Abstract
The Early Responsive to Dehydration (ERD) genes are defined as those genes that are rapidly activated during drought stress. The encoded proteins show a great structural and functional diversity, with a particular class of proteins acting as connectors of stress response pathways. Recent studies have shown that ERD15 proteins from different species of plants operate in cross-talk among different response pathways. In this mini-review, we show the recent progress on the functional role of this diverse family of proteins and demonstrate that a soybean ERD15 homolog can act as a connector in stress response pathways that trigger a programmed cell death signal.
Keywords: ER stress, ERD15, N-rich proteins, osmotic stress
As sessile organisms, plants have developed sophisticated signaling pathways that are responsible for the detection of and rapid adaptation to variations in the environment. Abiotic stresses, such as drought, salt and freezing, lead to the disruption of the plant water status that, in turn, provokes similar physiological consequences, such as osmotic stress and the excessive accumulation of reactive oxygen species (ROS) in plant cells.1 A major common response to these environmental stressors is the upregulation of transcriptional factors that induce the expression of genes involved in the synthesis of osmoprotectants, and encoding lipid desaturases and detoxification proteins that act to minimize the deleterious effects of the stress signals.2 Such stress-induced genes have been classified according to their pattern of expression, including RD (responsive to dehydration), ERD (early responsive to dehydration), COR (cold regulated), LTI (low-temperature induced), and KIN (cold inducible).3
The Early Responsive to Dehydration Genes
The ERD genes have been identified by their capacity to be rapidly induced by dehydration.4 The functional characterization of ERD genes (ERD1 through 16) has demonstrated that they exhibit diverse and heterogeneous biochemical functions and are present in different subcellular compartments. For instance, ERD1 encodes a chloroplast ATP-dependent protease,5 ERD2 encodes a cytosolic HSP70,4 and ERD4 encodes a membrane protein. Whereas the functions of ERD3 and ERD7 are unknown, ERD5 and ERD6 encode a mitochondrial proline dehydrogenase and a carbohydrate transporter, respectively,6,7 ERD8 produces Hsp81,4 and ERD16 encodes a ubiquitin extension protein 1, UBQ1.4 ERD9, ERD11 and ERD13 belong to the glutathione-S-transferase family,8 and ERD10 and ERD14 are members of the family of late embryogenesis abundant (LEA) proteins.9 Lastly, ERD15 is a hydrophilic protein10 that possesses a PAM2 domain that interacts with polyA-binding proteins (PABP11). We have recently identified a soybean ERD15 homolog, GmERD15 (Glycine max ERD15), which is induced by osmotic and endoplasmic reticulum stresses and activates the osmotic and ER stress-induced N-rich protein (NRP)-mediated cell death response.12
A Soybean ERD15 Homolog (GmERD15) Connects Endoplasmic Reticulum Stress with an Osmotic Stress-induced Cell Death Signal
NRP-mediated cell death signaling has been described in soybean as a novel branch of the ER-stress signaling pathway that diverges from the molecular chaperone-inducing branch of the UPR (unfolded protein response) and transduces a programmed cell death signal.13,14 This pathway integrates the ER-stress and osmotic-stress signals to increase the synergistic expression of N-rich proteins (NRP-A and NRP-B) and an NAC domain-containing protein, GmNAC6, which are critical mediators of stress-induced cell death in plants.13-16 This cell death integrated pathway has emerged as a relevant adaptive response of plant cells to multiple environmental stimuli.16
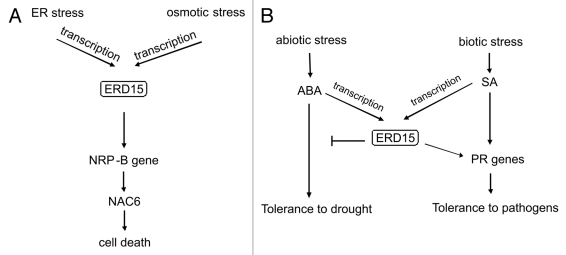
The soybean GmERD15 homolog has been described as a new ER stress- and osmotic stress-induced transcription factor that binds to the promoter and induces the expression of the NRP-B gene. In fact, GmERD15 was isolated by its capacity to associate stably with the promoter of NRP-B in yeast cells using the one-hybrid system.12 The GmERD15 binding site in the NRP-B promoter was mapped to a 12-bp palindromic sequence that resembles binding sites for ssDNA binding proteins.17 Furthermore, GmERD15 is located in the nucleus, and chromatin immunoprecipitation (ChIP) assays revealed that it binds to the NRP-B promoter in vivo. The ectopic expression of GmERD15 in soybean cells activates the NRP-B promoter and induces NRP-B expression. Collectively, these results indicate that GmERD15 functions as an upstream component of the NRP-mediated cell death signaling pathway that is induced by ER stress and osmotic stress (Fig. 1A).
