Abstract
Chromosome 22q11.2 deletion syndrome (22q11DS) is the most common microdeletion syndrome in humans. It is typified by highly variable symptoms, which might be explained by epigenetic regulation of genes in the interval. Using computational algorithms, our laboratory previously predicted that DiGeorge critical region 6 (DGCR6), which lies within the deletion interval, is imprinted in humans. Expression and epigenetic regulation of this gene have not, however, been examined in 22q11DS subjects. The purpose of this study was to determine if the expression levels of DGCR6 and its duplicate copy DGCR6L in 22q11DS subjects are associated with the parent-of-origin of the deletion and childhood psychopathologies. Our investigation showed no evidence of parent-of-origin-related differences in expression of both DGCR6 and DGCR6L. However, we found that the variability in DGCR6 expression was significantly greater in 22q11DS children than in age and gender-matched control individuals. Children with 22q11DS who had anxiety disorders had significantly lower DGCR6 expression, especially in subjects with the deletion on the maternal chromosome, despite the lack of imprinting. Our findings indicate that epigenetic mechanisms other than imprinting contribute to the dysregulation of these genes and the associated childhood psychopathologies observed in individuals with 22q11DS. Further studies are now needed to test the usefulness of DGCR6 and DGCR6L expression and alterations in the epigenome at these loci in predicting childhood anxiety and associated adult-onset pathologies in 22q11DS subjects.
Keywords: DGCR6, DGCR6L, DiGeorge syndrome, epigenetics, schizophrenia, 22q11DS syndrome
Introduction
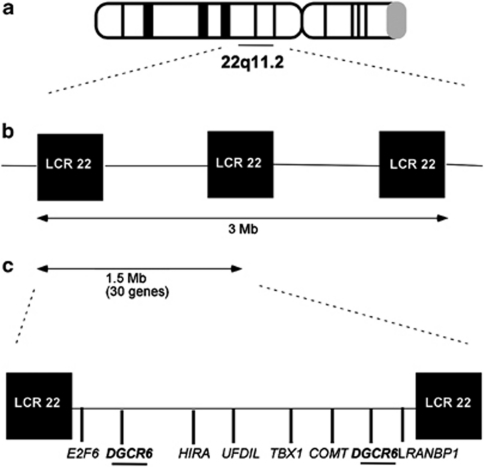
Chromosome 22q11.2 deletion syndrome (22q11DS), also known as velocardiofacial or DiGeorge syndrome, is a common hemizygous microdeletion syndrome occurring in 1 in 1600–4000 live births.1 Approximately 85% of subjects carry a 3-Mb deletion (Figure 1). A minority with a smaller 1.5-Mb deletion still show all the characteristics of the disorder, delimiting this as the 22q11DS minimal DiGeorge critical region (DGCR). Individuals with 22q11DS display variable conotruncal heart defects, atypical facial features, velopharyngeal insufficiency, and cognitive and psychiatric abnormalities.2, 3, 4
Figure 1.
Genomic rearrangements in 22q11.2 deletion syndrome (22q11DS) and the DiGeorge Critical Region (DGCR6)/DGCR6L locus. (a) 22q11DS patients bear a well-defined 3-Mb microdeletion at 22q11.2, mediated by low copy repeats (LCR22s) flanking the breakpoints. (b) The DGCR is a nested 1.5-Mb deletion that exhibits all the characteristic psychological and psychiatric symptoms of the syndrome. (c) Approximately 30 genes lie within the critical region, and some of these that are believed to be implicated in the phenotype are shown, including DGCR6 and its duplicate copy DGCR6L.
The cognitive abnormalities include borderline IQ to mild intellectual impairment, poor sustained attention, executive dysfunction and visual–spatial skills.5, 6, 7, 8 Minor psychiatric manifestations are common during childhood, with as many as 50% experiencing an anxiety disorder and/or ADHD.9, 10, 11, 12, 13 In late adolescence and early adulthood, major psychotic disorders, such as schizophrenia, bipolar illness and major depression, develop in 25–40% of the affected individuals.14, 15, 16 These cognitive and psychiatric manifestations vary in their severity and frequency.
Moreover, differential brain effects have been reported between 22q11DS subjects with a maternal or paternal deletion. One magnetic resonance imaging study and another involving the characterization of language reported that in children with 22q11DS gray-matter volume is more reduced and language disabilities are more severe when the deletion is on the maternal chromosome.17, 18 These findings suggest that the parent-of-origin of the deletion may differentially affect neurodevelopmental abnormalities. How the parental origin of a deletion can affect gene expression and psychological outcomes in individuals with 22q11DS has not been determined.
Genomic imprinting is a parent-of-origin-dependent epigenetic mechanism that results in the monoallelic silencing of genes. Epigenetic factors, such as DNA methylation and histone modifications, result in the monoallelic silencing of these genes. Monoallelic silencing not only occurs in a parent-of-origin-dependent manner, as observed in genomic imprinting, but can also occur in a random parental manner.19 This novel form of gene regulation is necessary for appropriate development, but it renders loci functionally haploid, thereby increasing their vulnerability to disorders caused by both genetic and epigenetic changes. In fact, disrupted genomic imprinting patterns cause several clearly defined syndromes,20, 21, 22 and are also implicated in complex conditions like schizophrenia.23 In complex disorders, like 22q11DS, epigenetic dysregulation could similarly lead to variable features such as age of onset, severity and parental effects.24 Nevertheless, direct evidence for monoallelic silencing of genes or metastable epialleles25 in the DGCR has not yet been demonstrated.
