Abstract
Glioblastoma multiforme (GBM) is the most lethal primary brain tumor. Extensive proliferation and dispersal of GBM tumor cells within the brain limits patient survival to approximately 1 year. Hence, there is a great need for the development of better means to treat GBM. Receptor protein tyrosine phosphatase (PTP)µ is proteolytically cleaved in GBM to yield fragments that promote dispersal of GBM cells. While normal brain tissue retains expression of full-length PTPµ, low-grade human astrocytoma samples have varying amounts of full-length PTPµ and cleaved PTPµ. In the highest-grade astrocytomas (i.e., GBM), PTPµ is completely proteolyzed into fragments. We demonstrate that short hairpin RNA mediated knockdown of full-length PTPµ and PTPµ fragments reduces glioma cell growth and survival in vitro. The reduction in growth and survival following PTPµ knockdown is enhanced when cells are grown in the absence of serum, suggesting that PTPµ may regulate autocrine signaling. Furthermore, we show for the first time that reduction of PTPµ protein expression decreases the growth and survival of glioma cells in vivo using mouse xenograft flank and i.c. tumor models. Inhibitors of PTPµ could be used to reduce the growth and survival of GBM cells in the brain, representing a promising therapeutic target for GBM.
Keywords: cell growth, cell survival, glioma, PTPmu (PTPµ), receptor protein tyrosine phosphatase
Gliomas, the most common primary brain tumors, arise from the supportive glial cells of the CNS.1,2 Astrocytomas (a type of glioma) are classified into 4 grades (I to IV) by the World Health Organization (WHO) based on histopathological features.3 Low-grade astrocytomas (LGAs), corresponding to WHO grades I–III, have a better prognosis and survival time than do grade IV astrocytomas, known as glioblastoma multiforme (GBM). GBM is the most common, aggressive, and lethal form of glioma, with an overall prognosis of 12–14 months survival.3,4 Treatment for GBM includes maximal surgical resection, chemotherapy, and radiotherapy.5
GBM is characterized by complex cell heterogeneity.6 Recent therapeutic strategies are aimed at targeting glioma stem-like cells and the molecular signaling pathways they affect.7,8 These glioma stem-like cells constitute a small proportion of the bulk of tumor cells in the brain and often express surface markers, including CD133, CD15/SSEA, and A2B5.7–11 Having higher self-renewal capacity than other tumor cells, glioma stem-like cells are believed to contribute to radioresistance and chemoresistance that are responsible for tumor recurrence after initial surgical resection and therapy.7,10,12
The malignancy of GBM can be attributed to various genetic alterations of components involved in critical signaling pathways contributing to cell survival, growth, migration, and dispersal.6,13,14 Altered receptor tyrosine kinase (RTK) signaling pathways are commonly found in GBM.6,13,14 Aberrant activation of RTK signaling allows tumor cells to stimulate their own growth (autocrine signaling), resulting in enhanced cell growth and survival contributing to GBM initiation and progression.6,13,14 Hence, current treatment strategies aimed at combination therapies involving multiple targets in cell growth and survival signaling are being investigated for GBM.5,15–21
Tyrosine phosphorylation is a reversible process, catalyzed by both protein tyrosine kinases and protein tyrosine phosphatases (PTPs).22–25 Several PTPs have been identified as tumor suppressors in various cancers,24–26 including GBM.22,26 The disruption of PTPs could synergize with RTK signaling to promote tumor growth, survival, and dispersal.22,24,26
PTPµ is a transmembrane receptor-like PTP with 2 intracellular tyrosine phosphatase domains. PTPµ is a member of the type IIb subfamily, which has an extracellular segment with a meprin-A5 antigen-PTPµ (MAM) domain, an immunoglobulin domain, and 4 fibronectin type III repeats. PTPµ mediates homophilic cell–cell adhesion via its extracellular segment and transduces signals from extracellular cues to the intracellular environment via its phosphatase activity.25,27–29 PTPµ protein expression is upregulated at high cell density in primary astrocytes30 and may play a role in regulating contact inhibition of growth.31 One of the early events in tumor progression is an inability of cells to undergo contact inhibition of growth and movement in response to external cues. Interestingly, low-grade human astrocytoma tumors express varying levels of full-length PTPµ protein.30 Similar to LGAs, the U-87 malignant glioma (MG) human glioma cell line expresses full-length PTPµ protein in tissue culture.30 In contrast, in the LN-229 human glioma cell line and higher-grade human GBM tumors, the full-length PTPµ protein is proteolytically processed by a series of proteases.30,32 The full-length PTPµ protein is normally cleaved in the Golgi complex by a furin-like protease giving rise to an extracellular segment (the E-subunit) that is noncovalently attached to the remainder of the protein, which includes a small, residual portion of the extracellular segment, the transmembrane domain, and intracellular segment (the P-subunit).32 In gliomas, the P-subunit is further cleaved at the membrane by an α-secretase (ADAM [A Disintegrin And Metalloprotease] or matrix metalloproteinase) generating a shed extracellular segment and a PΔE fragment that lacks the extracellular segment but retains the transmembrane domain and therefore remains at the plasma membrane.32 This α-secretase cleavage primes PΔE for cleavage by γ-secretase within the membrane, resulting in a membrane-free fragment (the intracellular domain [ICD]) that can translocate to the nucleus.32 Human GBM tumors and GBM xenograft tumors in nude mice retain the expression of the extracellular-domain (ECD) and intracellular (PΔE and ICD) fragments.32,33 The extracellular fragment of PTPµ is a novel biomarker of GBM that can be used as a diagnostic tool to noninvasively detect GBM in vivo.33
We previously demonstrated that PTPµ fragments positively regulate the migration of glioma cells in vitro.32 Phosphatase activity of the fragments is likely important in regulating cell migration, as inhibition of PTPµ phosphatase activity using a peptide inhibitor targeting the wedge motif in the ICD of PTPµ blocked glioma cell migration in vitro.32 In addition, PTPµ fragments are required for LN-229 glioma cell survival in vitro.32 It is not known if PTPµ regulates glioma cell survival in vivo.
