Summary
Variable Lymphocyte Receptors (VLRs) are the adaptive immune receptors of jawless fish, which evolved adaptive immunity independent of other vertebrates. In lieu of the immunoglobulin-fold based T- and B-cell receptors, lymphocyte-like cells of jawless fish express VLRs (A, B or C) composed of leucine-rich repeats and are similar to toll-like receptors (TLRs) in structure, but antibodies (VLRB) and T cell receptors (VLRA, C) in function. Here we present the structural and biochemical characterization of VLR4, a VLRB, in complex with BclA, the immunodominant glycoprotein of Bacillus anthracis spores. Using a combination of crystallography, mutagenesis and binding studies, we delineate the mode of antigen recognition and binding between VLR4 and BclA, examine commonalities in VLRB recognition of antigens, and demonstrate the potential of VLR4 as a diagnostic tool for the identification of B. anthracis spores.
Introduction
All vertebrate animals have evolved an adaptive immune system in order to generate a diverse repertoire of highly specific anticipatory receptors against novel antigens (Cooper and Alder, 2006; Pancer et al., 2004). Fish of the phylum agnatha are the most ancient lineage of vertebrates that persist today. These jawless fish produce immune cells that are remarkably similar in function to the B-cells and T-cells of other vertebrates. However, these lymphocyte-like cells lack receptors carrying the conventional immunoglobulin-fold (Ig-fold) of B- and T-cell receptors (BCRs and TCRs). Instead, jawless fish have independently evolved an adaptive immune system using leucine-rich repeat motifs (LRR) as the protein scaffold rather than the immunoglobulin domain (Pancer et al., 2004). Like conventional immune receptors, these variable lymphocyte receptors (VLR) have both humoral (VLRB) and cell-mediated functions (VLRA and VLRC), which are produced by discrete populations of lymphocyte-like cells (Guo et al., 2009; Kasamatsu et al., 2010).
The germline lamprey VLR loci contain conserved regions for the signal peptide, portions of the N- and C-terminal capping domains, and the invariant stalk region. In lymphocyte-like cells, LRRs are copied from cassettes that flank the incomplete germline gene via a mechanism thought to resemble gene conversion with putative roles for two cytidine deaminases that are expressed in specific populations of lymphocyte-like cells (Alder et al., 2005; Rogozin et al., 2007). VLRA and VLRC are expressed as membrane-bound receptors thought to resemble T-cell receptors in function (Deng et al., 2010; Guo et al., 2009; Kasamatsu et al., 2010). VLRBs resemble B-cell receptors and antibodies in that they can be either membrane-bound or secreted from VLRB+ cells. The affinity of a single VLRB protomer for its antigen is usually in the low micromolar range (Herrin et al., 2008; Velikovsky et al., 2009). However, secreted VLRB form large, disulfide linked octamers or decamers that show very high affinity for antigen by using multivalency to increase avidity (Herrin et al., 2008).
VLR antigen binding domains have a motif organization that is highly similar to that of other LRR-containing proteins, such as TLRs. The N-terminal capping region (LRRNT) serves to stabilize the protein and covers what would otherwise be the exposed end of the N-terminal hydrophobic core of the LRR solenoid (Bella et al., 2008). The LRRNT is followed by three or more LRR motifs: LRR1, LRRV and LRRVe and the connecting peptide (CP) that create the LRR solenoid (Pancer et al., 2004). The C-terminal capping region (LRRCT) acts to bury the C-terminal hydrophobic core of the LRR solenoid using a long α-helix (Bella et al., 2008). The LRRCT also contains a highly variable insert, which forms an extended loop that is crucial for contacting antigen (Rogozin et al., 2007). The β-sheet created by the LRRNT and LRR motifs forms a concave surface, the variable residues of which, along with the LRRCT-loop, compose the antigen-binding surface of VLRs (Deng et al., 2010; Han et al., 2008; Velikovsky et al., 2009). In addition, VLRs have an invariant C-terminal Thr/Pro stalk, a putative GPI attachment site and a Cys-rich region (Herrin et al., 2008; Pancer et al., 2004), which is responsible for the multimerization of VLRB into disulfide-linked oligomers and crucial for high avidity VLRB.
In previous work, VLR4, a monoclonal VLRB specific for BclA, was isolated from lampreys that had been immunized with purified exosporium from spores of Bacillus anthracis, the causative agent of anthrax (Herrin et al., 2008). The exosporium is the outermost layer of the B. anthracis spore, and BclA (the Bacillus collagen-like protein of anthracis) makes up the hair-like nap that is covalently attached to the exosporium basal layer (Tan et al. 2011). BclA is expressed on spores of a limited number of Bacillus species, all from the Bacillus cereus group. This trimeric protein has a 38-amino acid N-terminal domain containing sequences required for basal layer attachment, a central collagen-like domain, and a 134-residue C-terminal domain that drives trimerization (Boydston et al., 2005). The collagen-like domain is polymorphic in length ranging from 51-228 amino acids (Sylvestre et al., 2003; Boydston et al., 2005) and is heavily O-glycosylated (Daubenspeck et al., 2004). Trimers of the C-terminal domain form the tips of the spore’s hair-like nap and are the immunodominant target of vertebrate antibodies (Steichen et al., 2003; Boydston et al., 2005; Swiecki et al., 2006). B. anthracis is an uncommon human pathogen; however, infections from inhalation of bacterial spores have high mortality unless promptly treated with antibiotics and intensive care (Spencer, 2003). Unfortunately, B. anthracis spores have been used as biological weapons and in the bioterrorism attacks of 2001 in the United States, highlighting the need for rapid diagnostics to identify B. anthracis spores from spores of highly related, but more benign species (Higgins et al., 2003). Here, we describe the crystal structure and molecular interactions of VLR4 with BclA and show VLR4 to be a unique reagent in its ability to discriminate B. anthracis spores.
