et al. A new type of bacteriophytochrome acts in tandem with a classical bacteriophytochrome to control the antennae synthesis in Rhodopseudomonas palustris. J Biol Chem. 2005;280:32389–97. doi: 10.1074/jbc.M506890200. [DOI] [PubMed] [Google Scholar]
- 26.Yeh KC, Wu SH, Murphy JT, Lagarias JC. A cyanobacterial phytochrome two-component light sensory system. Science. 1997;277:1505–8. doi: 10.1126/science.277.5331.1505. [DOI] [PubMed] [Google Scholar]
- 27.Li H, Zhang J, Vierstra RD, Li H. Quaternary organization of a phytochrome dimer as revealed by cryoelectron microscopy. Proc Natl Acad Sci U S A. 2010;107:10872–7. doi: 10.1073/pnas.1001908107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson S, Srajer V, Moffat K. Structural heterogeneity of cryotrapped intermediates in the bacterial blue light photoreceptor, photoactive yellow protein. Photochem Photobiol. 2004;80:7–14. doi: 10.1562/2004-03-15-RA-115.1. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt M, et al. Protein kinetics: structures of intermediates and reaction mechanism from time-resolved x-ray data. Proc Natl Acad Sci U S A. 2004;101:4799–804. doi: 10.1073/pnas.0305983101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rockwell NC, et al. A second conserved GAF domain cysteine is required for the blue/green photoreversibility of cyanobacteriochrome Tlr0924 from Thermosynechococcus elongatus. Biochemistry. 2008;47:7304–16. doi: 10.1021/bi800088t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 32.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–42. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–54. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 34.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 35.Giraud E, Lavergne J, Vermeglio A. Characterization of bacteriophytochrome from photosynthetic bacteria: histidine kinase signaling triggered by light and redox sensing. Methods in Enzymology. 2010;471:135–159. doi: 10.1016/S0076-6879(10)71009-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.