Abstract
Actin filaments, an essential part of the cytoskeleton, drive various cell processes, during which they elongate, disassemble and form different architectures. Over the past 30 years, the study of actin dynamics has relied mainly on bulk solution measurements, which revealed the kinetics and thermodynamics of actin self-assembly at barbed and pointed ends, its control by ATP hydrolysis and its regulation by proteins binding either monomeric actin or filament ends and sides. These measurements provide quantitative information on the averaged behavior of a homogeneous population of filaments. They have been complemented by light microscopy observations of stabilized individual filaments, providing information inaccessible using averaging methods, such as mechanical properties or length distributions. In the past ten years, the improvement of light microscopy techniques has allowed biophysicists to monitor the dynamics of individual actin filaments, thus giving access to the length fluctuations of filaments or the mechanism of processive assembly by formins. Recently, in order to solve some of the problems linked to these observations, such as the need to immobilize filaments on a coverslip, we have used microfluidics as a tool to improve the observation, manipulation and analysis of individual actin filaments. This microfluidic method allowed us to rapidly switch filaments from polymerizing to depolymerizing conditions, and derive the molecular mechanism of ATP hydrolysis on a single filament from the kinetic analysis of its nucleotide-dependent disassembly rate. Here, we discuss how this work sets the basis for future experiments on actin dynamics, and briefly outline promising developments of this technique.
Keywords: actin assembly dynamics, microfluidics, single filament, TIRF microscopy
Power and Limitations of Bulk Solution Studies of Actin Assembly Dynamics
Since 1981, the change in fluorescence of pyrenyl-labeled actin,1 and to a lesser extent of NBD-labeled actin,2 has proven instrumental in the quantitative analysis of actin self-assembly parameters at barbed and pointed ends. The size of the nucleus (a trimer) was derived from the analysis of spontaneous assembly curves;3-5 the assembly and disassembly rate parameters at barbed and pointed ends were derived from seeded assembly assays using spectrin-actin seeds and gelsolin-actin seeds, and dilution-induced depolymerization assays. These methods were powerful, in addition to standard sedimentation and other biochemical assays, to quantitatively characterize the activities of G-actin sequesterers and of filament capping, severing, stabilizing or destabilizing factors.6 Bulk solution measurements actually measure the reactivity of filament ends. On the other hand, these averaging methods were blind to the length distribution of filaments. How many nuclei were formed, and how the number of filaments is affected by fragmentation and reannealing reactions was derived from kinetic modeling, not directly measured.5 Bulk solution studies provide no information on fluctuations in length and conformations of filaments in solution, or on any heterogeneity in dynamics of the filaments that compose the population, which could result from possible structural changes or cooperative binding of some regulators. Finally, reactions like filament branching appear in bulk solution as the autocatalytic generation of ends by a molecular mechanism that can be specified, but ignoring the branched structure. Bulk solution methods evidently do not allow to monitor processive assembly by formins. Quantifying all the reactions that regulate filament assembly at the level of individual filaments is important since these processes are essential aspects of their function in vivo.
Light Microscopy Live Imaging of Individual Filaments: New Insights and Limitations of TIRFM
Bulk measurements have often been complemented with epifluorescence (or electron) microscopy techniques, which have first provided images of individual filaments, stabilized by regulatory proteins, drugs, or by the presence of unlabeled actin monomers. This has brought information on the mechanical properties of the filament in various ATP hydrolysis states and in the presence of various stabilizing or destabilizing proteins.7-10 The branched filament structure was generated by WASP proteins with the Arp2/3 complex,11 or their fragmentation and reannealing were visualized.12
Over the past decade, the improvements of microscopy techniques, and Total Internal Reflection Microscopy (TIRFM) in particular, have enabled the observation of the dynamics of individual actin filaments in real time.13 It has become possible to monitor the elongation of filaments at their barbed and pointed ends individually,14 and to verify that the method provided assembly rate parameters identical to those derived from solution studies. Filament severing by ADF/cofilin15 and processive assembly by formin16 are typical examples of novel information provided by TIRF microscopy. In addition, the observation of individual filaments should also offer the possibility to monitor different subpopulations of filaments, for instance gelsolin-capped and non-capped, a situation similar to what takes place in living cells, where different filament structures coexist.
