Abstract
The transcriptomic response of A9:u-ATP9 and apetala3:u-ATP9 lines carrying a mitochondrial dysfunction in flower tissues has been characterized. Both lines showed an alteration in the transcription of several genes involved in carbon and nitrogen metabolism, stress responses, transcription factors and DNA binding proteins. Interestingly, several transcripts of photosynthetic-related genes were also affected in their expression such as the mRNAs encoding for chlorophyllase, chlorophyll binding proteins and a PSII. Moreover, chlorophyll levels were reduced and the Mg-dechelatase activity was increased, indicating an alteration in chlorophyll metabolism. Our results suggest that the mitochondrial dysfunction may also affect chloroplastic functions, and that our model could be useful to uncover retrograde signaling mechanisms operating between the three different plant genomes.
Keywords: Arabidopsis, chloroplasts, mitochondria, mitochondrial dysfunction
Plants require the contribution of three different genomes found in separate compartments. Chloroplasts and mitochondria, which are of endosymbiotic origin, contain only relatively few proteins encoded by their own genomes, following the transfer of a great part of the genetic material from the prokaryotic ancestors into the nucleus of the host. Consequently, most of the mitochondrial and chloroplast proteins are nuclear-encoded, synthesized in the cytoplasm and imported into organelles.1-3 Given that several cellular functions are performed by proteins encoded in different compartments, the existence of mechanisms that coordinate the expression of nuclear and organellar genes should be necessary. One important question concerns the character (identity) of the signals responsible for interorganellar cross-talk able to direct the expression of a set of nuclear genes.
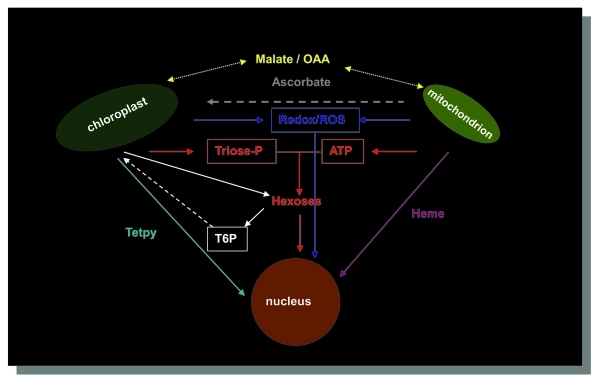
The intercompartment cross-talk includes anterograde (nucleus-to-organelle) and retrograde (organelle-to-nucleus) controls. Anterograde mechanisms are responsive to endogenous and environmental signals received by the kernel and coordinate the expression of genes in chloroplasts and mitochondria. Retrograde signaling regulates the expression of nuclear genes in response to the physiological state of organelles. Besides the cross-talk between chloroplasts/mitochondria and nucleus, interactions between chloroplast and mitochondria has been established during the evolution of plants to coordinate the activities of the two organelles which exhibit a high degree of metabolic independence4 (Fig. 1).
Figure 1.
Communication between the different compartments in higher plants. Ascorbate has been proposed to act as a mitochondrial signal for plastids. OAA; oxaloacetate; Tetpy: tetrapyrroles. T6P: threalose-6P. Adapted from4,25,26
Profound changes in gene expression of nuclear-encoded genes were observed in Arabidopsis plants exhibiting impaired mitochondrial function.5-12 The recent study of the transcriptome in Arabidopsis plants carrying a mitochondrial dysfunction, revealed important modifications in the expression of some genes from the carbon metabolic pathways with inhibition of glycolysis and the induction of the MDH alternative pathways.13 This model, where the mitochondrial flaw was induced by the expression of the unedited form of the ATP synthase subunit 9 (u-ATP9),8 is useful to uncover the interactions between organelles in plant cells.
