Abstract
The extraordinary evolutionary success of transposable elements (TEs) invites us to question the nature of the co-evolutionary dynamics between TE and host. Although sometimes assumed to be wholly parasitic, TEs have penetrated and spread throughout eukaryotic genomes at a rate unparalleled by other parasites. This near-ubiquity, occurring despite the potentially deleterious effects of insertional mutagenesis, raises the possibility that a counterbalancing benefit exists for the host. Such a benefit may act at the population level to generate genomic diversity within a species and hence greater adaptability under new selective pressures, or at the level of primary gain for the individual. Recent studies have highlighted the occurrence of retrotransposition events in the germline and discovered a surprisingly high rate of mobilization in somatic cells. Here we examine the available evidence for somatic retrotransposition and discuss how this phenomenon may confer a selective advantage upon an individual or species.
Keywords: Alu, LINE-1, parasitism, retrotransposon, somatic retrotransposition, SVA, symbiotism, transposable element
Transposable elements are a prominent feature of our genetic heritage. In addition to providing nearly half of the human genome,1,2 TEs have generated numerous sequences that distinguish our DNA from that of other primates and more distant relatives.3,4 Whether these differences are a cause or effect of evolution, and whether TEs are parasitic or symbiotic mobile genetic elements, is the subject of long-term debate.5,6
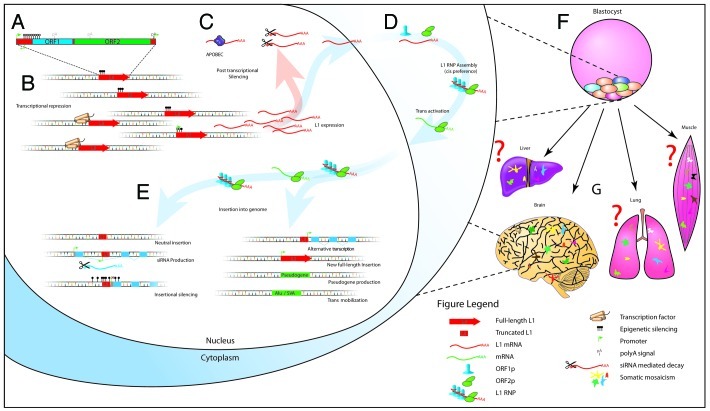
Three retrotransposon families remain mobile in the human genome: L1, Alu and SVA.7,8 Of these, L1 is considered the main driver of retrotransposition (Fig. 1A). Proteins translated from its two open reading frames mobilize L1 RNAs in cis9 as well as Alu, SVA and other RNAs incorporating a polyA tail in trans10-12 (Fig. 1D). Approximately 3,000 retrotransposons (~100 L1, ~3,000 Alu, < 100 SVA) are transposition-competent per individual,13 in contrast to the millions of immobile sequences produced by ancestral TEs.1
Figure 1. L1 is the main driver of retrotransposition in human cells. (A) L1 structure. ORF1 encodes an RNA-packaging protein and ORF2 encodes a protein (ORF2p) with endonuclease and reverse transcriptase domains.50,51 (B) Expression of L1 is limited by transcriptional repression and (C) post transcriptional regulation. (D) L1 ORF1p and ORF2p form an RNP with a marked cis preference,9 but ORF2p can also mobilize other RNAs with a polyA tail in trans. (E) Diverse effects of L1 insertional mutagenesis on gene expression. (F) L1 is known to be highly active during embryogenesis, and in neural cells (G), resulting in somatic mosaicism. Somatic retrotransposition in other adult tissues may also occur.
Other than a common pattern of near-exclusion from exons,14 the genomic distributions of L1, Alu and SVA are markedly different. L1 sequences are depleted in introns1 and very recent L1 insertions are more likely to be excluded from protein-coding genes than older insertions,14,15 suggesting that these events are strongly selected against.16,17 By contrast, recent Alu insertions are almost randomly distributed in the genome and SVA insertions are enriched in protein-coding genes.14 As noted above, the L1 machinery mediates L1, Alu and SVA mobilization, implying that each family is inserted in a similar genomic pattern and then redacted from the genome by natural selection depending on their impact. It is also possible that insertion site preference is modulated by unknown host factor interactions specific to each family.
