Abstract
In order to adapt to environmental changes of light species and intensity, higher plants furnish complicate signaling systems such as the UVR/COP/HY5 cascade which links several diverse classes of photoreceptors. In addition UV-B light provokes accelerated production of UV-B protectants such as flavonoids and vitamins. Following intensive research efforts, genes in the UV-B signaling cascade have been characterized via forward genetics approaches following mutant screens relying on sensitivity to UV-B irradiation. However detailed processes of the linkage between light signaling and the upregulation of metabolite accumulation remain unclear. Here we review both the light signal cascades and metabolite pathways responding to UV-B exposure. Finally we generate co-expression network analysis using published data in order to find novel candidate genes which link light signaling and transcriptional regulation to metabolic biosynthesis in attempt to describe how these processes are interlinked.
Keywords: Arabidopsis thaliana, Flavonoid, light responses, Metabolic responses, network analysis, Transcriptome analysis, UV-B protectant, UV-B signaling cascade
Introduction
Sunlight irradiation is one of the most critical factors for energy metabolism and the regulation of plant growth and development. Accordingly, sunlight as an environment factor is not only for energy source for photosynthesis in higher plants but also an informational signal influencing their entire life cycle. Despite the fact that sunlight has a wide range of different light fractions, limited light of wavelength above ~290nm reaches to the surface of the earth,1 because sunlight is filtrated by the atmosphere such as ozone phase. This atmospheric filtration can, however, not eliminate all toxic light fractions, such as UV-B light (UV-B, 280 to 315nm). UV-B is long known to damage DNA, inhibit photosynthesis and arrest the cell cycle.2
In order to survive the broad range of physiological responses caused by toxic light fractions, higher plants possess several classes of photoreceptors. The model plant Arabidopsis, utilizes more than three independent photoreceptor systems, perceiving the red/far-red (phytochromes, phyA-E), blue/UV-A (cryptochromes cry1 and 2) and UV-B (UVR8, UVB-RESISTANCE 8) fractions.3 Genes of the member in UV-B signaling cascade have been characterized via forward genetics approaches following mutant screens relying on sensitivity to UV-B irradiation. The principal genes identified as mediators of UV-B photomorphogenic responses were COP1 (CONSTITUTIVE PHOTOMORPHOGENIC 1)1 and HY5 (ELONGATED HYPOCOTYL 5),4 while recently UVR8 which has been identified as a UV-B receptor.3 Detailed gene expression and phenotypic analysis using gene knockout mutant analysis with UV-B treatment revealed the structure and functional interplay of a complex UV-B signaling cascade. Taking a different approach knockout mutants that were deficient in metabolites which are known to exhibit photoprotective function such as flavonoid mutants,5-7 phenylpropanoid mutants,7,8 ascorbate mutants9 and tocopherol mutants10 have been characterized. These mutants displayed UV-B sensitive phenotypic changes alongside an enhancement of gene expression of UV-B responsive genes. In this mini-review we will attempt both to summarize both the signal transduction events and metabolic shifts which follow perception of UV-B stress and where possible to synthesize their action to form a common model of response.
Although the UV-B signaling cascade and the pathways of light protectant have been long studied and are at least partially identified, the combined network of signaling cascade to protectant metabolism has received relatively little attention. HY5 regulation of PFG1 (PRODUCTION OF FLAVONOL GLYCOSIDES)/AtMYB12 gene expression which controls flavonol metabolism has been uncovered.6 However, the roles of other, more minor, players in the UV-B responsive network remain unclear, potentially simply because mutation of such genes do not reveal severe phenotypic variance. That said a recent combined metabolite and transcript profiling analysis of phenolic deficient mutants following UV-B irradiation revealed the relative importance of metabolic priming and induction of flavonol production in response to this treatment7 In addition, since considerable transcriptome data sets concerning UV-B stress are publicly available11 advanced in silico gene expression analysis and co-expression network analysis can be used in order to further characterize the response to this stress.12 Here, we show an example of in silico gene expression analysis based on UV-B experiments, for finding novel candidate genes out of transcriptional network between signaling cascade to phenolic protectant metabolism.