To explore the role that epigenetic regulation has in the etiology of 22q11DS, one mouse study investigated the imprint status of 25 genes within the DGCR and found them all to be bi-allelically expressed in developing and adult brains. Thus, the authors concluded that imprinting or allelic biasing could not explain the phenotypic features of 22q11DS.26 Nevertheless, computational approaches in our laboratory predicted that the gene DGCR6, lying in the DGCR, is imprinted in the human,27 but not in the mouse.28
DGCR6 is thought to contribute to the manifestations of 22q11DS, although its exact function has not been clearly defined. It is implicated in neural crest cell migration,29 pharyngeal arch development, and in the regulation of other genes implicated in 22q11DS (for example, TBX-1 and UFD1L).30 DGCR6 is duplicated in the primate lineage (DGCR6L), with both genes residing in the DGCR.29 DGCR6 and DGCR6L share 97% identity at the cDNA level, similar expression profiles, redundant functions and have only seven amino-acid differences between them.29 The duplication of DGCR6 is particularly interesting, as gene duplication could lead to dosage compensation by random silencing of one allele of each paralog that later could have evolved into parental allele-specific silencing present in genomically imprinted genes.31
In this study, we set out to examine whether specific deletion of one of the parental alleles on chromosome 22 directs null expression of DGCR6/DGCR6L, as would be expected from an imprinted gene expressed only from that allele. Our first hypothesis was that 22q11DS children with paternally derived deletions would demonstrate null expression of DGCR6 as our computational analyses had predicted that DGCR6 would be expressed from the paternal allele.27 Our second hypothesis was that such alterations of DGCR6 gene-expression patterns based on parent-of-origin of the deletion would be correlated with the neuropsychological findings in children with 22q11DS. We also examined the expression of DGCR6L as it is a duplicate of DGCR6 and thus its expression and association with the neuropsychological findings in children with 22q11DS could be similar. Thus, our aims were to determine the expression patterns of DGCR6 and DGCR6L in subjects with 22q11DS compared with age- and gender-matched control subjects. We also wanted to know whether the expression of the two genes could be attributed to differential methylation at their promoter regions. We further wanted to examine the relationship between the parent-of-origin of the deletion, expression of DGCR6/DGCR6L and neuropsychological findings in children with 22q11DS.
Materials and methods
Sample collection
Blood was collected from 38 subjects (males=23, females=15; mean age=10.4±2.6 years) carrying a 22q11.2 microdeletion confirmed by fluorescent in situ hybridization and 16 controls who were age and gender matched to the subjects (males=7, females=9; mean age=11.6±2.0 years). Samples were obtained from the Duke University Medical Center and Wake Forest University Health Sciences under the protocols approved by the Institutional Review Boards of these institutions. There were no significant differences in age (t statistic=1.7, P>0.05), gender (χ2=1.3, P>0.2) and parental socio-economic status (t statistic=0.006, P>0.9) between the two groups. Thirty-four of the 22q11DS subjects were Caucasian, two were African American and two were Hispanic. Of the control subjects, eight were Caucasian and eight were African American, resulting in a significant difference in race between the two groups (χ2=15.2, P<0.001). The subjects were all non-psychotic. A three-generation pedigree was drawn to ascertain developmental or genetic disorders, mental illness, learning disabilities and other cognitive defects in the families of the 22q11DS subjects as well as the control subjects. Personal or family histories of cognitive defects, psychotic illness or congenital anomalies in first-degree relatives were used as exclusion criteria for the control subjects. Children with 22q11DS who had an IQ <50 were excluded from the study, as were control subjects with an IQ >115. This minimized the intellectual disparities between the two groups and ensured optimal performance of 22q11DS children on the neurocognitive battery.
Determining parental origin of the 22q11.2 deletion
DNA was extracted from the blood samples of 22q11DS subjects and their parents (at least from one or both when available) using the Gentra Puregene Kit (Qiagen Sciences, Valencia, CA, USA). These subjects were then genotyped for 450 single-nucleotide polymorphisms (SNPs) corresponding to the 1.5-Mb deleted region using the iPLEX assay from Sequenom (Sequenom, Inc., San Diego, CA, USA). SNPs were selected using the Tagger program built into the Haploview software for the HapMap data, which utilizes linkage disequilibrium to identify a minimal set of SNPs for this region. We compared parental and child genotypes across 50 unlinked SNPs spanning the 1.5-Mb DGCR interval. In instances where only one parent was available, the parent-of-origin could be determined with certainty when the deletion was inherited from the parent whose genotype was available. In subjects in whom the deletion was thought to be on the chromosome from the unavailable parent, the hemizygous genotypes in the subject would match those of the available parent. A probability analysis was then performed with the SNPs to determine the likelihood that the other parent could have had identical genotypes across the interval; a probability of at least 0.95 of the parent-of-origin of the deletion was the threshold at which the determination was made. This was the case for four of the subjects with 22q11DS. The parent-of-origin of the deletion for the rest was easily determined.