Nonetheless, insight derived from single filament observations suffers from various limitations. Single filament techniques, whether performed in TIRFM or epifluorescence microscopy, often rely on the anchoring of filaments to the microscope coverslip via side-binding proteins.13 In this situation, the filaments interact strongly with the surface, and this constraint has been suspected to cause artifacts in the observed dynamics.14 In particular, changes in structure of the filament linked to binding of regulators like ADF/cofilin or tropomyosin, or to filament branching cannot be considered to occur with the same freedom as in a 3D environment. To minimize this problem, the density of anchoring sites can be reduced, but the filaments are then very mobile which can make their analysis cumbersome and inaccurate. The frequent use of methylcellulose to confine the filament in a 2D geometry, while avoiding anchorage, has an impact on filaments (e.g., bundling) due to excluded volume interactions and possible charge screening. Molecular confinement has non-trivial effects.17 The influence of methylcellulose on the interaction of actin filaments with regulatory proteins should be considered with caution. These effects are difficult to test.
Changing the composition of the medium in which filaments are observed is possible using an open flow cell,18 however the often long dead time (typically of the order of one minute19) needed to switch to the new conditions precludes kinetic studies.
As we shall see, a standard fluorescence microscope can be fitted out with microfluidics to go beyond these limits.
Assets of Microfluidics for the Observation of Single Actin Filaments
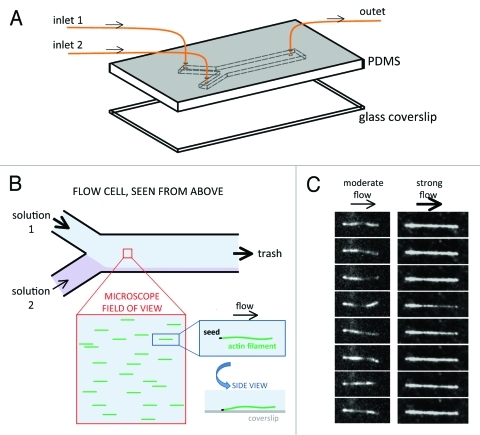
The microfluidic setup we have used to observe single actin filaments20 is very simple and similar to setups used for the study of individual DNA polymers.22 Actin filaments are grown from spectrin-actin seeds, which are adsorbed on the coverslip. The filaments fluctuate away from the coverslip surface when the flow rate is low, while they are maintained close to the surface and aligned with the flow when it is faster (Fig. 1). Today’s commercially available microfluidics apparatus allow a fast and reliable control of pressures and flow rates in various ranges. It is therefore possible to vary the flows in the flow-cell at will, and in real time.
Figure 1. Schematic principle of the microfluidics setup used for the study of individual actin filaments. (A) A structured Poly Dimethyl Siloxane (PDMS) block and a coverslip are assembled to create a flow cell, with typically 2 or 3 inlets and one outlet. Each inlet is connected to a reservoir (not shown) in which the pressure is regulated in order to control the flow in the corresponding channel. The chamber height is in the 10–100 μm range. (B) The entry channels merge into one channel. Filaments in a field located a few millimeters downstream of the junction are exposed to the incoming solution with the strongest flow. The actin filaments are grown from spectrin-actin seeds anchored to the coverslip surface. If the flow rate is high enough, the filaments align with the flow, and remain in the vicinity of the coverslip. (C) TIRFM images of two different actin filaments, roughly 10 μm long, exposed to a moderate or a strong flow of solution in the flow cell. In each series, the time interval between consecutive images is 1 sec.