Our previous results raise the question of whether proteolysis of PTPµ plays a role in regulating cell survival in vivo by rendering the cell refractory to normal extracellular cues. In our present study, we used the U-87 MG human glioma cell line that expresses full-length PTPµ and the LN-229 human glioma cell line in which full-length PTPµ is constitutively proteolyzed into PTPµ fragments.32 We demonstrate that PTPµ undergoes proteolysis in U-87 MG cells in postconfluent states in vitro and in glioma xenograft tumors of U-87 MG cells in vivo. The pattern of PTPµ fragments generated in U-87 MG cells under these conditions is similar to the pattern of fragments present in the LN-229 cells, where PTPµ undergoes constitutive proteolysis. We demonstrate that CD133+ glioma stem-like cells contain predominantly PTPµ fragments, unlike CD133− cells (which do not have stem-cell properties in our model). Knockdown of PTPµ in U-87 MG results in a decrease in cell number and viability in growth curves in vitro and in colony formation assays. We demonstrate that the cell growth and viability defect observed upon PTPµ knockdown in U-87 MG glioma cells is enhanced in the absence of exogenous growth factors, suggesting a role for PTPµ in regulating autocrine signaling mechanisms. Finally, we show that knockdown of PTPµ in human glioma cells reduces GBM tumor growth in vivo in heterotopic and orthotopic xenograft tumors in nude mice. Taken together, our studies suggest that proteolytic fragments of PTPµ promote GBM tumor growth and survival in vivo. Hence, PTPµ could represent a promising target for novel therapeutics against GBM.
Materials and Methods
Cell Culture
The U-87 MG and LN-229 human glioma cell lines were obtained from American Type Culture Collection. Cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with either 10% (for U-87 MG) or 5% (for LN-229) fetal bovine serum (FBS; HyClone) and antibiotic-antimycotic (Invitrogen) in a humidified atmosphere of 5% CO2 at 37°C. For studying the growth of cells in the absence of exogenous growth factors, cells were cultured in DMEM without the addition of FBS.
Isolation and Characterization of Glioma Stem-like Cells
Glioma stem-like cells were isolated from primary human brain tumor specimens from patients or human glioblastoma xenografts as previously described34 in accordance with a protocol approved by the Case Comprehensive Cancer Center Institutional Review Board and concurrent with national regulatory standards, with patients signing informed consent. Briefly, specimens from tumor CW421 were disaggregated by a Papain Dissociation System (Miltenyi Biotech) and filtered with a 70-µm cell strainer according to the manufacturer's instructions. Cells were then cultured in Neurobasal media (Invitrogen), supplemented with the following: B27 lacking vitamin A (Invitrogen), 2 mM l-glutamine (Invitrogen), 1 mM sodium pyruvate (Invitrogen), 10 ng/mL basic fibroblast growth factor (R&D Systems), and 10 ng/mL epidermal growth factor (R&D Systems) for at least 4 hours to recover surface antigens. Isolated cells were separated with a magnetic sorting column using microbead-conjugated CD133 antibodies (Miltenyi). CW421 CD133+ cells formed neurospheres in serum-free medium, were self-perpetuating, differentiated into neuronal and glial phenotypes when exposed to serum, were tumorigenic in 7 of 8 nude mice, and were thus deemed “stem-like.” CW421 CD133− cells were not self-perpetuating or tumorigenic and were thus nonstem glioma cells.
Lentiviral Infection
The lentiviral short hairpin (sh)RNA construct V2LHS_171008 (Open Biosystems) or TRCN0000 273327 (Sigma-Aldrich), targeting a region in the extracellular domain or the 3′ untranslated region of human PTPµ mRNA, respectively, was used to suppress PTPµ expression. Both shRNA constructs were equally efficient in reducing PTPµ protein levels as tested by immunoblotting. A lentiviral shRNA construct (control for V2LHS_171008) was a gift from Drs E. Johnson and R. Keri (Case Western Reserve University). Another lentiviral shRNA construct, pLKO.1 (control for TRCN0000273327), was purchased from Sigma-Aldrich. A lentiviral construct encoding green fluorescent protein (GFP) was generated in our lab by cloning the insert sequence for GFP35 into the pCDH1-MCS (multiple cloning site) 2 Cloning and Expression Vector (System Biosciences). Production of lentiviral particles and infection of the U-87 MG cell line were performed as previously described.30 Lentiviruses were titered using the Retro-Tek HIV-1 p24 Antigen Enzyme-Linked Immunosorbent Assay (ZeptoMetrix Corporation). The concentration of HIV-1 p24 antigen and the number of transducing units (TUs) per milliliter for every picogram of p24 antigen present were calculated using a protocol from Sigma-Aldrich. The number of transducing units (TU) of PTPµ shRNA and control shRNA lentivirus were 2.68 × 106 TU and 3.35 × 106 TU, respectively, for U-87 MG and LN-229, plated at a density of 2 × 105 cells. This TU for PTPµ shRNA was sufficient to reduce PTPµ expression as assessed by immunoblot.
