Abstract
During seed development, endosperm cells of highly productive cereals, including rice, synthesize disulfide-rich proteins in large amounts and deposit them into storage organelles. Disulfide bond formation involves electron transfer and generates H2O2 as a by-product. To ensure proper development and maturation of seeds, the endosperm cells must supply large amounts of oxidizing equivalents to dithiols in nascent proteins in a controlled manner. This review compares multiple oxidative protein folding systems in yeast, cultured human cells, and rice endosperm. We discuss possible roles of ERO1, other sulfhydryl oxidases, and the protein disulfide isomerase family in the formation of disulfide bonds in storage proteins and the development of protein bodies. Rice prolamins, encoded by a multigene family, are divided into Cys-rich and Cys-depleted subgroups. We discuss the potential importance of disulfide bond formation in the evolution of the prolamin family in japonica rice.
Keywords: electron transfer, ERO1, hydrogen peroxide, protein disulfide bond, protein disulfide isomerase, ROS, sulfhydryl oxidase
Introduction
Prokaryotes and eukaryotes have evolved distinct electron transfer systems for disulfide bond formation, such as DsbA-DsbB in the periplasm of Escherichia coli,1 ERO1-PDI in the endoplasmic reticulum (ER),2 and Erv1-Mia40 in the intermembrane space of mitochondria.3 Protein disulfide isomerase (PDI), which is a ubiquitous housekeeping enzyme found in all eukaryotic cells, is the principal catalyst for the formation of native disulfide bonds, functioning as the direct donor of disulfides to polypeptides in dithiol-disulfide exchange reactions.4 The genomes of rice (Oryza sativa), Arabidopsis (Arabidopsis thaliana), and maize (Zea mays) each encode about 20 members of the PDI family.5 However, it remains unknown how the plant cells establish an ER redox environment favorable for oxidation of PDI and various substrates that are physicochemically and structurally diverse.
Because rice endosperm synthesizes large amounts of disulfide-rich storage proteins during seed development, it provides an excellent system for studying how the cell can maintain an oxidizing environment in the ER. This review briefly summarizes the function of multiple systems for oxidative protein folding and focuses in particular on the functions of the oxidoreductase ERO1 and specific members of the PDI family. We then discuss the possible relationship between the formation of disulfides and the evolution of the prolamin family in japonica rice.
Rice Seed Proteins and Development of Protein Storage Organelles
During seed development, rice endosperm cells synthesize large amounts of disulfide-bonded storage proteins which differ considerably in structure and physicochemical properties: glutelins (acid- or alkaline-soluble 11S-type globulins), prolamins (highly hydrophobic and alcohol-soluble), and α-globulin (saline-soluble).6
Disulfide bond formation plays a critical role in accumulating storage proteins in protein bodies (PBs). Prolamins are polymerized through intermolecular disulfide bonds and packed into the ER-derived type-I PB (PB-I; spherical structure with a diameter of 1–2 μm).7-9 The 10-kDa Cys-rich prolamins (Os03 g0766100) are concentrated at the center core region of PB-I and the 13-kDa Cys-poor prolamins (Os05 g0329100) are distributed mainly to the outer layers of PB-I.10,11 In contrast, glutelin precursors (proglutelins) and α-globulin acquire intramolecular disulfide bonds and then exit the ER for delivery to the protein storage vacuole type-II PB (PB-II; crystalloid structure with a diameter of 2–4 μm). Proglutelins that are targeted to PB-II are processed into acidic and basic subunits by a vacuolar processing enzyme (VPE) and accumulate in the form of a higher-order complex through intermolecular disulfide bonds and hydrophobic interactions.7,12-14 Inside PB-II, mature glutelins and α-globulin are predominantly segregated to the crystalloids and matrix, respectively.9 Recent studies indicated that the processing of proglutelins is necessary for the formation of crystalloids14 and that lowering the α-globulin level leads to malformed PB-II.15
In addition to oxidative protein folding, RNA targeting and intracellular trafficking systems play important roles in the regulation of PB development in rice endosperm. mRNAs encoding glutelins and prolamins are targeted to discrete subdomains of the ER.16,17 Proglutelins are targeted from the ER to PB-II via the Golgi apparatus.18 Recent studies showed that a small GTPase protein, Rab5a, plays a critical role in the vesicular trafficking of proglutelins to PB-II.19,20
The ERO1-Dependent Electron Transfer Pathway: Roles in Disulfide Bond Formation and Development of Protein Bodies in Rice Endosperm
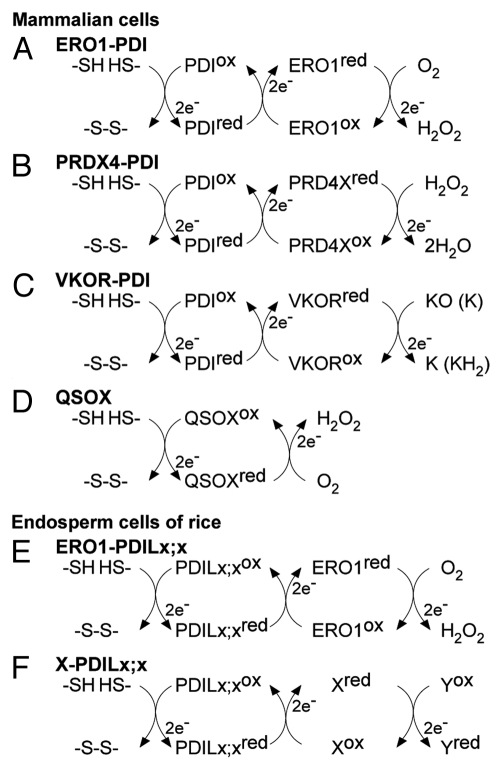
Oxidative protein folding of secretory proteins in the ER has been extensively studied in the yeast Saccharomyces cerevisiae and in cultured human cells.2 As shown in Figure 1A, the oxidizing power for the oxidation of a pair of sulfhydryls in a nascent secretory protein is directly supplied by oxidized PDI.4 The reduced form of PDI is then reoxidized by the FAD-containing sulfhydryl oxidase ERO1.21,22 The net products of the electron transfer are one disulfide bond and one molecule of H2O2. Structural studies on yeast ERO1 have identified the active and shuttle Cys pairs.23 Besides these catalytic Cys residues, ERO1 proteins contain regulatory Cys residues, which play an important role in redox-dependent feedback regulation of ERO1 activity in response to fluctuations in the ER redox environment.24,25
Figure 1.
