Abstract
Microsporidia are obligate intracellular parasites whose genomes have been shaped by an extreme lifestyle. Specifically, their obligate intracellular parasitism has resulted in the loss of many genes and biochemical pathways, but these reductive processes have been often offset by the acquisition of several genes by means of horizontal gene transfer (HGT). Until recently, these HGTs were all found to have derived from prokaryotic donors, but a recent study suggests that some species took advantage of this mechanism to acquire one gene from an animal, which they maintained in their genome for metabolic purposes. The gene encodes for a purine nucleoside phosphorylase, and shows a strong phylogenetic signal of arthropod origin. Here, we briefly review our current knowledge of HGTs discovered across microsporidian genomes and discuss the implications of the most recent findings in this research area for understanding the origin and evolution of this highly adapted group of intracellular parasites. A novel gene potentially transferred by means of HGT to one microsporidia is also reported.
Keywords: ATP transporters, catalase, encephalitozoon, folylpolyglutamate synthase, horizontal gene transfer, microsporidia, purine nucleoside phosphorylase, superoxide dismutase
Introduction
Microsporidia represent a group of ultra-adapted, obligate intracellular parasites that are found to infect virtually every known animal lineage; from worms to humans. Microsporidian cells are very simplistic in form, lacking conventional mitochondria, and harbouring an atypical Golgi apparatus and “prokaryote-like” rRNA molecules (rRNA).1 The presence of these unconventional features in microsporidia have long been thought to reflect their primitive eukaryotic nature.2-5 However, it is now well recognized that these result from their adaptation for an atypical lifestyle, and these curious unicellular organisms are currently best described as representing one of many offshoots of the fungal kingdom.6-10
The genomes of microsporidia are also compelling mirrors of their specialized way of living—they are small, and simplistic in both form and content. Until now, sequencing efforts have revealed that the genomes of all members of the group are characterized by at most 2,500 genes, which are usually involved in a few, reduced metabolic pathways.11 Obviously, this reduction in many biochemical pathways has resulted in microsporidia being unable to produce a number of cellular compounds, so members of this lineage strongly depend on their host’s metabolism for many cytoplasmic supplies; including nucleotides and amino acids.12,13 In the most extreme case, one microsporidia appears to have lost the capacity to produce ATP altogether;14 resulting in its dependence on the host for even the most basic source of energy.
Undergoing massive gene losses may not appear as an ideal solution to evolve a successful lifestyle, but microsporidian parasites have managed to offset the miniaturization of their proteome by acquiring a battery of genomic tools; which they now use to protect themselves from environmental insults and to profit more fully from their hosts.
Role of Horizontal Gene Transfers in the Evolution of Microsporidia
The compelling reductive processes that characterize microsporidia have often overshadowed what’s abundant in their genomes. Indeed, all species with surveyed genomes are all surprisingly rich in “transporter” proteins (e.g., ATP transporters, Folate transporters), that are typically used by these parasites to steal metabolites13-15 and energy from the hosts they invade16 (Fig. 1A). In the case of ATP/ADP transporters, these appear to have been acquired by microsporidian genomes from a number of donors by means of HGT, and it is now increasingly accepted that such transfers have likely originated from co-infecting bacteria; such as Chlamydia.15,17,18
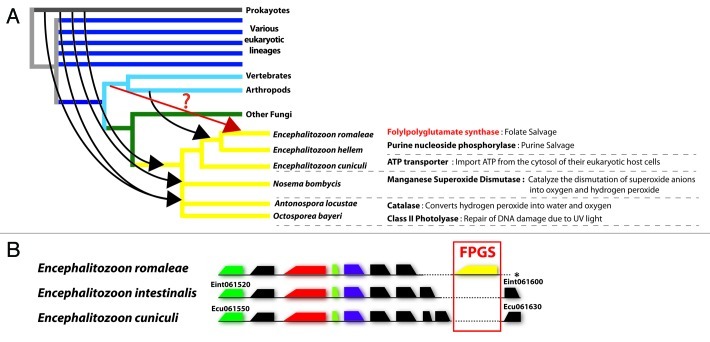
Figure 1. (A) Events of horizontal gene transfer (HGT) known to have occurred in microsporidia based on published sequence data. A schematic representation of major evolutionary lineages is shown. Arrows depicts the evolutionary origin (e.g., gene donor) of different HGTs acquired by different microsporidian genera. The red arrow highlights the gene transfer of the FPGS gene, whose origin is currently unclear. (B) Genomic location of the FPGS gene in the genome of E. romaleae. Alignment of a homologous region of approximately 15kb region between the from E. romaleae, E. intestinalis and E. cuniculi. This region is located on the chromosome 6 of E. intestinalis and E. cuniculi. The location of the folylpolyglutamate synthase (FPGS) genes in E. romaleae is shown in yellow. Conserved hypothetical proteins are shown as black rectangles, whereas genes encoding for proteins with known functions are shown as colored rectangles. Dashed lines indicate portions of the genome sequence that are missing a particular gene. * end of the E. romaleae contig currently available.