Immunoblotting
Cells were lysed in a buffer containing 100 mM Tris pH 7.5, 1% Triton-X-100, 0.1% sodium dodecyl sulfate (SDS), 140 mM NaCl, 0.5% sodium deoxycholate, 5 mM EDTA, protease inhibitors (1 mM benzamidine, 5 µg/mL aprotinin, 5 µg/mL leupeptin, 1 µg/mL pepstatin A), and phosphatase inhibitors (1 mM sodium fluoride [NaF], 1 mM sodium orthovanadate [NaVO3]) on ice for 30 min. Cell lysates were spun at 10 000 rpm for 3 min, and clarified lysates were resolved by SDS-polyacrylamide gel electrophoresis (PAGE). Proteins were transferred to nitrocellulose membranes and blocked using Tris/Tween-buffered saline (TTBS) containing 5% (weight/volume [w/v]) nonfat dry milk. All antibodies were diluted in the same blocking buffer. Full-length and fragments of PTPµ protein were detected on immunoblots using monoclonal antibodies to PTPµ (SK18) that recognize the membrane-proximal phosphatase domain in the intracellular segment of PTPµ.36 SK18 and anti-vinculin antibodies (Sigma-Aldrich) were detected using goat anti-mouse secondary antibodies conjugated to horseradish peroxidase (Jackson Immuno Research Laboratories). Secondary antibodies were detected by chemiluminescence using an Image Quant LAS 4000 camera system (General Electric Healthcare) capable of capturing digital images. Densitometry values were calculated using ImageJ software.37 Vinculin was used as a control for equal protein loading. To reprobe for vinculin, blots were stripped by rocking in Re-Blot Plus Strong Solution (Millipore) diluted in distilled water for 15 min at room temperature. Stripped blots were washed twice with blocking buffer (TTBS containing 5% [w/v] nonfat dry milk) and then incubated with anti-vinculin antibodies.
To collect lysates for pre- and postconfluent states, cells were harvested either at less than100% or at 100% confluency in the lysis buffer described above. To collect lysates of cells growing in an in vivo 3D environment, U-87 MG glioma cells suspended in Matrigel (BD Biosciences) were injected subcutaneously into the right flank of female athymic nude mice (National Institutes of Health [NIH]). Three-week-old tumors were excised, snap-frozen on dry ice for transport, and stored at −80°C. Frozen tissue was thawed, ground using a Pro 200 tissue tearor (PRP Scientific), and homogenized using a dounce homogenizer in lysis buffer containing 20 mM Tris pH 7.0, 0.5% Triton-X-100, 5 mM EDTA, 2 mM ethylene glycol tetraacetic acid (EGTA), protease inhibitors (1 mM phenylmethanesulfonylfluoride, 3 mM benzamidine, 5 µg/mL aprotinin, 5 µg/mL leupeptin, 1 µg/mL pepstatin A), and phosphatase inhibitors (30 mM NaF, 2 mM NaVO3, 40 mM glycerol 2-phosphate). Cell lysates were then centrifuged at 10 000 rpm for 6 min. The cleared supernatant was resolved by SDS-PAGE and immunoblotted as described above.
Collection of human GBM tumor tissue and nontumor normal tissue was in accordance with an approved protocol from the University Hospitals Case Medical Center Institutional Review Board, as previously described, along with lysate preparation of the tissue samples.30,32
To collect lysates of glioma stem-like cells, the cells were washed once with 1X phosphate buffered saline (PBS) and harvested with sample buffer (10% SDS, 1 M dithiothreitol, 1 M Tris pH 6.8 and glycerol) preheated to 100°C. Harvested cells were sonicated and centrifuged at 10 000 rpm for 5 min. The cleared supernatant was collected, boiled for 5 min, resolved by SDS-PAGE, and immunoblotted as described above.
Cell Growth Assay
To study growth, uninfected U-87 MG cells and cells infected with lentivirus encoding either control shRNA or PTPµ shRNA were trypsinized and plated at a density of 2 × 105 cells 1 day post-infection. Cell counts of adherent cells were taken in triplicate for 11 days by counting cells on a hemocytometer. Prior to counting, cells were imaged using 10× and 40× objectives on a TE-200 inverted microscope (Nikon Instruments) with a SPOT–Real Time camera and SPOT software version 3.2 (Diagnostic Instruments). Growth medium was replenished every day by removing half of the old medium and adding half fresh medium. Cell viability of adherent cells was determined using trypan blue to label dead cells. Data were analyzed from 3 independent experiments, and error bars represent standard error.
Colony Formation Assay
The colony formation assay using crystal-violet staining to assess growth factor–independent cell survival was performed as previously described.32 Data were quantitated from 4 independent experiments performed in triplicate and were analyzed for statistical significance using an unpaired Student's t-test.
Heterotopic Xenograft Flank Tumors
Animals were maintained in the Animal Core Facility at Case Western Reserve University according to institutional policies. All animal procedures were in accordance with approved protocols from the Institutional Animal Care and Use Committee. U-87 MG cells that were uninfected or infected with lentivirus encoding either control or PTPµ shRNA were trypsinized 3 days after infection and resuspended at a concentration of 2 × 104 cells/µL in PBS. A total of 250–300 µL of a 1:1 suspension of cells plus Matrigel (BD Biosciences) was injected subcutaneously into the right flank of NIH athymic nude female mice (6–8 weeks old). After 21 days, the animals were sacrificed and imaged using the Maestro FLEX In Vivo Imaging system (Caliper Life Sciences), followed by tumor removal. The length, width, and height of tumors were measured using calipers (Fine Science Tools) and then the tumors were snap-frozen in dry ice for transport and processed for preparing lysates for immunoblotting or for cryosectioning and immunohistochemistry. Data were collected from at least 3 animals injected per condition, and error bars denote standard error. Statistical significance was determined using an unpaired Student's t-test.