Multiple electron transfer pathways for oxidative protein folding in the ER of mammalian and endosperm cells. Listed pathways are (A) ERO1-PDI, (B) peroxiredoxin IV (PRDX4)-PDI, (C) vitamin K epoxide reductase (VKOR)-PDI, and (D) quiescin sulfhydryl oxidase (QSOX), which all facilitate oxidative protein folding in the ER of mammalian cells. In the ER of rice endosperm, it is unlikely that OsERO1 directly oxidizes the active sites of OsPDIL1;1. Instead, it is plausible that OsERO1 oxidizes as-yet-unidentified members of the PDI family, indicated by PDILx;x, to promote the oxidative folding of storage proteins (E). Although there is no direct evidence, other oxidoreductases (X) and some members of the PDI family, indicated by PDILx;x, may also operate in oxidative folding of storage proteins (F). Yox represents unidentified electron acceptors, such as O2, H2O2, and quinone (F). KO, vitamin K epoxide; K, quinone; KH2, hydroquinone.
Whole-genome sequencing has identified members of sulfhydryl oxidase families, including ERO1 and PDI, in the genomes of rice, Arabidopsis, and maize. When RNAi knockdown of rice ERO1 (OsERO1) is induced under the control of an endosperm-specific promoter, the formation of native disulfide bonds in proglutelins is inhibited, and instead the formation of nonnative intermolecular disulfide bonds is promoted, leading to the formation of proglutelin-containing aggregates.26 Endosperm cells with severely reduced levels of OsERO1 exhibit defects in the development of PB-I and PB-II, and instead form abnormal aggregates of small particles.26
ERO1 proteins are tightly associated with the luminal face of the ER membrane in yeast27 through the C-terminal tail of 127-aa residues,28 whereas human ERO1α and ERO1β are retained in the ER lumen by covalent interactions with PDI and ERp44, another member of the PDI family.29 In rice endosperm, OsERO1 is an ER-localized integral membrane protein, and the ER localization of OsERO1 depends on a hydrophobic sequence present in the N-terminal region.26 The mechanism by which OsERO1 is retained in the ER membrane remains unclear.
Distinct but Coordinated Functions of PDI Family Members in Rice Endosperm
The PDI family members vary widely in the organization of the catalytic thioredoxin (TRX)-fold domains (a and a′), which have redox-active Cys-x-x-Cys motifs, and the noncatalytic TRX-like domains (b and b′). The genomes of the higher plants rice, Arabidopsis, and maize, each contain at least seven members of the PDI family with two redox-active sites, PDIL1;1−1;4 and PDIL2;1−2;3.5 PDIL1;1−1;4 members have an a-b-b′-a′ domain organization, similar to that of human PDI and ERp57 and yeast Pdi1p, a U-shaped molecule in which the two active-site Cys pairs face each other.30 PDIL2;1−2;3 members, however, have an a-a′-b domain organization, similar to that of human ER protein 5 (called P5), in which two redox-active TRX-like domains repeat in tandem followed by a single redox-inactive TRX domain.31 Members of the PDI and P5 subfamilies are likely to have evolved to acquire distinct redox activities. Human P5 shows lower reductase activity (using insulin as substrate) and chaperone activity than human PDI.32,33 Rice OsPDIL2;3, which belongs to the P5 subfamily, exhibits lower activity for oxidative folding of α-globulin in vitro than does OsPDIL1;1, a PDI subfamily member.10 However, recombinant OsPDIL2;3 exhibits higher sulfhydryl oxidase activity for the formation of nonnative disulfide bonds between α-globulin Cys79Phe mutant proteins.10
Multiple lines of evidence support specific functions of the PDI family members. In human cells, PDI prevents neurotoxicity associated with ER stress and with protein misfolding in Parkinson disease.34 Human P5 promotes tumor immune evasion by reducing disulfide bonds in the major histocompatibility complex class I–related ligand MICA on the surface of tumor cells35 and promotes breast cancer progression by activating ErbB2 and phosphoinositide-3-kinase signaling.36 In Arabidopsis, AtPDIL1;1 regulates the timing of programmed cell death in the endothelial cells,37 whereas AtPDIL2;1 plays a role in embryo sac development.38 Although the distinct expression patterns and catalytic activities of the PDI members suggest a potential for specific functions, the actual specificity or redundancy of the individual PDI members in vivo remains an open question.