DGCR6 and DGCR6L expression analysis
RNA was extracted from control and subject blood samples (white blood cells) using the PAXgene Blood RNA Kit (Qiagen Sciences), and then reverse transcribed using oligo dT primer and Superscript II (Invitrogen, Carlsbad, CA, USA). The expression levels were quantified using Taqman real-time PCR on the Applied Biosystems 7900HT Fast Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA). Custom-designed ABI gene expression assays for DGCR6 (Hs00606390mH) and DGCR6L (Hs00819920mH) were utilized, and each reaction was performed in triplicate with β-actin as an internal control. Real-time data were processed as reported previously to calculate the expression relative to that of β-actin, to avoid errors related to amounts of mRNA used in the reactions, a standard procedure in expression experiments.32
To analyze the effect that the parent-of-origin of the deletion has on DGCR6 and DGCR6L expression, parametric and non-parametric analyses were performed. Children with 22q11DS were divided into three groups, based on the expression of DGCR6 and DGCR6L relative to control subjects: those showing underexpression (<2 s.e.m. of the controls), average expression (expression within 2 s.e.m. of the controls) or overexpression (>2 s.e.m. of the controls). Analysis of variance (ANOVA) was performed to ascertain expression differences in the three groups. The χ2 tests were used to determine if the distributions of low, average and high expressers were significantly different between the controls, and maternally and paternally deleted 22q11DS subjects (GraphPad Prism; GraphPad Software, La Jolla, CA, USA).
Correlation of DGCR6 and DGCR6L expression with methylation in 22q11DS blood samples
Genomic DNA from 16 subjects with 22q11DS and 3 control subjects' blood samples were bisulfite treated as reported previously.33 Primers were designed to amplify bisulfite-treated DNA in the promoter regions of DGCR6 and DGCR6L (Supplementary Table 1) using Epidesigner-beta (http://www.epidesigner.com) from Sequenom (Sequenom, Inc.). Primer specificity was checked by BLAST against the entire bisulfite-treated genome (http://bisearch.enzim.hu). Regions were PCR-amplified and methylation was quantified using the Sequenom Massarray system (Sequenom).34, 35 The results were analyzed using Sequenom's Epityper software and statistical tests were done using repeated-measures ANOVA, with multiple CpG sites being evaluated.
Association of DGCR6 and DGCR6L expressions with neuropsychological data
Neuropsychological data were collected from the subjects and controls. Assessments were made for vigilance/attention, verbal learning and reasoning, and executive function based on task-force recommendations developed by the NIMH Measurement and Treatment Research to Improve Cognition in Schizophrenia (MATRICS).36 Executive function was measured with the Wisconsin Card Sort Test (WCST), which involves matching of cue cards containing different shapes and colors.37 Sustained attention was assessed with the identical pairs and AX conditions of the continuous performance test, which are based on recognition of identical numbers and a pattern of numbers, respectively.38, 39 In addition, the California verbal learning test40 was used to assess verbal learning and memory. The Wechsler Intelligence Scale for children (WISC) was used for intelligence testing. The computerized diagnostic interview schedule for children was administered to ascertain psychiatric disorders, such as anxiety, based on DSM-IV criteria.
Statistical analyses were performed using the SPSS Version 18.0 (SPSS, Chicago, IL, USA). Independent two-sample t-tests and ANOVA were performed to evaluate group differences on continuous variables and Fisher's exact test and χ2 for categorical variables. Pearson correlations were performed to assess associations between the psychological and gene-expression data. All of our analyses were performed on the basis of a priori hypotheses. Thus, we did not correct for multiple testing, an approach adopted by other investigators in the field of 22q11DS research.41
Results
Parental origin of 22q11.2 deletion
Parental genotypes were available in 35/38 subjects with 22q11DS. Of these, 22 deletions were present on the maternal chromosome while 13 were on the paternal chromosome. One child with 22q11DS had an inherited deletion of the 22q11.2 region from his mother; the others were de novo deletions. Complete psychological data as well as gene-expression data were available for 19 subjects with a maternal deletion and 11 subjects with a paternal deletion. Correlational analyses with the neuropsychological data were performed with this subset of patients.