Orthotopic Xenograft Brain Tumors
U-87 MG and LN-229 glioma cells were infected with lentivirus to express GFP and either control or PTPµ shRNA. Cells were trypsinized 2 days after infection and resuspended to a concentration of 1.25 × 104 cells/µL of PBS. Cells were injected i.c. into NIH athymic nude female mice using a stereotactic device (David Kopf Instruments). Animals were anesthetized using 50 mg/kg of a 1:1 ketamine/xylazine solution by an i.p. injection. An incision was made and a burr hole (0.7 mm) was drilled in the skull 2 mm to the right and 0.5 mm anterior to the bregma. Cells were injected into the right striatum of the brain to a depth of 4.5 mm at a rate of 1 µL/min. The incision was closed with sutures and the animals were allowed to heal. After 21 days, the animals were sacrificed and the brains were removed and imaged using the Maestro FLEX In Vivo Imaging System with appropriate filters for GFP (tumor; excitation = 445–490 nm, emission = 515 nm long-pass filter, acquisition settings = 500–720 in 10-nm steps). Tissue autofluorescence spectra were collected from nontumor regions of the brain. The multispectral fluorescence images were background-subtracted and unmixed using Maestro software (Caliper Life Sciences). The brains were then frozen for sectioning followed by immunohistochemistry. Data were collected from at least 3 animals per condition.
Immunohistochemistry
Flank tumors or brains injected with GFP-labeled tumor cells were excised and fixed in 4% paraformaldehyde in PEM buffer (80 mM Pipes, 5 mM EGTA, 1 mM Magnesium chloride, 3% sucrose), pH 7.4. The fixative was then sequentially replaced with 10% sucrose in PBS and 25% sucrose in PBS before embedding in Tissue Freezing Medium (TFM Electron Microscopy Sciences). For flank tumors, 7-µm-thick sections were stained for anti-human vimentin (NeoMarkers Clone SP20) to detect human tumor cells and were counterstained with hematoxylin and eosin (H&E) to study tissue morphology. TFM was washed off the section with water. The section was incubated in block buffer containing PBS, 2% goat serum, 0.25% saponin, and 0.5% bovine serum albumin to permeabilize the cells and block nonspecific binding. Blocked tissue was incubated overnight at 4°C with anti-human vimentin antibodies (NeoMarkers) that were then detected using AlexaFluor 568 goat anti-rabbit (Invitrogen) secondary antibodies by incubation for 2 h at room temperature. Fluorescent images were captured using a 40× objective on a DMI 6000B automated inverted microscope (Leica Microsystems) fitted with a Retiga EXi camera (QImaging). For brain tumors, fixed brain tissue was cryosectioned at 5µm intervals from the anterior end of the brain until the GFP-expressing tumor cells were no longer visible under a microscope equipped with fluorescence. Sections 5 µm thick were examined and imaged for GFP-expressing tumor cells using a 10× objective on a Leica DMI 6000B automated inverted microscope, prior to staining with H&E. H&E staining was done second to prevent loss of GFP signal during staining.
Results
Cleavage of PTPµ Increases at Postconfluence and in In Vivo Conditions
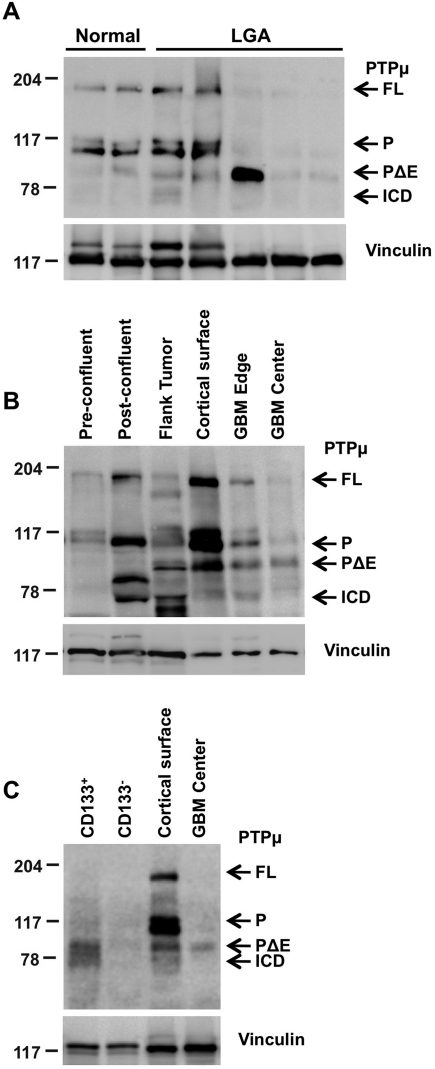
Full-length PTPµ was expressed as a 200-kDa protein and as 2 noncovalently associated fragments of ∼100 kDa each, the E- and P-subunits (Fig. 1A).29,38,39 At the time of our original study, in which we examined PTPµ protein expression in LGAs,30 we did not know that PTPµ was further proteolyzed into biologically active fragments. Therefore, when we examined PTPµ protein expression in LGAs by immunoblotting, samples were run on 6% gels to detect the 200-kDa and 100-kDa bands. However, the smaller fragments of PTPµ were undetectable due to migration off the gels.30 To understand the importance of proteolysis of PTPµ in glioma progression, we characterized the protein levels of both full-length PTPµ and PTPµ fragments in human samples from normal brain tissue, LGAs, and GBM tumors by running them on 8% gels to detect the fragments on an immunoblot (Fig. 1A and B). LGA samples from different patients express varying levels of full-length PTPµ protein30 and PTPµ fragments (Fig. 1A). GBM tissue from the edge of a tumor has both full-length PTPµ and PTPµ fragments, because the edge sample also contains some normal tissue (Fig. 1B), whereas GBM tissue from the center of the tumor almost exclusively expresses PTPµ fragments (Fig. 1B). In some samples, additional cleavage fragments are observed, but we have yet to identify these fragments. Hence, proteolysis of PTPµ may be one step in the progression of gliomas from low grade to high grade.