Results
Engineering of BclA-CTD
In agreement with multiple studies, we found that the BclA C-terminal domain (CTD) tended to form crystals in nearly every condition commonly used for protein crystallization (Boydston et al., 2005; Liu et al., 2008; Nuttall et al., 2011). To prevent crystallization of BclA-CTD alone and facilitate formation of VLR4-BclA complex crystals, we introduced a mutation in BclA-CTD to disrupt crystal packing based on a previously determined BclA-CTD structure from our laboratory (2R6Q.pdb). We mutated Thr185, a key residue in forming the BclA-CTD crystal lattice, to an Asn with the expectation that this conservative mutation would obstruct BclA-CTD crystal growth without drastically altering the structure of BclA. The mutant BclA-CTD-Thr185Asn was expressed and purified from E. coli and mixed with a monomeric VLR4 expressed from insect cells using a baculovirus expression system. The VLR4 construct was C-terminally truncated to remove the Thr/Pro stalk and Cys-rich regions that multimerize VLRB. The strategy was highly successful and allowed crystals of the VLR4-BclA complex to be grown in space group I222. X-ray diffraction data were collected to 2.55 Å resolution (Table 1). Comparison of the mutated BclA-CTD to wild-type structures in the Protein Data Bank (PDB) indicated no major changes, including the loop containing the mutated amino acid.
Table 1.
Data Collection and Refinement Statistics
| VLR4-BclA-Thr185Asn 3TWI |
BclA-Pro159Ser 3TYJ |
|
|---|---|---|
| Data Collection | ||
| Wavelength (Å) | 1.033 | 1.000 |
| Resolution (Å) | 50-2.55 (2.64-2.55) | 40-2.15 (2.19-2.15) |
| Space group | I222 | P6322 |
| Cell dimensions | ||
| a, b, c (Å) | 74.8 146.5 209.5 | 68.2 68.2 163.7 |
| α, β, γ (°) | 90 90 90 | 90 90 120 |
| No. observations | 116,551 | 144,923 |
| Unique reflections | 37,042 | 12,588 |
| Redundancy | 3.1 (2.9) | 11.5 (7.8) |
| Completeness (%) | 97.3 (99.1) | 96.9 (97.9) |
| Rsym(%)b | 0.122 (0.581) | 0.168 (0.647) |
| I/σ | 9.5 (2.0) | 11.7 (3.2) |
| Refinement statistics | ||
| Resolution | 41.9-2.55 (2.61-2.55) | 34-2.15 (2.21-2.15) |
| No. reflections (working) | 33,162 (2,559) | 11,338 (840) |
| No. reflections (test) | 1,869 (131) | 619 (40) |
| Rcryst(%)c | 0.19 (0.29) | 0.21 (0.25) |
| Rfree(%)d | 0.23 (0.33) | 0.23 (0.25) |
| No. mols./ asu | 3 | 1 |
| No. protein atoms | 6,731 | 1,072 |
| No. protein chains | 6 | 1 |
| Overall B values (Å2) | ||
| BclA | 32.6 | 15.0 |
| VLR4 (Chains D, E, F) | 38.6, 52.2, 98.0 | |
| Waters | 39.1 | 32.8 |
| Wilson B-value (Å2) | 32.0 | 16.6 |
| Ramachandran plot (%)e | ||
| Favored | 96.4 | 100.0 |
| Allowed | 3.6 | 0.0 |
| Disallowed | 0.0 | 0.0 |
| rmsd | ||
| Bond length (Å) | 0.011 | 0.009 |
| Angle (°) | 1.21 | 1.22 |
Values in parentheses are for the outer shell
Rsym=∑hkl ∣I-<I>∣/∑hkl∣I∣
Rcryst=∑hkl ∣Fobs-Fcalc∣/∑hkl Fobs
Rfree is the same as Rcryst except for 5% of the data excluded from refinement
Ramachandran statistics were calculated with MolProbity.
We also used biolayer interferometry to quantify VLR4 binding to modified and wild-type BclA proteins. Binding of truncated monomeric VLR4 containing the antigen-binding motifs (residues 22-188) to wild-type BclA-CTD resulted in a Kd of 2.6 μM (Table 2) in agreement with relatively weak binding by monomeric VLRB (Herrin et al., 2008; Velikovsky et al., 2009). The BclA-CTD-Thr185Asn mutant surprisingly showed a four-fold increase in affinity, with a Kd of 0.6 μM, indicating that the mutation did not disrupt the epitope for VLR4 and may have even slightly enhanced the VLR4-BclA interaction.
Table 2. Binding of monomeric VLR4 to recombinant BclA-CTD variants.
VLR4 has similar binding affinities for BclA despite amino-acid changes corresponding to differences in the sequences of BclA of B. anthracis Sterne, B. cereus T and B. thruingiensis Kurstaki. The BclA mutation created to disrupt crystal packing, Thr185Asn, shows a slight increase in affinity for VLR4. See also supplemental Figure S3.
| Sample | Kd (M) |
|---|---|
| BclA-CTD-B. anthracis Sterne | 2.6×10−6 |
| BclA-CTD-Thr185Asn | 0.6×10−6 |
| BclA-CTD-Ser129Ala | 3.2×10−6 |
| BclA-CTD-Glu130Gln | 2.4×10−6 |
| BclA-CTD-Pro159Ser | 3.5×10−6 |
| BclA-CTD-Ser129Ala-Pro159Ser | 4.2×10−6 |
| BclA-CTD-Glu130Gln-Pro159Ser | 3.2×10−6 |
| BclA-CTD-B. cereus T | 3.5×10−6 |
| BclA-CTD-B. thruingiensis Kurstaki | 3.4×10−6 |
Structure of VLR4-BclA
The asymmetric unit (asu) of the crystal contains three VLR4 head domain monomers (chains D,E,F) bound to the trimeric BclA (chains A,B,C) such that each VLR4 contacts a single BclA monomer using its concave surface (Figure 1). VLR4 binds lower down BclA than perhaps expected near the proximal end of BclA with respect to the surface of the spore, and the C-terminus of the VLR4 head domain points away from the spore where it would normally connect to the decameric VLR4. Although three VLR4 head domains are bound multivalently to the BclA trimer using identical binding surfaces, one of the VLR4 domains shows a significant amount of disorder, as reflected by increased B-values and substantially fewer contacts in the crystal lattice. Nevertheless, the concave surface of this less-ordered VLR4 (chain F) in contact with BclA has lower B-values and better electron density than its more distal region.