DGCR6 and DGCR6L expression
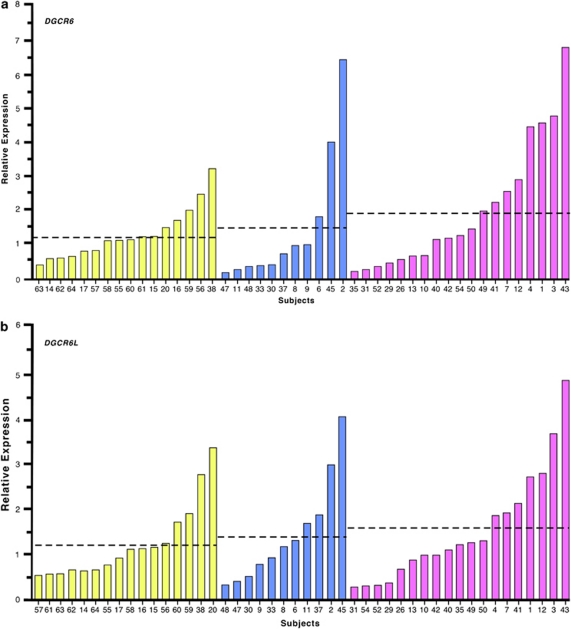
DGCR6 and DGCR6L expressions were highly correlated in the 22q11DS subjects (r=0.9; P<0.01) and controls (r=0.9; P<0.01). There were no significant differences in expression of both DGCR6 and DGCR6L between the 22q11DS and control groups. The average relative expression of DGCR6 for 22q11DS subjects was 1.7±0.3 (mean and s.e.m.) (n=38) while that in controls was 1.2±0.2 (n=16) (F=1.07, P=not significant, Cohen's d=0.4). The average relative expression of DGCR6L for 22q11DS subjects was 1.5±0.2 (n=38) while that in controls was 1.2±0.2 (n=16) (F=0.677, P=not significant, Cohen's d=0.3). However, the variability of DGCR6 expression was significantly greater among the subjects with 22q11DS than in the controls (Levene's test for homogeneity of variance, F=7.68, P<0.05). DGCR6 expression did not vary significantly between the control subjects (1.2±0.2) and 22q11DS subjects with maternal (2.0±0.4) and paternal deletions (1.4±0.6) (F=1.00, P=not significant). Similarly, for DGCR6L, there were no differences in expression between controls (1.2±0.2), maternal (1.5±0.3) and paternal deletions (1.4±.3) F=0.40, P=not significant. When we trichotomized the 22q11DS subjects into normal, high and low expressers based on their expressions relative to controls, as described in the methods (Figures 2a and b), we found that for DGCR6 expression there was a trend toward more high and low expressers in the 22q11DS subjects with a maternally derived deletion (χ2=8.45, P=0.07). The distribution of high, low and average expressers was not significant for DGCR6L (χ2=3.0, P>0.05).
Figure 2.
Relative gene expression of human DiGeorge Critical Region (DGCR6) (a) and DGCR6L (b). The fold expression levels relative to the average mean of the controls are displayed for controls (yellow), 22q11DS subjects with paternal deletions (blue) and 22q11DS subjects with maternal deletions (pink). The average relative expression level of each group is marked with a dashed line.
As there was a significant difference in race in the control group and the 22q11DS group, with half the control group being African American, we looked for differences in expression in the Caucasian controls compared with the African-American controls and there were no significant differences for DGCR6 (F=0.008, P>0.05) and DGCR6L (F=0.136, P>0.05). Thus, we did not covary for race in further analyses.
DGCR6 and DGCR6L promoter methylation levels in 22q11DS subjects
We compared the DNA methylation levels at the promoter regions of DGCR6 and DGCR6L in 16 22q11DS subjects, which included eight maternally and eight paternally deleted subjects; three controls were also investigated. As the expense associated with the methylation analyses were prohibitive, we selected a representative set of maternal and paternally deleted subjects with 22q11DS and control subjects for this analyses. We observed that this region was primarily unmethylated in all controls and subjects, regardless of the parental origin of the deletion. The average methylation across 16 CpG sites at −284 to −122 upstream of the DGCR6 promoter was 3.1±1.7% for maternally deleted subjects, 3.1±1.6% for paternally deleted subjects and 2.2±1.9% for controls (ANOVA, P>0.05, F=0.66). The average methylation across 50 CpG sites at +206 to −772 bp upstream of the DGCR6L promoter was 7.5±1.3% for maternally deleted subjects, 8.0±1.8% for paternally deleted subjects and 6.2±1.9% for controls (ANOVA, P>0.05, F=0.85).
Relationship between DGCR6 and DGCR6L expression and neuropsychological symptoms
Consistent with our previous report, the subjects with 22q11DS performed worse than the control subjects on all the psychological tests.6, 11 In the 22q11DS group, DGCR6 expression was significantly lower (F=5.42, P<0.05) in those with anxiety disorders (n=17/37) (Table 1). Similarly, the maternally deleted subjects with an anxiety disorder (n=6/19) showed a significantly lower expression of both DGCR6 (F=7.8, P<0.01) and DGCR6L (F=5.37, P<0.05) (Table 2). This significant difference in the expression levels was not present in the paternally deleted subjects or the control subjects. Upon dividing the 22q11 subjects into low, average and high expressers, there was a significantly higher incidence of anxiety disorders in the low expressers (χ2=6.55, P<0.05). Higher internalizing behaviors, which are indicative of anxiety, were also significantly correlated with lower expression of DGCR6L (Table 1). No such relationships between the level of expression and anxiety disorders were seen in the control subjects.
Table 1. Relationship between DGCR6 and DGCR6L gene expression and neuropsychological performance in 22q11DS subjects (n=36).
| Test | DGCR6 | DGCR6L |
|---|---|---|
| IQ (WISC)—verbal comprehension | −0.215 | −0.183 |
| IQ—perceptual organization | −0.049 | −0.030 |
| IQ—working memory | 0.12 | 0.2 |
| IQ—processing speed | −0.090 | −0.025 |
| Executive function (WCST)—PE | 0.244 | 0.194 |
| Sustained attention—CPT AX | −0.307 | −0.245 |
| Sustained attention—CPT IP | −0.271 | −0.288 |
| Parent CBCL internalizing T-score | −0.116 | −0.326* |
| Anxiety disorders (C-DISC) | F=5.42* | F=0.85 |
*P<0.05.
