Abstract
Synaptic function in the central nervous system (CNS) is highly dependent on a dynamic actin cytoskeleton in both the pre- and the postsynaptic compartment. Remodelling of the actin cytoskeleton is controlled by tropomyosins, a family of actin-associated proteins which define distinct actin filament populations. Here we show that TPM3 and TPM4 gene products localize to the postsynaptic region in mouse hippocampal neurons. Furthermore our data confirm previous findings of isoform segregation to the pre- and postsynaptic compartments at CNS synapses. These data provide fundamental insights in the formation of functionally distinct actin filament populations at the pre- and post-synapse.
Keywords: actin cytoskeleton, central nervous system, postsynapse, tropomyosin
Introduction
The actin cytoskeleton is the predominant cytoskeletal structure in the postsynaptic compartment of excitatory synapses in the mammalian brain. The majority of postsynaptic compartments are formed at the distal ends of dendritic spines,1 small protrusions that form along the length of dendrites. Constant remodelling of actin filaments is crucial to support the structural and functional changes that occur at CNS synapses during memory and learning. On the molecular level, changes of the structure and dynamics of actin filaments is required for the trafficking and recycling of neurotransmitter receptors such as 2-amino-3-(5-methyl-3-oxo-1,2- oxazol-4-yl)propanoic acid (AMPA) and N-Methyl-d-aspartic acid or N-Methyl-D-aspartate (NMDA) receptors (AMPAR and NMDAR respectively). These neurotransmitter receptors are integrated in the spine head and linked to actin filaments by a number of scaffolding proteins. Recent studies have shown the presence of distinct actin filament populations in dendritic spines which are characterized by different structural and dynamic properties.2,3 On the structural level (for a review see3), a meshwork made of short, straight actin filaments has been observed. These filaments have a diameter of 4–6 nm and a length of 20 nm, are part of the post-synaptic density (PSD) and extend into the spine head. Throughout the spine head an additional network of actin filaments is present of which only few are making contact with the PSD. A third population of filaments with a diameter of 5–7 nm has been localized in the neck of the spine. Analysis of actin dynamics at the post-synapse has identified three types of actin filament pools, namely a dynamic pool, an enlargement pool and a stable pool.2 The dynamic pool is thought to allow for glutamate sensitivity and the regulation of spine volume while the enlargement pool is required for processes of long-term potentiation involving long-term enlargement of the spines. The stable pool on the other hand is thought to be more involved in giving the spines stability.
Previous functional studies on the actin filament system at CNS synapses have focused on the role of known actin filament regulators that have been characterized in other cell types and subcellular compartments. These include the actin filament nucleator Arp2/3,4,5 the filament severing protein cofilin6,7 and actin-associated motor proteins such as myosin IIb.8 In contrast, little is known about tropomyosins, key regulators of actin filament function at synapses.9 Tropomyosins have been suggested to play a role at CNS synapses based on their identification in synaptosome preparations and immunogold electon microscopy of rat brain tissue showing an isoform specific segregation of tropomyosins at synapses in the cerebellum.10-12 However the question of whether different tropomyosin isoforms co-exist in the same sub-cellular compartment at synapses has so far not been addressed.
Tropomyosin is a coiled-coil dimer which forms a head-to-tail polymer lying in the major groove of the actin-filament. Over 40 isoforms of tropomyosin are generated in mammals by alternative splicing from four different genes (TPM1–4).9 This diversity is reflected in remarkable spatial segregation of isoforms13 and their capacity to regulate actin filament function in an isoform specific manner.9,14
Tropomyosin expression is developmentally regulated in both a tissue and cell type specific manner.9,15 In neuronal tissue products from three of the four Tm genes have been found: TmBr1, TmBr2 and TmBr3 (TPM1); Tm5NM1–11 (TPM3); Tm4 (TPM4).16
Tropomyosins regulate actin filament function at least in part by controlling the access and recruitment of other actin binding proteins to the filaments thereby differentially slowing the ‘off-rate’ of actin monomers from actin filament ends and increasing actin filament stiffness.17 Previous studies have demonstrated that tropomyosins can protect filaments from the severing action of ADF/cofilin18,19 and influence myosin mechanochemistry.20 A number of actin filament-associated proteins that have been shown to interact with actin filaments in a tropomyosin dependent way are enriched at the postsynapse of excitatory synapses in the CNS. These include the actin motor protein myosin IIb which is important in regulating actin dynamics during synapse function.21 We have previously shown that in B35 neuroblastoma cells the TPM1 product Tm5NM1 is able to recruit myosin IIb to actin filaments.22 The localization of Tm5NM1 in mature neuronal networks has not yet been addressed.