HGTs have also affected microsporidian lineages in other ways. For instance, species in genus Antonospora, Nosema and Octosporea have experienced additional gene transfers that provided a welcomed protection for their cells.15,19,20 One of these genes encodes for a superoxide dismutase that catalyzes the conversion of superoxide anions into oxygen and hydrogen peroxide, while the other encodes for a catalase; a protein responsible for the decomposition of hydrogen peroxide to water and oxygen. Both genes are, therefore, critical in the detoxification of cytoplasmic environment from reactive oxygen species. Interestingly, the catalase is found in the peroxysome in other organisms, an organelle that is lacking from microsporidian cells; and both sequences harbor a strong prokaryotic signal.19,20 Some of these latter genera have also gained protection by acquiring a “photolyase,” which is required for the repair of UV-induced DNA damage; an essential tool to avoid cell death as a consequence of mutagenic factors.21 These genes are all absent in more diverged lineages of the group (i.e., Encephalitozoon spp), leading to the prediction that spores from Antonospora, Nosema and Octosporea may be better protected from environmental factors than those of other species.21
An Animal Gene in a Microsporidia
For some time, questions have remained as to whether microsporidia have also acquired genes from more closely related donors and, perhaps, used these HGTs to rebuild broken biochemical pathways. Interestingly, a recent study based on ongoing genome sequencing projects suggests that these processes are likely to have happened in two species.
Inspections of the genome draft of Encephalitzoon romaleae, an insect-infecting microsporidia, have revealed the presence of a protein encoding gene that was previously unknown from other members of this group; a purine nucleoside phosphorylase (PNP).22 This enzyme is known to be involved in the salvage of purines,23-25 so its presence is likely to benefit the metabolic power of E. romaleae. Surprisingly, the amino acid sequence of the E. romaleae PNP gene did not show significant similarities with homologs from prokaryotes, or from closely related unicellular eukaryotes, but shared instead a strong identity with homologs from arthropods. This unexpected finding was further confirmed using a variety of methods and models for phylogenetic analyses, and resulted in the discovery of the first compelling case of HGT between a microsporidian parasite and an animal (Fig. 1A). Because E. romaleae is an obligate intracellular parasite of insects, this HGT seems better explained if it had derived from its host, rather than from another unknown source. It is also notable that the PNP appears to be somehow amenable to HGTs, since similar gene acquisitions have been reported in other parasites, such as Giardia lamblia26 Borrelia hermsii27 and Cryptosporidium parvum28; although never from eukaryotic donors. Certainly, the unusual frequency at which PNP has been moved across many distantly related lineages suggests that the acquisition of this gene represents a strong selective advantage for many parasites. One may, therefore expect other metabolically relevant genes to have been moved around in the same way.
A Novel HGT Candidate in the Genome of Encephalitozoon Romaleae
Interestingly, more recent inspections of the genome of E. romaleae in our lab have identified the presence of another HGT candidate. This latter genome has been sequenced using the Illumina technology, and assembled and annotated as described in22 As for PNP, this gene is absent from other relatives with published genomes,12,13 and encodes for a metabolically important protein; the folylpolyglutamate synthase (FPGS). This protein represents an ATP-dependent enzyme that plays a key role in the salvage of folate and in its cellular retention,29 so its presence in the genome is another strong indication that the metabolism of this species may be more elaborated than that of other microsporidia. The gene is located on a large contig that resembles subtelomeric regions of sibling species (Fig. 1B), and its presence in the genome was confirmed by PCR and conventional DNA sequencing. Verification of the location of folylpolyglutamate synthase within E. romaleae was performed using the following set of primers; 5′-FPGS_F1: GGATCGATGTTCGTGACTAAAAGGGT and FPGS_R1: 5′ TTCCATCTTCAAAGCGCCTTAGATCCT-3′. PCR reactions were performed in 25μl containing a final concentration of 1X EconoTaq® DNA Polymerase (Lucigen, WI, USA), 0.5mM of each primer and 0.3μl of DNA template; and the products were sequenced using conventional Sanger sequencing.
