Abstract
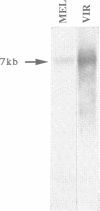
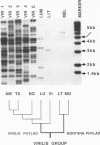
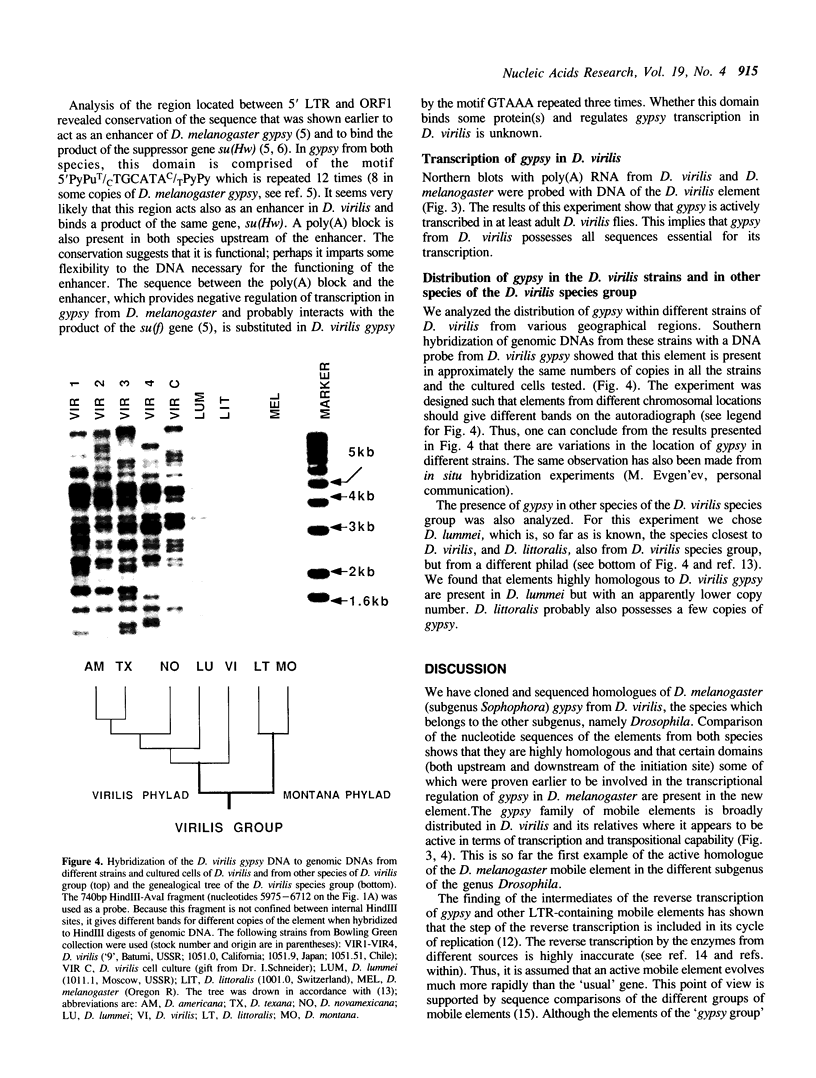
The homologue of the Drosophila melanogaster mobile element gypsy was cloned from the distantly related species D. virilis. It has three ORFs highly homologous to those of the element from D. melanogaster. gypsy from D. virilis appears to be actively transcribed and is capable of transposition. Comparison of the untranslated regions of both elements revealed conserved sequences including those which had previously been demonstrated to be important in transcription regulation. Distribution of gypsy among the different strains of D. virilis and different species within the D. virilis group was analyzed. Possible involvement of horizontal transmission in the process of spreading and evolution of gypsy is discussed.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arkhipova I. R., Mazo A. M., Cherkasova V. A., Gorelova T. V., Schuppe N. G., Llyin Y. V. The steps of reverse transcription of Drosophila mobile dispersed genetic elements and U3-R-U5 structure of their LTRs. Cell. 1986 Feb 28;44(4):555–563. doi: 10.1016/0092-8674(86)90265-5. [DOI] [PubMed] [Google Scholar]
- Bender W., Akam M., Karch F., Beachy P. A., Peifer M., Spierer P., Lewis E. B., Hogness D. S. Molecular Genetics of the Bithorax Complex in Drosophila melanogaster. Science. 1983 Jul 1;221(4605):23–29. doi: 10.1126/science.221.4605.23. [DOI] [PubMed] [Google Scholar]
- Blackman R. K., Meselson M. Interspecific nucleotide sequence comparisons used to identify regulatory and structural features of the Drosophila hsp82 gene. J Mol Biol. 1986 Apr 20;188(4):499–515. doi: 10.1016/s0022-2836(86)80001-8. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., Corces V. G. Transcription and reverse transcription of retrotransposons. Annu Rev Microbiol. 1989;43:403–434. doi: 10.1146/annurev.mi.43.100189.002155. [DOI] [PubMed] [Google Scholar]
- Colot H. V., Hall J. C., Rosbash M. Interspecific comparison of the period gene of Drosophila reveals large blocks of non-conserved coding DNA. EMBO J. 1988 Dec 1;7(12):3929–3937. doi: 10.1002/j.1460-2075.1988.tb03279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels S. B., Peterson K. R., Strausbaugh L. D., Kidwell M. G., Chovnick A. Evidence for horizontal transmission of the P transposable element between Drosophila species. Genetics. 1990 Feb;124(2):339–355. doi: 10.1093/genetics/124.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle R. F., Feng D. F., Johnson M. S., McClure M. A. Origins and evolutionary relationships of retroviruses. Q Rev Biol. 1989 Mar;64(1):1–30. doi: 10.1086/416128. [DOI] [PubMed] [Google Scholar]
- Heberlein U., Rubin G. M. Structural and functional comparisons of the Drosophila virilis and Drosophila melanogaster rough genes. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5916–5920. doi: 10.1073/pnas.87.15.5916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassis J. A., Poole S. J., Wright D. K., O'Farrell P. H. Sequence conservation in the protein coding and intron regions of the engrailed transcription unit. EMBO J. 1986 Dec 20;5(13):3583–3589. doi: 10.1002/j.1460-2075.1986.tb04686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konsolaki M., Komitopoulou K., Tolias P. P., King D. L., Swimmer C., Kafatos F. C. The chorion genes of the medfly, Ceratitis capitata, I: Structural and regulatory conservation of the s36 gene relative to two Drosophila species. Nucleic Acids Res. 1990 Apr 11;18(7):1731–1737. doi: 10.1093/nar/18.7.