Figure 1.
ERD15 is involved in multiple stress response signaling pathways. (A) The induction of GmERD15 gene expression by ER or osmotic stress leads to the activation of the NRP-mediated cell death signaling pathway. (B) The induction of AtERD15 gene expression by abiotic or biotic stress attenuates ABA signaling and induces PR genes, leading to the enhanced resistance to pathogens.
Functional Diversity of the ERD15 Family
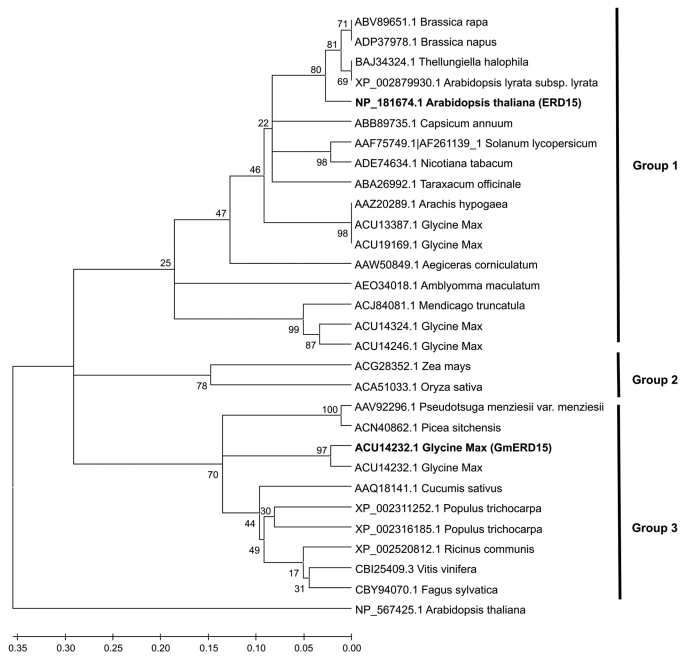
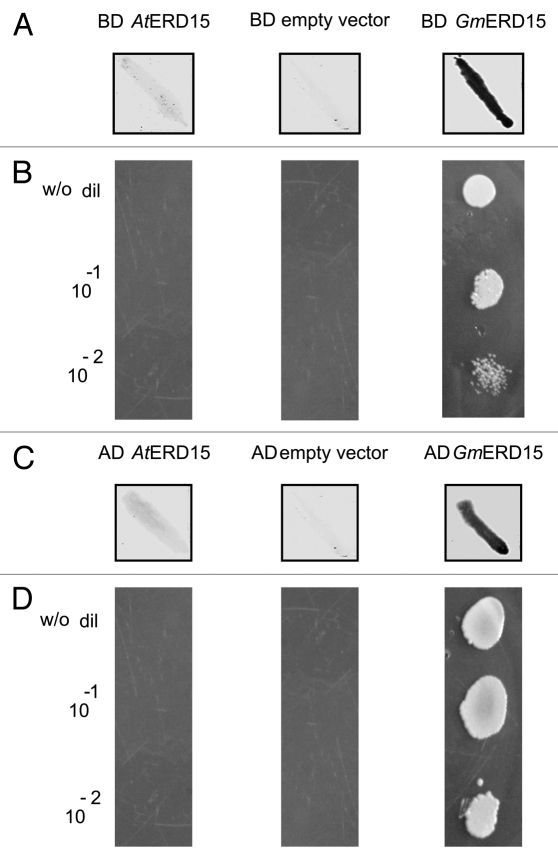
A sequence analysis of the primary structure of the ERD family in soybean and Arabidopsis thaliana indicates that GmERD15 is represented by two copies in the genomes of both of these plant species (Fig. 2). The analyzed sequences are distributed into four distinct groups, and the soybean GmERD15 protein (group 2) is clustered separately from the Arabidopsis ERD15 protein (group 1), as described by Kariola et al.11 In contrast to GmERD15, Arabidopsis ERD15 does not exhibit a transactivation activity, as evaluated by transactivation assays in yeast (Fig. 3). In fact, Arabidopsis ERD15 does not associate with the NRP-B promoter in yeast and, more importantly, does not transactivate a GAL4-dependent promoter when fused to the GAL-4 binding domain (Fig. 3). Functional diversity has also been described among other putative ERD15 homologs. The ERD15 homologs from Solanum licopersicum and from Solanum pennelli are 98% identical and belong to the same group as AtERD15, indicating a possible analogous function.18 However, the tomato protein is clearly located in the nucleus and confers tolerance to freezing when ectopically expressed in transgenic tomato plants, which is in marked contrast to the phenotypes displayed by ERD15-overexpressing Arabidopsis lines.11 These contrasting results in transgene overexpression studies suggest that the Arabidopsis and tomato ERD15 homologs may be functionally diverse.