Our present study reports for the first time the presence of different tropomyosin gene products from the TPM3 and TPM4 genes within the same synaptic compartment at CNS synapses. Furthermore our data obtained using dissociated cultures of mouse hippocampal neurons confirm previous findings of the segregation of TPM1 and TPM4 products to the pre- and postsynaptic compartment respectively. Taken together with data on the functional diversity of tropomyosins we can conclude that tropomyosins are instrumental in the formation of actin filament populations with different functional properties at the pre- and postsynapse. This will have important implications for future studies that aim to understand actin filament dependent mechanisms at CNS synapses.
Results
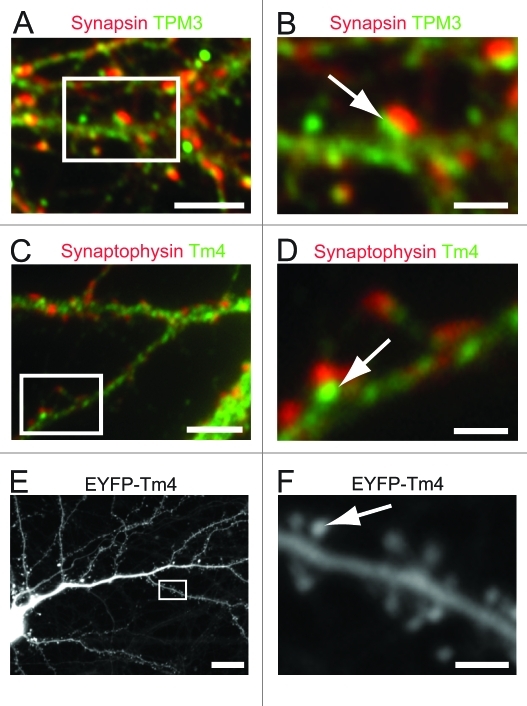
Previous studies using immunogold electron microscopy of cerebellar tissue from rats revealed the localization of TPM1 products to the pre- and TPM4 products to the postsynaptic compartment. For our study we used primary neuronal cultures from mouse embryonal hippocampi, harvested at 16.5 d after gestation as previously described.23 Immunocytochemical staining using an antibody directed against exon 9c of the TPM1 gene shows the localization of TmBr1/3 at the presynapse as determined by co-localization with the synaptic marker protein synapsin (Fig. 1). Using an antibody directed against exon 1b of the TPM3 gene, detecting all non-muscle products of the TPM3 gene we demonstrate the postsynaptic localization of these products as indicated by the close proximity but lack of overlap with the presynaptic marker synapsin (Fig. 2A and B). Similarly we show the localization of Tm4 to the postsynaptic compartment with only little immunostaining of the presynaptic compartment which is detected using an antibody against the presynaptic marker synaptophysin (Fig. 2C and D). To further confirm the localization of Tm4 at the postsynapse we decided to use a non-antibody dependent approach. We tested the localization of exogenously expressed EYFP-Tm4 in mature cultures of hippocampal neurons using lentivirus-mediated gene-transfer techniques which allows the identification of individual dendritic trees in a neuronal network. Analysis of infected cells demonstrated the presence of EYFP-tagged Tm4 in dendritic spines (Fig. 2E and F) supporting a postsynaptic localization of TPM4 gene products.
