Abstract
The root endodermis is the cylindrical boundary that separates the inner vascular tissue from the outer cortex and functions as an apoplasmic barrier for selective nutrient uptake. Recent developmental and cell biological studies have started to reveal the mechanisms by which this single cell layer serves as a key regulatory module of root growth, tissue patterning and nutrient flow, which in concert support the plant’s ability to survive in a terrestrial habitat. This review provides an overview of the key factors that contribute to the functioning of the root endodermis and discusses how this single cell layer dictates root growth and tissue patterning.
Keywords: casparian strip, Endodermis, gibberellin, GRAS, HD-ZIP III, microRNA, pattern formation, root
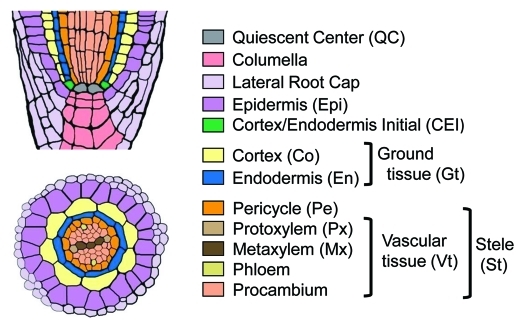
The roots of vascular plants are composed of functionally distinct tissues organized into a concentric pattern of cell layers.1 Although both the number and morphology of each tissue vary with plant species, the overall organization of root tissue patterns are well conserved, indicating their importance for basic root functions, such as anchorage of the plant and nutritional uptake. Figure 1 shows a schematic view of the Arabidopsis root tissue organization.1,2 The roots of certain plant species, such as rice, maize and tobacco, exhibit more complex patterns, mainly due to the presence of additional cortical layers.3
Figure 1.
Schematic representation of Arabidopsis root tissue organization (reproduced from ref2). Cell types are shown in different colors, and their abbreviations are in parentheses.
Laser ablation and mosaic analyses indicated that stem cell maintenance and tissue patterning are regulated by cell-cell communication.4-6 Recent molecular genetic studies using the Arabidopsis root as a model system have started to reveal the molecular identity of the intercellular signals involved in root growth and tissue patterning, and highlight the importance of the endodermis as a central regulator of these processes. Moreover, a search for genes specifically expressed in the endodermis has led to the identification of the elusive components of the Casparian strip (CS), the cell wall structure that serves as an impermeable apoplasmic barrier to nutrients and water. This minireview introduces recent advances in our understanding of the mechanisms by which the root endodermis differentiates and how this single-celled layer controls root growth and tissue patterning.
Formation of the Root Endodermis
In Arabidopsis, the developmental origin of the root endodermis can be traced back to triangular-stage embryos, where the ground meristem, the progenitor of the ground tissue (endodermis and cortex), divides periclinally to form two-layered ground tissue primordia.7 After germination, this two-layered ground tissue organization is maintained by the stereotypical cell division of the cortex/endodermis initials (CEIs) and their immediate daughter cells (CEIDs) (Fig. One and 2D).1 An unidentified signal that possibly emanates from the adjacent quiescent center (QC) maintains the pluripotent stem cell capacity of the CEI.6 The CEI divides transversely to give rise to a CEID at the distal position from the QC, whereas the daughter cell abutting the QC maintains its CEI identity. The CEID divides periclinally to give rise to two daughter cells, each of which differentiates into either an endodermis or cortex cell, depending on its position relative to the stele.