Amino acid sequences of publicly available FPGS’s homologs from other taxa were acquired from RefSeq GenBank, ESTdb, as well as from complete eukaryotic genome databases deposited in the Broad institute and DOE Joint Genome Institute databases, and aligned using Muscle 3.7.30 to carry phylogenetic analyses as described in.22 BLAST searches (Table 1) and phylogenetic analyses all pointed toward an animal origin for this gene but, as opposed to the PNP gene, statistical support for most analyses was low and phylogenetic reconstructions sometimes failed to support the monophyly of natural lineages (e.g., the fungi). So, while the FPGS gene has undoubtedly been acquired by E. romaleae by means of HGT, its donor cannot be identified with certainty. Whatever its origin, however, FPGS represents another outstanding addition to the biochemical repertoire of E. romaleae, and highlights how remarkably well this species has benefited from these stochastic events compared with any other member of the group. The contig containing the FPGS gene is available in GenBank under the following accession number JN859606.
Table 1. Protein with closest homology to FPGS gene from E. romaleae.
| Evalue | % Pairwise Identity | Query coverage | Accession | Organism |
|---|---|---|---|---|
|
1.49E-100 |
43.00% |
95.24% |
NP_998602 |
Danio rerio |
|
1.03E-98 |
43.20% |
95.24% |
XP_003440209 |
Oreochromis niloticus |
|
9.84E-96 |
42.70% |
97.14% |
EFX89284 |
Daphnia pulex |
|
1.51E-94 |
40.00% |
96.43% |
XP_003230625 |
Anolis carolinensis |
|
2.77E-91 |
42.00% |
96.90% |
XP_003440208 |
Oreochromis niloticus |
|
3.89E-91 |
38.80% |
96.90% |
XP_003470799 |
Cavia porcellus |
|
6.21E-91 |
38.80% |
97.14% |
AAH05484 |
Mus musculus |
|
1.49E-90 |
40.80% |
97.14% |
XP_001365989 |
Monodelphis domestica |
|
3.60E-90 |
38.70% |
96.90% |
NP_001230938 |
Cricetulus griseus |
|
8.49E-90 |
38.50% |
96.67% |
NP_001019651 |
Bos taurus |
|
2.58E-89 |
38.50% |
96.90% |
XP_851481 |
Canis lupus |
|
4.68E-89 |
37.40% |
96.90% |
XP_002915221 |
Ailuropoda melanoleuca |
|
5.78E-89 |
42.00% |
95.48% |
XP_002109877 |
Trichoplax adhaerens |
|
8.69E-89 |
42.30% |
96.19% |
XP_002430516 |
Pediculus humanus |
| 1.00E-88 | 39.50% | 92.38% | DAA24139 | Bos Taurus |
An Insect Gene in a Parasite of Humans: Underpinning the Frequency of “Host-Switch” in Microsporidian Parasites
The identification of an arthropod gene in E. romaleae (PNP) was certainly surprising, but not inconceivable given that this species is an intimate intracellular parasite of insects. More intriguing, however, was the fact that the gene could also be found in the genome of a notorious human pathogen, Encephalitozoon hellem.31 So, how can an insect gene move into the genome of parasite of vertebrates? This question was partly answered using phylogenetic analyses, which demonstrated that E. hellem and E. romaleae are closely related, sister species22,32 sharing a recent common ancestor from which they have likely both acquired the PNP gene. Whether E. hellem can also infect insects is currently unknown, but the discovery of an insect gene in this species indicates that “host-switching” must have occurred at one point in this particular lineage. Importantly, infection of both insects and vertebrate hosts by microsporidia has already been documented for other species, e.g., Anncaliia algerae,33 Trachipleistophora hominis34 and Trachipleistophora extenrec35; so having an ancestor that could infect both mammals and insects is a likely event.