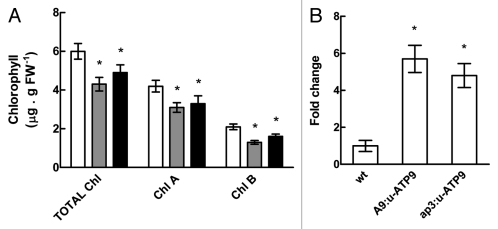
A striking fact was the changes observed in the expression of genes related to carbon and nitrogen metabolism, and some genes involved in stress responses in plants. Particularly interesting is the behavior of some photosynthesis-related genes. To explore this prospect, we quantified chlorophylls from A9:u-ATP9 and apetala3:u-ATP9 transgenic plants according to the method of Moran.14 It was noticed that u-ATP9 transgenic plants showed a decrease of about 25% in the levels of chlorophyll A and B, and about 30% for total chlorophylls, per gram of fresh weight compared with untransformed control plants, suggesting that the production of chlorophyll is impaired or that there is an increase in its degradation (Fig. 2A).
Figure 2.
(A) Determination of total chlorophyll, chlorophyll A and B levels in wt (white bars), A9:u-ATP9 (gray bars) and ap3:u-ATP9 (black bars) flowers (stage 12). Values are the mean ± SD of four independent replicates. (B) qRT-PCR analysis of AtCLH2 gene (At5g43860) in flowers (stage 12) from wt, A9:u-ATP9 and ap3:u-ATP9 lines. Columns represent mean values (error bars ± SD) of three independent experiments. Relative expression levels are shown as fold change values with respect to β-actin mRNA levels. The asterisk signals a statistically different result from the control value (p < 0.05).
It has been described that the biosynthesis and the degradation of chlorophylls occur through different processes.15 Among relevant enzymes in chlorophyll catabolism, chlorophyllase (CLH) which catalyzes the hydrolysis of ester bond to yield chlorophyllide and phytol, was found affected in the microarray experiments of u- ATP9 plants, particularly AtCLH2 (At5g43860) one of two CLHs from Arabidopsis.13 Previous studies indicate that AtCLH2 is constitutively expressed throughout leaf development, and that its expression is unaffected by stress or senescence.16-18 In contrast with these observations, we found an increase of 6 and 5 times in AtCLH2 mRNA levels in A9:u-ATP9 and ap3:u-ATP9 plants respectively, compared with wild type plants (Fig. 2B) which agrees with the results of microarray data.13
An indicator of the chlorophyll breakdown is the release of Mg atoms from these molecules. The Mg-dechelatase activity is performed either by heat stable low-molecular weight metal chelating substances (MCS) or by Mg-releasing proteins (MRP), which differs by their substrate specificity.19,20 We found that A9:u-ATP9 and ap3:u-ATP9 plants presented an increase of 25 and 40% of the Mg-dechelatase activity respectively compared with wild-type plants (Fig. 3).
Figure 3.
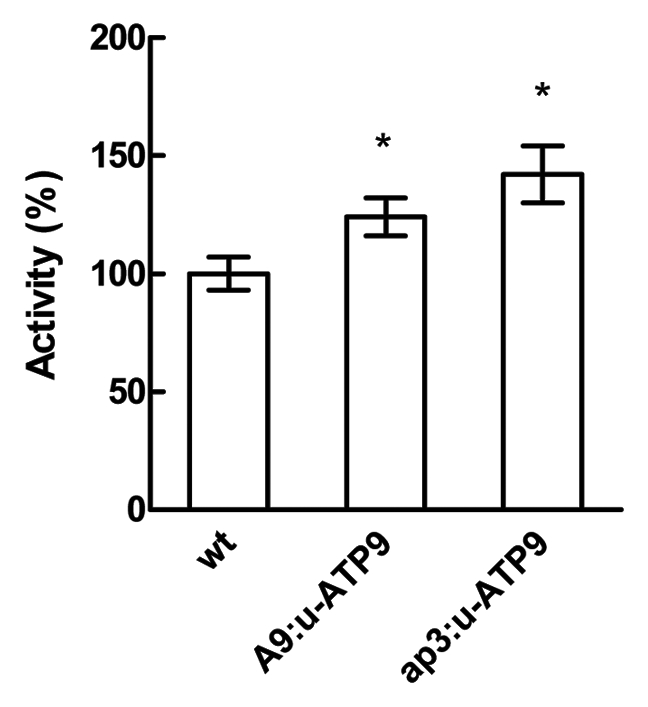
Magnesium dechelatase activity from wild type (wt), A9:u-ATP9 and ap3:u-ATP9 lines analyzed in flowers extracts (stage 12). 100% of activity represents 0.03 ∆Abs686 . g−1 FW s−1. Values are the mean ± SD of four independent replicates. The asterisk indicates values statistically different from the control (p < 0.05).