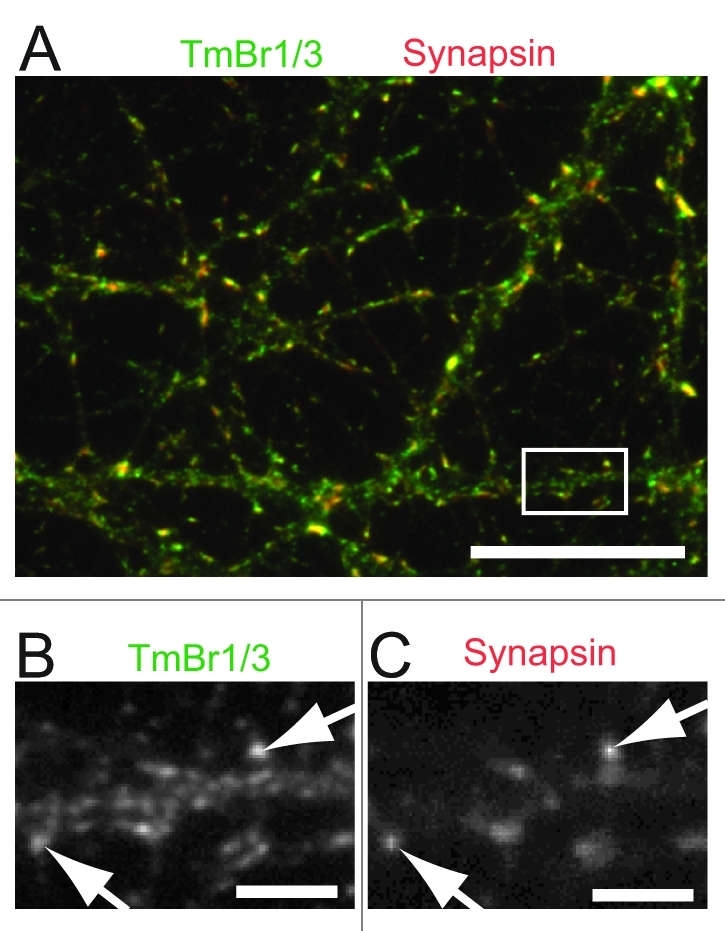
Figure 1. Localization of TPM3 gene products at the post-synapse of hippocampal neurons using immmunocytochemistry. Hippocampal neurons were co-stained using an antibody detecting exon 9c of TPM3 gene products (TmBr1,3)24 (A–E) together with anti-synapsin to identify the pre-synaptic compartment. Note the co-localization of TmB1/3 with the presynaptic marker synapsin (arrows). Scale Bars = 25 μm (A), 5 μm (B–E).
Figure 2. Localization of TPM1 and TPM4 gene products at the post-synapse of hippocampal neurons using immmunocytochemistry. Hippocampal neurons were co-stained using an antibody detecting the TPM1 gene products (A and B) or TPM3 gene products (C and D) together with anti-synapsin (A and B) or anti-synaptophysin (C and D) to identify the presynaptic compartment. Note the close localization but lack of overlap of tropomyosins (arrow) and synapsin or synaptophysin signal which demonstrates the postsynaptic enrichment of these tropomyosins. (E and F) Infection of 20 d in vitro cultured hippocampal neurons with lentiviral particles expressing EYFP-tagged Tm4. Note the localization of Tm4 at the spine heads along the dendrites of the imaged neuron. Scale Bars = 10 μm (A, C and E), 5 μm (B, D and F).