An obvious consequence of insertional mutagenesis is genetic disease; TEs are associated with more than 75 human disorders.13,18 Likewise there are numerous documented cases of alternative transcripts and chimeric genes produced by TE insertions, often leading to expression of a host gene in a new spatiotemporal context19,20 (Fig. 1E). Several L1 sequence features, including a long polyA tail and strong internal 5′ and 3′ promoters19,21 can also dramatically alter the expression of a host gene in cases of intronic integration,17 while the epigenetic marks associated with L1 and other retrotransposons22 can modify chromatin state at integration sites and thereby drive rapid shifts in gene expression (Fig. 1E).
Given the multiple routes by which TEs can deleteriously alter the functional landscape of a genome, it is perhaps surprising that the global human population presents such a large number of dimorphic insertions.23 Recent studies using high-throughput sequencing (for reviews, see refs. 13, 24 and 25) have yielded a wealth of new insertion sites in healthy and diseased individuals, suggesting the full catalog of dimorphic and private insertions has been vastly underestimated and that roughly 1/20 live births harbor de novo retrotransposition events.
Most of these new insertions are thought to be neutral and are ultimately lost or fixed through genetic drift. The overall impact of the remaining insertions is likely to be overwhelmingly deleterious, raising the question of why retrotransposition is allowed to continue at an apparently high rate. More effective TE suppression would prevent harmful mutations, both in the germline and during somatic development.26 A model of successful parasitism would suggest that we have simply failed; that somehow despite a clear selective advantage to the host in silencing retrotransposons, L1 has managed to evade all attempts to prevent its activity. However, suppression of L1 has been effective during our recent evolution: less than 0.002% of human L1 copies are transposition-competent, and even fewer are frequently active or “hot”7,27. While the current state of L1 activity is a snapshot of a dynamic system, this could nonetheless suggest that it is evolutionarily advantageous to limit retrotransposition but not to totally eradicate it. For example, the South American rat genus Oryzomys28,29 has won this evolutionary arms race, apparently achieving L1 quiescence but, interestingly, this outcome coincides with a notable increase in karyotypic instability.30
This leads us to consider the position that regulated germline retrotransposition confers a benefit upon a host population. L1 provides clues for how this system may have co-evolved with the governing transcriptional programs of the host. Paradoxically, the canonical L1 promoter has retained motifs necessary for its transcriptional suppression in the germline and throughout development (e.g., SOX2 binding sites31,32) while new, usually 5′ truncated, L1 insertions are rapidly inactivated despite breaking free of the suppression inherent to the canonical L1 promoter33,34 (Fig. 1B and C). Thus, L1 maintains its own suppression but is not entirely silenced, leading to a tolerable rate of insertional mutagenesis while maintaining increased genomic malleability and genetic diversity that may be selected on when a population is strongly pressured (e.g., in the cases of pandemic or famine). For example, an L1-mediated TRIM5-CypA gene fusion35 following the divergence of Old and New World primates provides owl monkeys with HIV resistance not seen in other New World monkeys.
Nonetheless, a model founded exclusively upon observations of germline retrotransposition may be critically incomplete. We propose that L1 activity during ontogenesis36-38 (Fig. 1F and G) may serve to accelerate TE and host co-evolution. Recent reports suggest that the brain is a hotspot of somatic mosaicism caused by L1 mobilization during neurogenesis.31,39-41 If calculations of 80 somatic L1 insertions per neuron, of which there are ~1011 present in the human body,42 are even approximately accurate,31 then a single human individual may have more somatic L1 insertions than the total number of private germline L1 insertions in the global population. Informing this scenario further, we recently developed a technology to map somatic L1 insertions in human cells.39 Our principal conclusions were that these events preferentially impacted protein-coding genes expressed in the brain, that the hippocampus—as seen previously31—was particularly enriched for somatic retrotransposition and that neural cells indeed present43 a remarkable degree of somatic genome mosaicism. Despite this advance, numerous questions are yet to be answered, including (1) the timing of somatic L1 mobilization throughout life; (2) how many events occur per individual, organ or cell; (3) whether certain population groups are particularly affected; (4) which transcription factors govern L1 activation in somatic cells other than neurons and (5) whether the same rules that apply in germ cells (e.g., a limited number of “hot” donor elements and families7,27) also apply to somatic cells.