Fig. 1.
PTPµ proteolysis varies with astrocytoma grade, growth conditions, and in glioma stem-like cells. Lysates were separated by 8% SDS-PAGE. Proteins were transferred onto nitrocellulose membranes and immunoblotted using antibodies against the intracellular segment of PTPµ (SK18). Immunoblots were stripped and reprobed for vinculin to control for loading. Full-length PTPµ (FL) is detected at 200 kDa on an immunoblot. Full-length PTPµ is normally cleaved into 2 ∼100-kDa fragments that remain noncovalently associated. The intracellular segment (P) is detected at ∼100 kDa and is further proteolytically cleaved into PΔE and ICD fragments (∼83 and 74 kDa, respectively) in gliomas. (A) Relative expression of full-length PTPµ and PTPµ intracellular fragments from noncancerous brain tissue (epilepsy), a cortical surface sample from noncancerous brain tissue of a GBM patient, and LGAs are shown. (B) Expression of full-length PTPµ and PTPµ fragments in parental U-87 MG cells growing either in vitro in 2D tissue culture dishes at pre- and postconfluent states or in vivo in flank tumor xenografts. Matched normal cortical surface sample from nontumor brain tissue of a GBM patient (cortical surface) and cancerous tissue from the edge and center of a tumor (GBM edge and center) are shown as controls for the expression of full-length PTPµ and PTPµ fragments. (C) Expression of PTPµ fragments in CD133+ cells compared with CD133− cells.
Since the extracellular domain, or ectodomain, shedding of molecules at the cell surface increases at postconfluence,40 we hypothesized that there would be an increase in the cleavage of PTPµ protein with an increase in cell confluency. In standard, 2D tissue culture at approximately 80%–90% confluence, U-87 MG glioma cells express full-length PTPµ and noncovalently associated E- and P-subunits of PTPµ, with little to no PΔE or ICD fragments (Fig. 1B). Postconfluent U-87 MG cells had a high level of full-length and noncovalently associated fragments of PTPµ compared with the preconfluent state but also had a high level of ICD fragment (Fig. 1B), supporting our hypothesis. To further test the hypothesis that PTPµ proteolysis is density dependent, we collected lysates of U-87 MG cells grown in a nude mouse flank tumor, an in vivo 3D environment akin to an in vitro high-density culture. Full-length PTPµ was almost completely proteolyzed into PTPµ fragments in flank xenograft tumors (Fig. 1B). This confirms our hypothesis that there is increased PTPµ cleavage into PTPµ fragments at high cell density in vitro and in vivo.
Previous studies have identified a small proportion of human brain tumor cells to be glioma stem-like cells that express CD133 and have high self-renewal capacity compared with other cells in the tumor.10 In our studies, CD133+ glioma stem-like cells express predominantly PTPµ fragments similar to tissue derived from the center of GBM tumor from human patients (Fig. 1C). In contrast, PTPµ fragment expression was absent in CD133− cells (Fig. 1C). Matched normal, nontumor cortical surface tissue and GBM tumor tissue from the center of the tumor from the same patient are shown as a reference for the migration of full-length PTPµ and PTPµ fragments (Fig. 1C).