Plant light signaling cascades
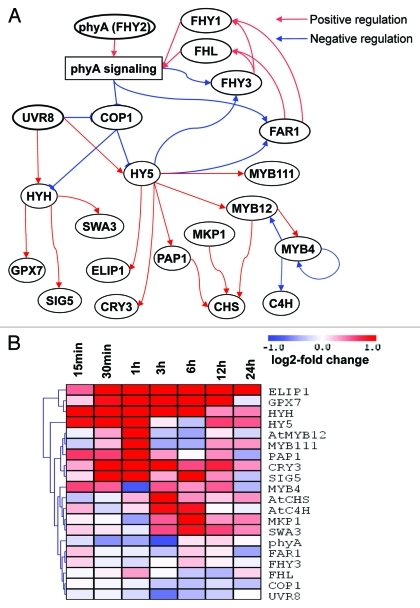
Higher plants are known to adapt a range of strategies in response to the incidence of UV-B light including increases in leaf thickness, UV-B reflective properties and in the cellular levels of UV-B absorbing metabolites. In recent years, considerable advances in our understanding of UV-B perception have been made. This process involves phytochrome-mediated photoprotection, UV-B invoked cell-death and UV-B signaling responses.13 In the UV-B signaling transduction cascade, the bZIP transcription factor, HY5, a mediator of several photomorphogenic pathways, is required for UV-B-mediated gene expression.1 The multi-functional E3-ubiquitin ligases, (COP1)14 and HY5-HOMOLOG (HYH)15 transcription factors have also been demonstrated to play important roles in UV-B signaling. In addition, UVR8 was recently elegantly demonstrated to be a UV-B receptor mediating key photomorphogenic responses3 Here we intend to broaden the focus beyond the specific signal transduction cascades which fire in response to varying light conditions to additionally encompass more general UV-light response Hypothetical signaling transduction and transcriptional regulation network of light response in higher plants are summarized in Figure 1A. Within this cascade, the UV-B photoreceptor, UVR8, acts to upregulate HY5 and HYH, and downregulate expression of COP1. This regulation system can adjust balance of photomorphogenesis and high light responsive pathways. The transcription factors, HY5 and HYH, control several important genes and pathways. For example EARLY LIGHT-INDUCABLE PROTEIN 1 (ELIP1) is one of the major light responsive genes which leads to tolerance to photoinhibition and photooxidative stress.16 PRODUCTION OF ANTHOCYANIN PIGMENT 1 (PAP1, AtMYB75) and MYB12 are regulators of flavonol/anthocyanin pathways.6,17 MYB4 is known as negative regulator of phenylpropanoids, because MYB4 downregulates C4H expression.18 These regulators are all under the control of HY5 suggesting that the complex structure of light signaling cascade can adapt to changes of light intensity to survive under more severe environments.
Figure 1.
Hypothetical light signaling cascades. (A) Light signaling transduction and transcriptional regulation network of light response in higher plants. (B) Hieratical Clustering Analysis (HCA) of gene expression profiling of light signaling related gene under UV-B irradiation. Abbreviations: AtCHS, naringenin-chalcone synthase, At5g13930; AtMYB12, At2g47460; AtC4H, cinnamate-4-hydroxylase, At2g30490; COP1, CONSTITUTIVE PHOTOMORPHOGENIC 1, At2g32950; CRY3, CRYPTOCHROME 3, At5g24850; ELIP1, EARLY LIGHT-INDUCABLE PROTEIN 1, At3g22840; FAR1, FAR-RED IMPAIRED RESPONSE 1, At4g15090; FHL, FAR-RED-ELONGATED HYPOCOTYL1-LIKE, At5g02200; FHY1, FRY1, PAT3, FAR-RED ELONGATED HYPOCOTYLS 1, At2g37678; FHY3, FAR-RED ELONGATED HYPOCOTYLS 3, At3g22170; GPX7, GLUTATHIONE PEROXIDASE 7, At4g31870; HY5, ELONGATED HYPOCOTYL 5, At5g11260; HYH, HY5-HOMOLOG, At3g17609; MKP1, MITOGEN-ACTIVATED PROTEIN KINASE PHOSPHATASE 1, At3g55270; MYB111, At5g49330; MYB4, At4g38620; PAP, MYB75, At1g56650; phyA, FHY2, hy8, PHYTOCHROME A, At1G09570; SIG5, SIGMA FACTOR 5, At5g24120; SWA3, WALK8, SLOW WALKER 3, At1g16280; UVR8, UV REPAIR DEFECTIVE 8, At5g63860.