In a recent study, we demonstrated that OsPDIL1;1 and OsPDIL2;3, when expressed in the same rice cell, play distinct roles in the formation of the two types of PBs. OsPDIL1;1 is evenly distributed within the dilated ER and plays an important role in the formation of disulfide bonds in the PB-II-targeted storage proteins, including proglutelins and α-globulin.26 In contrast, OsPDIL2;3 is mostly localized on the surface of PB-I and has a specific function in the intermolecular disulfide bond–assisted assembly of prolamin polypeptides in PB-I.10,26 In addition to these distinct roles of OsPDIL1;1 and OsPDIL2;3, their coordinated functions are required for the development of PB-I and PB-II in the endosperm. The activity of OsPDIL1;1 indirectly supports the assembly of prolamin species in the ER.10 Additionally, OsPDIL1;1 may be involved in the formation of a structural disulfide bond in the b domain of OsPDIL2;3.10
In yeast, among the active sites of PDI family members present in the ER, the CxxC active site in the N-terminal TRX domain of Pdi1p is preferentially oxidized by ERO1.39 Surprisingly, complementation analysis suggests that the a and a′ domains of OsPDIL1;1 may independently assist the oxidative folding of proglutelins through reduction and/or isomerization reactions.10 Because ERO1 oxidizes only the defined active sites of the PDI family members in yeast and mammalian cells, it is unlikely that OsERO1 directly oxidizes the active sites of OsPDIL1;1 (Fig. 1E). Alternatively, if OsERO1 does oxidize the active sites of OsPDIL1;1, it is likely that other oxidases also provide oxidizing power for proglutelins via electron transfer pathways that do not involve OsPDIL1;1 (Fig. 1F).
ERO1-Independent Disulfide Formation and Other Candidate Sulfhydryl Oxidases
In simple eukaryotes, such as yeast, the lack of ERO1 impairs cell viability.27,40 Mammalian and plant ERO1 proteins also function in the process of disulfide bond formation in the ER. Nevertheless, when the functions of two mouse paralogues of human ERO1, ERO1α and ERO1β, are simultaneously impaired, the mice are still viable although having a mild diabetic phenotype.41 ERO1-knockdown endosperm cells of rice also produce a reduced, but substantial, amount of disulfide bonds in storage proteins.
In addition to ERO1, other sulfhydryl oxidases have emerged as candidates for providing oxidizing power for disulfide bond formation in mammalian cells. Peroxiredoxin IV (PRDX4) oxidizes PDI to promote the formation of disulfide bonds in substrates, using H2O2 as the source of oxidizing power (Fig. 1B).42 Plants also have the peroxiredoxin family, but no ER-localized peroxiredoxin has been identified yet.43 Another system is the vitamin K epoxide reductase (VKOR)–dependent electron transfer system (Fig. 1C). Using oxidized vitamin K, VKOR interacts specifically with membrane-anchored PDI family members to transfer an oxidizing equivalent to PDI for disulfide bond formation in substrates.44-46 Homologs of VKOR are present in a variety of organisms, spanning from bacteria, archaea, vertebrates, and plants.47 Additionally, quiescin sulfhydryl oxidase (QSOX) functions as a sulfhydryl oxidase in mammalian cells (Fig. 1D).48 It is important to note that QSOX can directly oxidize dithiols of substrate proteins49 and is localized in structures involved in the secretory pathway, including the Golgi and secretory granules.50,51
In rice endosperm, the redox-active form of OsPDIL2;3 is localized mainly on the PB-I surface, whereas the redox-inactive form dissociates from PB-I and is localized in the ER lumen.10 These results indicate that not only the activity of OsPDIL2;3 but also the specific localization in the ER is regulated by the redox status at the active sites. As mentioned above, because ERO1 oxidizes only the defined active sites of PDI family members in yeast and mammalian cells, and because S. cerevisiae does not have an apparent P5 ortholog in its genome, it seems likely that an oxidase other than OsERO1 serves as a source of oxidizing power for OsPDIL2;3. Further studies are needed to identify the direct donor of oxidizing power to OsPDIL2;3 and to elucidate the distinct and overlapping roles of OsERO1 and other sulfhydryl oxidases (Fig. 1F).