PTPµ Regulates Growth and Viability of Glioma Cells In Vitro
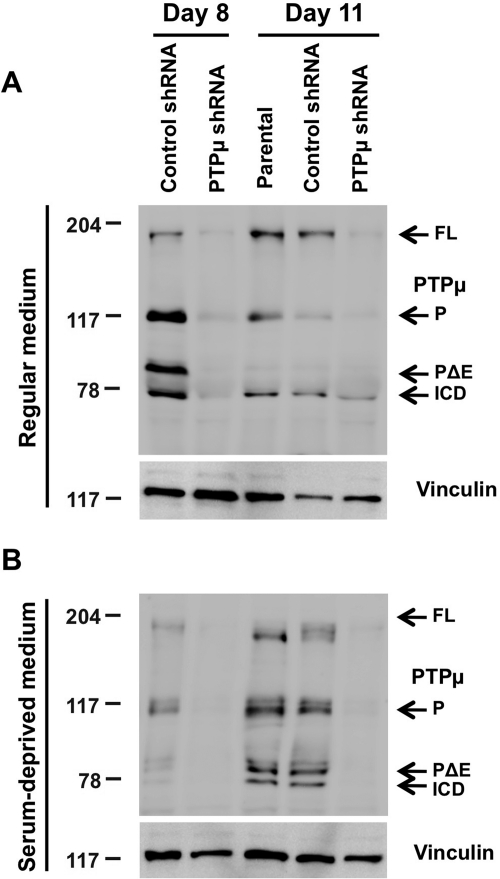
To study the role of full-length PTPµ and PTPµ fragments in regulating growth of U-87 MG human glioma cells, we compared growth curves of cells infected with lentivirus encoding control or PTPµ shRNA for 11 days. To mimic the growth factor independence of GBM tumor cells in vivo, growth curves were performed in the presence and absence of exogenous serum growth factors. In our experiments, viable adherent and floating cells were counted separately every day, and cell lysates were collected for immunoblot analysis on days 8 and 11, when cells were considered to be at a postconfluent state (Fig. 2A and B, data from adherent cells are shown). U-87 MG glioma cells infected with lentivirus encoding PTPµ shRNA showed a reduction in PTPµ protein levels when compared with cells infected with control shRNA lentivirus in serum-containing and serum-deprived growth conditions (Fig. 2A and B). Based on densitometry analysis of U-87 MG samples infected with either control shRNA or PTPµ shRNA on days 8 and 11, there was a 56.18% ± 6.86% knockdown of protein levels of both full-length PTPµ (200 kDa) and proteolytically processed forms of PTPµ (P, PΔE, and ICD) in cells infected with PTPµ shRNA compared with controls in serum-containing conditions (Fig. 2A). PTPµ shRNA–infected cells grown in serum-deprived conditions had a 67.03% ± 8.93% reduction in full-length PTPµ and PTPµ fragment expression compared with controls (Fig. 2B). Therefore, PTPµ shRNA reduced the expression of both full-length PTPµ and PTPµ fragments (Fig. 2A and B).
Fig. 2.
shRNA-mediated reduction of PTPµ protein levels. (A) Parental U-87 MG cells or those infected with lentivirus encoding either control shRNA or PTPµ shRNA were grown in medium containing serum. Cells were lysed on days 8 and 11, separated by 8% SDS-PAGE, and immunoblotted for antibodies against the intracellular segment of PTPµ (SK18). Immunoblots were stripped and reprobed for vinculin to control for loading. (B) Lysates of parental U-87 MG cells or cells expressing control shRNA or PTPµ shRNA growing in serum-deprived medium for 8 and 11 days were separated by 8% SDS-PAGE, immunoblotted using antibodies against the intracellular segment of PTPµ (SK18), and reprobed for vinculin.
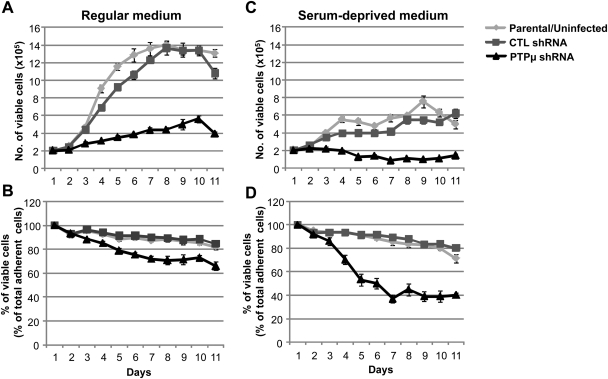
Control shRNA–infected U-87 MG cells grew exponentially in tissue culture medium containing serum until day 8, after which they reached saturation density and entered into a stationary phase, defined as confluence (Fig. 3A). In contrast, PTPµ shRNA–infected U-87 MG cells proliferated at a significantly reduced rate in regular culture medium (Fig. 3A), resulting in an 18.65% ± 3.68% decrease (P< .001) in viable cell number on day 8 compared with control-infected cells (Fig. 3B). In the absence of serum, U-87 MG cells infected with lentivirus encoding control shRNA were able to grow and survive, albeit at much reduced levels compared with the serum-containing cultures (Fig. 3C). Moreover, in the absence of serum, U-87 MG cells infected with lentivirus encoding PTPµ shRNA had a 43.18% ± 5.27% reduction (P< .0001) in viable cell number after 8 days in culture compared with controls (Fig. 3C and D). This suggests that while the control shRNA–infected cells have autocrine survival signals, the cells with reduced PTPµ expression lacked a robust autocrine signaling mechanism.
Fig. 3.
Knockdown of PTPµ reduces growth and viability of glioma cells in vitro. U-87 MG cells infected with lentivirus encoding either control shRNA or PTPµ shRNA were grown in media with serum (A and B) or without serum (C and D). Total number of viable adherent cells in both types of growth conditions are shown in (A) and (C). The percentage of viable adherent cells (as a percentage of total adherent cells) in both growth conditions is plotted in (B) and (D). Counts of adherent cells were taken every day for 11 days, and cell viability was assessed by trypan blue exclusion. The number and percentage of viable adherent cells (as a percentage of total adherent cells) are plotted. The data represent mean ± SE of 3 independent experiments.