Figure 1. The VLR4-BclA Structure.
VLR4 binds BclA using its concave surface (green) and LRRCT-loop (red). The VLR4 binding site lies on the proximal end of BclA with respect to the spore, near the termini. A) Stereo view of VLR4 interacting with a BclA monomer. B) Depiction of trimeric BclA decorated by VLR4.
VLR4 uses its six-stranded β-sheet of the concave surface, as well as LRRCT, to bury 480 Å2 of surface area on BclA. The VLR4 concave surface lies along the proximal end of each BclA monomer and primarily contacts the loop regions connecting strands of the two β-sheets. The extended antigen-contacting loop of LRRCT lies across a shallow groove between two of these loops. Six hydrogen bonds are formed between VLR4 and BclA, four of which are contributed by the LRRCT (Figure 2). Because the LRRCT-loop contributes a greater fraction of the surface buried on VLR4 than any single LRR motif and provides the most hydrogen bonds, this loop is likely responsible for most of the antigen binding specificity of VLR4.
Figure 2. Molecular Interactions of VLR4 with BclA.
Detailed view of molecular interactions between VLR4 and BclA-CTD. A) The concave surface of VLR4 (green and blue) contributes two hydrogen bonds (red dashes) and buries hydrophobic surface on BclA (yellow). B) The LRRCT (pink) makes interactions with BclA (yellow) using both the LRRCT-loop and Asn164, which lies between the loop and structurally conserved Cys167. The LRRCT makes four hydrogen bonds to BclA and buries more surface area than any other single LRR motif from the concave surface. See also supplemental Figure S1.
VLR4 Mutagenesis and Binding to BclA
In a previous study (Herrin et al., 2008), several VLRBs with highly similar sequences were isolated as binders of BclA-CTD, including VLR4 (Figure S1), and contain highly identical LRRCT-loops, bind to BclA with similar affinities as VLR4, and display significant species specificity for the spores of Bacillus anthracis. Although several VLR mutagenesis experiments were described, with the VLR4-BclA structure now in hand, we can now ask more specific questions about the contributions of particular residue positions to differences in antigen recognition among these VLRBs. For instance, position 55 of VLR4 is a His, whereas an Arg in highly related VLRBs is important for BclA binding. However, His55 does not contact BclA in the VLR4-BclA complex structure and mutation to Arg in VLR4 did not significantly alter binding to BclA (Table 3). We also probed position 106 at the edge of the VLR4-BclA interface that is Ile in VLR4, but Thr, Asn, Asp or Ile in other BclA-binding VLRBs. Mutation of Ile106 to Thr reduced binding of VLR4 for BclA by approximately eight-fold (Table 3).
Table 3. Binding of monomeric VLR4 variants to recombinant B. anthracis Sterne BclA-CTD.
The VLR4 His55Arg mutation has no significant impact on BclA binding, but Ile106Thr shows greatly reduced affinity suggesting that other BclA-binding VLRB recognize the BclA epitope differently.
| Sample | Kd (M) |
|---|---|
| VLR4 | 3.0×10−6 |
| VLR4-His55Arg | 2.7×10−6 |
| VLR4-Ile106Thr | 24.0×10−6 |
BclA Mutagenesis and VLR4 Specificity
VLR4 shows remarkable specificity for B. anthracis Sterne (Ba) spores, but does not bind Bacillus cereus T (Bc) or Bacillus thuringiensis Kurstaki (Bt) spores (Herrin et al., 2008). This finding is in contrast to many conventional vertebrate antibodies against BclA, which show some amount of cross reactivity among Bacillus species (Liu et al., 2008; Nuttall et al. 2011). To better understand this discrimination, we compared sequence differences of the non-binding bacterial species with BclA from Ba (Supplemental Figure S2) and mapped them onto the BclA structure. Very few differences are found in or near the VLR4 binding site (Figure 3) and only one difference in a contact residue arises in Bc at Ser129Ala, with a nearby sequence difference in Bt at Glu130Gln. The two non-binding species also share a different residue at nearby Pro159Ser and we hypothesized that mutation of this Pro to Ser might alter the conformation of a loop that is contacted by VLR4 and, hence, alter binding to the VLR4 LRRCT-loop. We, therefore, determined the crystal structure of the BclA-CTD-Pro159Ser mutant (Table 1) and were surprised to find that mutation at this position had no effect on the loop conformation. We also tested binding of monomeric VLR4 to Ser129Ala, Glu130Gln and Pro159Ser BclA-CTD single and double mutants, as well as binding to recombinant BclA-CTD from Bc and Bt (Table 2). Again to our surprise, no significant differences were found in binding affinity among any of these mutants compared to Ba wild-type BclA-CTD. In addition, with multimeric VLR4, we observed strong binding to recombinant BclA-CTD from each species. However, we were able to confirm the previous observation that VLR4 is specific for Ba spores, as distinct from recombinant Ba BclA, and little, if any, binding to Bc spores (Supplemental Figure S3) (Herrin et al., 2008). We also tested VLR4 binding to modified Ba Sterne spores with a shortened central collagen-like region of 51 residues (strain CLT314) that is much shorter than Ba BclA (228 residues), Bc BclA (123 residues), or Bt BclA (117 residues). VLR4 binds this modified BclA on Ba spores nearly as well as wild type (Supplemental Figure S3). We interpret these results to mean that VLR4 recognizes and discriminates among BclAs only in the context of bacterial spores, independent of the length of the collagen-like region, and not with regard to the respective recombinant proteins, when expressed in Escherichia coli.