To understand time dependent expression of UV-B signaling cascade related genes during UV-B irradiation, in silico gene expression analysis was performed (Fig. 1B). At the gene expression level, half of genes related to light signaling cascade were highly induced following application of 24h UV-B irradiation. The key positive regulators of the UV-B response -HY5 and HYH- displayed early responses (within the first 6h) following their increased expression genes downstream of HY5 and HYH such as MYB111, PAP1, cryptochrome 3 (CRY3) and RNA polymerase sigma subunit E (SIG5), were upregulated. Furthermore, after increasing of gene expression of MYB111 and PAP1, enzymatic gene such as AtCHS and AtC4H were slightly induced as a consequence of MYB111 and PAP1 being direct regulators of AtCHS expression. Far-red specific signaling related genes (FHZ1, FHL and FHY3) were unchanged by UV-B irradiation. The COP1 gene, however, which is a negative regulator of UV-B light-dependent development displayed a slight decreased in expression onwards from 3h. However, the UV-B receptor UVR8 displayed unaltered gene expression under short-term UV-B irradiation.
Metabolic protection against UV-B irradiation
Increasing evidence for the protective function of various compounds against UV-B light has been provided by the analysis of mutants in a range of biosynthetic pathways. In order to elucidate the function of putative protectant metabolites several gene expression and phenotypic analysis using knockout mutants exposed to UV-B treatment have been performed. Flavonoids, one of the major polyphenol compounds derived from phenylalanine, are arguably the foremost compounds for UV-B protection in plants. Li et at (1993) reported that flavonoid-less mutants, tt4 (CHS, chalcone synthase) and tt5 (CHI, chalcone isomerase) mutants revealed hypersensitive phenotypic responses to UV-B irradiation. Subsequently, gene expression of CHS has been used as one of the major markers of UV-B stress.1,13,19 High accumulation of anthocyanin has additionally been observed in higher plants under UV-B treatment. In addition, obvious evidence for the importance of flavonoid in protection against UV-B has been obtained in experiments to confirm the regulation of CHS and MYB12 by HY5.1,6 Phenylpropanoids which are also derived from phenylalanine are regarded as major UV-B protectants, because knockout mutant of MYB4 gene which negatively regulates cinnamate 4-Hydroxylase gene (AtC4H) expression has a reduced tolerance to UV-B light.20 Furthermore, ascorbate, carotenoids, tocopherol and vitamin B6 are also considered as key compounds for protection of UV-B light.9,10 UV-B protectant metabolites can thus be categorised to three different families according to their chemical structural functions, (i) light protection in proportion to aromatic ring, (ii) antioxidant activity corresponding to reduction moieties such as phenolic moiety and unsaturated carbon-carbon bonds, and (iii) increasing osmotic pressure cased by compound accumulation in specific location for sample in vacuole. In our recent study we identified that flavonoid-less mutants were strongly effected by UV-B, even more so than mutants deficient in other phenylpropanoids.7 Transcriptional regulation is, furthermore, much better characterized for these metabolites than for those vitamins mentioned above. For these reasons we focused here on flavonoids. Function of flavonoid accumulation following UV-B irradiation has been investigated with flavonoid deficient mutants (see examples, Jin et al., 2000; Kusano et al., 2011). In flavonoid-less mutants, large metabolic changes and enhancement of UV-B associated transcriptional programs inducing senescence were observed.7 These changes are likely the consequence of an increased effective dose of UV-B perceived in the cell due to the deficiency of protection afforded by aromatic ring structures.