Roles of O2 and H2O2 in Oxidative Protein Folding
The source of the oxidizing power in the yeast ERO1-PDI system is O2. ERO1 accepts electrons from the active site of PDI and transfers them to O2 via the FAD cofactor, producing H2O2 as a by-product (Fig. 1A).52 By using an H2O2-specific fluorescence probe (BES-H2O2) in the living cells of developing endosperm of rice, we showed that the oxidation of sulfhydryl groups is accompanied by the production of H2O2 in the ER. The homozygous mutant seeds of the rice EM49 line have fewer sulfhydryl groups for disulfide bonds than the wild-type because they accumulate Cys-rich prolamins at markedly reduced levels.26,53 Homozygous EM49 mutant seeds produce H2O2 in the ER at a substantially reduced level compared with the wild-type.26
High concentrations of H2O2, a reactive oxygen species (ROS), may cause peroxidation of the membrane lipid and gradually deteriorate the membrane integrity, leading to the leakage of small molecules, including water.54 Conversely, EM49 homozygous mutant seeds, which produce less H2O2, exhibit markedly slower desiccation and maturation than the wild-type.26 Although ROS, including H2O2, are cytotoxic, they also play key roles as signal transduction messengers in eukaryote cells.55 The 3n endosperm cells, but not the 2n embryo, are destined for programmed cell death during the late stages of seed maturation. The generated H2O2 in the ER may have a role as a signal for inducing programmed endosperm cell death and subsequent seed desiccation/maturation. If so, uncontrolled formation of disulfide bonds could lead to premature cell death in the endosperm; therefore, the quantity and timing of disulfide bond formation in storage proteins may affect the seed maturation, and hence the seed quality.
Disulfide Bond Formation and Evolution of the Cys-Depleted Prolamin Subfamily in japonica Rice
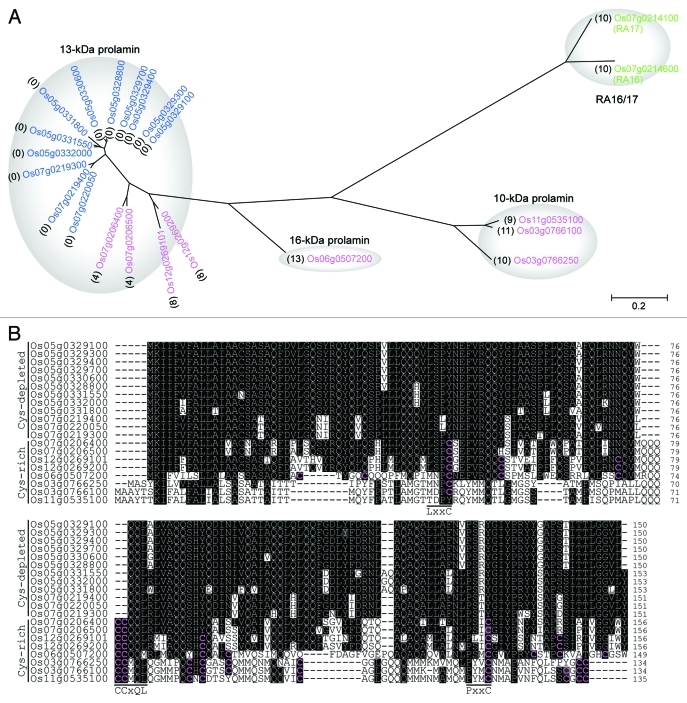
Rice prolamins are encoded by a multigene family and have been conventionally divided into Cys-rich and Cys-poor prolamins.56 We searched the genome of japonica rice (Nipponbare) and conducted a phylogenetic analysis of prolamins together with protein allergens of RA16/1757 as an outgroup. RA16/17 are water-soluble monomers containing intramolecular disulfide bonds.10 Surprisingly, although prolamins and RA16/17 proteins are substantially different in physicochemical properties, RA16/17 proteins are more closely related to prolamins than expected. The phylogenetic tree indicates that prolamins are clustered into three subgroups: (1) 13-kDa prolamins containing 0 to 8 Cys residues, (2) 10-kDa prolamins containing 9 to 11 Cys residues, and (3) a 16-kDa prolamin containing 13 Cys residues (Fig. 2A). These findings suggest that the 10-kDa and the 13-Da/16-kDa subgroups diverged before the 13-kDa and the 16-kDa subgroups branched away from each other, and that the evolution and diversification of prolamins are relatively recent events.
Figure 2.
Comparison of the amino acid sequences of prolamins and protein allergens from rice seeds. (A) Phylogenetic analysis of Cys-rich prolamins, Cys-depleted prolamins, and protein allergens of RA16/17. The phylogenetic tree was constructed by the neighbor-joining method using MEGA version 458 (http://www.megasoftware.net). The reliability of different phylogenetic groupings was evaluated by the bootstrap test (1000 replicates). Magenta, blue, and green letters indicate Cys-rich prolamins, Cys-depleted prolamins, and RA16/17 proteins, respectively. Numbers in parentheses indicate the numbers of Cys residues in the predicted mature-sized proteins. The NCBI accession numbers are Os03 g0766100, NP_001051380; Os03 g0766250, NP_001173654; Os05 g0328800, NP_001055211; Os05 g0329100, NP_001055213; Os05 g0329300, NP_001055214; Os05 g0329400, NP_001055215; Os05 g0329700, NP_001055216; Os05 g0330600, NP_001055218; Os05 g0331550, NP_001174359; Os05 g0331800, NP_001055221; Os05 g0332000, NP_001055222; Os06 g0507200, NP_001057724; Os07 g0206400, NP_001059151; Os07 g0206500, NP_001059152; Os07 g0219300, NP_001059197; Os07 g0219400, NP_001059198; Os07 g0220050, NP_001175103; Os11 g0535100, NP_001068024; Os12 g0269101, NP_001176891; Os12 g0269200, NP_001066544. (B) The amino acid sequences of Cys-rich and Cys-depleted prolamins were aligned with the CLUSTALW program. The genes of Os03 g0766100 and Os05 g0329100 encode the Cys-rich 10-kDa prolamin (crP10) and Cys-depleted 13-kDa prolamin (cpP13), respectively, analyzed in our recent paper.10 Magenta-on-black letters indicate Cys residues in the predicted mature-sized proteins. White-on-black letters indicate amino acid residues conserved in more than 10 of the sequences analyzed.