Microfluidics offers many features that overcome the technical drawbacks of standard light microscopy. The artifacts that may arise from the flow of fluid, in particular the resulting drag force on the filament, can be circumscribed easily—as well as used as a tool in further studies. In the following, we list improvements and resulting achievements brought by this method.
Because the filaments are aligned parallel to the coverslip by the flow, a single attachment point at the pointed end (spectrin-actin seeds) or at the barbed end (gelsolin-actin seeds) is sufficient and the constraints in filament structure linked to several side-attachment points are avoided.
Protein interactions might be affected by the flow, either because of the flow velocity of the proteins in solution, or because of the tension applied to the anchored filaments. To circumvent this potential problem and appreciate its extent, control assays were performed. For example, filament dynamics were monitored comparatively in a strong constant flow and at a very low flow rate, except during the acquisition of images (when a strong flow facilitates the imaging of the filaments). We verified that a large range of flow rates, up to a few mm/s on average in the chamber, can be used without affecting actin assembly/disassembly dynamics. Higher flow rates, which we have not tested, may affect protein interactions or filament structure, and this could be studied using a similar setup (see below).
An essential feature offered by microfluidics is the possibility to rapidly change the medium to which the filaments are exposed while continuously monitoring them, by switching the flow rate of the incoming solutions from different inlets. Rapid kinetics can thus be performed on single filaments like in a stopped-flow apparatus. This flexibility in the control of filament history also allows the creation of various assays where specific filament compositions are achieved. For example, an artificial solid ADP-Pi-actin cap can be built to mimic vectorial Pi release.20 One can also build filaments with an embedded non-fluorescent segment, hereby getting insight into the dynamics of non-labeled actin, which so far could not be observed under the microscope.23
The continuous flow of fresh medium in the flow cell ensures that reactions are monitored at constant protein concentration (in contrast with uncontrolled changes in G/F-actin ratio with time in standard microscopy assays). This improvement allows quantitative interpretation of the collected data.
The alignment of the filaments with the flow, giving them a nearly straight contour, makes the measurement of filament length and position of the end straightforward and accurate.
The accumulation of statistically relevant data at the individual filament level is made considerably easier using microfluidics because a field of view typically contains a hundred parallel filaments, which are exposed to identical conditions and have the same history. Taken together, these features contribute to making the data accurate, and reliable.
In our recent work,20 the performance of microfluidics-assisted microscopy has revealed the molecular mechanism of inorganic phosphate release in actin filaments. The kinetics of change in the depolymerizing rate of actin filament barbed-ends, rapidly switched from an assembly to a disassembly regime, actually reflects the ADP-Pi profile in a growing filament. The data provide clear evidence for a random mechanism, while bulk solution measurements provided the same global slow rate constant of Pi release but could not discriminate, due to averaged measurements, between a vectorial and a random mechanism.21
The same microfluidics setup was used to monitor filament disassembly and analyze in deeper detail the recently reported “dynamic stabilization” of actin filaments.19 In this study, the spatial and temporal resolution provided by microfluidics was crucial to determine the molecular mechanism responsible for pauses during the depolymerization of filaments and rule out a stabilizing structural change of the polymer upon aging.23 How the random nature of reactions on individual actin filaments are used, modified, enhanced or limited by actin regulatory proteins will thus be easily addressed using the microfluidics approach combined with fluorescence microscopy. The method opens promising perspectives into the molecular mechanisms by which the specificity and efficiency of actin-based processes are achieved.
Analysis of length fluctuations of filaments has been proposed to provide insight into the mechanism of ATP cleavage in actin filaments.24 Such measurements have been difficult to achieve so far, and should be facilitated in the future by the improved accuracy provided by the microfluidics setup. As we discuss in the following, the present simple setup can be modified and made more sophisticated to broaden the range of applications, in particular addressing the mechanism of regulators of actin dynamics.