Integrative analysis of signaling cascades and metabolism
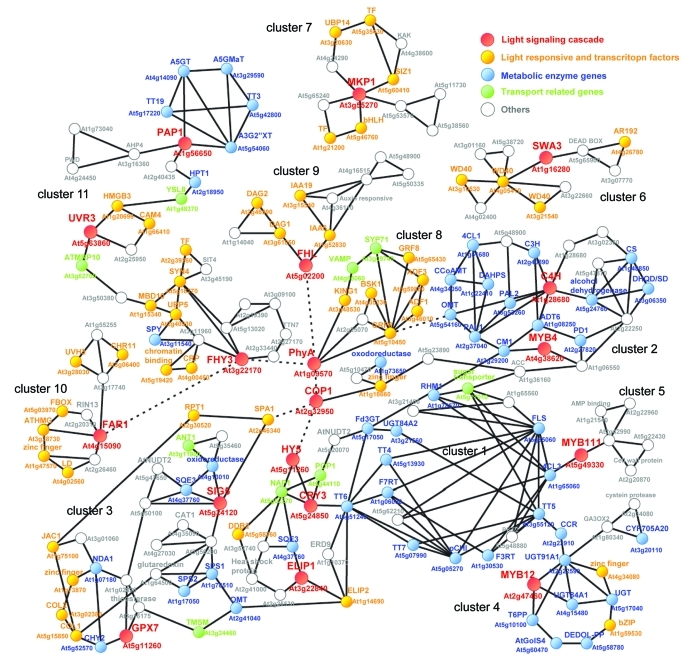
Although normally studied in isolation the relationship between cascade networks and protectant biosynthesis is likely highly intertwined. In recent years, given the ever accumulating amount of transcriptome data has been possible to perform novel approaches for facilitate annotation of plant gene function. Co-expression network analysis based on similarity/distance measure calculated from the vast publically available microarray data are one of the new approach for elucidation of transcriptionally regulated gene network.12,21 To construct UV-B responsive gene network, co-expression network analysis was performed using 21 genes of UV-B signaling cascade (Fig. 2). In an attempt to predict the framework of UV-B responsive networks, data obtained from ATTED-II version 622 (http://atted.jp/) were used in the construction of a co-expression network. Network analysis queried the 21 light signaling cascade related genes shown in Figure 1A.
Figure 2.
Co-expression network analysis of light signaling related genes. Co-expression data obtained from ATTED-II version 6 (http://atted.jp/) was used in the construction of a co-expression network. Network analysis queried the 20 one light signaling cascade related genes shown in Figure 1A. Solid edges indicate gene co-expression, and red dotted edges indicate known protein-protein-interaction. Red node indicates light signaling cascade genes, yellow indicates putative light responsive and transcription factor genes, blue indicates genes encoding protectant metabolism, green indicates transport related genes.
This correlation analysis revealed 12 specific sub-clusters which were connected by the known signaling cascade related genes, COP1, HY5, PhyA, FHL, FHY3, SIG5 and CRY3. COP1 which is a negative regulator of light-dependent development played a central role in the network being connected to HY5, PhyA and FHL. These connections have been experimentally confirmed by identification of protein-protein-interactions between them.22 Cluster 1 contributed of HY5, CRY3, ELIP3 and chalcone synthase (TT4, AtCHS). This sub-cluster revealed a network with flavonol biosynthesis, because it included major flavonol related genes such as 4-coumarate-CoA ligase 3 (4CL3), chalcone isomerase (CHS, TT5), flavanone 3-hydroxylase (F3H, TT6), flavonoid 3′-hydroxylase (F3′H, TT7), flavonol synthase (FLS), flavonol 7-O-rhamnosyltransferse (F7RT), flavonol 3-O-rhamnosyltransferse F3RT and UDP-rhamnose synthase (RHM1). In the cluster 1 network, some genes -putatively related to light signaling cascade- such as ELIP2 and DDB2 and transport related genes (ATP-binding cassette A21, POP1; ATP-binding cassette I20, NAP9; sugar transporter, At5g17010) were observed. Subsequently, cluster 1 was connected to second larger sub-network named cluster 2 which was contributed shikimate-phenylpropanoid biosynthetic genes such as chorismate mutase 1 (CM1), arogenate dehzdratase 6 (ADT6), phenylalanine ammonia-lyase 1 (PAL1), phenylalanine ammonia-lyase 2 (PAL2), cinnamate-4-hydroxylase (C4H), -O-methyltransferase (OMT1), 4CL1, quinate O-hydroxycinnamoyltransferas (HCT) and prephenate dehydratase 1 (PD1). In addition, cluster 1 also well connected to cluster 3 which included Sigma factor 5 (SIG5) and glutathione peroxidase 7 (GPX7). In cluster 3, genes of the carotenoid related MEP pathway (β-carotene hydroxylase 2, CHY2; solanesyl diphosphate synthase 1, SPS1 and SPS2) and squalene epoxidase 3 (SQE3) were correlated. The connections between clusters 1, 2 and 3 revealed the strong connections from light signaling cascade related genes (HY5, CRY3, ELIP1, GPX7 and SIG5) to light irradiation protectant biosynthetic genes of flavonol, phenylpropanoid, epoxysqualene and carotenoid. Since the HYH gene was not included in the ATTED-II database, connections between HYH could not be observed in this network. However, connection between HY5 to