Moderate and large effect sizes are in bold (small=0.1, medium=0.3 and large=0.5 for Pearson correlations and small=0.10, medium=0.25 and large=0.40 for ANOVA).
Please note that neuropsychological data for correlational analyses were available in 36/38 subjects with 22q11DS.
Pearson's correlations were computed for all variables, except for anxiety disorders, wherein ANOVA was computed, to examine differences in expression of DGCR6 and DGCR6L in those with and without an anxiety disorder.
Table 2. Relationship between maternally derived deletions in the 22q11DS subjects and neuropsychological performance (data available for 19 subjects with maternally derived deletions).
| Test | DGCR6 | DGCR6L |
|---|---|---|
| IQ (WISC)—verbal comprehension | −0.191 | −0.236 |
| IQ—perceptual organization | −0.120 | −0.185 |
| IQ—working memory | 0.170 | 0.171 |
| IQ—processing speed | −0.184 | −0.228 |
| Executive function (WCST)—PE | 0.183 | 0.251 |
| Sustained attention—CPT AX | −0.337 | −0.331 |
| Sustained attention—CPT IP | −0.380 | −0.332 |
| Parent CBCL internalizing T-score | −0.441 | −0.461 |
| Anxiety disorders | F=7.8* | F=5.37* |
*P<0.05.
**P<0.01.
Moderate and large effect sizes are in bold (small=0.1, medium=0.3 and large=0.5 for Pearson's correlations and small=0.10, medium=0.25 and large=0.40 for ANOVA).
Pearson's correlations were computed for all variables, except for anxiety disorders, wherein ANOVA was computed, to examine differences in expression of DGCR6 and DGCR6L in those with and without an anxiety disorder.
Discussion
22q11DS is characterized by a multitude of neuropsychological abnormalities.3 Two studies reported that maternal inheritance of the deletion was associated with a greater reduction in cortical gray matter and increased language-learning disabilities.17, 18 This parental effect suggested a possible role for epigenetic regulation, such as genomic imprinting. Although mouse studies ruled out parent-of-origin-specific expression of the 25 genes lying within the 22q11DS critical region,26 computational analysis in our laboratory predicted DGCR6 to be monoallelically expressed in humans, with expression from the paternal allele.27 It was not predicted to be imprinted in mice.28 DGCR6 is believed to be required for neural crest cell migration during development30 and GABAB-receptor localization.42 It is also linked to schizophrenia susceptibility in completely independent association studies.43 Thus, we postulated that DGCR6 could be imprinted and that functional dysregulation of DGCR6 would be correlated with the psychological abnormalities observed in subjects with 22q11DS, wherein one allele is deleted.
We first compared the expression levels of DGCR6 and its duplicate copy DGCR6L between 22q11DS subjects with maternal and paternal 22q11.2 deletions, as well as with normal age- and sex-matched controls. Contrary to the expectations based on the expected hemizygous state of DGCR6 and DGCR6L in 22q11DS subjects, a large number of 22q11DS subjects showed dysregulated rather than reduced/null-gene expression when compared with those in the control individuals. This finding excludes the possibility of genomic imprinting of these genes in peripheral blood, wherein all subjects deleted from one particular parental allele would be expected to have null expression. It is, however, possible that imprinting could occur in other tissues that were not examined in this study. There were three groups that could be distinguished in the 22q11DS subjects; those with DGCR6 and DGCR6L expression levels markedly lower than controls, those with the expression levels similar to the controls and the individuals who exhibited gross overexpression of the genes. One possible explanation for the dysregulated expression of DGCR6 and DGCR6L in children with 22q11DS is that the other epigenetically regulated imprinted genes residing in the 22q11DS interval on the intact chromosome 22 could influence the expression of DGCR6 and DGCR6L in the affected individuals. Other deleted genes are also likely to contribute to the psychopathological manifestations of 22q11DS, as this is a contiguous microdeletion syndrome.44 Thus, it is now critical to determine the imprint status of all the genes in the human that reside in the 22q11DS minimum-deleted region.