It has been reported that the mRNA levels of some antenna proteins were increased in plants with mitochondrial dysfunction.13 Particularly, the LCHI type II gene (At1g19150) coding for the chlorophyll A-B binding protein, which transfers the energy absorbed by chlorophylls to photochemical reaction centers. The qRT-PCR analysis of LCHI mRNA levels showing an increase of about 5-fold in both, A9:u-ATP9 and ap3:u-ATP9 lines compared with wild type (Fig. 4) confirms the upregulation observed in microarray experiments. An induction of about 3-fold was also detected for the mRNAs of PSBQ2 (AT4g05180). This gene encodes the PsbQ subunit of the oxygen evolving complex of photosystem II, and this transcriptional response indicates that photosynthesis is affected (Fig. 4).
Figure 4.
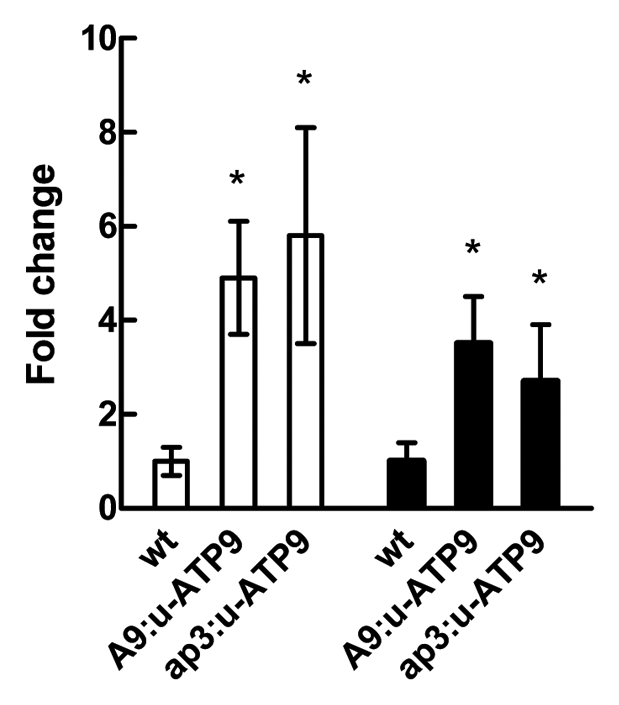
qRT-PCR analysis of LCHI gene (white bars) and PSBQ2 gene (black bars) in flowers (stage 12) from wt, A9:u-ATP9 and ap3:u-ATP9 lines. Columns represent mean values (error bars ± SD) of three independent experiments. Relative expression levels are shown as fold change values with respect to β-actin mRNA levels. The asterisk signals a statistically different result from the control value (p < 0.05).
Since, u-ATP9 transgenic plants present altered levels of ROS and ascorbic acid concomitant with the increase in the mRNA levels of PER50 (At4g37520) and PER57 (At5g17820), it is plausible to consider that the reduced levels of chlorophyll might result from bleaching by peroxidases. These enzymes are found in several subcellular compartments, including chloroplasts. Although the role of peroxidases in chlorophyll degradation is controversial, several studies have supported their possible participation in chlorophyll catabolism.21
An interesting point of our work is the fact that mitochondrial dysfunction in transgenic plants, induced by an “unedited” version of the ATPase subunit 9 gene can affect photosynthesis by reducing the chlorophyll levels.13 The chlorophyll metabolic pathway has been associated with nuclear expression control.22 It has been postulated that the tetrapyrrole intermediate Mg-protoporphyrin IX acts as a signal molecule in one of the signaling pathways between the chloroplast and the nucleus and chloroplasts and mitochondria, and the accumulation of this metabolite is necessary to regulate the expression of several nuclear genes encoding proteins associated with photoshythesis.23,24 In fact, the degradation of these pigments may be due to both, the increase of Mg- dechelatase activity (nuclear response to mitochondrial dysfunction) and ROS accumulation (mitochondrial product acting on chloroplasts), causing dysfunction of the light-harvesting (antenna) complex. A possible consequence of the chloroplasts dysfunction is the induction of a nuclear response that causes increased expression of LCHI and PBSQ2 genes.