Figure 2.
Relatedness of ERD15 proteins from different plant species. The multiple alignment was made using ClustalW, and the dendrogram was built with the MEGA5 software using the UPGMA method (the numbers at the nodes indicate the bootstrap scores). The ERD15 proteins were separated into at least 3 groups, as indicated. The Arabidopsis thaliana ERD15 described by Kariola et al.11 and the Glycine max ERD15 described by Alves et al.12 are in bold. The proteins accession numbers are indicated.
Figure 3.
Arabidopsis thaliana ERD15 protein does not exhibit transactivation activity or DNA binding activity in yeast. The transactivation activities of ERD15 proteins were revealed through the expression of the lacZ reporter gene (β-galactosidase activity), detected as a blue color (shown here in dark gray/black; panels A and C), or by prototrophic growth (activation of the HIS3 reporter gene) in medium lacking histidine (His; panels B and D). Yeast AH109 cells were transformed with the indicated GAL4-BD (DNA-binding domain) chimeric constructs (panels A and B). The chimeric BD-AtERD15 neither activates β-galactosidase gene expression nor induces His prototrophy, whereas BD-GmERD15 strongly activates the expression of both reporter genes, indicating that GmERD15, but not AtERD15, exhibits transactivation activity. The same results were observed when the reporter genes were under the control of the NRP-B promoter, and the chimeras AD-AtERD15 and AD-GmERD15 were used (Panels C and D) to demonstrate that AtERD15 does not bind to the NRP-B promoter.
ERD15 as a Linker of Multiple Stress Signaling Pathways
ERD15 from Arabidopsis has been functionally characterized as a common regulator of the abscisic acid (ABA) response and the salicylic acid (SA)-dependent defense pathway.11 The overexpression of ERD15 reduced the sensitivity to ABA, as the transgenic plants were less tolerant to drought and were impaired in increasing their freezing tolerance in response to ABA. In contrast, the loss of the ERD15 function caused a hypersensitivity to ABA, and the silenced plants displayed enhanced tolerance to both drought and freezing. The negative effect of ERD15 on ABA signaling enhances the SA-dependent defense pathway because the overexpression of ERD15 was associated with an increased resistance to the bacterial necrotroph Erwinia carotovora and the enhanced induction of marker genes for systemic acquired resistance. The authors of the study also addressed the antagonistic effect of ABA on SA-mediated defense by demonstrating the enhanced expression of marker genes for systemic acquired resistance in the knockouts abi1–1 and abi2–1, which are defective for ABA metabolism. Together, these results implicate ERD15 from Arabidopsis as a shared component of the ABA- and SA-mediated responses (Fig. 1B). Although AtERD15 from Arabidopsis and GmERD15 from soybean are clustered into different groups (Fig. 2) and exhibit distinct biochemical activities (Fig. 3), the ERD15 proteins are believed to operate as connectors of signaling pathways, as shown in Figure 1. The analysis of GmERD15 expression in response to treatments with ABA, SA and JA, cold acclimation and wounding may provide insights into the function of GmERD15 as a linker of the multiple stress signaling pathways that may transduce a programmed cell death signal in plants.
Acknowledgments
This research was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico CNPq and Fundação de Amparo a Pesquisa do Estado de Minas Gerais (FAPEMIG).