Future Developments of the Technique
The first results obtained by implementing microfluidics in TIRF microscopy to monitor filament dynamics open a large variety of perspectives on issues that can easily be addressed, by combining the technological refinements of microfluidics, that have been developed in other fields,22,25 with the biochemical tools that proved instrumental in solution studies of actin.
For instance, the filaments grown from spectrin-actin seeds anchored to the glass coverslip have a free barbed end and a stabilized pointed end. This configuration can be used to characterize the kinetic and thermodynamic aspects of interaction of filament barbed ends, in various bound nucleotide states, with a large number of barbed end regulators like capping protein, formins or barbed end trackers like VASP. As an example, we characterized the interaction of ADP- and ADP-Pi bound barbed ends with profilin, to quantify its impact on assembly dynamics.20
In order to study the dynamics of the pointed end, one can anchor filaments from stabilized barbed ends, while their pointed ends are free to elongate and depolymerize. This can be achieved by anchoring biotinylated gelsolin to a streptavidin-coated surface.26 The dynamics of the pointed ends can thus be analyzed as was done for barbed ends. The idea that the structure and reactivity of the filament is determined by the protein bound to its end, like formin27 or gelsolin,28 can be easily addressed by analyzing the effect of either ADF, tropomyosin, myosin or other filament side-binding protein on actin assembly dynamics when the filaments are anchored by various end-binding proteins.
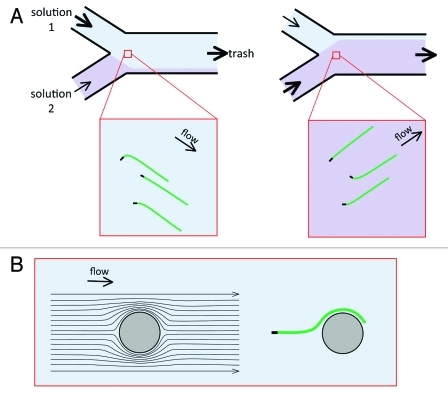
Comparison between various effectors can be performed using micropatterned chambers, so as to bind different anchoring proteins in different spatially defined regions, thus mimicking a cellular environment in which different filament arrays co-exist and turnover in a coordinated fashion. Additionally, the complexity of the medium flowing over the filaments can be varied at will, to investigate how the observed filaments, in a defined state (with free or bound barbed ends for instance), react to signal-mimicking cues, or to changes in the steady-state of filament assembly. In other words, biomimetics should experience new developments using microfluidics. Alternatively, micropatterning can be used to anchor proteins of interest in specific regions, so that growing filaments could interact with them as they fluctuate in the vicinity of the surface. In that frame of mind, one could take advantage of the microfluidic flows to orient the filaments (Fig. 2A) in order to control the interaction of growing filaments with different functionalized microdomains of the coverslip. Whether filament branching by WASP proteins with Arp2/3 complex occurs via barbed end branching or side branching mechanism can similarly be addressed in a straightforward fashion.
Figure 2. Using the flow to orient and bend the filaments. (A) In a region close to the junction of the entry channels, the filaments will have different orientations, depending on the dominant incoming flow. This provides another possibility for the manipulation of the filaments. It could be used to orient the filaments toward protein-coated patterns, for example. In addition, the spectrin-actin seeds have random orientations, and the filaments bend near the seed when they align with the flow. This bending can be modulated by changing the orientation of the filaments. (B) A fixed obstacle (e.g., a PDMS pillar) will deform the flow lines. This can also be used to bend actin filaments, with a radius of curvature that will be close to the obstacle radius.
One limitation of the microfluidics setup presented in Figure 1 is the time taken for the solutions to transit from the reservoirs to the flow cell. A serious drawback results when the protein solution undergoes chemical reactions after mixing. For example, maintaining a high concentration of G-actin in high ionic strength polymerizing conditions is hampered by the spontaneous nucleation of filaments. This limitation could be overcome by mixing reagents (G-actin and salts in this case) rapidly enough at the entrance of the flow cell, or directly within the flow cell. Different types of rapid mixing devices (e.g., using multiple adjacent flows of solutions to establish a rapid mix) have been developed.25 Another possibility is to establish controlled gradients to investigate different protein concentrations simultaneously.