We also observed a significant relationship between the increased frequency of anxiety disorders and low DGCR6 and DGCR6L expression in children with 22q11DS. Additionally, we found moderate correlations between low expression level and higher parent ratings of internalizing symptoms, which is an indirect indicator of anxiety symptoms. The importance of this observation is strengthened further by the observation that when analyses were performed after dividing the 22q11 subjects into low, average and high expressers, there was a significantly higher incidence of anxiety disorders in the low expressers. The association between childhood anxiety disorders, broader internalizing symptoms and DGCR6 expression may have implications for psychosis risk later in life, as 40–60% of children with 22q11DS have high levels of anxiety in childhood, and anxiety disorders are frequently seen in association with psychotic conditions such as schizophrenia.45 Most recently, the DGCR6 protein was shown to interact with GABAB receptor subunit, GABAB1, and aid its localization to the endoplasmic reticulum.42 This finding is intriguing as the GABAB receptor may be involved in schizophrenia. In fact, completely independent geneticlinkage studies implicate DGCR6 in schizophrenia susceptibility.43, 46 Prospective psychological/psychiatric follow-up studies in our cohort are underway and will help determine the relationship between anxiety, psychosis, and DGCR6 and DGCR6L expression. The correlation between DGCR6 and DGCR6L expression and neurocognition must also be further investigated as 22q11DS subjects with lower sustained attention scores tended to have a higher expression of DGCR6. Impaired sustained attention is an integral part of the neurocognitive phenotype in 22q11DS.11 Additionally, impaired sustained attention is a hallmark of schizophrenia, with the 22q11DS subjects showing decreased sustained attention with the onset of schizophrenia.47
In order to explain the dysregulated expression pattern of DGCR6 and DGCR6L in subjects with 22q11DS, we determined the level of DNA methylation of the promoters in both the 22q11DS and control individuals. We found the promoter regions to be unmethylated in all the cases. Nevertheless, it is likely there are epigenetic control regions at other genomic locations that regulate the expression of these genes, resulting in the variability of DGCR6 and DGCR6L expression that we found in the 22q11DS group. Our findings indicate that DNA methylation and histone modifications need to be mapped for the entire minimum-deleted region in subjects with 22q11DS, as it is possible that hemizygosity for the 22q11.2 interval could result in epigenetic dysregulation of the genes in the corresponding interval on the intact chromosome 22.
The limitation of this study is its small sample size. However, owing to the difficulty in ascertaining affected individuals with 22q11DS and obtaining the samples and measures required for a study as this, our sample size would not be unusually small. Another limitation is that we did not use a correction for multiple comparisons, such as a Bonferroni, although we chose all of our measures based on a priori hypotheses.
In conclusion, our investigations reveal that low expression levels of the vital genes DGCR6 and DGCR6L are associated with the observed variability in anxiety disorders in children with 22q11DS. This expression pattern could serve as a biomarker to predict anxiety and the development of associated schizophrenia if further studies confirm our findings. We also show here that expression of these genes is likely controlled by complex epigenetic mechanisms that when disrupted lead to dosage aberration, rather than simple reduction in gene expression, and the variability in symptoms seen in individuals with 22q11DS. Understanding their regulation and function directly in humans will be the key to understanding the role of epigenetics in complex disorders such as 22q11DS. Such knowledge could also prove to be of value in predicting those subjects who will develop psychopathologies and in developing novel therapies based on modifying the epigenome.
Acknowledgments
We thank the families for their participation in this study.This work was supported by NIH grants to VS (R01MH078015) and RLJ (R01ES015165, R01ES13053, DOE DE-0FG02-05ER64101, the Ester B. O'Keeffe Foundation Award, and Fred and Alice Stanback).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
Supplementary Material
References
- Shprintzen RJ. Velo-cardio-facial syndrome: a distinctive behavioral phenotype. Ment Retard Dev Disabil Res Rev. 2000;6:142–147. doi: 10.1002/1098-2779(2000)6:2<142::AID-MRDD9>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Driscoll DA, Budarf ML, Emanuel BS. A genetic etiology for DiGeorge syndrome: consistent deletions and microdeletions of 22q11. Am J Hum Genet. 1992;50:924–933. [PMC free article] [PubMed] [Google Scholar]
- Shprintzen RJ. Velo-cardio-facial syndrome: 30 Years of study. Dev Disabil Res Rev. 2008;14:3–10. doi: 10.1002/ddrr.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shprintzen RJ, Goldberg RB, Young D, Wolford L. The velo-cardio-facial syndrome: a clinical and genetic analysis. Pediatrics. 1981;67:167–172. [PubMed] [Google Scholar]