Figure 3. Species Specificity of VLR4.
Comparison of VLR4 bound to BclA (A) with Bacillus sequence polymorphisms mapped onto the BclA structure (B) shows that VLR4 recognizes a surface on BclA that is largely devoid of sequence differences seen in the different Bacillus strains. Differences from B. anthracis Sterne that are found in B. cereus T are colored in red, B. thuringiensis Kurstaki in blue and those that differ from B. anthracis Sterne in both strains in purple. See also supplemental Figure S2.
On the spore, BclA is heavily O-glycosylated on its central collagen-like region (Daubenspeck et al., 2004), and it is possible that additional glycosylation occurs on the C-terminal domain in Bc and Bt BclA, although we do not observe this in Ba BclA (C. Turnbough, unpublished data). VLR4 binds across a short stretch of threonine residues (amino acids 183-185, Supplemental Figure S2) and would be prevented from binding if these residues were sites of O-linked glycosylation. To test this hypothesis, we treated Ba and Bc spores with sodium meta-periodate, which partially removed the glycans from the spore surface. Deglycosylation of Bc spores significantly enhances the VLR4 binding to this strain supporting our hypothesis that the VLR4 epitope on BclA is a site of O-glycosylation on Bc spores (Figure 4). Binding of VLR4 to deglycosylated Ba spores was unaltered compared to non-treated spores indicating that treatment did not disrupt or enhance VLR4 interactions with the BclA epitope. While we expect this treatment removed the majority of glycans, a partial adduct likely remains attached to the protein that would reduce binding to the treated Bc spores compared to the Ba spores.
Figure 4. Glycosylation of B. cereus Spores Blocks VLR4 Binding.
Bacillus spores were treated with periodate to remove the majority of the glycans from the surface of the spores. VLR4 is unable to bind the non-treated B. cereus T spores. However, after treatment with periodate, VLR4 recognizes B. cereus T spores to a much greater extent than unmodified spores suggesting that glycosylation of the B. cereus T spores prevents VLR4 binding. Periodate treatment has no effect on the strong binding of VLR4 to B. anthracis Sterne spores.
Discussion
The crystal structure of VLR4-BclA complex marks only the third VLRB-antigen complex to be determined thus far, and common themes for VLR-antigen recognition are now beginning to emerge. VLRs use their concave surface created by the LRR motifs to interact with antigens, as distinct from TLRs, which can use this concave surface, but also other surface regions and faces for primary interactions with antigen. Although the number of LRRs that contact antigen in VLRs is variable, antigen does not contact the entire concave surface in much the same way that antibodies do not usually use their entire combining site surface for contacting antigen. The LRRCT-loop is key to the binding of antigens by VLRB, similar to the focused role of the variable heavy chain and the complementarity-determining region H3 in conventional vertebrate antibodies, especially for smaller antigens. VLR binding of antigen seems to be consistently focused on the C-terminal LRRs (Figure 5) proximal to the extended loop of LRRCT. This bias is most evident in the minimal use of the LRRNT. Although this motif contains variable residues at potential antigen-contacting positions, VLR4 is the only VLRB so far that has been demonstrated to bind to antigen using LRRNT but, even so, only makes minimal contacts to BclA. Although this may change as more examples become available, the evidence so far suggests that VLRs bind antigens primarily using their C-terminal repeats, with the N-terminal repeats being used less frequently, perhaps reflecting the importance of LRRCT in binding and specificity.
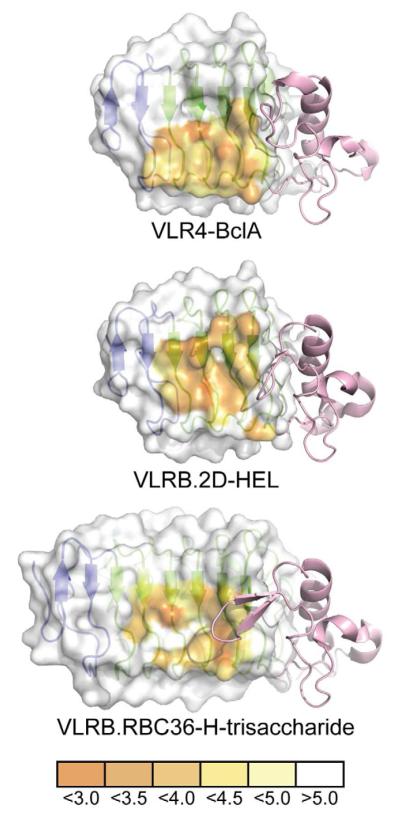
Figure 5. Comparison of VLRB Binding Interfaces with Different Antigens.
The concave surface of each VLRB is represented as a surface highlighting the proximity of each amino acid to its respective antigen in angstroms. The surface used by VLRB to contact antigens favors the use of the C-terminal LRR motifs and VLR4 is heavily biased towards the used of the C-terminal ends of the ß-strands of these motifs. The LRRCT motif (pink) of each VLRB shows dramatically different lengths and conformations in their LRRCT-loops.