Two GRAS-type transcription factors, SHORTROOT (SHR) and SCARECROW (SCR), play key roles in the formation and maintenance of the root endodermis. SHR proteins are produced in the stele and move to the adjacent cell layer, which is composed of the QC, CEI, CEID and endodermis (Fig. 2D).8,9 In this layer, SHR activates the transcription of several target genes, including SCR, and the SCR protein thus formed physically interacts with SHR, likely forming a transcription activation complex.10,11 In the CEID, the SHR-SCR complex activates the transcription of a cell cycle regulator, Cyclin D6;1, which in turn is required for the periclinal division of CEID (Fig. 2D).12 Thus, loss-of-function shr and scr mutants are unable to form the two-layered ground tissue. The ground tissue of the shr mutant does not possess the differentiated attributes of the endodermis, whereas that of scr contains at least partial characteristics of the endodermis.8,13 Therefore, the functioning of SHR, but not of SCR, is required for the differentiation of the endodermis. Consistent with this observation, the forced expression of SHR outside the stele results in the ectopic manifestation of endodermal characteristics.9
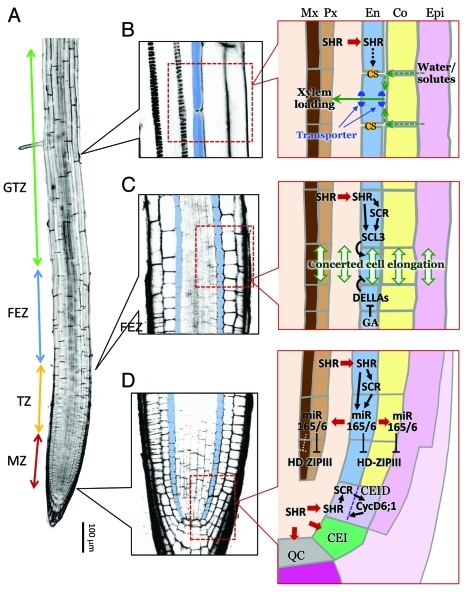
Figure 2.
Developmental zones of the Arabidopsis root and the signal transduction pathways and nutrient flow in the region of the root endodermis. (A) A confocal section of an Arabidopsis root. The four developmental zones, i.e., the meristematic zone (MZ), transition zone (TZ), fast elongation zone (FEZ), and growth terminating zone (GTZ) are labeled. (B-D) Magnified views of selected root regions (left; blue color indicates endodermis) and schematic representations of water/solute flow and developmental signaling in the endodermis (right). (B) Formation of the CS (represented as orange dots) and its role in regulating water and solute uptake. (C) GA- and SCL3-mediated control of endodermal cell elongation and its effect on the elongation of adjacent cell layers. (D) SHR- and miR165/6-mediated intercellular signaling between the endodermis and surrounding tissues. Arrows indicate activation/promotion, and T bars represent inhibition/suppression. Thick red arrows indicate cell-cell trafficking.
Whereas the number of cortical layers varies with plant species and the age of the root, the endodermis is typically composed of a single cell layer. This evolutionarily and developmentally conserved property of the root endodermis is likely due to the restricted cell-cell movement of SHR to one cell distance.10 Loss of SCR protein results in increased SHR movement and ectopic periclinal divisions, indicating that SCR captures SHR protein in the nucleus of the recipient cell layer, and thereby inhibits further cell-cell movement of SHR. Interestingly, this regulatory mechanism seems to be conserved in rice, indicating that the mechanism that ensures the formation of the single endodermis layer is evolutionarily conserved.10
The Endodermis Emits microRNAs for Tissue Patterning
Whereas functional studies of SHR and SCR demonstrated that a mobile transcription factor specifies endodermal cell fate in a position-dependent manner, the signaling pathway that controls root radial patterning also involves other non-cell-autonomous factors that function downstream of the SHR-SCR module. Recently, SHR was shown to activate the transcription of three MICRORNA165/6 genes, MIR165A, MIR166A and MIR166B, specifically in the single cell layer composed of the QC, CEI, CEID and endodermis (Fig. 2D),2,14 an expression pattern precisely coinciding with that of SCR.8,13 As expected, levels of mature miR165/6 were dramatically reduced in both shr and scr mutants, suggesting that MIR165/6 expression is regulated by the SHR/SCR transcription factor complex.2,14 Consistent with this hypothesis, chromatin immunoprecipitation (ChIP) analysis indicated that SHR binds to the 5′ upstream regions of MIR165A and MIR166B.14