Members of the prolamin superfamily, which includes the 2S albumin, contain conserved ABC peptide regions, and each region contains conserved sequence motifs: LxxC in the A region, CCxQL in the B region, and PxxC in the C region.9 Interestingly, the 13-kDa Cys-poor prolamins of rice contain replacements or deletion of the Cys residues in these conserved sequence motifs: LxxY in the A region (Cys is substituted by Tyr),–xQL in the B region (Cys pair is deleted), and PxxY in the C region (Cys is substituted by Tyr) (Fig. 2B). We therefore hereafter call these Cys-poor prolamins Cys-depleted prolamins to reflect that the Cys residues were selectively mutated within otherwise conserved motifs. Ushijima et al.59 identified a group of Cys-depleted prolamin genes on chromosomes 5 and 7 of japonica rice and elegantly showed that the cluster of Cys-depleted prolamin genes on chromosome 5 of japonica rice (Kinmaze) is not present in the corresponding chromosomal segment of indica rice (Kasalath). The phylogenetic tree indicates that the amino acid sequences of Cys-depleted prolamins show higher homology to the Cys-rich prolamins Os07 g0206400 and Os07 g0206500 than to the other Cys-rich prolamins (Fig. 2A). Although speculative, it is plausible that the evolution of Cys-depleted prolamins in japonica rice involved a series of mutations at the Cys residues on an ancestral Cys-rich prolamin closely related to Os07 g0206400 and Os07 g0206500. It is also likely that the resulting Cys-depleted, mutated prolamin gene was subsequently multiplied on chromosomes 7 and 5.
Because the critical mutations at the Cys residues that led to the generation of the Cys-depleted prolamin subfamily occurred at otherwise conserved regions in the Cys-rich prolamin (Fig. 2B), it is conceivable that these mutations were under strong selective pressure. The Cys-depleted prolamins constitute a high proportion of the total prolamin fraction, and their knockdown causes a reduction in the PB-I size.60 The mutations at the Cys residues in prolamins and a rapid increase in copy numbers of Cys-depleted prolamin genes could reduce the concentration of H2O2 in the ER and increase storage substances in PB-I. The evolution of the Cys-depleted prolamin subfamily in japonica rice could have reduced the total amount of disulfide bonds in storage proteins, which may have contributed to the establishment of an appropriate redox environment in the secretory system for the development and maturation of seeds. The domestication of highly productive crops, including rice, has involved genetic modifications to meet the needs of humans.61 We speculate that the dynamic genetic modifications that led to the generation of the Cys-depleted prolamin subfamily played a role in the natural or artificial selection of japonica rice. Further studies on the molecular evolution of prolamin in Oryza sativa subspecies, as well as the ancestors of rice, should provide new insight into how disulfide bond formation and H2O2 generation impact the development and quality of seeds and the natural or artificial selection of rice.
Acknowledgments
We thank T. W. Okita (Washington State University) and T. Masumura (Kyoto Prefectural University) for commenting on the manuscript. This work was supported by a Grant-in-Aid for Young Scientists from the Japan Society for the Promotion of Science (23780101 to Y.O.), by the Ministry of Education, Culture, Sports, Science and Technology of Japan (to Y.O.), and by the Ministry of Agriculture, Forestry, and Fisheries of Japan (Genomics for Agricultural Innovation, IPG-0023 to Y.K.).