Perhaps, the ability of some microsporidia to infect drastically different lineages (i.e. arthropods and vertebrates) is correlated with the metabolic complexity of some species. In particular, an increased metabolism may reduce the dependency of intracellular parasites for supplies from their host, facilitating their autonomy and their capacity to invade new environments (e.g., new hosts). This hypothesis is consistent with the recently discovered, improved metabolic capabilities of E. romaleae and E. hellem (i.e., the FPGS was also recently found in the unpublished genome of E. hellem; J.F. Pombert and Patrick Keeling, personal communication), but genome data from other species that are prone to radical host switches (i.e. A. algerae, T. hominis, T. extenrec) will be required to fully test this hypothesis.
Conclusion
HGT can be very profitable for their recipients, enabling parasites to invade new environments and adapt to conditions prevailing within animals.21,36-39 Microsporidia seem to have mastered this type of gene acquisitions, having received and maintained a number of genes from different donors by HGT that they now use for protection, or to fuel their metabolism. Microsporidian genomes have been studied for some time, but only recently sequencing efforts have started to focus on species that are not exclusively medically or ecologically relevant. Such efforts just started to pay off with the recent identification of an animal gene in these parasites. This outstanding finding has many implications for the field of evolutionary biology, and may result in an increased interest by many researchers to search for similar events in other, currently overlooked microsporidian species.
Acknowledgments
N.C. is a scholar of the Integrated Microbial Diversity program of the Canadian Institute for Advanced Research (CIFAR). Work in our lab is supported by grants from the Natural Sciences and Engineering Research Council of Canada (NSERC), the Canadian fund for Innovation, the Ontario ministry of research and the RDP program from the University of Ottawa. We thank Jean-François Pombert for useful comments on a previous version of this manuscript
Glossary
Keywords:
- horizontal gene transfer
microsporidia, encephalitozoon, purine nucleoside phosphorylase, catalase, superoxide dismutase, folylpolyglutamate synthase, ATP transporters
Footnotes
Previously published online: www.landesbioscience.com/journals/mge/article/18611
References
- 1.Keeling PJ, Fast NM. Microsporidia: biology and evolution of highly reduced intracellular parasites. Annu Rev Microbiol. 2002;56:93–116. doi: 10.1146/annurev.micro.56.012302.160854. [DOI] [PubMed] [Google Scholar]
- 2.Cavalier-Smith T. Eukaryote kingdoms: seven or nine? Biosystems. 1981;14:461–81. doi: 10.1016/0303-2647(81)90050-2. [DOI] [PubMed] [Google Scholar]
- 3.Cavalier-Smith T. Eukaryotes with no mitochondria. Nature. 1987;326:332–3. doi: 10.1038/326332a0. [DOI] [PubMed] [Google Scholar]
- 4.Vossbrinck CR, Maddox JV, Friedman S, Debrunner-Vossbrinck BA, Woese CR. Ribosomal RNA sequence suggests microsporidia are extremely ancient eukaryotes. Nature. 1987;326:411–4. doi: 10.1038/326411a0. [DOI] [PubMed] [Google Scholar]
- 5.Brown JR, Doolittle WF. Root of the universal tree of life based on ancient aminoacyl-tRNA synthetase gene duplications. Proc Natl Acad Sci USA. 1995;92:2441–5. doi: 10.1073/pnas.92.7.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gill EE, Lee RC, Corradi N, Grisdale CJ, Limpright VO, Keeling PJ, et al. Splicing and transcription differ between spore and intracellular life stages in the parasitic microsporidia. Mol Biol Evol. 2010;27:1579–84. doi: 10.1093/molbev/msq050. [DOI] [PubMed] [Google Scholar]