HYH can be assumed as a function of the connection between HY5 to GPX7 and SIG5. In cluster 3, we also observed some candidate genes which are putatively related to the light signaling cascade (suppressor of PhyA, SPA1; root phototropism 2, RPT1; J-domain protein required for chloroplast accumulation response, JAC1) and transcriptional regulation (constans-like 1, COL1; COL2; damaged DNA-binding 2, DDB2). By contrast, clusters 5 through 10 contained few metabolite associated genes. Some putative signaling related genes (calmodulin 4, CAM4; ubiquitin-specific protease 5, UBP5; cryptic precocious, CPR), transcription factors (methyl-CpG binding 10, MBD10; At2g39260) and chromatin regulation related genes (high mobility mobility group B3, HMGB3; sister chromatid cohesion 1 SYN4; At5g19420) were observed in cluster 10 which was constructed by three light signaling cascade related genes UVR8, FHY3 and PAP1. Interestingly, the homogentisate phytyltransferase (HPT1) gene which encodes homogentisate phytyltransferase involved in tocopherol biosynthesis, was included in this UVR-anthocyanin network. Despite the fact that the UV-B receptor UVR8 was unaltered following shorten UV-B irradiation, it was correlated closely to the the cluster of anthocyanin and tocopherol biosynthesis.
Toward application in crop species
Given the twin problems of environmental deterioration and a booming global population efforts to maintain or improve crop productivity are becoming ever more important. In view of the fact that UV-B strongly affects plant growth and development, understanding of UV-B response and adaptation is very important in crops on account of the potential effect of UV-B on food security. In recent years, several crop species have been investigated based on wide-target analyses such as transcriptomics, proteomics and metabolomics with respect to their UV-B tolerance and response including studies on maize leaves,23 tomato fruits,24 carrot culture25 and barley seedlings.26 The majority of these investigations focused on flavonoid biosynthesis which is conserved across higher plants. Metabolite profiling of maize leaves, tomato fruits, barley seedlings and carrot culture under UV-B treatment revealed a common increase of flavonoid related gene expression and indeed of the metabolites themselves. This fact reveals that flavonoid related compounds play an important role for the response to UV-B irradiation across both species and tissue boundaries. Thus metabolic engineering of flavonoid production within crop species is beneficial not only for health27 but also for ensuring food security. The use of recently developed tools and methodologies for translational research (see28-31) will likely greatly extend the number of species for which the knowledge we have acquired to date can be applied to.
In summary, in this review we suggest that gene expression of UV-B signaling cascade and the production of UV-B protectants are transcriptionally regulated under the same gene expression network controlled by the UVR/COP/HY5 system. Plant metabolites which can protect against UV-B light damage, for example, flavonol, phenylpropanoid, anthocyanin and carotenoids, have a transcriptional linkage to the UV-B response. These transcriptional linkages thus facilitate identification of novel associated genes via co-expression analysis. Given the genes increasing availability of genome information many tools for translational biology are becoming available. Therefore in silico network analysis based on Arabidopsis can be extended to other important crop species, a construction of framework between UV-B signaling and important metabolisms will be important targets for metabolic engineering and/or breeding strategies for the maintenance of plant performance in the future global climate.
Acknowledgments
Research activity of TT is supported by the Alexander von Humboldt Foundation. Funding from the Max-Planck-Society (to TT and ARF) is gratefully acknowledged.