It was noted in both the VLRB.2D-hen-egg lysozyme (HEL) and VLR.RBC36-H-trisaccharide complex structures that the LRRCT-loop contains an aromatic residue that makes important contacts with antigen. For VLR.RBC36, a Trp residue sandwiches the H-trisaccharide against the concave surface of the VLR (Han et al., 2008) and shields the LRRCT-loop from further interaction with the ligand. For VLRB.2D, the LRRCT-loop is inserted into the active site of HEL, burying Leu and Asn residues and positioning a Tyr near other aromatic residues in the active site (Velikovsky et al., 2009). In contrast, VLR4 lacks an aromatic residue in the LRRCT-loop. However, similar to VLRB.2D, VLR4 uses both side-chain and main-chain atoms of LRRCT for hydrogen bonding with ligand. VLR4 also makes use of the region between the LRRCT-loop and the structurally conserved Cys167, which is located on the C-terminal side of the loop, where Asn164 hydrogen bonds to BclA (Figure 2 and Supplemental Figure S1). Whereas VLRB.2D inserts its LRRCT-loop directly into a deep pocket, the LRRCT-loop of VLR4 meanders across a shallow groove on BclA such that Gly158 at the loop apex does not contact BclA. This latter mode of interaction is reminiscent of VLRA.R2.1, which uses the LRRCT-loop to bind a flat surface on the antigen rather than in a pocket (Deng et al., 2010).
VLR4 mutagenesis experiments and the work by Herrin et al. leads us to hypothesize that, although the set of BclA-binding VLRBs (Herrin et al., 2008) are highly similar in sequence, they likely recognize BclA with subtle differences. The exceptional degree of similarity, particularly in the highly variable LRRCT-loop, would suggest that they all recognize a similar epitope on BclA. However, the binding differences observed by our mutagenesis seem to indicate that the other identified VLRB clones could rotate on BclA (Figure 6). The proposed rotation would bring the more N-terminal variable positions on the concave surface ß-strands into greater contact with antigen, while the C-terminal positions would have a diminished role in antigen binding. Such structural variation in antigen recognition among these VLRBs would account for the observed variation among the isolated VLRB at position 106 and allow an Arg55 to contact Glu130 on BclA accounting for the previously ascribed importance of this position. In fact, comparison of the available VLRB-antigen complex structures demonstrates that VLR4 is much more shifted towards the use of the C-terminal ends of the ß-strands for antigen recognition than seen previously (Han et al., 2008; Velikovsky et al., 2009). The proposed rotation of sequence-related BclA-binding VLRBs on the surface of BclA would allow contact with VLRB positions that interact with antigen in the other structures (Figure 5).
Figure 6. BclA-binding VLRBs Likely Recognize Subtly Different Epitopes.
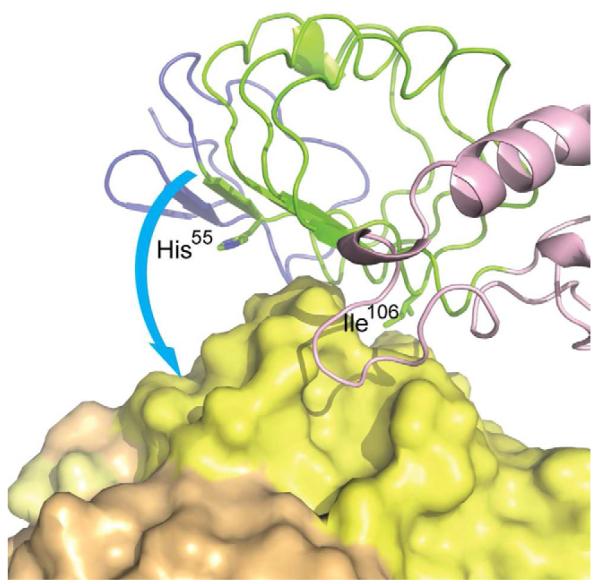
VLRBs with high amino-acid sequence homology to VLR4 likely bind a similar epitope on BclA. Nevertheless, an Arg at position 55 of other VLRBs was determined to be important for binding and position 106 can be Ile, Asn, Asp or Thr, suggesting that these VLRB recognize BclA slightly differently than VLR4. The proposed rotation of VLR4 on BclA is indicated as a hypothesized recognition mode for these VLRBs.
Although VLR4 shows remarkable specificity for the spores of Ba over Bc and Bt, the epitope on BclA is relatively conserved across these strains. While some neighboring sequence differences exist on the surface of BclA, mutation of these residues singly or in combination does not abrogate binding to VLR4 (Table 3). Thus, we conclude that VLR4 discrimination of Bacillus species only occurs in the context of the bacterial spore. Glycosylation of Bc spores blocks recognition by VLR4 and likely is the mode of species discrimination by VLR4. VLR4 binding to deglycosylated Bc spores was less than to Ba spores, which we attribute to partial residual sugar adducts on BclA. However, an additional possibility is the differential packing of proteins in the exosporium limiting the accessibility to the VLR4 epitope on the underside of BclA in these species (Sylvestre et al., 2003; Ball et al., 2008). Indeed, it is possible that the VLR4 epitope is not accessible to the Fab regions of conventional Ig-antibodies, for example, owing to their much larger size (Nuttall et al., 2011). These data on fine discrimination of BclA on spores, therefore, suggest that VLR4 would make an excellent diagnostic tool for the identification B. anthracis spores and encourages testing of VLR4 against a greater panel of Bacillus cereus group strains. No natural polymorphisms in BclA-CTD are found in B. anthracis species, but are found among strains of B. cereus and B. thuringiensis species. Sequences identical to the Ba, Bc or Bt BclA-CTD amino-acid sequences can be found in both B. cereus, and B. thuringiensis species. Because VLR4 discriminates on the basis of glycosylation of a conserved protein epitope, use of VLR4 to recognize B. anthracis spores overcomes the issue of recognition of BclA having amino-acid sequences identical to the other Bacillus species. Use of VLR4 as a diagnostic tool able to identify B. anthracis spores despite naturally occurring aminoacid polymorphisms would be invaluable for the identification of bioterrorism agents with obvious biodefense applications.