Moreover, as somatic events are by definition non-heritable, it is the propensity for L1 mobilization, rather than its consequences, on which natural selection may apply. If true, this may suggest that the brain is enriched for somatic mobilization as an innocent bystander in an evolutionary arms race occurring primarily in the germline. A large percentage of genes expressed in the brain are also expressed in the testis (the “brains and balls” phenomenon44), meaning that L1 transcription may be activated in somatic cells as an accident of evolution. The mutagenic effects of these insertions may then be simply tolerated by somatic cells; in addition to a reduction in impact due to heterozygosity, each mutation is expected to affect only a small sub-population of mature cells.
Another, more striking, possibility is that somatic retrotransposition confers some primary gain upon the individual host. As noted by others,45 Barbara McClintock’s celebrated discovery of transposition-derived kernel variegation in maize46 was also the first description of somatic mosaicism caused by a transposable element. Singer et al.43 more recently provided a compelling case for the potential action of L1 in producing somatic mosaicism in neural cells, resulting in greater genetic diversity and thus a greater variety of behavioral phenotypes in isogenic animals. As at the population level, genetic diversity may be beneficial at the cellular level. One classic example, driven by RAG proteins domesticated from an ancient transposon,47,48 is V(D)J recombination, where somatic rearrangements in immunoglobins and T-cell receptors49 provide genetic diversification crucial for the adaptive immune system.
The contribution of TEs to the fitness and success of species may not be limited to their well-documented effects on the genome mediated through germline retrotransposition. Their potential role in driving genetic diversity both within and between individuals adds yet another layer to the complex relationship between TEs and their hosts. Characterization of the regulation and functional impact of somatic retrotransposition is now feasible,39 and may soon settle debate on whether TEs are merely globally successful parasites, or diverse genomic symbiotes.
Acknowledgments
G.J.F. receives the support of a C.J. Martin Overseas Based Biomedical Fellowship from the Australian NHMRC (575585). J.K.B. is supported by a Wellcome Trust Clinical Fellowship (090385/Z/09/Z) through the Edinburgh Clinical Academic Track (ECAT). G.J.F. is funded by an Institute Strategic Programme Grant and a New Investigator Award from the British BBSRC (BB/H005935/1) and an EU FP7 Collaborative Project research grant (FP7-HEALTH-2010–259743).
Glossary
Abbreviations:
- TE
transposable element
- LINE- 1 or L1
long interspersed nuclear element
- ORF
open reading frame
Footnotes
Previously published online: www.landesbioscience.com/journals/mge/article/18422
References
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science. 2001;291:1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 3.Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 4.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–62. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 5.Britten RJ, Davidson EH. Gene regulation for higher cells: a theory. Science. 1969;165:349–57. doi: 10.1126/science.165.3891.349. [DOI] [PubMed] [Google Scholar]
- 6.Doolittle WF, Sapienza C. Selfish genes, the phenotype paradigm and genome evolution. Nature. 1980;284:601–3. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- 7.Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, et al. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci USA. 2003;100:5280–5. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mills RE, Bennett EA, Iskow RC, Devine SE. Which transposable elements are active in the human genome? Trends Genet. 2007;23:183–91. doi: 10.1016/j.tig.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, et al. Human L1 retrotransposition: cis preference versus trans complementation. Mol Cell Biol. 2001;21:1429–39. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 2003;35:41–8. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 11.Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat Genet. 2000;24:363–7. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 12.Hancks DC, Goodier JL, Mandal PK, Cheung LE, Kazazian HH., Jr Retrotransposition of marked SVA elements by human L1s in cultured cells. Hum Mol Genet. 2011;20:3386–400. doi: 10.1093/hmg/ddr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faulkner GJ. Retrotransposons: mobile and mutagenic from conception to death. FEBS Lett. 2011;585:1589–94. doi: 10.1016/j.febslet.2011.03.061. [DOI] [PubMed] [Google Scholar]