Glossary
Abbreviations:
- CHI
chalcone isomerase
- CHS
chalcone synthase
- COP
constitutively photomorphogenic
- HYH
HY5 homolog
- HY
elongated hypocotyl
- phy
phytochrome
- UV
ultra violet
- UVR
UV resistance locus
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/18240
REFERENCES
- 1.Oravecz A, Baumann A, Mate Z, Brzezinska A, Molinier J, Oakeley EJ, et al. Constitutively photomorphogenic1 is required for the uv-b response in arabidopsis. Plant Cell. 2006;18:1975–90. doi: 10.1105/tpc.105.040097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang L, Wang Y, Björn LO, Li S. Does cell cycle arrest occur in plant under solar uv-b radiation? Plant Signal Behav. 2011;6:892–4. doi: 10.4161/psb.6.6.15317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rizzini L, Favory J-J, Cloix C, Faggionato D, O'Hara A, Kaiserli E, et al. Perception of uv-b by the arabidopsis uvr8 protein. Science. 2011;332:103–6. doi: 10.1126/science.1200660. [DOI] [PubMed] [Google Scholar]
- 4.Ulm R, Baumann A, Oravecz A, Mate Z, Adam E, Oakeley EJ, et al. Genome-wide analysis of gene expression reveals function of the bzip transcription factor hy5 in the uv-b response of arabidopsis. Proc Natl Acad Sci USA. 2004;101:1397–402. doi: 10.1073/pnas.0308044100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J, Oulee TM, Raba R, Amundson RG, Last RL. Arabidopsis flavonoid mutants are hypersensitive to uv-b irradiation. Plant Cell. 1993;5:171–9. doi: 10.1105/tpc.5.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stracke R, Favory J-J, Gruber H, Bartelniewoehner L, Bartels S, Binkert M, et al. The arabidopsis bzip transcription factor hy5 regulates expression of the pfg1/myb12 gene in response to light and ultraviolet-b radiation. Plant Cell Environ. 2010;33:88–103. doi: 10.1111/j.1365-3040.2009.02061.x. [DOI] [PubMed] [Google Scholar]
- 7.Kusano M, Tohge T, Fukushima A, Kobayashi M, Hayashi N, Otsuki H, et al. Metabolomics reveals comprehensive reprogramming involving two independent metabolic responses of arabidopsis to ultraviolet-b light. Plant J. 2011;67:354–69. doi: 10.1111/j.1365-313X.2011.04599.x. [DOI] [PubMed] [Google Scholar]
- 8.Brown BA, Headland LR, Jenkins GI. Uv-b action spectrum for uvr8-mediated hy5 transcript accumulation in arabidopsis. Photochem Photobiol. 2009;85:1147–55. doi: 10.1111/j.1751-1097.2009.00579.x. [DOI] [PubMed] [Google Scholar]
- 9.Page M, Sultana N, Paszkiewicz K, Florance H, Smirnoff N. The influence of ascorbate on anthocyanin accumulation during high light acclimation in arabidopsis thaliana: Further evidence for redox control of anthocyanin synthesis. Plant Cell Environ. 2011 doi: 10.1111/j.1365-3040.2011.02369.x. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 10.Harvaux M, Kloppstech K. The protective functions of carotenoid and flavonoid pigments against excess visible radiation at chilling temperature investigated in arabidopsis npq and tt mutants. Planta. 2001;213:953–66. doi: 10.1007/s004250100572. [DOI] [PubMed] [Google Scholar]
- 11.Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, et al. The atgenexpress global stress expression data set: Protocols, evaluation and model data analysis of uv-b light, drought and cold stress responses. Plant J. 2007;50:347–63. doi: 10.1111/j.1365-313X.2007.03052.x. [DOI] [PubMed] [Google Scholar]
- 12.Tohge T, Fernie AR. Combining genetic diversity, informatics and metabolomics to facilitate annotation of plant gene function. Nat Protoc. 2010;5:1210–27. doi: 10.1038/nprot.2010.82. [DOI] [PubMed] [Google Scholar]
- 13.Jenkins GI. Signal transduction in responses to uv-b radiation. Annu Rev Plant Biol. 2009;60:407–31. doi: 10.1146/annurev.arplant.59.032607.092953. [DOI] [PubMed] [Google Scholar]