MicroRNA165/166 (miR165/6) target the mRNA of five Class III HOMEODOMAIN LEUCINE ZIPPER (HD-ZIPIII) genes, PHABULOSA (PHB), PHAVOLUTA (PHV), REVOLUTA/INTERFASCICULAR FIBERLESS1 (REV/IFL1), CORONA/INCURVATA4 (CNA/ICU4) and ATHB8.15-20 In Arabidopsis roots, the expression of all HD-ZIPIII mRNA is confined to the vascular tissue by the action of miR165/6-dependent regulation.14 Loss-of-function mutants of a single HD-ZIP III gene exhibit no discernible defect in root tissue pattern,14 whereas miR-resistant gain-of-function alleles, especially those of PHB (phb-d alleles), exhibit severe patterning defects in a broad range of root cell layers, including the cortex, endodermis, pericycle and xylem vessels.2,14 Most notably, differentiation of the two xylem cell types, protoxylem (Px) and metaxylem (Mx), is disturbed in phb-d mutants, with Mx occupying the region where Px normally forms. This same phenotype is also observed in shr and scr roots, where the level of miR165/6 is reduced and hence HD-ZIPIII expression is slightly expanded relative to the wild type. In contrast, quadruple loss-of-function mutants of the HD-ZIPIII genes, as well as transgenic plants overexpressing miR165 in the stele, form supernumerary Px files at the expense of Mx.14 These data suggest that the differentiation of the two xylem cell types is determined by the dosage of HD-ZIPIII TFs in the central stele, which in turn is defined in a non-cell-autonomous fashion by the miR165/6 derived from the endodermis.
The mode of non-cell-autonomous miR165 action has been characterized quantitatively by manipulating the level of miR165 production in the ground tissue and correlating it with PHB expression patterns and xylem differentiation in the stele.2 The level of miR165 in the ground tissue was indeed found to regulate the graded distribution of PHB across the stele, as well as the differentiation of Px and Mx. Moreover, this study revealed that the ground tissue-derived miR165 (and possibly also miR166) suppresses the expression of PHB in the pericycle and cortex, and that this suppression is essential for the correct differentiation of the pericycle and cortex. Therefore, SHR/SCR-dependent activation of miR165/6 production in the endodermis not only specifies xylem cell types in the stele, but also controls a broader range of cell differentiation in Arabidopsis roots.2
It is noteworthy that the miR165/6-dependent suppression of HD-ZIPIII has been suggested to play a key role in establishing apical-basal polarity during embryogenesis, dorsoventral patterning of leaf primordia and shoot vascular organization.17,21,22 A particularly interesting research question is whether the non-cell-autonomous function of miR165/6 also operates during these developmental events, or whether it is a specific feature of the root endodermis.
The Endodermis as an Integrator of Growth-regulating Hormonal Signals
Root growth is a consequence of two distinct processes that take place in spatially distinct regions along the root; i.e., (1) the production of new cells in the meristematic zone (MZ) at the root tip and (2) the directional cell expansion that initiates in the transition zone (TZ) and accelerates in the fast elongation zone (FEZ) (Fig. 2A).23The exit of root cells from the MZ is thought to be controlled by the interplay between the two plant hormones, auxin and cytokinin.24 The concerted action of several PIN-FORMED (PIN) auxin efflux carriers establishes a cyclic flow of auxin in the root tip region encompassing the MZ and TZ.23,25 A high concentration of auxin is thus maintained in the MZ and promotes root cell division.25 Cytokinin antagonizes this auxin-mediated cell division activity by promoting the expression of SHY2, an inhibitor of auxin signaling in the TZ. This leads to the repression of PIN expression, and hence promotes the transition from cell division to cell elongation.24
Recently, another class of plant hormone, the gibberellins (GAs), emerged as a player in the hormonal network that controls root growth.26 A key event in the GA signal transduction pathway is the proteasome-dependent degradation of DELLA repressor proteins (see ref27 for a review). DELLA proteins are members of the GRAS family of transcription factors, and suppress GA-responsive gene expression by interacting with other transcription regulators. The Arabidopsis genome encodes five DELLA proteins, i.e., GAI, RGA, RGL1, RGL2 and RGL3. The conserved DELLA motif mediates the interaction between these proteins and the GA-bound form of the soluble GA receptor, GA INSENSITIVE DWARF1 (GID1), and thereby targets DELLA proteins for proteasome-dependent degradation. Deletion of the DELLA motif renders DELLA proteins non-degradable and hence constitutively blocks GA signaling.