Glossary
Abbreviations:
- ER
endoplasmic reticulum
- PDI
protein disulfide isomerase
- PB
protein body
- QSOX
quiescin sulfhydryl oxidase
- TRX
thioredoxin
- VKOR
vitamin K epoxide reductase
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/17967
References
- 1.Ito K, Inaba K. The disulfide bond formation (Dsb) system. Curr Opin Struct Biol. 2008;18:450–8. doi: 10.1016/j.sbi.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Sevier CS, Kaiser CA. Ero1 and redox homeostasis in the endoplasmic reticulum. Biochim Biophys Acta. 2008;1783:549–56. doi: 10.1016/j.bbamcr.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 3.Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–44. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hatahet F, Ruddock LW, Ahn K, Benham A, Craik D, Ellgaard L, et al. Protein disulfide isomerase: a critical evaluation of its function in disulfide bond formation. Antioxid Redox Signal. 2009;11:2807–50. doi: 10.1089/ars.2009.2466. [DOI] [PubMed] [Google Scholar]
- 5.Houston NL, Fan CZ, Xiang QY, Schulze JM, Jung R, Boston RS. Phylogenetic analyses identify 10 classes of the protein disulfide isomerase family in plants, including single-domain protein disulfide isomerase-related proteins. Plant Physiol. 2005;137:762–78. doi: 10.1104/pp.104.056507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shewry PR, Napier JA, Tatham AS. Seed storage proteins: structures and biosynthesis. Plant Cell. 1995;7:945–56. doi: 10.1105/tpc.7.7.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamagata H, Sugimoto T, Tanaka K, Kasai Z. Biosynthesis of storage proteins in developing rice seeds. Plant Physiol. 1982;70:1094–100. doi: 10.1104/pp.70.4.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogawa M, Kumamaru T, Satoh H, Iwata N, Omura T, Kasai Z, et al. Purification of protein body-I of rice seed and its polypeptide composition. Plant Cell Physiol. 1987;28:1517–27. [Google Scholar]
- 9.Kawagoe Y, Suzuki K, Tasaki M, Yasuda H, Akagi K, Katoh E, et al. The critical role of disulfide bond formation in protein sorting in the endosperm of rice. Plant Cell. 2005;17:1141–53. doi: 10.1105/tpc.105.030668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Onda Y, Nagamine A, Sakurai M, Kumamaru T, Ogawa M, Kawagoe Y. Distinct roles of protein disulfide isomerase and P5 sulfhydryl oxidoreductases in multiple pathways for oxidation of structurally diverse storage proteins in rice. Plant Cell. 2011;23:210–23. doi: 10.1105/tpc.110.079509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagamine A, Matsusaka H, Ushijima T, Kawagoe Y, Ogawa M, Okita TW, et al. A role for the cysteine-rich 10 kDa prolamin in protein body I formation in rice. Plant Cell Physiol. 2011;52:1003–16. doi: 10.1093/pcp/pcr053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krishnan HB, Okita TW. Structural relationship among the rice glutelin polypeptides. Plant Physiol. 1986;81:748–53. doi: 10.1104/pp.81.3.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Zhu S, Liu S, Jiang L, Chen L, Ren Y, et al. The vacuolar processing enzyme OsVPE1 is required for efficient glutelin processing in rice. Plant J. 2009;58:606–17. doi: 10.1111/j.1365-313X.2009.03801.x. [DOI] [PubMed] [Google Scholar]
- 14.Kumamaru T, Uemura Y, Inoue Y, Takemoto Y, Siddiqui SU, Ogawa M, et al. Vacuolar processing enzyme plays an essential role in the crystalline structure of glutelin in rice seed. Plant Cell Physiol. 2010;51:38–46. doi: 10.1093/pcp/pcp165. [DOI] [PubMed] [Google Scholar]
- 15.Ashida K, Saito Y, Masumura T, Iida S. Ultrastructure of protein bodies in mature rice (Oryza sativa L.) with altered storage protein composition. Breed Sci. 2011;61:201–7. doi: 10.1270/jsbbs.61.201. [DOI] [Google Scholar]
- 16.Li X, Franceschi VR, Okita TW. Segregation of storage protein mRNAs on the rough endoplasmic reticulum membranes of rice endosperm cells. Cell. 1993;72:869–79. doi: 10.1016/0092-8674(93)90576-C. [DOI] [PubMed] [Google Scholar]
- 17.Choi SB, Wang C, Muench DG, Ozawa K, Franceschi VR, Wu Y, et al. Messenger RNA targeting of rice seed storage proteins to specific ER subdomains. Nature. 2000;407:765–7. doi: 10.1038/35037633. [DOI] [PubMed] [Google Scholar]
- 18.Krishnan HB, Franceschi VR, Okita TW. Immunochemical studies on the role of the Golgi-complex in protein-body formation in rice seeds. Planta. 1986;169:471–80. doi: 10.1007/BF00392095. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Ren Y, Liu X, Jiang L, Chen L, Han X, et al. OsRab5a regulates endomembrane organization and storage protein trafficking in rice endosperm cells. Plant J. 2010;64:812–24. doi: 10.1111/j.1365-313X.2010.04370.x. [DOI] [PubMed] [Google Scholar]
- 20.Fukuda M, Satoh-Cruz M, Wen L, Crofts AJ, Sugino A, Washida H, et al. The small GTPase Rab5a is essential for intracellular transport of proglutelin from Golgi apparatus to the protein storage vacuole and endosomal membrane organization in develiping rice endosperm. Plant Physiol. 2011;157:632–44. doi: 10.1104/pp.111.180505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frand AR, Kaiser CA. Ero1p oxidizes protein disulfide isomerase in a pathway for disulfide bond formation in the endoplasmic reticulum. Mol Cell. 1999;4:469–77. doi: 10.1016/S1097-2765(00)80198-7. [DOI] [PubMed] [Google Scholar]