Glossary
Abbreviations:
- ERD
early responsive to dehydration
- NRP
asparagine rich protein
- ER
endoplasmic reticulum
- UPR
unfolded protein response
- PR
pathogenesis related genes
- ABA
abscisic acid
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/18268
References
- 1.Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59:651–81. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- 2.Yamaguchi–Shinozaki K, Shinozaki K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005;10:88–94. doi: 10.1016/j.tplants.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 3.Shinozaki K, Yamaguchi-Shinozaki K. Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol. 2000;3:217–23. [PubMed] [Google Scholar]
- 4.Kiyosue T, Yamaguchi–Shinozaki K, Shinozaki K. Cloning of cDNAs for genes that are early-responsive to dehydration stress (ERDs) in Arabidopsis thaliana L.: identification of three ERDs as HSP cognate genes. Plant Mol Biol. 1994;25:791–8. doi: 10.1007/BF00028874. [DOI] [PubMed] [Google Scholar]
- 5.Soitamo AJ, Piippo M, Allahverdiyeva Y, Battchikova N, Aro E. Light has a specific role in modulating Arabidopsis gene expression at low temperature. BMC Plant Biol. 2008;8:13. doi: 10.1186/1471-2229-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakashima K, Satoh R, Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K. A gene encoding proline dehydrogenase is not only induced by proline and hypoosmolarity, but is also developmentally regulated in the reproductive organs of Arabidopsis. Plant Physiol. 1998;118:1233–41. doi: 10.1104/pp.118.4.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiyosue T, Abe H, Yamaguchi-Shinozaki K, Shinozaki K. ERD6, a cDNA clone for an early dehydration-induced gene of Arabidopsis, encodes a putative sugar transporter. Biochim Biophys Acta. 1998;1370:187–91. doi: 10.1016/S0005-2736(98)00007-8. [DOI] [PubMed] [Google Scholar]
- 8.Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K. Characterization of two cDNAs (ERD11 and ERD13) for dehydrationinducible genes that encode putative glutathione S-transferases in Arabidopsis thaliana L. FEBS Lett. 1993;335:189–92. doi: 10.1016/0014-5793(93)80727-C. [DOI] [PubMed] [Google Scholar]
- 9.Kiyosue T, Yamaguchi–Shinozaki K, Shinozaki K. Characterization of two cDNAs (ERD10 and ERD14) corresponding to genes that respond rapidly to dehydration stress in Arabidopsis thaliana. Plant Cell Physiol. 1994;35:225–31. [PubMed] [Google Scholar]
- 10.Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K. ERD15, a cDNA for a dehydration-inducible gene from Arabidopsis thaliana. Plant Physiol. 1994;106:1707. doi: 10.1104/pp.106.4.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kariola T, Brader G, Helenius E, Li J, Heino P, Palva ET. EARLY RESPONSIVE to DEHYDRATION 15, a negative regulator of abscisic acid responses in Arabidopsis. Plant Physiol. 2006;142:1559–73. doi: 10.1104/pp.106.086223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alves MS, Reis PAB, Dadalto SP, Faria JAQA, Fontes EPB, Fietto LG. A novel transcription factor, ERD15 (Early Responsive to Dehydration 15), connects endoplasmic reticulum stress with an osmotic stress-induced cell death signal. J Biol Chem. 2011;286:20020–30. doi: 10.1074/jbc.M111.233494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irsigler AST, Costa MDL, Zhang P, Reis PAB, Dewey RE, Boston RS, et al. Expression profiling on soybean leaves reveals integration of ER- and osmotic-stress pathways. BMC Genomics. 2007;8:431. doi: 10.1186/1471-2164-8-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costa MDL, Reis PAB, Valente MAS, Irsigler AST, Carvalho CM, Loureiro ME, et al. A new branch of endoplasmic reticulum stress signaling and the osmotic signal converge on plant specific asparagine-rich proteins to promote cell death. J Biol Chem. 2008;283:20209–19. doi: 10.1074/jbc.M802654200. [DOI] [PubMed] [Google Scholar]
- 15.Pinheiro GL, Marques CS, Costa MDBL, Reis PAB, Alves MS, Carvalho CM, et al. Complete inventory of soybean NAC transcription factors: Sequence conservation and expression analysis uncover their distinct roles in stress response. Gene. 2009;444:10–23. doi: 10.1016/j.gene.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 16.Reis PAB, Soares-Ramos JRL, Fontes EPB. Endoplasmic Reticulum Stress Response. Soybean - Biochemistry, Chemistry and Physiology, In:TECH 2011; ISBN 978-953-307-219-7. [Google Scholar]
- 17.Desveaux D, Després C, Joyeux A, Subramaniam R, Brisson N. PBF-2 is a novel single-stranded DNA binding factor implicated in PR-10 gene activation in potato. Plant Cell. 2000;12:1477–89. doi: 10.1105/tpc.12.8.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ziaf K, Loukehaich R, Gong P, Liu H, Han Q, Wang T, et al. A multiple stress responsive gene ERD15 from Solanum pennelli confers stress tolerance in tobacco. Plant Cell Physiol. 2011;52:1055–67. doi: 10.1093/pcp/pcr057. [DOI] [PubMed] [Google Scholar]