Ubeda-Tomas et al. (2008) analyzed the effect of blocking GA signaling in selected root tissues by the targeted expression of non-degradable GAI (designated gai).28 They found that expression of gai in tissues that included the endodermis (or the endodermis alone) resulted in root growth retardation, whereas gai expression in other root tissues had no effect. The endodermis-specific effect of gai expression on the growth of the entire root is likely due to the highly responsive nature of endodermal cells to modulation of GA signaling. Because root cell layers are interconnected by cell walls, reduced endodermal cell elongation should retard growth of the entire root. Based on these observations, the authors proposed that GA signaling in the endodermis plays a key role in the regulation of whole root growth (Fig. 2C).28
These same authors later reported another role of GA signaling in the endodermis, namely the promotion of cell division in the MZ, which is required to enlarge the MZ during the first few days after germination.29 An independent study by Achard et al. (2009) reached a similar conclusion as to the role of GA signaling in the control of root meristem size.30 This latter study further reported that two types of Cyclin-dependent kinase (CDK) inhibitors (CDKIs), Kip-related protein2 (KRP2) and a SIAMESE (SIM) family protein, were upregulated in the GA-deficient ga1 mutants, and that this phenotype was cancelled by quadruple-DELLA mutations.30 It is currently unknown whether blocking GA signaling in the endodermis affects the local expression of CDKI in the endodermis or whether it modulates CDKI levels in broader cell layers in the MZ.
It is not clear why the endodermis exhibits increased sensitivity to the inhibition of GA signaling. Two recent studies identified a GRAS transcription factor, SCARECROW-LIKE3 (SCL3), as a key modulator of the GA response in the endodermis (Fig. 2C).31,32 Transcription of SCL3 is spatially restricted to the endodermis by the SHR/SCR transcription complex,10,11 whereas the level of SCL3 is controlled by GA through a DELLA protein, RGA.33 Genetic evidence indicated that SCL3 is a positive regulator of the GA response and acts as an attenuator of DELLA repressors; loss-of-function scl3 and SCL3-overexpressing roots were, respectively, more susceptible to and resistant against endodermis-specific gai expression than were wild-type roots. Although a detailed mechanistic understanding of the downstream signaling pathways that translate endodermal GA signaling to growth of the entire root awaits future studies, the preferential expression of a DELLA attenuator, SCL3, in the endodermis suggests that the root endodermis has increased sensitivity to GA-mediated growth-regulating signaling, and is capable of integrating the hormone signaling pathways that govern root growth.