Discussion
The cytoskeletal architecture at the post-synapse of synaptic connections in the CNS plays a pivotal role in supporting and mediating processes of memory and learning. To understand the mechanisms that control the actin cytoskeleton at the CNS synapse we studied the segregation of TPM1, 3 and 4 gene products to the pre- and postsynaptic compartments of cultured mouse hippocampal neurons. Our data show for the first time the localization of products from the TPM3 gene at synapses of CNS neurons with an enrichment at the post-synaptic site. The localization of TPM1 and TPM4 products at the pre- and post-synapse of CNS neurons is consistent with previous findings by Had et al. which showed an enrichment of TmBr1/3 at the presynaptic terminals of parallel fibers in the cerebellum and an enrichment of Tm4 at the post-synapse of synapses in the molecular layer.11
The spatial segregation of tropomyosins at the CNS synapse is intriguing due to the central role of tropomyosins in defining different actin filament populations with distinct dynamic and mechanical properties. Functional characterization of the TPM1 gene product TmBr3 and the TPM3 gene product Tm5NM1 in B35 neuroblastoma cells allowed us to formulate a current model of the formation of distinct actin filament populations. While the association of TmBr3 with actin filaments promotes the formation of shorter less stiff filaments, the association of Tm5NM1 results in longer and stiffer filaments by blocking ADF/cofilin activity and the recruitment of the actin motor protein myosin IIb.22 The functional properties of Tm4 have not yet been fully assessed in this cellular model. However in a fluorescence recovery after photobleaching (FRAP) approach YFP-Tm4 showed a different dynamic association with actin filaments in human osteosarcoma cells (U2OS) as compared with YFP-tagged TPM3 gene products with YFP-Tm4 displaying a more rapid fluorescence recovery at actin structures.25 These data are suggestive for specific functional roles for Tm5NM1 and Tm4 in the same subcellular compartments.
The confirmation of the segregation of tropomyosins to the the pre- and post-synapse and the finding of different tropomyosin isoforms at the post-synapse in hippocampal neurons is significant since cultures of primary hippocampal neurons are an easily accessible system for genetic manipulation and electrophysiological analyses to study the actin cytoskeleton at the CNS synapse. Elevated expression of Tm5NM1 leads to an increase in the filamentous actin pool and the size of growth cones of hippocampal and cortical neurons.26 Changes in the size of dendritic spines are associated with long-term potentiation (LTP),27,28 in which strengthening of individual synapses occurs through presynaptic input on depolarized postsynaptic compartments, an essential process for learning and memory. Increased dendritic spine head size as seen during LTP is also associated with an increased pool of filamentous actin in the spine heads.29 Therefore the localization of TPM3 gene products in the post-synapse provides a regulatory component to control actin filament pool size in this compartment which eventually can control learning and memory.
The presence of different tropomyosin isoforms provides a potential mechanism for the formation of the structurally and functionally distinct actin filament populations that have been observed at the postsynapse.2,3 LTP involves the increased expression and insertion of AMPARs into the postsynaptic plasma membrane30,31 and it is this AMPAR trafficking that is believed to be controlled by a dynamic actin cytoskeleton.32,33 Gu et al. showed that increased ADF/cofilin activity enhanced insertion of AMPARs at the postsynaptic plasma membrane after tetraethylammonium (TEA) induced LTP.33
This emphasizes the importance of understanding how changes in the actin network at the post-synapse are mediated during LTP. NMDA receptor activation leads to the phosphorylation and activation of Myosin Light Chain (MLC)21 and the phosphorylation and deactivation of ADF/cofilin.34 A product from the TPM3 gene has been shown to direct these functional outcomes.22 Both the phosphorylation of MLC and the phosphorylation of ADF/cofilin are essential for stable LTP.21,32,35 Based on the role of tropomyosins in controlling access of actin-associated proteins to distinct actin filaments, we speculate that TPM3 and TPM4 may be instrumental in mediating the effect of NMDAR receptor induced changes downstream or in parallel to the phosphorylation of MLC and ADF/cofilin.