- Gerdes M, Solot C, Wang PP, Moss E, LaRossa D, Randall P, et al. Cognitive and behavior profile of preschool children with chromosome 22q11.2 deletion. Am J Med Genet. 1999;85:127–133. [PubMed] [Google Scholar]
- Shashi V, Kwapil TR, Kaczorowski J, Berry MN, Santos CS, Howard TD, et al. Evidence of gray matter reduction and dysfunction in chromosome 22q11.2 deletion syndrome. Psychiatry Res. 2010;181:1–8. doi: 10.1016/j.pscychresns.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swillen A, Devriendt K, Legius E, Eyskens B, Dumoulin M, Gewillig M, et al. Intelligence and psychosocial adjustment in velocardiofacial syndrome: a study of 37 children and adolescents with VCFS. J Med Genet. 1997;34:453–458. doi: 10.1136/jmg.34.6.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodin M, Wang PP, Aleman D, McDonald-McGinn D, Zackai E, Moss E. Neuropsychological profile of children and adolescents with the 22q11.2 microdeletion. Genet Med. 2001;3:34–39. doi: 10.1097/00125817-200101000-00008. [DOI] [PubMed] [Google Scholar]
- Bearden CE, Woodin MF, Wang PP, Moss E, McDonald-McGinn D, Zackai E, et al. The neurocognitive phenotype of the 22q11.2 deletion syndrome: selective deficit in visual-spatial memory. J Clin Exp Neuropsychol. 2001;23:447–464. doi: 10.1076/jcen.23.4.447.1228. [DOI] [PubMed] [Google Scholar]
- Feinstein C, Eliez S, Blasey C, Reiss AL. Psychiatric disorders and behavioral problems in children with velocardiofacial syndrome: usefulness as phenotypic indicators of schizophrenia risk. Biol Psychiatry. 2002;51:312–318. doi: 10.1016/s0006-3223(01)01231-8. [DOI] [PubMed] [Google Scholar]
- Lewandowski K, Shashi V, Berry P, Kwapil T. Schizophrenic-like neurocognitive deficits in children and adolescents with 22q11 deletion syndrome. Am J Med Genet B Neuropsychiatr Genet. 2007;144:27–36. doi: 10.1002/ajmg.b.30379. [DOI] [PubMed] [Google Scholar]
- Shashi V, Howard T, Keshavan M, Berry M, Schoch K, Spence E, et al. COMT and anxiety and cognition in children with chromosome 22q11.2 deletion syndrome. Psychiatry Res. 2010;178:433–436. doi: 10.1016/j.psychres.2010.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swillen A, Vandeputte L, Cracco J, Maes B, Ghesquiere P, Devriendt K, et al. Neuropsychological, learning and psychosocial profile of primary school aged children with the velo-cardio-facial syndrome (22q11 deletion): evidence for a nonverbal learning disability. Child Neuropsychol. 1999;5:230–241. doi: 10.1076/0929-7049(199912)05:04;1-R;FT230. [DOI] [PubMed] [Google Scholar]
- Bassett AS, Chow EW, Husted J, Weksberg R, Caluseriu O, Webb GD, et al. Clinical features of 78 adults with 22q11 deletion syndrome. Am J Med Genet A. 2005;138:307–313. doi: 10.1002/ajmg.a.30984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KC, Jones LA, Owen MJ. High rates of schizophrenia in adults with velo-cardio-facial syndrome. Arch Gen Psychiatry. 1999;56:940–945. doi: 10.1001/archpsyc.56.10.940. [DOI] [PubMed] [Google Scholar]
- Shprintzen RJ, Goldberg R, Golding-Kushner KJ, Marion RW. Late-onset psychosis in the velo-cardio-facial syndrome. Am J Med Genet. 1992;42:141–142. doi: 10.1002/ajmg.1320420131. [DOI] [PubMed] [Google Scholar]
- Eliez S, Antonarakis SE, Morris MA, Dahoun SP, Reiss AL. Parental origin of the deletion 22q11.2 and brain development in velocardiofacial syndrome: a preliminary study. Arch Gen Psychiatry. 2001;58:64–68. doi: 10.1001/archpsyc.58.1.64. [DOI] [PubMed] [Google Scholar]
- Glaser B, Mumme D, Blasey C. Language skills in children with velocardiofacial syndrome (deletion 22q11.2) J Pediatr. 2002;140:753–758. doi: 10.1067/mpd.2002.124774. [DOI] [PubMed] [Google Scholar]
- Tycko B. Allele-specific DNA methylation: beyond imprinting. Hum Mol Genet. 2010;19:R210–R220. doi: 10.1093/hmg/ddq376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy S, Scwartz S. Prader-Willi and angelman syndromes: disorders of genomic imprinting. Medicine. 1998;77:140–151. doi: 10.1097/00005792-199803000-00005. [DOI] [PubMed] [Google Scholar]
- Choufani S, Shuman C, Weksberg R. Beckwith-Wiedemann syndrome. Am J Med Genet C Semin Med Genet. 2010;154C:343–354. doi: 10.1002/ajmg.c.30267. [DOI] [PubMed] [Google Scholar]
- Das R, Hampton DD, Jirtle RL. Imprinting evolution and human health. Mamm Genome. 2009;20:563–572. doi: 10.1007/s00335-009-9229-y. [DOI] [PubMed] [Google Scholar]
- Crespi B. Genomic imprinting in the development and evolution of psychotic spectrum conditions. Biol Rev Camb Philos Soc. 2008;83:441–493. doi: 10.1111/j.1469-185X.2008.00050.x. [DOI] [PubMed] [Google Scholar]
- Petronis A. Human morbid genetics revisited: relevance of epigenetics. Trends Genet. 2001;17:142–146. doi: 10.1016/s0168-9525(00)02213-7. [DOI] [PubMed] [Google Scholar]
- Rakyan VK, Blewitt ME, Druker R, Preis JI, Whitelaw E. Metastable epialleles in mammals. Trends Genet. 2002;18:348–351. doi: 10.1016/s0168-9525(02)02709-9. [DOI] [PubMed] [Google Scholar]