The structure presented here shows that VLR4 interacts with BclA using its concave surface and LRRCT-loop similar to other VLRB-antigen complexes. This structure combined with the mutagenesis and binding studies of VLR4 reveals that sequence-related VLRBs likely recognize the BclA epitope with slight differences. It is interesting to speculate that BclA binding by sequence-related VLRBs (Herrin et al., 2008) may share a common precursor which then diversified to bind the BclA epitope in subtly different ways or may, instead, have arisen by parallel evolution. However, this determination will have to wait for a clearer understanding of VLRB assembly, diversification and selection. Importantly, our studies of VLR4 species specificity have revealed that VLR4 can be used as an excellent diagnostic tool for the identification of B. anthracis spores.
EXPERIMENTAL PROCEDURES
Monomeric VLR4 Expression and Purification
Amino acids 22-167 of VLR4 were cloned into pBAC6 using the SacII and BamHI restriction sites putting the gene in frame with an N-terminal gp64 signal peptide and 6xHis-tag. This construct lacks the C-terminal 82 amino acids containing the Thr/Pro stalk, putative GPI linkage site and Cys-rich region used for multimerization. The VLR4-pBAC6 transfer vector was co-transfected with ProFold-ER1 (AB Vector) into Sf9 cells using Cellfectin (Invitrogen). Recombinant baculovirus was amplified to a titer of approximately 109 pfu/mL. This virus was used to infect Sf9 cells at a density of 2.0 × 106 cells/mL at a multiplicity of infection of six. Cultures were harvested after two days of shaking at 28°C. Culture supernatants were buffer exchanged into 10 mM TrisCl pH 8.0, 300 mM sodium chloride, 20 mM imidazole and passed over Ni-NTA (Qiagen). The resin was washed with 10 mM HEPES pH 6.9, 300 mM NaCl, 30 mM imidazole, and protein was eluted in the same buffer with 300 mM imidazole. Protein fractions were concentrated and run on a Superdex 75 size exclusion column in 50 mM HEPES pH 6.9, 100 mM NaCl. The protein eluted at the predicted molecular weight of the monomeric species. The protein was finally purified by cation exchange on a MonoS column in 50 mM Bis-TrisCl pH 6.5 and eluted with a gradient of 0-0.5 M NaCl. Purified protein was buffer exchanged into 50 mM HEPES pH 6.8 and concentrated to an A280 of 8.1.
BclA Mutagenesis, Expression and Purification
The recombinant B. anthracis BclA protein was from the Sterne strain, which has 400 amino acids (Boydston et al., 2005). The C-terminal 138 amino acids (residues 263-400, which includes the CTD) were cloned into pET28b with an N-terminal His6-tag and TEV cleavage site. The numbering of the amino acids in BclA varies among strains due to length polymorphism of the collagen-like region. The numbering of BclA presented here corresponds to B. anthracis strain Ceb 9732 for ease of comparison with previously determined structures of the BclA-CTD (residues 77-214). Mutagenesis of this construct to disrupt a common crystal packing interaction of apo BclA-CTD (Boydston et al., 2005; Liu et al., 2008) was accomplished by MPIPE (Klock et al., 2008) and resulted in Thr185 to Asn mutation. This construct also contains a C-terminal cloning artifact encoding the sequence FRFRN. Vectors containing the BclA-CTD coding sequences were transformed into E. coli BL21(DE3) (Novagen). Cultures were induced with IPTG at an OD600 of 0.8 and expression was carried out overnight at 16°C. Cells were lysed using a continuous flow cell-disruptor in 50 mM sodium phosphate pH 7.5, 500 mM NaCl, 30 mM imidazole. Cleared lysates were bound to Ni-NTA and washed in the same buffer. The protein was eluted in 50 mM sodium phosphate, 500 mM NaCl, 300 mM imidazole, concentrated and run on a Superdex 200 size exclusion column in 50 mM TrisCl pH 8.0, 150 mM NaCl where the protein eluted as a trimeric species. These protein fractions were concentrated to an A280 of 9.0. The BclA-CTD-wt protein was treated similarly; however, the expressed protein was recalcitrant towards being further concentrated.
VLR4-BclA Crystallization
VLR4-BclA-CTD-Thr185Asn crystals grew in 15% PEG 6000, 1.2 M LiCl, 0.1 M sodium citrate pH 5.26, as clusters of thin plates after 3-7 days and were cryoprotected in 25% glycerol for storage and data collection. Diffraction data were collected at APS beamline 23ID-D. We initially located three BclA monomers and a single VLR4 by molecular replacement (MR) using 2R6Q.pdb for BclA and 2R9U.pdb for a VLRB as MR models, and placed an additional two VLR4 molecules by hand into residual electron density after the initial MR solution.
Crystals of BclA-CTD-Pro159Ser grew in 0.7 M ammonium phosphate monobasic, 0.1 M sodium citrate pH 5.0 at room temperature in a hexagonal plate morphology and were cryoprotected in a 1:1 mixture of 5 M sodium malonate pH 6.4 and mother liquor. Diffraction data were collected at ALS beamline 8.2.1 and indexed and processed with HKL2000 (Otwinoski and Minor, 1997). Phasing was done by MR with Phaser (McCoy et al., 2007) using 2R6Q.pdb as a MR model for BclA. We located a single BclA-CTD monomer in the asymmetric unit with identical crystal packing to 2R6Q.pdb.
Rigid body and restrained refinements were performed in Refmac (Murshudov et al., 1997) and Buster-TNT (Blanc et al., 2004) with the final refinements performed in Refmac. A summary of the data processing and refinement can be found in Table 1. The VLR4-BclA structure was analyzed using HBPLUS (McDonald and Thornton, 1994), MS (Connolly, 1993), Contacsym (Sheriff et al., 1987), and Molprobity (Davis et al., 2007) to assess hydrogen-bonding, buried surface areas, van der Waals contacts and structure quality, respectively.