- 22.Mezghrani A, Fassio A, Benham A, Simmen T, Braakman I, Sitia R. Manipulation of oxidative protein folding and PDI redox state in mammalian cells. EMBO J. 2001;20:6288–96. doi: 10.1093/emboj/20.22.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gross E, Kastner DB, Kaiser CA, Fass D. Structure of Ero1p, source of disulfide bonds for oxidative protein folding in the cell. Cell. 2004;117:601–10. doi: 10.1016/S0092-8674(04)00418-0. [DOI] [PubMed] [Google Scholar]
- 24.Sevier CS, Qu H, Heldman N, Gross E, Fass D, Kaiser CA. Modulation of cellular disulfide-bond formation and the ER redox environment by feedback regulation of Ero1. Cell. 2007;129:333–44. doi: 10.1016/j.cell.2007.02.039. [DOI] [PubMed] [Google Scholar]
- 25.Appenzeller-Herzog C, Riemer J, Christensen B, Sorensen ES, Ellgaard L. A novel disulphide switch mechanism in Ero1α balances ER oxidation in human cells. EMBO J. 2008;27:2977–87. doi: 10.1038/emboj.2008.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Onda Y, Kumamaru T, Kawagoe Y. ER membrane-localized oxidoreductase Ero1 is required for disulfide bond formation in the rice endosperm. Proc Natl Acad Sci USA. 2009;106:14156–61. doi: 10.1073/pnas.0904429106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frand AR, Kaiser CA. The ERO1 gene of yeast is required for oxidation of protein dithiols in the endoplasmic reticulum. Mol Cell. 1998;1:161–70. doi: 10.1016/S1097-2765(00)80017-9. [DOI] [PubMed] [Google Scholar]
- 28.Pagani M, Pilati S, Bertoli G, Valsasina B, Sitia R. The C-terminal domain of yeast Ero1p mediates membrane localization and is essential for function. FEBS Lett. 2001;508:117–20. doi: 10.1016/S0014-5793(01)03034-4. [DOI] [PubMed] [Google Scholar]
- 29.Otsu M, Bertoli G, Fagioli C, Guerini-Rocco E, Nerini-Molteni S, Ruffato E, et al. Dynamic retention of Ero1α and Ero1β in the endoplasmic reticulum by interactions with PDI and ERp44. Antioxid Redox Signal. 2006;8:274–82. doi: 10.1089/ars.2006.8.274. [DOI] [PubMed] [Google Scholar]
- 30.Tian G, Xiang S, Noiva R, Lennarz WJ, Schindelin H. The crystal structure of yeast protein disulfide isomerase suggests cooperativity between its active sites. Cell. 2006;124:61–73. doi: 10.1016/j.cell.2005.10.044. [DOI] [PubMed] [Google Scholar]
- 31.Ferrari DM, Soling HD. The protein disulphide-isomerase family: unravelling a string of folds. Biochem J. 1999;339:1–10. doi: 10.1042/0264-6021:3390001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kikuchi M, Doi E, Tsujimoto I, Horibe T, Tsujimoto Y. Functional analysis of human P5, a protein disulfide isomerase homologue. J Biochem. 2002;132:451–5. doi: 10.1093/oxfordjournals.jbchem.a003242. [DOI] [PubMed] [Google Scholar]
- 33.Akama K, Horikoshi T, Sugiyama A, Nakahata S, Akitsu A, Niwa N, et al. Protein disulfide isomerase-P5, down-regulated in the final stage of boar epididymal sperm maturation, catalyzes disulfide formation to inhibit protein function in oxidative refolding of reduced denatured lysozyme. Biochim Biophys Acta. 1804. 2010:1272–84. doi: 10.1016/j.bbapap.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 34.Uehara T, Nakamura T, Yao D, Shi ZQ, Gu Z, Ma Y, et al. S-nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature. 2006;441:513–7. doi: 10.1038/nature04782. [DOI] [PubMed] [Google Scholar]
- 35.Kaiser BK, Yim D, Chow IT, Gonzalez S, Dai Z, Mann HH, et al. Disulphide-isomerase-enabled shedding of tumour-associated NKG2D ligands. Nature. 2007;447:482–6. doi: 10.1038/nature05768. [DOI] [PubMed] [Google Scholar]
- 36.Gumireddy K, Sun FX, Klein-Szanto AJ, Gibbins JM, Gimotty PA, Saunders AJ, et al. In vivo selection for metastasis promoting genes in the mouse. Proc Natl Acad Sci USA. 2007;104:6696–701. doi: 10.1073/pnas.0701145104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andème Ondzighi C, Christopher DA, Cho EJ, Chang SC, Staehelin LA. Arabidopsis protein disulfide isomerase-5 inhibits cysteine proteases during trafficking to vacuoles before programmed cell death of the endothelium in developing seeds. Plant Cell. 2008;20:2205–20. doi: 10.1105/tpc.108.058339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H, Boavida LC, Ron M, McCormick S. Truncation of a protein disulfide isomerase, PDIL2-1, delays embryo sac maturation and disrupts pollen tube guidance in Arabidopsis thaliana. Plant Cell. 2008;20:3300–11. doi: 10.1105/tpc.108.062919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vitu E, Kim S, Sevier CS, Lutzky O, Heldman N, Bentzur M, et al. Oxidative activity of yeast Ero1p on protein disulfide isomerase and related oxidoreductases of the endoplasmic reticulum. J Biol Chem. 2010;285:18155–65. doi: 10.1074/jbc.M109.064931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pollard MG, Travers KJ, Weissman JS. Ero1p: a novel and ubiquitous protein with an essential role in oxidative protein folding in the endoplasmic reticulum. Mol Cell. 1998;1:171–82. doi: 10.1016/S1097-2765(00)80018-0. [DOI] [PubMed] [Google Scholar]