The Establishment of Endodermal Cell Polarity and Casparian Strip
The most distinctive feature of the root endodermis is the presence of the Casparian strip (CS), a focused deposition of cell wall materials that prevents apoplasmic flow of water-soluble substances between the cortex and stele (Fig. 2B).3 Although the chemical constituents of the CS have been known for over six decades, little is known about the mechanism by which this cell wall structure is formed along the equatorial position in the endodermis.3 A recent study provided the first molecular clue as to how this process is controlled. Roppolo et al. (2011)34 sought to identify the genes that are specifically required for the development of the root endodermis by consulting the gene expression map of the Arabidopsis root.35,36 This approach identified five genes (later designated CASP1-CASP5) that encode members of an uncharacterized plant-specific protein family (UPF0497).34 This gene family consists of 38 members in Arabidopsis, among which only CASP1–5 are expressed in a pattern consistent with their specific involvement in root endodermal function. CASP1–5 are predicted to be transmembrane proteins and share a conserved amino acid sequence in their putative extracellular domains. GFP fusion proteins of CASP1–5 localized to the membrane domain precisely beneath the CS (designated CS membrane domain, CSD), and their expression preceded the establishment of the apoplasmic barrier of the endodermis, indicating that CASPs not merely mark the preformed CS, but actively participate in CS formation. In accordance with this view, casp1 casp3 double mutants failed to accumulate CS constituents at the predicted CSD positions.34
What is the function of CASPs in CS formation? CASPs appear to cycle between the plasma membrane and endomembranes via BFA-sensitive trafficking pathways during the early phases of endodermis differentiation, but are later immobilized at the CSD. Once immobilized, CASPs become insolubilized at the CSD. At least some CASPs physically interact with each other, and ectopic expression of CASP5-GFP altered the morphology of the ER membrane. Roppolo et al. (2011) proposed that CASPs form a scaffold at the CSD that serves as a platform for the localization of the cell wall biosynthetic machinery that is required for the formation of the CS.34 The chemical nature of the predicted CASP scaffold and the components of the wall-producing complex, however, have yet to be identified.
The mechanisms by which CASPs are targeted to the CSD are also unknown. When ectopically expressed outside the endodermis, CASPs spread over the entire plasma membrane, indicating that unknown endodermis-specific factors specify the CSD and/or recruit CASPs.34 Earlier, the group that conducted this study had demonstrated that endodermal cells are intrinsically polarized, by visualizing the polar localization of two boron transporters, NIP5;1 and BOR1, to the outer and inner PM domains of the endodermis, respectively.37 Whereas these transporters already exhibit partial polar localization in embryos and root meristematic cells, which exist prior to CS formation, their polar localization becomes more tightly regulated as the CS forms.34,37 These observations suggest that the CSD not only functions as a structural platform for the accumulation of cell wall materials, but also serves as a membrane boundary that separates the inner and outer surfaces of the endodermal cell.
Conclusions and Perspectives
Since the identification of SHR and SCR transcription factors as master regulators of root ground tissue patterning,8,13 considerable progress has been made in our understanding of how these transcription factors and their downstream components act in the formation of the endodermis, and in the specification of remote cell types, such as xylem vessels. These studies revealed the existence of a non-cell-autonomous signaling pathway that is mediated by a mobile transcription factor and microRNAs. In addition, the highly specific expression of SCR contributed to the construction of the reliable gene expression map of Arabidopsis roots from which Roppolo et al. (2011) identified CASPs, the first structural components of the CS.34 However, the molecular mechanisms that link the SHR/SCR module to the expression of CASPs as well as the identity of the other genes required for the functional specialization of the root endodermis are unknown. As reported in the CASP study,34 structural proteins are likely encoded by highly redundant genes. Therefore, extensive reverse genetic studies are necessary to identify these genes. The large number of downstream factors identified for the SHR/SCR module and the public availability of the Arabidopsis root expression map are useful tools for the identification of novel factors required for the functional specialization of the root endodermis.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/18079