Application to Mechanical Studies
We found that over a large range of flow rates, the filaments are aligned close to the surface with no interference of the flow with assembly dynamics. On the other hand, the flowing fluid could be used as a tool to exert a significant mechanical force on the filaments. Then the method would become instrumental to address important issues regarding the interplay between the mechanical and biochemical properties of actin filaments, under tensile strengths similar to those experienced in living cells. Recent reports actually point to a modulation of the structure of the filament29 and to the regulation of filament side-binding by mechanical constraints.30,31 A few examples of possible assays follow.
When adsorbing on the glass surface, spectrin-actin seeds or short filaments used as seeds orient randomly. In the presence of G-actin in the flow, filaments initially grow in the direction imposed by the seeds, before bending and aligning in the direction of the flow (Fig. 2A). When the flow is fast, the filaments are bent over a very small region, and appear straight.20 When the flow rate is lower, or when polymers are stiffer (e.g., actin bundles, or microtubules) the curvature is lower and detectable. Alternatively, fixed obstacles placed downstream from the filament anchor would bend the filament as it follows the deviated flow lines (Fig. 2B). Flow-induced bending of filaments could be used to study the impact of local filament curvature on the affinity of actin-binding proteins, or on filament severing31 and branching.32
The flowing fluid exerts a friction force along the contour of the filament. This force is maximal at the anchoring point, decreases progressively along the filament, and is null at the growing end. This force straightens the filament, and puts it under mechanical tension. In the range of flow rates that we have used (up to a few tens of microliters per minute, for a flow cell cross-section of 600 × 40 microns), this force is smaller than one pico-Newton and does not provoke the detachment of the spectrin-actin seeds from the coverslip. However, these flow rates are sufficient to greatly reduce the lateral fluctuations of the filaments. The microfluidics approach hereby offers a way to study filament fluctuations, which may have physiological relevance, in a way that differs from classical setups used to manipulate individual filaments by holding then from both ends, such as optical or magnetic tweezers.
More significant forces could be applied to filaments either by increasing the flow rate, or by anchoring the filaments a few micrometers above the coverslip surface (for example, by growing the filaments from micro-beads anchored to the bottom of the flow cell23), where the local flow velocity is higher. Actin filaments could thus be put under significant mechanical tension (in the pico-Newton range and beyond). The gradient of force along the filament, resulting from the flow, can be used as a tool to analyze how proteins like ADF or myosin bind more or less well (in a gradient) along filaments maintained under a variable tension. Quantitative correlations could then be established between protein binding and local tension.
Alternatively, a quasi-constant tension might be applied along the filament by attaching small beads to the filament in a region downstream the flow. In this setup, the drag force applied by the flow to the bead largely predominates over the variable weak force applied along the filament. The ability to perform measurements for tens of filaments in parallel, represents an important advantage of the microfluidics approach as opposed to tweezers techniques in which only one filament is manipulated.
Concluding Remarks
Adding microfluidics to fluorescence microscopy enhances the control of biochemical and mechanical conditions when monitoring the dynamics of individual actin filaments, while eliminating potential sources of artifacts. The resulting improved reliability and accuracy, in space and time, opens broad perspectives for future studies in the actin field. These perspectives will certainly be expanded by further developments of the method, inspired by forthcoming biological issues.
Acknowledgments
GRL acknowledges support from the Human Frontier Science Program (grant RGY0067/2008). MFC acknowledges support from ANR-PCV program, the Ligue Nationale contre le Cancer (équipe labelisée) a EU 241548 FP7 grant and an ERC advanced grant (ERC 2009-249982-Forcefulactin).