While our study established the localization of TPM3 and TPM4 gene products at the post-synapse, further studies will be required to analyze the sub-compartment specific distribution of products from these two genes. For this, recent advances in super-resolution microscopy applications such as STED, STORM and PALM microscopy will be very valuable and will help to characterize the role of these product in synapse function.36-38
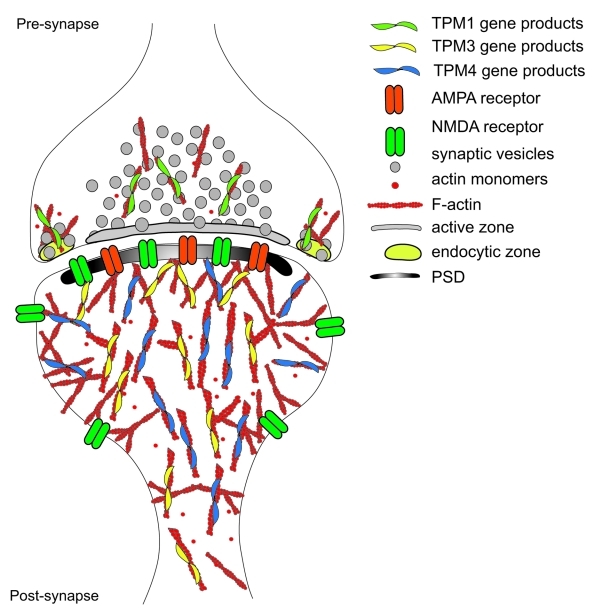
Our findings of the presence of products from both the TPM3 and TPM4 gene (illustrated in Fig. 3) adds significantly to our current view of actin filament populations at the post-synapse. The identification of two different tropomyosins at the post-synapse provides a potential mechanism to create and regulate different actin filament populations.
Figure 3. Schematic of an excitatory synapse in the CNS. The actin filament organization and relative localization of Tropomyosins at the CNS synapse are depicted. Note the localization of TPM1 gene products at the presynapse while TPM3 and TPM4 gene products localize to the postsynapse.
Material and Methods
Hippocampal-cortical co-cultures were prepared as described previously.23 In brief, hippocampi and cortices were dissected from embryonal day 16.5 C57Bl6 mice and triturated in plating medium containing DMEM/10% FBS (Gibco) using firepolished glass Pasteur pipettes. 1x103 hippocampal cells were plated on poly d-lysine (0.1 mg/ml; Sigma)-coated 12-mm glass coverslips (Menzel) mounted in the center of a 12-well culture plate well and 1 x 105 cortical cells around the edge of the wells. After 2 h, the plating medium was replaced by 1 ml of Neurobasal medium containing B27 supplement and Glutamax (Gibco). For immunofluorescence staining, cells were fixed after 21 d with 4% paraformaldehyde in PBS. Cells were permeabilized with 0.1% Triton in PBS solution and stained with the following antiobodies: monoclonal CG3, directed against all non-muscle TPM3 products,24 rabbit anti Tm4,24 anti-synapsin (AB1543, Millipore) and anti-synaptophysin (Millipore, MAB5258). All fluorophore-conjugated antibodies were obtained from Molecular Probes. Catalogue numbers are given in brackets. The following secondary antibodies were used: Alexa 647-conjugated donkey anti-rabbit (A31573), Alexa 555-conjugated donkey anti-rabbit (A31572), Alexa 488 donkey anti-mouse (A21202), Alexa 555 donkey anti-mouse (A31570). Images of neuronal cultures were taken on an IX81 (Olympus) and an Axioskop 40 (Carl Zeiss) microscope.
A EYFP-Tm4 expression construct was prepared by sub-cloning of EYFP-Tm4 from pEYPF-Tm425 into a pRRLsin18.PPT.CMV.Wpre39 resulting in the EYFP-Tm4 expressing vector pCMV-EYFP-Tm4. For the preparation of Lenti-viral particles 293 cells were plated in T75 flasks at a density of 4 × 106 cells per flask. Cells were co-transfection with pCMV-YFP-Tm4, pREV, pRRE and pVSVg using CaCl2 transfection as previously described.40 The transfected cells were incubated at 37˚C and 5% CO2 in DMEM, 10% FBS, the medium was changed for fresh medium and incubated for 28–32 h. After the incubation, supernatant was collected and the titer determined using a FACSCalibur flow cytometer (BD Biosciences).