Binding of VLR4 and BclA-CTD
Binding of VLR4 to mutant BclA-CTD was determined using biolayer interferometry on an OctetRED (ForteBio). DNA encoding the peptide MSGLNDIFEAQKIEWHE was inserted N-terminal of the BclA-CTD coding sequence for biotin ligation. Protein constructs were expressed as above and prepared in 100 mM TrisCl pH 8.1 to which 10X reaction buffer was added (final concentrations 10 mM ATP disodium salt, 10 mM magnesium acetate and 50 μM D-biotin) and 100 μg of BirA, biotin ligase. The reaction was incubated overnight at 37°C and then buffer exchanged on a Superdex 200 16/60 column. The BclA proteins were not concentrated. Biotinylated protein was loaded onto streptavidin biosensors and incubated with a dilution series of monomeric VLR4 produced as described above. Curve fitting was done using the OctetRED analysis software.
For surface plasmon resonance experiments, wild-type BclA-CTD proteins were prepared as above without the biotinylation peptide or reaction with BirA. VLR4 mutants were generated by MPIPE and expressed and purified as the monomeric wild-type protein. VLR4 affinity was analyzed using a Biacore 3000 (GE) instrument. Recombinant BclA was coupled to a Biacore CM5 chip using amine chemistry. A reference channel was created by capping off one channel of the EDC/NHS activated CM5 chip with ethanolamine. Recombinant monomeric VLR4 was injected over the biosensor surface in duplicate at a flow rate of 30 μl/min in 10 mM MES, 150 mM NaCl, 0.05% Tween-20, 3 mM EDTA, pH 6.5. MES buffer without VLR4 was injected during the course of the experiment for double referencing. The surface was regenerated after each cycle with a 30 sec pulse of 50 mM NaOH. Sensorgrams were processed using BiaEvaluation (GE) and Scrubber2 software (BioLogic software, Australia). Sensorgrams were zeroed on the y-axis and bulk refractive index changes were removed by subtracting the reference channel.
Periodate treatment of Bacillus spores
B. anthracis and B. cereus spores (1 × 106) were washed twice with 0.1 M sodium acetate buffer, pH 5.5, then incubated with 20 mM sodium meta-periodate (Thermo Scientific) in acetate buffer at room temperature for 1 to 6 hr . After incubation, the spores were washed three times with 0.1 M Tris buffer, pH 8.0, before staining with purified, multivalent VLR4 or a non-specific control VLRB (C4.39) in FACS buffer. The spore samples were acquired using an Accuri C6 flow cytometer and analyzed with CFLOW software.
Supplementary Material
Highlights.
VLRBs use their C-terminal LRRs and the LRRCT-loop to interact with antigen Sequence-related VLRBs exhibit differential recognition of their BclA epitopes VLR4 binds a conserved protein epitope, yet is specific for B. anthracis spores
ACKNOWLEDGEMENTS
We gratefully acknowledge Robyn Stanfield for help with data processing, the Joint Center for Structural Genomics (www.jcsg.org) for help with protein crystallization and use of their quality control server, and Sylvia A. McPherson for preparation of Bacillus spores for the FACS experiments. Support is acknowledged from NIH AI042266 to IAW, AI081777 to CLT, and AI072435 to MDC, and the Basic Science Research Program (2011-0027449) and the Global Frontier (NRF-M1AXA002-2010-0029770) through the National Research Foundation of Korea to BWH. GM/CA CAT has been funded in whole or in part with Federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Sciences (Y1-GM-1104). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract No. DE-AC02-06CH11357. The Berkeley Center for Structural Biology is supported in part by the National Institutes of Health, National Institute of General Medical Sciences, and the Howard Hughes Medical Institute. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Footnotes
ACCESSION NUMBERS Coordinates for VLR4-BclA-CTD-Thr185Asn and BclA-CTD-Pro159Ser structures have been deposited in the Protein Data Bank with accession codes 3TWI and 3TYJ, respectively.
SUPPLEMENTAL INFORMATION Supplemental Information includes three figures and can be found with this article online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing financial interests. This is manuscript 21227 from TSRI.