References
- 1.Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, et al. Cellular organisation of the Arabidopsis thaliana root. Development. 1993;119:71–84. doi: 10.1242/dev.119.1.71. [DOI] [PubMed] [Google Scholar]
- 2.Miyashima S, Koi S, Hashimoto T, Nakajima K. Non-cell-autonomous microRNA165 acts in a dose-dependent manner to regulate multiple differentiation status in the Arabidopsis root. Development. 2011;138:2303–13. doi: 10.1242/dev.060491. [DOI] [PubMed] [Google Scholar]
- 3.Esau K. The root: Primary state of growth. Anatomy of Seed Plants. New York: John Wiley & Sons, 1977:215-42. [Google Scholar]
- 4.Kidner C, Sundaresan V, Roberts K, Dolan L. Clonal analysis of the Arabidopsis root confirms that position, not lineage, determines cell fate. Planta. 2000;211:191–9. doi: 10.1007/s004250000284. [DOI] [PubMed] [Google Scholar]
- 5.van den Berg C, Willemsen V, Hage W, Weisbeek P, Scheres B. Cell fate in the Arabidopsis root meristem determined by directional signalling. Nature. 1995;378:62–5. doi: 10.1038/378062a0. [DOI] [PubMed] [Google Scholar]
- 6.van den Berg C, Willemsen V, Hendriks G, Weisbeek P, Scheres B. Short-range control of cell differentiation in the Arabidopsis root meristem. Nature. 1997;390:287–9. doi: 10.1038/36856. [DOI] [PubMed] [Google Scholar]
- 7.Scheres B, Wolkenfelt H, Willemsen V, Terlouw M, Lawson E, Dean C, et al. Embryonic origin of the Arabidopsis primary root and root meristem initials. Development. 1994;120:2475–87. [Google Scholar]
- 8.Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, et al. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell. 2000;101:555–67. doi: 10.1016/S0092-8674(00)80865-X. [DOI] [PubMed] [Google Scholar]
- 9.Nakajima K, Sena G, Nawy T, Benfey PN. Intercellular movement of the putative transcription factor SHR in root patterning. Nature. 2001;413:307–11. doi: 10.1038/35095061. [DOI] [PubMed] [Google Scholar]
- 10.Cui H, Levesque MP, Vernoux T, Jung JW, Paquette AJ, Gallagher KL, et al. An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science. 2007;316:421–5. doi: 10.1126/science.1139531. [DOI] [PubMed] [Google Scholar]
- 11.Levesque MP, Vernoux T, Busch W, Cui H, Wang JY, Blilou I, et al. Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol. 2006;4:e143. doi: 10.1371/journal.pbio.0040143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sozzani R, Cui H, Moreno-Risueno MA, Busch W, Van Norman JM, Vernoux T, et al. Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature. 2010;466:128–32. doi: 10.1038/nature09143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, et al. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell. 1996;86:423–33. doi: 10.1016/S0092-8674(00)80115-4. [DOI] [PubMed] [Google Scholar]
- 14.Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, Zhou J, et al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature. 2010;465:316–21. doi: 10.1038/nature08977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baima S, Nobili F, Sessa G, Lucchetti S, Ruberti I, Morelli G. The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development. 1995;121:4171–82. doi: 10.1242/dev.121.12.4171. [DOI] [PubMed] [Google Scholar]
- 16.Green KA, Prigge MJ, Katzman RB, Clark SE. CORONA, a Member of the Class III Homeodomain Leucine Zipper Gene Family in Arabidopsis, Regulates Stem Cell Specification and Organogenesis. Plant Cell. 2005;17:691–704. doi: 10.1105/tpc.104.026179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature. 2001;411:709–13. doi: 10.1038/35079635. [DOI] [PubMed] [Google Scholar]
- 18.Ochando I, Jover-Gil S, Ripoll JJ, Candela H, Vera A, Ponce MR, et al. Mutations in the microRNA complementarity site of the INCURVATA4 gene perturb meristem function and adaxialize lateral organs in arabidopsis. Plant Physiol. 2006;141:607–19. doi: 10.1104/pp.106.077149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prigge MJ, Otsuga D, Alonso JM, Ecker JR, Drews GN, Clark SE. Class III Homeodomain-Leucine Zipper Gene Family Members Have Overlapping, Antagonistic, and Distinct Roles in Arabidopsis Development. Plant Cell. 2005;17:61–76. doi: 10.1105/tpc.104.026161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Talbert PB, Adler HT, Parks DW, Comai L. The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development. 1995;121:2723–35. doi: 10.1242/dev.121.9.2723. [DOI] [PubMed] [Google Scholar]