For infection, hippocampal neurons were plated as described above. Lenti-viral particle stocks were diluted in neurobasal medium to achieve a concentration of 3 × 105 particles per 10 µl. Infected cultures were fixed at two days post infection by applying equal volume of 8% PFA in PBS. After fixation, coverslips were washed with PBS and mounted on glass slides for imaging.
Acknowledgments
We wish to thank Prof. Ian Alexander for providing the viral plasmids for the generation of Lenti-viral infectious particles as well as Dr. Samantha Ginn and Dr. Belinda Kramer for their advice and assistance in virus stock preparation.
Ethics Statement
All procedures with animals were performed to avoid unnecessary discomfort, pain or injury to the animals and have been approved by the Animal Care and Ethics Committee of the Children’s Medical Research Institute and Children’s Hospital at Westmead, Australia.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/BioArchitecture/article/19336
References
- 1.Matus A, Ackermann M, Pehling G, Byers HR, Fujiwara K. High actin concentrations in brain dendritic spines and postsynaptic densities. Proc Natl Acad Sci U S A. 1982;79:7590–4. doi: 10.1073/pnas.79.23.7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Honkura N, Matsuzaki M, Noguchi J, Ellis-Davies GC, Kasai H. The subspine organization of actin fibers regulates the structure and plasticity of dendritic spines. Neuron. 2008;57:719–29. doi: 10.1016/j.neuron.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 3.Dillon C, Goda Y. The actin cytoskeleton: integrating form and function at the synapse. Annu Rev Neurosci. 2005;28:25–55. doi: 10.1146/annurev.neuro.28.061604.135757. [DOI] [PubMed] [Google Scholar]
- 4.Rácz B, Weinberg RJ. Organization of the Arp2/3 complex in hippocampal spines. J Neurosci. 2008;28:5654–9. doi: 10.1523/JNEUROSCI.0756-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hotulainen P, Llano O, Smirnov S, Tanhuanpää K, Faix J, Rivera C, et al. Defining mechanisms of actin polymerization and depolymerization during dendritic spine morphogenesis. J Cell Biol. 2009;185:323–39. doi: 10.1083/jcb.200809046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Racz B, Weinberg RJ. Spatial organization of cofilin in dendritic spines. Neuroscience. 2006;138:447–56. doi: 10.1016/j.neuroscience.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 7.Görlich A, Wolf M, Zimmermann AM, Gurniak CB, Al Banchaabouchi M, Sassoè-Pognetto M, et al. N-cofilin can compensate for the loss of ADF in excitatory synapses. PLoS One. 2011;6:e26789. doi: 10.1371/journal.pone.0026789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ryu J, Liu L, Wong TP, Wu DC, Burette A, Weinberg R, et al. A critical role for myosin IIb in dendritic spine morphology and synaptic function. Neuron. 2006;49:175–82. doi: 10.1016/j.neuron.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 9.Gunning P, O’Neill G, Hardeman E. Tropomyosin-based regulation of the actin cytoskeleton in time and space. Physiol Rev. 2008;88:1–35. doi: 10.1152/physrev.00001.2007. [DOI] [PubMed] [Google Scholar]
- 10.Blitz AL, Fine RE. Muscle-like contractile proteins and tubulin in synaptosomes. Proc Natl Acad Sci U S A. 1974;71:4472–6. doi: 10.1073/pnas.71.11.4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Had L, Faivre-Sarrailh C, Legrand C, Méry J, Brugidou J, Rabié A. Tropomyosin isoforms in rat neurons: the different developmental profiles and distributions of TM-4 and TMBr-3 are consistent with different functions. J Cell Sci. 1994;107:2961–73. doi: 10.1242/jcs.107.10.2961. [DOI] [PubMed] [Google Scholar]