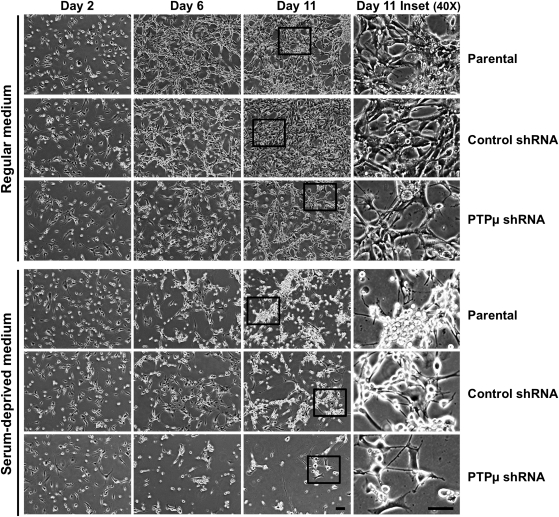
In standard tissue culture conditions, U-87 MG cells grew close to one another and formed a network. After reaching saturation density, U-87 MG cells did not occupy the entire space on a tissue culture plate as depicted in the photomicrographs of day 11 (Fig. 4, parental and control shRNA). Instead, the cells tended to grow in close proximity to one another (Fig. 4). As was observed in the growth curves (Fig. 3), serum deprivation resulted in an appreciable reduction in cell number in PTPµ shRNA–infected cells compared with controls observed in the photomicrographs after 6 and 11 days in culture (Fig. 4). Additionally, U-87 MG cells infected with either control or PTPµ shRNA-encoding lentivirus had a rounded-up morphology in the absence of serum (Fig. 4).
Fig. 4.
Knockdown of PTPµ reduces cell growth of glioma cells. (A) Parental U-87 MG cells or those infected with lentivirus encoding either control shRNA or PTPµ shRNA were grown in medium (with or without serum) for 11 days and imaged at 100× and 400× magnification. Representative images of cells on days 2, 6, and 11 are shown. The cells in the inset in day 11 images were imaged with the 40x objective and are shown at 400× magnification to depict cell morphology. Scale bar, 100 µm.
PTPµ Regulates Growth Factor–Independent Survival of Glioma Cells In Vitro
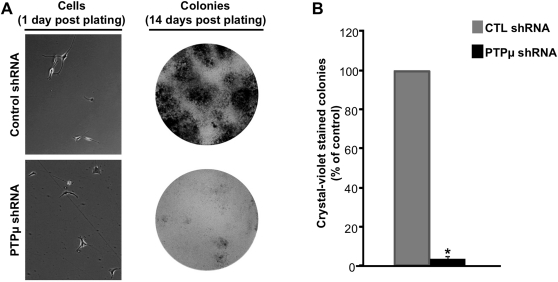
Our previous studies demonstrated that shRNA-mediated reduction of PTPµ in LN-229 cells decreased colony formation in vitro in colony formation assays.32 To test the ability of U-87 MG cells to survive in vitro under growth factor–independent conditions, we performed colony formation assays.41 In our studies, control- or PTPµ shRNA–infected U-87 MG cells were plated at low density and allowed to form colonies for 2 weeks. PTPµ shRNA–infected cells had a 96.93% ± 0.88% reduction (P< 0.0001) in colony formation compared with control cells (Fig. 5A and B). These data further suggest that PTPµ is involved in regulating autocrine responses in GBM cells to promote survival.
Fig. 5.
Knockdown of PTPµ reduces growth factor–independent survival of glioma cells in vitro. (A) U-87 MG cells infected with lentivirus encoding either control shRNA or PTPµ shRNA were plated at low density (representative images at 200× magnification are shown) and deprived of growth factors. Cells were allowed to form colonies for 14 days. Crystal violet–stained colonies are shown in (A). The number of colonies is graphically represented in (B). The data represent mean ± SE of 4 independent experiments performed in triplicate. *Statistically significant reduction in colony formation (P< .0001, n = 4).
PTPµ Regulates Glioma Cell Survival In Vivo
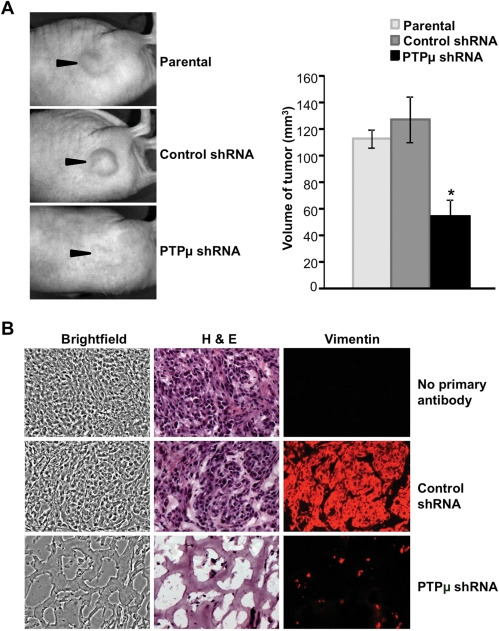
To determine whether the survival defect observed upon reduction of PTPµ protein expression can be translated to an in vivo environment, parental, control-infected or PTPµ shRNA–infected U-87 MG cells were mixed with Matrigel and injected subcutaneously into the flank region of nude mice. Twenty-one days post-injection, animals were sacrificed, and the tumors were excised. The tumors formed from cells infected with control shRNA lentivirus were similar in volume to the tumors formed from uninfected U-87 MG cells (Fig. 6A). In contrast, the volume of tumors from PTPµ shRNA–infected cells was significantly less (57.49% ± 12.7% reduction, n= 3, P< .05) than that of the tumors formed from control shRNA–infected cells (Fig. 6A). Cryosections of the flank tumors were immunolabeled with anti-human vimentin antibody to highlight human tumor cells. PTPµ shRNA–expressing tumors showed a marked reduction in the number of vimentin-positive cells surviving in the tumor compared with control shRNA–expressing tumors (Fig. 6B). Staining of the tumor cryosections with H&E to label cell nuclei and the extracellular matrix demonstrated that the few surviving cells in PTPµ shRNA–expressing tumors were scattered in the Matrigel that formed the bulk of the tumor (Fig. 6B).