- 21.Smith ZR, Long JA. Control of Arabidopsis apical-basal embryo polarity by antagonistic transcription factors. Nature. 2010;464:423–6. doi: 10.1038/nature08843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, et al. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol. 2003;13:1768–74. doi: 10.1016/j.cub.2003.09.035. [DOI] [PubMed] [Google Scholar]
- 23.Verbelen JP, De Cnodder T, Le J, Vissenberg K, Baluska F. The Root Apex of Arabidopsis thaliana Consists of Four Distinct Zones of Growth Activities: Meristematic Zone, Transition Zone, Fast Elongation Zone and Growth Terminating Zone. Plant Signal Behav. 2006;1:296–304. doi: 10.4161/psb.1.6.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dello Ioio R, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, Morita MT, et al. A genetic framework for the control of cell division and differentiation in the root meristem. Science. 2008;322:1380–4. doi: 10.1126/science.1164147. [DOI] [PubMed] [Google Scholar]
- 25.Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, et al. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature. 2005;433:39–44. doi: 10.1038/nature03184. [DOI] [PubMed] [Google Scholar]
- 26.Moubayidin L, Perilli S, Dello Ioio R, Di Mambro R, Costantino P, Sabatini S. The rate of cell differentiation controls the Arabidopsis root meristem growth phase. Curr Biol. 2010;20:1138–43. doi: 10.1016/j.cub.2010.05.035. [DOI] [PubMed] [Google Scholar]
- 27.Harberd NP, Belfield E, Yasumura Y. The angiosperm gibberellin-GID1-DELLA growth regulatory mechanism: how an “inhibitor of an inhibitor” enables flexible response to fluctuating environments. Plant Cell. 2009;21:1328–39. doi: 10.1105/tpc.109.066969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ubeda-Tomás S, Swarup R, Coates J, Swarup K, Laplaze L, Beemster GT, et al. Root growth in Arabidopsis requires gibberellin/DELLA signalling in the endodermis. Nat Cell Biol. 2008;10:625–8. doi: 10.1038/ncb1726. [DOI] [PubMed] [Google Scholar]
- 29.Ubeda-Tomás S, Federici F, Casimiro I, Beemster GT, Bhalerao R, Swarup R, et al. Gibberellin signaling in the endodermis controls Arabidopsis root meristem size. Curr Biol. 2009;19:1194–9. doi: 10.1016/j.cub.2009.06.023. [DOI] [PubMed] [Google Scholar]
- 30.Achard P, Gusti A, Cheminant S, Alioua M, Dhondt S, Coppens F, et al. Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr Biol. 2009;19:1188–93. doi: 10.1016/j.cub.2009.05.059. [DOI] [PubMed] [Google Scholar]
- 31.Heo JO, Chang KS, Kim IA, Lee MH, Lee SA, Song SK, et al. Funneling of gibberellin signaling by the GRAS transcription regulator scarecrow-like 3 in the Arabidopsis root. Proc Natl Acad Sci USA. 2011;108:2166–71. doi: 10.1073/pnas.1012215108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang ZL, Ogawa M, Fleet CM, Zentella R, Hu J, Heo JO, et al. Scarecrow-like 3 promotes gibberellin signaling by antagonizing master growth repressor DELLA in Arabidopsis. Proc Natl Acad Sci USA. 2011;108:2160–5. doi: 10.1073/pnas.1012232108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zentella R, Zhang ZL, Park M, Thomas SG, Endo A, Murase K, et al. Global analysis of della direct targets in early gibberellin signaling in Arabidopsis. Plant Cell. 2007;19:3037–57. doi: 10.1105/tpc.107.054999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roppolo D, De Rybel B, Tendon VD, Pfister A, Alassimone J, Vermeer JE, et al. A novel protein family mediates Casparian strip formation in the endodermis. Nature. 2011;473:380–3. doi: 10.1038/nature10070. [DOI] [PubMed] [Google Scholar]
- 35.Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, et al. A gene expression map of the Arabidopsis root. Science. 2003;302:1956–60. doi: 10.1126/science.1090022. [DOI] [PubMed] [Google Scholar]
- 36.Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–6. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- 37.Alassimone J, Naseer S, Geldner N. A developmental framework for endodermal differentiation and polarity. Proc Natl Acad Sci USA. 2010;107:5214–9. doi: 10.1073/pnas.0910772107. [DOI] [PMC free article] [PubMed] [Google Scholar]