- 12.Mello CF, Sultana R, Piroddi M, Cai J, Pierce WM, Klein JB, et al. Acrolein induces selective protein carbonylation in synaptosomes. Neuroscience. 2007;147:674–9. doi: 10.1016/j.neuroscience.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin C, Gunning P. Isoform sorting of tropomyosins. Adv Exp Med Biol. 2008;644:187–200. doi: 10.1007/978-0-387-85766-4_15. [DOI] [PubMed] [Google Scholar]
- 14.Gunning P, Hardeman E, Jeffrey P, Weinberger R. Creating intracellular structural domains: spatial segregation of actin and tropomyosin isoforms in neurons. Bioessays. 1998;20:892–900. doi: 10.1002/(SICI)1521-1878(199811)20:11<892::AID-BIES4>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 15.Schevzov G, Vrhovski B, Bryce NS, Elmir S, Qiu MR, O’neill GM, et al. Tissue-specific tropomyosin isoform composition. J Histochem Cytochem. 2005;53:557–70. doi: 10.1369/jhc.4A6505.2005. [DOI] [PubMed] [Google Scholar]
- 16.Curthoys N, Gunning P, Fath T. Tropomyosins in neuronal morphogenesis and development. Book Chapter 2011; (Eds) R.A. Nixon & A. Yuan (2011) XV, 411-446. [Google Scholar]
- 17.Adami R, Cintio O, Trombetta G, Choquet D, Grazi E. On the stiffness of the natural actin filament decorated with alexa fluor tropomyosin. Biophys Chem. 2003;104:469–76. doi: 10.1016/S0301-4622(03)00036-X. [DOI] [PubMed] [Google Scholar]
- 18.Bernstein BW, Bamburg JR. Tropomyosin binding to F-actin protects the F-actin from disassembly by brain actin-depolymerizing factor (ADF) Cell Motil. 1982;2:1–8. doi: 10.1002/cm.970020102. [DOI] [PubMed] [Google Scholar]
- 19.Ishikawa R, Yamashiro S, Matsumura F. Differential modulation of actin-severing activity of gelsolin by multiple isoforms of cultured rat cell tropomyosin. Potentiation of protective ability of tropomyosins by 83-kDa nonmuscle caldesmon. J Biol Chem. 1989;264:7490–7. [PubMed] [Google Scholar]
- 20.Fanning AS, Wolenski JS, Mooseker MS, Izant JG. Differential regulation of skeletal muscle myosin-II and brush border myosin-I enzymology and mechanochemistry by bacterially produced tropomyosin isoforms. Cell Motil Cytoskeleton. 1994;29:29–45. doi: 10.1002/cm.970290104. [DOI] [PubMed] [Google Scholar]
- 21.Rex CS, Gavin CF, Rubio MD, Kramar EA, Chen LY, Jia Y, et al. Myosin IIb regulates actin dynamics during synaptic plasticity and memory formation. Neuron. 2010;67:603–17. doi: 10.1016/j.neuron.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bryce NS, Schevzov G, Ferguson V, Percival JM, Lin JJ, Matsumura F, et al. Specification of actin filament function and molecular composition by tropomyosin isoforms. Mol Biol Cell. 2003;14:1002–16. doi: 10.1091/mbc.E02-04-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fath T, Ke YD, Gunning P, Götz J, Ittner LM. Primary support cultures of hippocampal and substantia nigra neurons. Nat Protoc. 2009;4:78–85. doi: 10.1038/nprot.2008.199. [DOI] [PubMed] [Google Scholar]
- 24.Schevzov G, Whittaker SP, Fath T, Lin JJ, Gunning PW. Tropomyosin isoforms and reagents. Bioarchitecture. 2011;1:135–64. doi: 10.4161/bioa.1.4.17897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tojkander S, Gateva G, Schevzov G, Hotulainen P, Naumanen P, Martin C, et al. A molecular pathway for myosin II recruitment to stress fibers. Curr Biol. 2011;21:539–50. doi: 10.1016/j.cub.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 26.Schevzov G, Fath T, Vrhovski B, Vlahovich N, Rajan S, Hook J, et al. Divergent regulation of the sarcomere and the cytoskeleton. J Biol Chem. 2008;283:275–83. doi: 10.1074/jbc.M704392200. [DOI] [PubMed] [Google Scholar]