Glossary
Abbreviations:
- F-actin
filamentous actin
- G-actin
globular (monomeric) actin
- TIRFM
Total Internal Reflection Fluorescence (or evanescent wave) Microscopy
Footnotes
Previously published online: www.landesbioscience.com/journals/BioArchitecture/article/19338
References
- 1.Kouyama T, Mihashi K. Fluorimetry study of N-(1-pyrenyl)iodoacetamide-labelled F-actin. Local structural change of actin protomer both on polymerization and on binding of heavy meromyosin. Eur J Biochem. 1981;114:33–8. doi: 10.1111/j.1432-1033.1981.tb06167.x. [DOI] [PubMed] [Google Scholar]
- 2.Detmers P, Weber A, Elzinga M, Stephens RE. 7-Chloro-4-nitrobenzeno-2-oxa-1,3-diazole actin as a probe for actin polymerization. J Biol Chem. 1981;256:99–105. [PubMed] [Google Scholar]
- 3.Tobacman LS, Korn ED. The kinetics of actin nucleation and polymerization. J Biol Chem. 1983;258:3207–14. [PubMed] [Google Scholar]
- 4.Wegner A, Engel J. Kinetics of the cooperative association of actin to actin filaments. Biophys Chem. 1975;3:215–25. doi: 10.1016/0301-4622(75)80013-5. [DOI] [PubMed] [Google Scholar]
- 5.Sept D, McCammon JA. Thermodynamics and kinetics of actin filament nucleation. Biophys J. 2001;81:667–74. doi: 10.1016/S0006-3495(01)75731-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bosch M, Le KH, Bugyi B, Correia JJ, Renault L, Carlier MF. Analysis of the function of Spire in actin assembly and its synergy with formin and profilin. Mol Cell. 2007;28:555–68. doi: 10.1016/j.molcel.2007.09.018. [DOI] [PubMed] [Google Scholar]
- 7.Isambert H, Venier P, Maggs AC, Fattoum A, Kassab R, Pantaloni D, et al. Flexibility of actin filaments derived from thermal fluctuations. Effect of bound nucleotide, phalloidin, and muscle regulatory proteins. J Biol Chem. 1995;270:11437–44. doi: 10.1074/jbc.270.19.11437. [DOI] [PubMed] [Google Scholar]
- 8.McGough A, Pope B, Chiu W, Weeds A. Cofilin changes the twist of F-actin: implications for actin filament dynamics and cellular function. J Cell Biol. 1997;138:771–81. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlier MF, Ressad F, Pantaloni D. Control of actin dynamics in cell motility. Role of ADF/cofilin. J Biol Chem. 1999;274:33827–30. doi: 10.1074/jbc.274.48.33827. [DOI] [PubMed] [Google Scholar]
- 10.McCullough BR, Grintsevich EE, Chen CK, Kang H, Hutchison AL, Henn A, et al. Cofilin-linked changes in actin filament flexibility promote severing. Biophys J. 2011;101:151–9. doi: 10.1016/j.bpj.2011.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blanchoin L, Amann KJ, Higgs HN, Marchand JB, Kaiser DA, Pollard TD. Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature. 2000;404:1007–11. doi: 10.1038/35010008. [DOI] [PubMed] [Google Scholar]
- 12.Husson C, Renault L, Didry D, Pantaloni D, Carlier MF. Cordon-Bleu uses WH2 domains as multifunctional dynamizers of actin filament assembly. Mol Cell. 2011;43:464–77. doi: 10.1016/j.molcel.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 13.Amann KJ, Pollard TD. Direct real-time observation of actin filament branching mediated by Arp2/3 complex using total internal reflection fluorescence microscopy. Proc Natl Acad Sci U S A. 2001;98:15009–13. doi: 10.1073/pnas.211556398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuhn JR, Pollard TD. Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys J. 2005;88:1387–402. doi: 10.1529/biophysj.104.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andrianantoandro E, Pollard TD. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol Cell. 2006;24:13–23. doi: 10.1016/j.molcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 16.Romero S, Le Clainche C, Didry D, Egile C, Pantaloni D, Carlier M-F. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell. 2004;119:419–29. doi: 10.1016/j.cell.2004.09.039. [DOI] [PubMed] [Google Scholar]