- 7.Lee SC, Corradi N, Byrnes EJ, 3rd, Torres-Martinez S, Dietrich FS, Keeling PJ, et al. Microsporidia evolved from ancestral sexual fungi. Curr Biol. 2008;18:1675–9. doi: 10.1016/j.cub.2008.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee SC, Corradi N, Doan S, Dietrich FS, Keeling PJ, Heitman J. Evolution of the sex-related locus and genomic features shared in microsporidia and fungi. PLoS ONE. 2010;5:e10539. doi: 10.1371/journal.pone.0010539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corradi N, Keeling PJ. Microsporidia: a journey through radical taxonomical revisions. Fungal Biol Rev. 2009;23:1–8. doi: 10.1016/j.fbr.2009.05.001. [DOI] [Google Scholar]
- 10.Gill EE, Fast NM. Assessing the microsporidia-fungi relationship: Combined phylogenetic analysis of eight genes. Gene. 2006;375:103–9. doi: 10.1016/j.gene.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 11.Corradi N, Slamovits CH. The intrigiung nature of microsporidian genomes. Brief Funct Genomics. 2011;10:115–24. doi: 10.1093/bfgp/elq032. [DOI] [PubMed] [Google Scholar]
- 12.Corradi N, Pombert JF, Farinelli L, Didier ES, Keeling PJ. The complete sequence of the smallest known nuclear genome from the microsporidian Encephalitozoon intestinalis. Nat Commun. 2010;1:77. doi: 10.1038/ncomms1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katinka MD, Duprat S, Cornillot E, Metenier G, Thomarat F, Prensier G, et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001;414:450–3. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- 14.Keeling PJ, Corradi N, Morrison HG, Haag KL, Ebert D, Weiss LM, et al. The reduced genome of the parasitic microsporidian Enterocytozoon bieneusi lacks genes for core carbon metabolism. Genome Biol Evol. 2010;2:304–9. doi: 10.1093/gbe/evq022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corradi N, Haag KL, Pombert JF, Ebert D, Keeling PJ. Draft genome sequence of the Daphnia pathogen Octosporea bayeri: insights into the gene content of a large microsporidian genome and a model for host-parasite interactions. Genome Biol. 2009;10:R106. doi: 10.1186/gb-2009-10-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsaousis AD, Kunji ER, Goldberg AV, Lucocq JM, Hirt RP, Embley TM. A novel route for ATP acquisition by the remnant mitochondria of Encephalitozoon cuniculi. Nature. 2008;453:553–6. doi: 10.1038/nature06903. [DOI] [PubMed] [Google Scholar]
- 17.Lee SC, Weiss LM, Heitman J. Generation of genetic diversity in microsporidia via sexual reproduction and horizontal gene transfer. Commun Integr Biol. 2009;2:414–7. doi: 10.4161/cib.2.5.8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richards TA, Hirt RP, Williams BA, Embley TM. Horizontal gene transfer and the evolution of parasitic protozoa. Protist. 2003;154:17–32. doi: 10.1078/143446103764928468. [DOI] [PubMed] [Google Scholar]
- 19.Fast NM, Law JS, Williams BA, Keeling PJ. Bacterial catalase in the microsporidian Nosema locustae: implications for microsporidian metabolism and genome evolution. Eukaryot Cell. 2003;2:1069–75. doi: 10.1128/EC.2.5.1069-1075.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiang H, Pan G, Vossbrinck CR, Zhang R, Xu J, Li T, et al. A tandem duplication of manganese superoxide dismutase in Nosema bombycis and its evolutionary origins. J Mol Evol. 2010;71:401–14. doi: 10.1007/s00239-010-9394-3. [DOI] [PubMed] [Google Scholar]
- 21.Slamovits CH, Keeling PJ. Class II photolyase in a microsporidian intracellular parasite. J Mol Biol. 2004;341:713–21. doi: 10.1016/j.jmb.2004.06.032. [DOI] [PubMed] [Google Scholar]
- 22.Selman M, Pombert JF, Solter L, Farinelli L, Weiss LM, Keeling P, et al. Acquisition of an animal gene by microsporidian intracellular parasites. Curr Biol. 2011;21:R576–7. doi: 10.1016/j.cub.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montgomery JA. Purine nucleoside phosphorylase: a target for drug design. Med Res Rev. 1993;13:209–28. doi: 10.1002/med.2610130302. [DOI] [PubMed] [Google Scholar]