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatowski J., Ayala F. J. Drosophila virilis Cu-Zn superoxide dismutase gene sequence. Nucleic Acids Res. 1989 Mar 11;17(5):2133–2133. doi: 10.1093/nar/17.5.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlor R. L., Parkhurst S. M., Corces V. G. The Drosophila melanogaster gypsy transposable element encodes putative gene products homologous to retroviral proteins. Mol Cell Biol. 1986 Apr;6(4):1129–1134. doi: 10.1128/mcb.6.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazo A. M., Mizrokhi L. J., Karavanov A. A., Sedkov Y. A., Krichevskaja A. A., Ilyin Y. V. Suppression in Drosophila: su(Hw) and su(f) gene products interact with a region of gypsy (mdg4) regulating its transcriptional activity. EMBO J. 1989 Mar;8(3):903–911. doi: 10.1002/j.1460-2075.1989.tb03451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael W. M., Bowtell D. D., Rubin G. M. Comparison of the sevenless genes of Drosophila virilis and Drosophila melanogaster. Proc Natl Acad Sci U S A. 1990 Jul;87(14):5351–5353. doi: 10.1073/pnas.87.14.5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizrokhi L. J., Georgieva S. G., Ilyin Y. V. jockey, a mobile Drosophila element similar to mammalian LINEs, is transcribed from the internal promoter by RNA polymerase II. Cell. 1988 Aug 26;54(5):685–691. doi: 10.1016/s0092-8674(88)80013-8. [DOI] [PubMed] [Google Scholar]
- Mizrokhi L. J., Mazo A. M. Evidence for horizontal transmission of the mobile element jockey between distant Drosophila species. Proc Natl Acad Sci U S A. 1990 Dec;87(23):9216–9220. doi: 10.1073/pnas.87.23.9216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizrokhi L. J., Obolenkova L. A., Priimägi A. F., Ilyin Y. V., Gerasimova T. I., Georgiev G. P. The nature of unstable insertion mutations and reversions in the locus cut of Drosophila melanogaster: molecular mechanism of transposition memory. EMBO J. 1985 Dec 30;4(13B):3781–3787. doi: 10.1002/j.1460-2075.1985.tb04148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modolell J., Bender W., Meselson M. Drosophila melanogaster mutations suppressible by the suppressor of Hairy-wing are insertions of a 7.3-kilobase mobile element. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1678–1682. doi: 10.1073/pnas.80.6.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. A., Coyne J. A. The genetics of postzygotic isolation in the Drosophila virilis group. Genetics. 1989 Mar;121(3):527–537. doi: 10.1093/genetics/121.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhurst S. M., Harrison D. A., Remington M. P., Spana C., Kelley R. L., Coyne R. S., Corces V. G. The Drosophila su(Hw) gene, which controls the phenotypic effect of the gypsy transposable element, encodes a putative DNA-binding protein. Genes Dev. 1988 Oct;2(10):1205–1215. doi: 10.1101/gad.2.10.1205. [DOI] [PubMed] [Google Scholar]
- Ricchetti M., Buc H. Reverse transcriptases and genomic variability: the accuracy of DNA replication is enzyme specific and sequence dependent. EMBO J. 1990 May;9(5):1583–1593. doi: 10.1002/j.1460-2075.1990.tb08278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey S. N., Lansman R. A., Brock H. W., Grigliatti T. A. Distribution and conservation of mobile elements in the genus Drosophila. Mol Biol Evol. 1986 Nov;3(6):522–534. doi: 10.1093/oxfordjournals.molbev.a040413. [DOI] [PubMed] [Google Scholar]
- Treier M., Pfeifle C., Tautz D. Comparison of the gap segmentation gene hunchback between Drosophila melanogaster and Drosophila virilis reveals novel modes of evolutionary change. EMBO J. 1989 May;8(5):1517–1525. doi: 10.1002/j.1460-2075.1989.tb03536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]