- 14.Favory JJ, Stec A, Gruber H, Rizzini L, Oravecz A, Funk M, et al. Interaction of cop1 and uvr8 regulates uv-b-induced photomorphogenesis and stress acclimation in arabidopsis. EMBO J. 2009;28:591–601. doi: 10.1038/emboj.2009.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown BA, Jenkins GI. Uv-b signaling pathways with different fluence-rate response profiles are distinguished in mature arabidopsis leaf tissue by requirement for uvr8, hy5, and hyh. Plant Physiol. 2008;146:576–88. doi: 10.1104/pp.107.108456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rossini S, Casazza AP, Engelmann ECM, Havaux M, Jennings RC, Soave C. Suppression of both elip1 and elip2 in arabidopsis does not affect tolerance to photoinhibition and photooxidative stress. Plant Physiol. 2006;141:1264–73. doi: 10.1104/pp.106.083055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tohge T, Nishiyama Y, Hirai MY, Yano M, Nakajima J, Awazuhara M, et al. Functional genomics by integrated analysis of metabolome and transcriptome of arabidopsis plants over-expressing an myb transcription factor. Plant J. 2005;42:218–35. doi: 10.1111/j.1365-313X.2005.02371.x. [DOI] [PubMed] [Google Scholar]
- 18.Hemm MR, Herrmann KM, Chapple C. Atmyb4: A transcription factor general in the battle against uv. Trends Plant Sci. 2001;6:135–6. doi: 10.1016/S1360-1385(01)01915-X. [DOI] [PubMed] [Google Scholar]
- 19.Jiang L, Wang Y, Bjorn LO, Li SS. Arabidopsis radical-induced cell death1 is involved in uv-b signaling. Photochem Photobiol Sci. 2009;8:838–46. doi: 10.1039/b901187k. [DOI] [PubMed] [Google Scholar]
- 20.Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, et al. Transcriptional repression by atmyb4 controls production of uv-protecting sunscreens in arabidopsis. EMBO J. 2000;19:6150–61. doi: 10.1093/emboj/19.22.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saito K, Hirai MY, Yonekura-Sakakibara K. Decoding genes with coexpression networks and metabolomics - 'majority report by precogs'. Trends Plant Sci. 2008;13:36–43. doi: 10.1016/j.tplants.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 22.Obayashi T, Kinoshita K, Nakai K, Shibaoka M, Hayashi S, Saeki M, et al. A database of co-expressed genes and cis elements for identifying co-regulated gene groups in arabidopsis. Nucleic Acids Res. 2007;35:D863–9. doi: 10.1093/nar/gkl783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Casati P, Walbot V. Gene expression profiling in response to ultraviolet radiation in maize genotypes with varying flavonoid content. Plant Physiol. 2003;132:1739–54. doi: 10.1104/pp.103.022871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giuntini D, Lazzeri V, Calvenzani V, Dall'Asta C, Galaverna G, Tonelli C, et al. Flavonoid profiling and biosynthetic gene expression in flesh and peel of two tomato genotypes grown under uv-b-depleted conditions during ripening. J Agric Food Chem. 2008;56:5905–15. doi: 10.1021/jf8003338. [DOI] [PubMed] [Google Scholar]
- 25.Maeda K, Kimura S, Demura T, Takeda J, Ozeki Y. Dcmyb1 acts as a transcriptional activator of the carrot phenylalanine ammonia-lyase gene (dcpal1) in response to elicitor treatment, uv-b irradiation and the dilution effect. Plant Mol Biol. 2005;59:739–52. doi: 10.1007/s11103-005-0910-6. [DOI] [PubMed] [Google Scholar]
- 26.Kaspar S, Matros A, Mock HP. Proteome and flavonoid analysis reveals distinct responses of epidermal tissue and whole leaves upon uv-b radiation of barley (hordeum vulgare l.) seedlings. J Proteome Res. 2010;9:2402–11. doi: 10.1021/pr901113z. [DOI] [PubMed] [Google Scholar]
- 27.Butelli E, Titta L, Giorgio M, Mock HP, Matros A, Peterek S, et al. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat Biotechnol. 2008;26:1301–8. doi: 10.1038/nbt.1506. [DOI] [PubMed] [Google Scholar]
- 28.Ogata Y, Suzuki H, Sakurai N, Shibata D. Cop: A database for characterizing co-expressed gene modules with biological information in plants. Bioinformatics. 2010;26:1267–8. doi: 10.1093/bioinformatics/btq121. [DOI] [PubMed] [Google Scholar]
- 29.Tohge T, Mettler T, Arrivault S, Carroll AJ, Stitt M, Fernie AR. From models to crop species: Caveats and solutions for translational metabolomics Front Physio. 2011;in press. [DOI] [PMC free article] [PubMed]
- 30.Huber SC. Grand challenges in plant physiology: The underpinning of translational research. Front Physio. 2011;2:1–5. doi: 10.3389/fpls.2011.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mutwil M, Klie S, Tohge T, Giorgi FM, Wilkins O, Campbell MM, et al. Planet: Combined sequence and expression comparisons across plant networks derived from seven species. Plant Cell. 2011;23:895–910. doi: 10.1105/tpc.111.083667. [DOI] [PMC free article] [PubMed] [Google Scholar]