- 17.Zhou HX, Rivas G, Minton AP. Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences. Annu Rev Biophys. 2008;37:375–97. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujiwara I, Vavylonis D, Pollard TD. Polymerization kinetics of ADP- and ADP-Pi-actin determined by fluorescence microscopy. Proc Natl Acad Sci U S A. 2007;104:8827–32. doi: 10.1073/pnas.0702510104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kueh HY, Brieher WM, Mitchison TJ. Dynamic stabilization of actin filaments. Proc Natl Acad Sci U S A. 2008;105:16531–6. doi: 10.1073/pnas.0807394105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jégou A, Niedermayer T, Orbánn J, Didry D, Lipowsky R, Carlier MF, et al. Individual actin filaments in a microfluidic flow reveal the mechanism of ATP hydrolysis and give insight into the properties of profilin. PLoS Biol. 2011;9:e1001161. doi: 10.1371/journal.pbio.1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carlier MF, Pantaloni D. Direct evidence for ADP-Pi-F-actin as the major intermediate in ATP-actin polymerization. Rate of dissociation of Pi from actin filaments. Biochemistry. 1986;25(24):7789–92. doi: 10.1021/bi00372a001. [DOI] [PubMed] [Google Scholar]
- 22.Brewer LR, Bianco PR. Laminar flow cells for single-molecule studies of DNA-protein interactions. Nat Methods. 2008;5:517–25. doi: 10.1038/nmeth.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niedermayer T, Jegou A, Chieze L, Helfer E, Romet-Lemonne G, Carlier MF, et al. Intermittent depolymerization of actin filaments caused by photo-induced dimerization of protomers. submitted. [DOI] [PMC free article] [PubMed]
- 24.Ranjith P, Mallick K, Joanny JF, Lacoste D. Role of ATP-hydrolysis in the dynamics of a single actin filament. Biophys J. 2010;98:1418–27. doi: 10.1016/j.bpj.2009.12.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tabeling P. Introduction to microfluidics. Oxford University Press, 2005. [Google Scholar]
- 26.Brangbour C, du Roure O, Helfer E, Démoulin D, Mazurier A, Fermigier M, et al. Force-velocity measurements of a few growing actin filaments. PLoS Biol. 2011;9:e1000613. doi: 10.1371/journal.pbio.1000613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bugyi B, Papp G, Hild G, Lõrinczy D, Nevalainen EM, Lappalainen P, et al. Formins regulate actin filament flexibility through long range allosteric interactions. J Biol Chem. 2006;281:10727–36. doi: 10.1074/jbc.M510252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prochniewicz E, Zhang Q, Janmey PA, Thomas DD. Cooperativity in F-actin: binding of gelsolin at the barbed end affects structure and dynamics of the whole filament. J Mol Biol. 1996;260:756–66. doi: 10.1006/jmbi.1996.0435. [DOI] [PubMed] [Google Scholar]
- 29.Shimozawa T, Ishiwata S. Mechanical distortion of single actin filaments induced by external force: detection by fluorescence imaging. Biophys J. 2009;96:1036–44. doi: 10.1016/j.bpj.2008.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uyeda TQ, Iwadate Y, Umeki N, Nagasaki A, Yumura S. Stretching actin filaments within cells enhances their affinity for the myosin II motor domain. PLoS One. 2011;6:e26200. doi: 10.1371/journal.pone.0026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayakawa K, Tatsumi H, Sokabe M. Actin filaments function as a tension sensor by tension-dependent binding of cofilin to the filament. J Cell Biol. 2011;195:721–7. doi: 10.1083/jcb.201102039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Risca V, Chaudhuri O, Chia J, Fletcher DA. Actin Branching Is Affected by Local Bending of the Mother Filament. Biophys J 2009; 96:122a-a.