There are several reports that address the relationship between mitochondrial respiration, photosynthesis and chloroplast functions. The respiration process provides energy for biosynthesis, and its balance with photosynthesis determines the rate of plant biomass accumulation. These interactions involve transcriptional control, co-localization of proteins, distribution of biochemical pathways between organelles, and the impact of substrate and product concentrations (metabolic shuttles).25 The increased levels of malate observed in our experiments suggest it like another metabolite involved in the mitochondria/chloroplast communication.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgements
We are grateful to Evelyne Lepage and Cindi Aknin for excellent technical assistance. This work was supported by grants from PICS-CNRS 3641, Universite Victor Segalen Bordeaux 2, ANPCyT (PICT 00614 and 0729) and Provincia de Santa Fe. MVB and DFGC are research members from CONICET.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/18050
References
- 1.Adams KL, Daley DO, Qiu YL, Whelan J, Palmer JD. Repeated, recent and diverse transfers of a mitochondrial gene to the nucleus in flowering plants. Nature. 2000;408:354–7. doi: 10.1038/35042567. [DOI] [PubMed] [Google Scholar]
- 2.Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476–81. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- 3.Kakizaki T, Inaba T. New insights into the retrograde signaling pathway between the plastids and the nucleus. Plant Signal Behav. 2010;5:196–9. doi: 10.4161/psb.5.2.11107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leister D. Genomics-based dissection of the cross-talk of chloroplasts with the nucleus and mitochondria in Arabidopsis. Gene. 2005;354:110–6. doi: 10.1016/j.gene.2005.03.039. [DOI] [PubMed] [Google Scholar]
- 5.Busi MV, Gomez-Casati DF, Perales M, Araya A, Zabaleta E. Nuclear-encoded mitochondrial complex I gene expression is restored to normal levels by inhibition of unedited ATP9 transgene expression in Arabidopsis thaliana. Plant Physiol Biochem. 2006;44:1–6. doi: 10.1016/j.plaphy.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 6.Busi MV, Maliandi MV, Valdez H, Clemente M, Zabaleta EJ, Araya A, et al. Deficiency of Arabidopsis thaliana frataxin alters activity of mitochondrial Fe-S proteins and induces oxidative stress. Plant J. 2006;48:873–82. doi: 10.1111/j.1365-313X.2006.02923.x. [DOI] [PubMed] [Google Scholar]
- 7.Dutilleul C, Garmier M, Noctor G, Mathieu C, Chetrit P, Foyer CH, et al. Leaf mitochondria modulate whole cell redox homeostasis, set antioxidant capacity, and determine stress resistance through altered signaling and diurnal regulation. Plant Cell. 2003;15:1212–26. doi: 10.1105/tpc.009464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gómez-Casati DF, Busi MV, Gonzalez-Schain N, Mouras A, Zabaleta EJ, Araya A. A mitochondrial dysfunction induces the expression of nuclear-encoded complex I genes in engineered male sterile Arabidopsis thaliana. FEBS Lett. 2002;532:70–4. doi: 10.1016/S0014-5793(02)03631-1. [DOI] [PubMed] [Google Scholar]
- 9.Gutierres S, Sabar M, Lelandais C, Chetrit P, Diolez P, Degand H, et al. Lack of mitochondrial and nuclear-encoded subunits of complex I and alteration of the respiratory chain in Nicotiana sylvestris mitochondrial deletion mutants. Proc Natl Acad Sci USA. 1997;94:3436–41. doi: 10.1073/pnas.94.7.3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.James AM, Sheard PW, Wei YH, Murphy MP. Decreased ATP synthesis is phenotypically expressed during increased energy demand in fibroblasts containing mitochondrial tRNA mutations. Eur J Biochem. 1999;259:462–9. doi: 10.1046/j.1432-1327.1999.00066.x. [DOI] [PubMed] [Google Scholar]