- 41.Zito E, Chin KT, Blais J, Harding HP, Ron D. ERO1-β, a pancreas-specific disulfide oxidase, promotes insulin biogenesis and glucose homeostasis. J Cell Biol. 2010;188:821–32. doi: 10.1083/jcb.200911086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zito E, Melo EP, Yang Y, Wahlander A, Neubert TA, Ron D. Oxidative protein folding by an endoplasmic reticulum-localized peroxiredoxin. Mol Cell. 2010;40:787–97. doi: 10.1016/j.molcel.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tripathi BN, Bhatt I, Dietz KJ. Peroxiredoxins: a less studied component of hydrogen peroxide detoxification in photosynthetic organisms. Protoplasma. 2009;235:3–15. doi: 10.1007/s00709-009-0032-0. [DOI] [PubMed] [Google Scholar]
- 44.Wajih N, Hutson SM, Wallin R. Disulfide-dependent protein folding is linked to operation of the vitamin K cycle in the endoplasmic reticulum - A protein disulfide isomerase-VKORC1 redox enzyme complex appears to be responsible for vitamin K-1 2,3-epoxide reduction. J Biol Chem. 2007;282:2626–35. doi: 10.1074/jbc.M608954200. [DOI] [PubMed] [Google Scholar]
- 45.Li W, Schulman S, Dutton RJ, Boyd D, Beckwith J, Rapoport TA. Structure of a bacterial homologue of vitamin K epoxide reductase. Nature. 2010;463:507–12. doi: 10.1038/nature08720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schulman S, Wang B, Li WK, Rapoport TA. Vitamin K epoxide reductase prefers ER membrane-anchored thioredoxin-like redox partners. Proc Natl Acad Sci USA. 2010;107:15027–32. doi: 10.1073/pnas.1009972107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goodstadt L, Ponting CP. Vitamin K epoxide reductase: homology, active site and catalytic mechanism. Trends Biochem Sci. 2004;29:289–92. doi: 10.1016/j.tibs.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 48.Kodali VK, Thorpe C. Oxidative protein folding and the quiescin-sulfhydryl oxidase family of flavoproteins. Antioxid Redox Signal. 2010;13:1217–30. doi: 10.1089/ars.2010.3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rancy PC, Thorpe C. Oxidative protein folding in vitro: a study of the cooperation between quiescin-sulfhydryl oxidase and protein disulfide isomerase. Biochemistry. 2008;47:12047–56. doi: 10.1021/bi801604x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tury A, Mairet-Coello G, Poncet F, Jacquemard C, Risold PY, Fellmann D, et al. QSOX sulfhydryl oxidase in rat adenohypophysis: localization and regulation by estrogens. J Endocrinol. 2004;183:353–63. doi: 10.1677/joe.1.05842. [DOI] [PubMed] [Google Scholar]
- 51.Chakravarthi S, Jessop CE, Willer M, Stirling CJ, Bulleid NJ. Intracellular catalysis of disulfide bond formation by the human sulfhydryl oxidase, QSOX1. Biochem J. 2007;404:403–11. doi: 10.1042/BJ20061510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gross E, Sevier CS, Heldman N, Vitu E, Bentzur M, Kaiser CA, et al. Generating disulfides enzymatically: reaction products and electron acceptors of the endoplasmic reticulum thiol oxidase Ero1p. Proc Natl Acad Sci USA. 2006;103:299–304. doi: 10.1073/pnas.0506448103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matsusaka H, Kumamaru T, Ogawa M, Satoh H. Biosynthesis of rice S-poor and S-rich prolamins is regulated by an independent genetic system. In: Khush GS, Brar DS, Hardy B, eds. Advances in Rice Genetics. Manila, Philippines: International Rice Research Institute, 2003:441-44. [Google Scholar]
- 54.Sattler SE, Mene-Saffrane L, Farmer EE, Krischke M, Mueller MJ, DellaPenna D. Nonenzymatic lipid peroxidation reprograms gene expression and activates defense markers in Arabidopsis tocopherol-deficient mutants. Plant Cell. 2006;18:3706–20. doi: 10.1105/tpc.106.044065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Veal EA, Day AM, Morgan BA. Hydrogen peroxide sensing and signaling. Mol Cell. 2007;26:1–14. doi: 10.1016/j.molcel.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 56.Mitsukawa N, Konishi R, Kidzu K, Ohtsuki K, Masumura T, Tanaka K. Amino acid sequencing and cDNA cloning of rice seed storage proteins, the 13kDa prolamins, extracted from type I protein bodies. Plant Biotechnol. 1999;16:103–13. [Google Scholar]
- 57.Tada Y, Akagi H, Fujimura T, Matsuda T. Effect of an antisense sequence on rice allergen genes comprising a multigene family. Breed Sci. 2003;53:61–7. doi: 10.1270/jsbbs.53.61. [DOI] [Google Scholar]
- 58.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 59.Ushijima T, Matsusaka H, Jikuya H, Ogawa M, Satoh H, Kumamaru T. Genetic analysis of cysteine-poor prolamin polypeptides reduced in the endosperm of the rice esp1 mutant. Plant Sci. 2011;181:125–31. doi: 10.1016/j.plantsci.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 60.Kawakatsu T, Hirose S, Yasuda H, Takaiwa F. Reducing rice seed storage protein accumulation leads to changes in nutrient quality and storage organelle formation. Plant Physiol. 2010;154:1842–54. doi: 10.1104/pp.110.164343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Doebley JF, Gaut BS, Smith BD. The molecular genetics of crop domestication. Cell. 2006;127:1309–21. doi: 10.1016/j.cell.2006.12.006. [DOI] [PubMed] [Google Scholar]