- Maynard TM, Meechan DW, Heindel CC, Peters AZ, Hamer RM, Lieberman JA, et al. No evidence for parental imprinting of mouse 22q11 gene orthologs. Mamm Genome. 2006;17:822–832. doi: 10.1007/s00335-006-0011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luedi PP, Dietrich FS, Weidman JR, Bosko JM, Jirtle RL, Hartemink AJ. Computational and experimental identification of novel human imprinted genes. Genome Res. 2007;17:1723–1730. doi: 10.1101/gr.6584707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luedi PP, Hartemink AJ, Jirtle RL. Genome-wide prediction of imprinted murine genes. Genome Res. 2005;15:875–884. doi: 10.1101/gr.3303505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann L, Stankiewicz P, Spiteri E, Pandita RK, Shaffer L, Lupski JR, et al. Two functional copies of the DGCR6 gene are present on human chromosome 22q11 due to a duplication of an ancestral locus. Genome Res. 2001;11:208–217. doi: 10.1101/gr.143101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hierck BP, Molin DG, Boot MJ, Poelmann RE, Gittenberger-de Groot AC. A chicken model for DGCR6 as a modifier gene in the DiGeorge critical region. Pediatr Res. 2004;56:440–448. doi: 10.1203/01.PDR.0000136151.50127.1C. [DOI] [PubMed] [Google Scholar]
- Walter J, Paulsen M. The potential role of gene duplications in the evolution of imprinting mechanisms. Hum Mol Genet. 2003;12 (Spec No. 2):R215–R220. doi: 10.1093/hmg/ddg296. [DOI] [PubMed] [Google Scholar]
- Livak K, Schmittgen T. Analysis of relative gene expression data using real-time quantitative PCR and the 2(Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Dolinoy D, Huang D, Jirtle R. Maternal nutrient supplementation counteracts bisphenol-A induced hypomethylation in early development. Proc Natl Acad Sci USA. 2007;104:13056–13061. doi: 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrich M, Field JK, Liloglou T, Xinarianos G, Oeth P, Nelson MR, et al. Cytosine methylation profiles as a molecular marker in non-small cell lung cancer. Cancer Res. 2006;66:10911–10918. doi: 10.1158/0008-5472.CAN-06-0400. [DOI] [PubMed] [Google Scholar]
- Hartmer R, Storm N, Boecker S, Rodi CP, Hillenkamp F, Jurinke C, et al. RNase T1 mediated base-specific cleavage and MALDI-TOF MS for high-throughput comparative sequence analysis. Nucleic Acids Res. 2003;31:e47. doi: 10.1093/nar/gng047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern RS, Green MF, Nuechterlein KH, Deng BH. NIMH-MATRICS survey on assessment of neurocognition in schizophrenia. Schizophr Res. 2004;72:11–19. doi: 10.1016/j.schres.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Chelune GJ, Baer RA. Developmental norms for the Wisconsin Card Sorting Test. J Clin Exp Neuropsychol. 1986;8:219–228. doi: 10.1080/01688638608401314. [DOI] [PubMed] [Google Scholar]
- Cornblatt BA, Erlenmeyer-Kimling L. Global attentional deviance as a marker of risk for schizophrenia: specificity and predictive validity. J Abnorm Psychol. 1985;94:470–486. doi: 10.1037//0021-843x.94.4.470. [DOI] [PubMed] [Google Scholar]
- Erlenmeyer-Kimling L, Rock D, Roberts SA, Janal M, Kestenbaum C, Cornblatt B, et al. Attention, memory, and motor skills as childhood predictors of schizophrenia-related psychoses: the New York high-risk project. Am J Psychiatry. 2000;157:1416–1422. doi: 10.1176/appi.ajp.157.9.1416. [DOI] [PubMed] [Google Scholar]
- Otto MW, Bruder GE, Fava M, Delis DC, Quitkin FM, Rosenbaum JF. Norms for depressed patients for the California verbal learning test: associations with depression severity and self-report of cognitive difficulties. Arch Clin Neuropsychol. 1994;9:81–88. [PubMed] [Google Scholar]
- da Silva Alves F, Boot E, Schmitz N, Nederveen A, Vorstman J, Lavini C, et al. Proton magnetic resonance spectroscopy in 22q11 deletion syndrome. PLoS One. 2011;6:e21685. doi: 10.1371/journal.pone.0021685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zunner D, Deschermeier C, Kornau H. GABA(B) receptor subunit 1 binds to proteins affected in 22q11 deletion syndrome. Biochem Biophys Res Commun. 2010;393:185–189. doi: 10.1016/j.bbrc.2009.12.120. [DOI] [PubMed] [Google Scholar]
- Liu H, Heath SC, Sobin C, Roos JL, Galke BL, Blundell ML, et al. Genetic variation at the 22q11 PRODH2/DGCR6 locus presents an unusual pattern and increases susceptibility to schizophrenia. Proc Natl Acad Sci USA. 2002;99:3717–3722. doi: 10.1073/pnas.042700699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard TM, Haskell GT, Lieberman JA, LaMantia AS. 22q11 DS: genomic mechanisms and gene function in DiGeorge/velocardiofacial syndrome. Int J Dev Neurosci. 2002;20:407–419. doi: 10.1016/s0736-5748(02)00050-3. [DOI] [PubMed] [Google Scholar]
- Dernovsek M, Sprah L. Comorbid anxiety in patients with psychosis. Psychiatr Danub. 2009;21:43–50. [PubMed] [Google Scholar]
- Karayiorgou M, Morris MA, Morrow B, Shprintzen RJ, Goldberg R, Borrow J, et al. Schizophrenia susceptibility associated with interstitial deletions of chromosome 22q11. Proc Natl Acad Sci USA. 1995;92:7612–7616. doi: 10.1073/pnas.92.17.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver H, Feldman P. Evidence for sustained attention and working memory in schizophrenia sharing a common mechanism. J Neuropsychiatry Clin Neurosci. 2005;17:391–398. doi: 10.1176/jnp.17.3.391. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.