- 14.Stewart C, Kural D, Stromberg MP, Walker JA, Konkel MK, Stutz AM, et al. A comprehensive map of mobile element insertion polymorphisms in humans. PLoS Genet. 2011;7:e1002236. doi: 10.1371/journal.pgen.1002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ewing AD, Kazazian HH., Jr High-throughput sequencing reveals extensive variation in human-specific L1 content in individual human genomes. Genome Res. 2010;20:1262–70. doi: 10.1101/gr.106419.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boissinot S, Entezam A, Furano AV. Selection against deleterious LINE-1-containing loci in the human lineage. Mol Biol Evol. 2001;18:926–35. doi: 10.1093/oxfordjournals.molbev.a003893. [DOI] [PubMed] [Google Scholar]
- 17.Han JS, Szak ST, Boeke JD. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004;429:268–74. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- 18.Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3:370–9. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- 19.Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, et al. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet. 2009;41:563–71. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- 20.Lamprecht B, Walter K, Kreher S, Kumar R, Hummel M, Lenze D, et al. Derepression of an endogenous long terminal repeat activates the CSF1R proto-oncogene in human lymphoma. Nat Med 2010; 16:571-9, 1p following 9. [DOI] [PubMed]
- 21.Swergold GD. Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol Cell Biol. 1990;10:6718–29. doi: 10.1128/mcb.10.12.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lunyak VV, Prefontaine GG, Nunez E, Cramer T, Ju BG, Ohgi KA, et al. Developmentally regulated activation of a SINE B2 repeat as a domain boundary in organogenesis. Science. 2007;317:248–51. doi: 10.1126/science.1140871. [DOI] [PubMed] [Google Scholar]
- 23.Ewing AD, Kazazian HH., Jr Whole-genome resequencing allows detection of many rare LINE-1 insertion alleles in humans. Genome Res. 2011;21:985–90. doi: 10.1101/gr.114777.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beck CR, Garcia-Perez JL, Badge RM, Moran JV. LINE-1 Elements in Structural Variation and Disease. Annu Rev Genomics Hum Genet. 2011;12:187–215. doi: 10.1146/annurev-genom-082509-141802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ray DA, Batzer MA. Reading TE leaves: new approaches to the identification of transposable element insertions. Genome Res. 2011;21:813–20. doi: 10.1101/gr.110528.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miki Y, Nishisho I, Horii A, Miyoshi Y, Utsunomiya J, Kinzler KW, et al. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 1992;52:643–5. [PubMed] [Google Scholar]
- 27.Beck CR, Collier P, Macfarlane C, Malig M, Kidd JM, Eichler EE, et al. LINE-1 Retrotransposition Activity in Human Genomes. Cell. 2010;141:1159–70. doi: 10.1016/j.cell.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Casavant NC, Scott L, Cantrell MA, Wiggins LE, Baker RJ, Wichman HA. The end of the LINE?: lack of recent L1 activity in a group of South American rodents. Genetics. 2000;154:1809–17. doi: 10.1093/genetics/154.4.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grahn RA, Rinehart TA, Cantrell MA, Wichman HA. Extinction of LINE-1 activity coincident with a major mammalian radiation in rodents. Cytogenet Genome Res. 2005;110:407–15. doi: 10.1159/000084973. [DOI] [PubMed] [Google Scholar]
- 30.Koop BF, Baker RJ, Genoways HH. Numerous chromosomal polymorphisms in a natural population of rice rats (Oryzomys, Cricetidae) Cytogenet Cell Genet. 1983;35:131–5. doi: 10.1159/000131854. [DOI] [PubMed] [Google Scholar]