- 24.Taylor EA, Rinaldo-Matthis A, Li L, Ghanem M, Hazleton KZ, Cassera MB, et al. Anopheles gambiae purine nucleoside phosphorylase: catalysis, structure, and inhibition. Biochemistry. 2007;46:12405–15. doi: 10.1021/bi7010256. [DOI] [PubMed] [Google Scholar]
- 25.Bzowska A, Kulikowska E, Shugar D. Purine nucleoside phosphorylases: properties, functions, and clinical aspects. Pharmacol Ther. 2000;88:349–425. doi: 10.1016/S0163-7258(00)00097-8. [DOI] [PubMed] [Google Scholar]
- 26.Morrison HG, McArthur AG, Gillin FD, Aley SB, Adam RD, Olsen GJ, et al. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science. 2007;317:1921–6. doi: 10.1126/science.1143837. [DOI] [PubMed] [Google Scholar]
- 27.Barbour AG, Putteet-Driver AD, Bunikis J. Horizontally acquired genes for purine salvage in Borrelia spp. causing relapsing fever. Infect Immun. 2005;73:6165–8. doi: 10.1128/IAI.73.9.6165-6168.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Striepen B, Pruijssers AJ, Huang J, Li C, Gubbels MJ, Umejiego NN, et al. Gene transfer in the evolution of parasite nucleotide biosynthesis. Proc Natl Acad Sci USA. 2004;101:3154–9. doi: 10.1073/pnas.0304686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun X, Bognar AL, Baker EN, Smith CA. Structural homologies with ATP- and folate-binding enzymes in the crystal structure of folylpolyglutamate synthetase. Proc Natl Acad Sci USA. 1998;95:6647–52. doi: 10.1073/pnas.95.12.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Didier ES, Didier PJ, Friedberg DN, Stenson SM, Orenstein JM, Yee RW, et al. Isolation and characterization of a new human microsporidian, Encephalitozoon hellem (n. sp.), from three AIDS patients with keratoconjunctivitis. J Infect Dis. 1991;163:617–21. doi: 10.1093/infdis/163.3.617. [DOI] [PubMed] [Google Scholar]
- 32.Johny S, Larson TM, Solter LF, Edwards KA, Whitman DW. Phylogenetic characterization of Encephalitozoon romaleae (Microsporidia) from a grasshopper host: relationship to Encephalitozoon spp. infecting humans. Infect Genet Evol. 2009;9:189–95. doi: 10.1016/j.meegid.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 33.Coyle CM, Weiss LM, Rhodes LV, 3rd, Cali A, Takvorian PM, Brown DF, et al. Fatal myositis due to the microsporidian Brachiola algerae, a mosquito pathogen. N Engl J Med. 2004;351:42–7. doi: 10.1056/NEJMoa032655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weidner E, Canning EU, Rutledge CR, Meek CL. Mosquito (Diptera: Culicidae) host compatibility and vector competency for the human myositic parasite Trachipleistophora hominis (Phylum Microspora) J Med Entomol. 1999;36:522–5. doi: 10.1093/jmedent/36.4.522. [DOI] [PubMed] [Google Scholar]
- 35.Vávra J, Kamler M, Modry D, Koudela B. Opportunistic nature of the mammalian microsporidia: experimental transmission of Trachipleistophora extenrec (Fungi: Microsporidia) between mammalian and insect hosts. Parasitol Res. 2011;108:1565–73. doi: 10.1007/s00436-010-2213-3. [DOI] [PubMed] [Google Scholar]
- 36.de Koning AP, Brinkman FS, Jones SJ, Keeling PJ. Lateral gene transfer and metabolic adaptation in the human parasite Trichomonas vaginalis. Mol Biol Evol. 2000;17:1769–73. doi: 10.1093/oxfordjournals.molbev.a026275. [DOI] [PubMed] [Google Scholar]
- 37.Huang J, Mullapudi N, Sicheritz-Ponten T, Kissinger JC. A first glimpse into the pattern and scale of gene transfer in Apicomplexa. Int J Parasitol. 2004;34:265–74. doi: 10.1016/j.ijpara.2003.11.025. [DOI] [PubMed] [Google Scholar]
- 38.Rogers M, Keeling PJ. Lateral transfer and recompartmentalization of Calvin cycle enzymes of plants and algae. J Mol Evol. 2004;58:367–75. doi: 10.1007/s00239-003-2558-7. [DOI] [PubMed] [Google Scholar]
- 39.Richards TA, Dacks JB, Jenkinson JM, Thornton CR, Talbot NJ. Evolution of filamentous plant pathogens: gene exchange across eukaryotic kingdoms. Curr Biol. 2006;16:1857–64. doi: 10.1016/j.cub.2006.07.052. [DOI] [PubMed] [Google Scholar]