REFERENCES
- Alder MN, Rogozin IB, Iyer LM, Glazko GV, Cooper MD, Pancer Z. Diversity and function of adaptive immune receptors in a jawless vertebrate. Science. 2005;310:1970–1973. doi: 10.1126/science.1119420. [DOI] [PubMed] [Google Scholar]
- Ball DA, Taylor R, Todd SJ, Redmond C, Couture-Tosi E, Sylvestre P, Moir A, Bullough PA. Structure of the exosporium and sublayers of spores of the Bacillus cereus family revealed by electron crystallography. Mol. Microbiol. 2008;68:947–958. doi: 10.1111/j.1365-2958.2008.06206.x. [DOI] [PubMed] [Google Scholar]
- Bella J, Hindle KL, McEwan PA, Lovell SC. The leucine-rich repeat structure. Cell. Mol. Life Sci. 2008;65:2307–2333. doi: 10.1007/s00018-008-8019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc E, Roversi P, Vonrhein C, Flensburg C, Lea SM, Bricogne G. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- Boydston JA, Chen P, Steichen CT, Turnbough CL., Jr Orientation within the exosporium and structural stability of the collagen-like glycoprotein BclA of Bacillus anthracis. J. Bacteriol. 2005;187:5310–5317. doi: 10.1128/JB.187.15.5310-5317.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly ML. The molecular surface package. J. Mol. Graph. 1993;11:139–141. doi: 10.1016/0263-7855(93)87010-3. [DOI] [PubMed] [Google Scholar]
- Cooper MD, Alder MN. The evolution of adaptive immune systems. Cell. 2006;124:815–822. doi: 10.1016/j.cell.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Daubenspeck JM, Zeng H, Chen P, Dong S, Steichen CT, Krishna NR, Pritchard DG, Turnbough CL., Jr Novel oligosaccharide side chains of the collagen-like region of BclA, the major glycoprotein of the Bacillus anthracis exosporium. J. Biol. Chem. 2004;279:30945–30953. doi: 10.1074/jbc.M401613200. [DOI] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Velikovsky CA, Xu G, Iyer LM, Tasumi S, Kerzic MC, Flajnik MF, Aravind L, Pancer Z, Mariuzza RA. A structural basis for antigen recognition by the T cell-like lymphocytes of sea lamprey. Proc. Natl. Acad. Sci. USA. 2010;107:13408–13413. doi: 10.1073/pnas.1005475107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P, Hirano M, Herrin BR, Li J, Yu C, Sadlonova A, Cooper MD. Dual nature of the adaptive immune system in lampreys. Nature. 2009;459:796–801. doi: 10.1038/nature08068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han BW, Herrin BR, Cooper MD, Wilson IA. Antigen recognition by variable lymphocyte receptors. Science. 2008;321:1834–1837. doi: 10.1126/science.1162484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrin BR, Alder MN, Roux KH, Sina C, Ehrhardt GR, Boydston JA, Turnbough CL, Jr., Cooper MD. Structure and specificity of lamprey monoclonal antibodies. Proc. Natl. Acad. Sci. USA. 2008;105:2040–2045. doi: 10.1073/pnas.0711619105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JA, Cooper M, Schroeder-Tucker L, Black S, Miller D, Karns JS, Manthey E, Breeze R, Perdue ML. A field investigation of Bacillus anthracis contamination of U.S. Department of Agriculture and other Washington, D.C., buildings during the anthrax attack of October 2001. Appl. Environ. Microbiol. 2003;69:593–599. doi: 10.1128/AEM.69.1.593-599.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasamatsu J, Sutoh Y, Fugo K, Otsuka N, Iwabuchi K, Kasahara M. Identification of a third variable lymphocyte receptor in the lamprey. Proc. Natl. Acad. Sci. USA. 2010;107:14304–14308. doi: 10.1073/pnas.1001910107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klock HE, Koesema EJ, Knuth MW, Lesley SA. Combining the polymerase incomplete primer extension method for cloning and mutagenesis with microscreening to accelerate structural genomics efforts. Proteins. 2008;71:982–994. doi: 10.1002/prot.21786. [DOI] [PubMed] [Google Scholar]
- Liu CQ, Nuttall SD, Tran H, Wilkins M, Streltsov VA, Alderton MR. Construction, crystal structure and application of a recombinant protein that lacks the collagen-like region of BclA from Bacillus anthracis spores. Biotechnol. Bioeng. 2008;99:774–782. doi: 10.1002/bit.21637. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nuttall SD, Wilkins ML, Streltsov VA, Pontes-Braz L, Dolezal O, Tran H, Liu CQ. Isolation, kinetic analysis, and structural characterization of an antibody targeting the Bacillus anthracis major spore surface protein BclA. Proteins. 2011;79:1306–1317. doi: 10.1002/prot.22971. [DOI] [PubMed] [Google Scholar]
- Otwinoski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pancer Z, Amemiya CT, Ehrhardt GR, Ceitlin J, Gartland GL, Cooper MD. Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature. 2004;430:174–180. doi: 10.1038/nature02740. [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Iyer LM, Liang L, Glazko GV, Liston VG, Pavlov YI, Aravind L, Pancer Z. Evolution and diversification of lamprey antigen receptors: evidence for involvement of an AID-APOBEC family cytosine deaminase. Nat. Immunol. 2007;8:647–656. doi: 10.1038/ni1463. [DOI] [PubMed] [Google Scholar]
- Sheriff S, Silverton EW, Padlan EA, Cohen GH, Smith-Gill SJ, Finzel BC, Davies DR. Three-dimensional structure of an antibody-antigen complex. Proc. Natl. Acad. Sci. USA. 1987;84:8075–8079. doi: 10.1073/pnas.84.22.8075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer RC. Bacillus anthracis. J. Clin. Pathol. 2003;56:182–187. doi: 10.1136/jcp.56.3.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steichen C, Chen P, Kearney JF, Turnbough CL., Jr Identification of the immunodominant protein and other proteins of the Bacillus anthracis exosporium. J. Bacteriol. 2003;185:1903–1910. doi: 10.1128/JB.185.6.1903-1910.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiecki MK, Lisanby MW, Shu F, Turnbough CL, Jr., Kearney JF. Monoclonal antibodies for Bacillus anthracis spore detection and functional analyses of spore germination and outgrowth. J. Immunol. 2006;176:6076–6084. doi: 10.4049/jimmunol.176.10.6076. [DOI] [PubMed] [Google Scholar]
- Sylvestre P, Couture-Tosi E, Mock M. Polymorphism in the collagen-like region of the Bacillus anthracis BclA protein leads to variation in exosporium filament length. J. Bacteriol. 2003;185:1555–1563. doi: 10.1128/JB.185.5.1555-1563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Li M, Turnbough CL., Jr An unusual mechanism of isopeptide bond formation attaches the collagen-like glycoprotein BclA to the exosporium of Bacillus anthracis. MBio. 2011;2:e00084–00011. doi: 10.1128/mBio.00084-11. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Velikovsky CA, Deng L, Tasumi S, Iyer LM, Kerzic MC, Aravind L, Pancer Z, Mariuzza RA. Structure of a lamprey variable lymphocyte receptor in complex with a protein antigen. Nat. Struct. Mol. Biol. 2009;16:725–730. doi: 10.1038/nsmb.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.