- 27.Yuste R, Bonhoeffer T. Morphological changes in dendritic spines associated with long-term synaptic plasticity. Annu Rev Neurosci. 2001;24:1071–89. doi: 10.1146/annurev.neuro.24.1.1071. [DOI] [PubMed] [Google Scholar]
- 28.Segal M. Dendritic spines and long-term plasticity. Nat Rev Neurosci. 2005;6:277–84. doi: 10.1038/nrn1649. [DOI] [PubMed] [Google Scholar]
- 29.Bosch M, Hayashi Y. Structural plasticity of dendritic spines. Curr Opin Neurobiol. [DOI] [PMC free article] [PubMed]
- 30.Malinow R, Malenka RC. AMPA receptor trafficking and synaptic plasticity. Annu Rev Neurosci. 2002;25:103–26. doi: 10.1146/annurev.neuro.25.112701.142758. [DOI] [PubMed] [Google Scholar]
- 31.Makino H, Malinow R. AMPA receptor incorporation into synapses during LTP: the role of lateral movement and exocytosis. Neuron. 2009;64:381–90. doi: 10.1016/j.neuron.2009.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rust MB, Gurniak CB, Renner M, Vara H, Morando L, Görlich A, et al. Learning, AMPA receptor mobility and synaptic plasticity depend on n-cofilin-mediated actin dynamics. EMBO J. 2010;29:1889–902. doi: 10.1038/emboj.2010.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gu J, Lee CW, Fan Y, Komlos D, Tang X, Sun C, et al. ADF/cofilin-mediated actin dynamics regulate AMPA receptor trafficking during synaptic plasticity. Nat Neurosci. 2010;13:1208–15. doi: 10.1038/nn.2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen LY, Rex CS, Casale MS, Gall CM, Lynch G. Changes in synaptic morphology accompany actin signaling during LTP. J Neurosci. 2007;27:5363–72. doi: 10.1523/JNEUROSCI.0164-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou Q, Homma KJ, Poo MM. Shrinkage of dendritic spines associated with long-term depression of hippocampal synapses. Neuron. 2004;44:749–57. doi: 10.1016/j.neuron.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 36.Tatavarty V, Kim EJ, Rodionov V, Yu J. Investigating sub-spine actin dynamics in rat hippocampal neurons with super-resolution optical imaging. PLoS One. 2009;4:e7724. doi: 10.1371/journal.pone.0007724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tønnesen J, Nadrigny F, Willig KI, Wedlich-Söldner R, Nägerl UV. Two-color STED microscopy of living synapses using a single laser-beam pair. Biophys J. 2011;101:2545–52. doi: 10.1016/j.bpj.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dani A, Huang B, Bergan J, Dulac C, Zhuang X. Superresolution imaging of chemical synapses in the brain. Neuron. 2010;68:843–56. doi: 10.1016/j.neuron.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Follenzi A, Ailles LE, Bakovic S, Geuna M, Naldini L. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat Genet. 2000;25:217–22. doi: 10.1038/76095. [DOI] [PubMed] [Google Scholar]
- 40.Ginn SL, Fleming J, Rowe PB, Alexander IE. Promoter interference mediated by the U3 region in early-generation HIV-1-derived lentivirus vectors can influence detection of transgene expression in a cell-type and species-specific manner. Hum Gene Ther. 2003;14:1127–37. doi: 10.1089/104303403322167975. [DOI] [PubMed] [Google Scholar]