- 11.Noctor G, Dutilleul C, De Paepe R, Foyer CH. Use of mitochondrial electron transport mutants to evaluate the effects of redox state on photosynthesis, stress tolerance and the integration of carbon/nitrogen metabolism. J Exp Bot. 2004;55:49–57. doi: 10.1093/jxb/erh021. [DOI] [PubMed] [Google Scholar]
- 12.Rius SP, Casati P, Iglesias AA, Gomez-Casati DF. Characterization of Arabidopsis thaliana lines deficient in GAPC-1, a cytosolic NAD-dependent glyceraldehyde 3-phosphate dehydrogenase. Plant Physiol. 2008;148:1655–67. doi: 10.1104/pp.108.128769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Busi MV, Gomez-Lobato ME, Rius SP, Turowski VR, Casati P, Zabaleta EJ, et al. Effect of mitochondrial dysfunction on carbon metabolism and gene expression in flower tissues of Arabidopsis thaliana. Mol Plant. 2011;4:127–43. doi: 10.1093/mp/ssq065. [DOI] [PubMed] [Google Scholar]
- 14.Moran R. Formulae for determination of chlorophyllous pigments extracted with n,n-dimethylformamide. Plant Physiol. 1982;69:1376–81. doi: 10.1104/pp.69.6.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matile P, Hortensteiner S, Thomas H, Krautler B. Chlorophyll Breakdown in Senescent Leaves. Plant Physiol. 1996;112:1403–9. doi: 10.1104/pp.112.4.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benedetti CE, Arruda P. Altering the expression of the chlorophyllase gene ATHCOR1 in transgenic Arabidopsis caused changes in the chlorophyll-to-chlorophyllide ratio. Plant Physiol. 2002;128:1255–63. doi: 10.1104/pp.010813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park SY, Yu JW, Park JS, Li J, Yoo SC, Lee NY, et al. The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell. 2007;19:1649–64. doi: 10.1105/tpc.106.044891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsuchiya T, Ohta H, Okawa K, Iwamatsu A, Shimada H, Masuda T, et al. Cloning of chlorophyllase, the key enzyme in chlorophyll degradation: finding of a lipase motif and the induction by methyl jasmonate. Proc Natl Acad Sci USA. 1999;96:15362–7. doi: 10.1073/pnas.96.26.15362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vicentini F, Iteen F, Matile P. Develoment of an assay for Mg-dechelatase of oilseed rape cotyledons using chlorophyllin as the substrate. Physiol Plant. 1995;94:57–63. doi: 10.1111/j.1399-3054.1995.tb00784.x. [DOI] [Google Scholar]
- 20.Kunieda T, Amano T, Shioi Y. Search for chlorophyll degradation enzymes, Mg-dechelatase, from extracts of Chenopodium album withy native and artificial substrates. Plant Sci. 2005;169:177–83. doi: 10.1016/j.plantsci.2005.03.010. [DOI] [Google Scholar]
- 21.Martinez GA, Civello PM, Chaves AR, Añon MC. Characterization of peroxidases-mediated chlorophyll bleaching in strawberry fruit. Phytochemistry. 2001;58:379–87. doi: 10.1016/S0031-9422(01)00266-7. [DOI] [PubMed] [Google Scholar]
- 22.Rintamaki E, Lepisto A, Kangasjarvi S. Implication of chlorophyll biosynthesis on chloroplast-to-nucleus retrograde signaling. Plant Signal Behav. 2009;4:545–7. doi: 10.4161/psb.4.6.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strand A, Asami T, Alonso J, Ecker JR, Chory J. Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature. 2003;421:79–83. doi: 10.1038/nature01204. [DOI] [PubMed] [Google Scholar]
- 24.Kanesaki Y, Kobayashi Y, Hanaoka M, Tanaka K. Mg-protoporphyrin IX signaling in Cyanidioschyzon merolae: multiple pathways may involve the retrograde signaling in plant cells. Plant Signal Behav. 2009;4:1190–2. doi: 10.4161/psb.4.12.10061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Millar AH, Whelan J, Soole KL, Day DA. Organization and regulation of mitochondrial respiration in plants. Annu Rev Plant Biol. 2011;62:79–104. doi: 10.1146/annurev-arplant-042110-103857. [DOI] [PubMed] [Google Scholar]
- 26.Pesaresi P, Schneider A, Kleine T, Leister D. Interorganellar communication. Curr Opin Plant Biol. 2007;10:600–6. doi: 10.1016/j.pbi.2007.07.007. [DOI] [PubMed] [Google Scholar]