- 31.Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, et al. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–31. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tchénio T, Casella JF, Heidmann T. Members of the SRY family regulate the human LINE retrotransposons. Nucleic Acids Res. 2000;28:411–5. doi: 10.1093/nar/28.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Athanikar JN, Badge RM, Moran JVA. YY1-binding site is required for accurate human LINE-1 transcription initiation. Nucleic Acids Res. 2004;32:3846–55. doi: 10.1093/nar/gkh698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang N, Zhang L, Zhang Y, Kazazian HH., Jr An important role for RUNX3 in human L1 transcription and retrotransposition. Nucleic Acids Res. 2003;31:4929–40. doi: 10.1093/nar/gkg663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sayah DM, Sokolskaja E, Berthoux L, Luban J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430:569–73. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- 36.Garcia-Perez JL, Marchetto MC, Muotri AR, Coufal NG, Gage FH, O'Shea KS, et al. LINE-1 retrotransposition in human embryonic stem cells. Hum Mol Genet. 2007;16:1569–77. doi: 10.1093/hmg/ddm105. [DOI] [PubMed] [Google Scholar]
- 37.Kano H, Godoy I, Courtney C, Vetter MR, Gerton GL, Ostertag EM, et al. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 2009;23:1303–12. doi: 10.1101/gad.1803909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van den Hurk JA, Meij IC, Seleme MC, Kano H, Nikopoulos K, Hoefsloot LH, et al. L1 retrotransposition can occur early in human embryonic development. Hum Mol Genet. 2007;16:1587–92. doi: 10.1093/hmg/ddm108. [DOI] [PubMed] [Google Scholar]
- 39.Baillie JK, Barnett MW, Upton KR, Gerhardt DJ, Richmond TA, De Sapio F, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534–7. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–10. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 41.Muotri AR, Marchetto MC, Coufal NG, Oefner R, Yeo G, Nakashima K, et al. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468:443–6. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mattick JS, Taft RJ, Faulkner GJ. A global view of genomic information–moving beyond the gene and the master regulator. Trends Genet. 2010;26:21–8. doi: 10.1016/j.tig.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 43.Singer T, McConnell MJ, Marchetto MC, Coufal NG, Gage FH. LINE-1 retrotransposons: mediators of somatic variation in neuronal genomes? Trends Neurosci. 2010;33:345–54. doi: 10.1016/j.tins.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Graves JA. Review: Sex chromosome evolution and the expression of sex-specific genes in the placenta. Placenta. 2010;31(Suppl):S27–32. doi: 10.1016/j.placenta.2009.12.029. [DOI] [PubMed] [Google Scholar]
- 45.Levin HL, Moran JV. Dynamic interactions between transposable elements and their hosts. Nat Rev Genet. 2011;12:615–27. doi: 10.1038/nrg3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McClintock B. Controlling elements and the gene. Cold Spring Harb Symp Quant Biol. 1956;21:197–216. doi: 10.1101/sqb.1956.021.01.017. [DOI] [PubMed] [Google Scholar]
- 47.Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell. 1998;94:463–70. doi: 10.1016/S0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- 48.Melek M, Gellert M, van Gent DC. Rejoining of DNA by the RAG1 and RAG2 proteins. Science. 1998;280:301–3. doi: 10.1126/science.280.5361.301. [DOI] [PubMed] [Google Scholar]
- 49.Hozumi N, Tonegawa S. Evidence for somatic rearrangement of immunoglobulin genes coding for variable and constant regions. Proc Natl Acad Sci USA. 1976;73:3628–32. doi: 10.1073/pnas.73.10.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Doucet AJ, Hulme AE, Sahinovic E, Kulpa DA, Moldovan JB, Kopera HC, et al. Characterization of LINE-1 ribonucleoprotein particles. PLoS Genet. 2010;6:e1001150. doi: 10.1371/journal.pgen.1001150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khazina E, Truffault V, Buttner R, Schmidt S, Coles M, Weichenrieder O. Trimeric structure and flexibility of the L1ORF1 protein in human L1 retrotransposition. Nat Struct Mol Biol. 2011;18:1006–14. doi: 10.1038/nsmb.2097. [DOI] [PubMed] [Google Scholar]