Fig. 6.
Knockdown of PTPµ reduces glioma cell survival in vivo at a heterotopic site. (A) Parental U-87 MG cells or cells infected with lentivirus encoding either control shRNA or PTPµ shRNA were resuspended in Matrigel and injected subcutaneously into the flank region of nude mice. Animals were sacrificed and images were taken (arrowheads point toward tumors in the flank). Volume of the excised tumor was calculated using length, width, and height measurements and plotted. The data represent mean ± SE of 3 animals per condition tested. *Statistically significant reduction in volume of the tumor growing from cells expressing either control shRNA or PTPµ shRNA (P< .05, n= 3). (B) Mouse flank tumor xenografts of parental U-87 MG cells or those expressing either control shRNA or PTPµ shRNA were cryosectioned and immunolabeled with antibodies against human vimentin protein to detect injected tumor cells, then histochemically stained with hematoxylin and eosin (H&E). Images of sections showing brightfield, H&E staining, and vimentin immunolabeling are shown at 400× magnification.
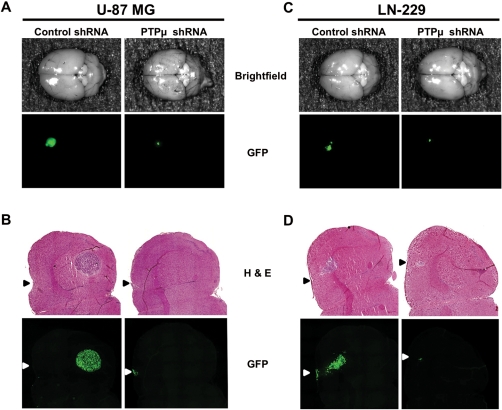
To assess glioma cell survival in vivo in an orthotopic environment, we performed i.c. injections of U-87 MG and LN-229 glioma cells coinfected with lentiviruses encoding GFP protein (to visualize tumor growth over time) and either control or PTPµ shRNA into the brains of nude mice. After 21 days, excised brains were imaged for GFP-expressing tumor cells. Control shRNA–expressing cells formed large tumors, as demonstrated by strong GFP signal intensity at the site of injection in the mouse brain striatum (Fig. 7A and C). In contrast, mice injected with PTPµ shRNA–expressing cells had a fainter GFP signal at the site of injection (Fig. 7A and C). Cryosections of the brains revealed that control shRNA–expressing cells formed large tumors in the striatum and close to the corpus callosum, as evidenced in both the H&E staining and GFP fluorescence (Fig. 7B and D). In contrast, no tumors were observed either with H&E staining or GFP fluorescence in the striatum or near the corpus callosum in the brains of mice injected with PTPµ shRNA–expressing cells (Fig. 7B and D). The small spot of fluorescence observed in brains injected with PTPµ shRNA–expressing cells appears to be tiny clusters of GFP-expressing cells present at the site of the injections (Fig. 7B and D). These results demonstrate that the absence of PTPµ protein in glioma cells has a dramatic effect on their ability to survive in the brain. Because U-87 MG and LN-229 cells preferentially express PTPµ fragments in vivo, these data suggest that proteolyzed PTPµ is required for tumor growth and survival in vivo.
Fig. 7.
Knockdown of PTPµ reduces orthotopic glioma cell survival in vivo. U-87 MG cells in (A) and (B) or LN-229 cells in (C) and (D) infected with lentivirus encoding either control shRNA or PTPµ shRNA were injected i.c. into nude mice and allowed to grow for 21 days. Images of the dorsal surface of whole brain showing GFP-expressing injected tumor cells are shown for U-87 MG (A) and LN-229 (C). Similar results were observed from 3 animals for each treatment. Intracranial mouse xenografts of U-87 MG (B) or LN-229 (D) cells infected with lentivirus encoding either control or PTPµ shRNA were cryosectioned and imaged for GFP fluorescence. Cryosections were then stained with H&E to depict the location of the tumor. Arrowheads represent the site of injection on the dorsal surface of the brain.
Discussion
The development of novel targeted therapeutics to treat GBM is essential due to the poor survival time postdiagnosis. Currently, small molecule inhibitors targeting specific components of growth factor–dependent survival signaling pathways deregulated in GBM are in clinical trials.5 For example, erlotinib (Tarceva, OSI-774, Genentech) and gefinitib (Iressa, ZD1839, AstraZeneca), specific small molecule inhibitors of the epidermal growth factor receptor tyrosine kinase, have been evaluated for GBM therapy without much improvement in survival of GBM patients.5 Like aberrant RTK signaling, PTPs are deregulated in GBM,22 implicating them as potential therapeutic targets. In the present article, we show that proteolyzed PTPµ is required for glioma cell survival in vitro and in vivo, suggesting that PTPµ is a promising therapeutic target for GBM.
In humans, normal brain tissue expresses full-length PTPµ, whereas LGAs express varying levels of full-length PTPµ and PTPµ fragments (Fig. 1A). In contrast, in GBM, full-length PTPµ is proteolyzed into PTPµ fragments (Fig. 1A and Burgoyne et al.32). Hence, we propose that as astrocytomas advance from low grade to high grade, they progressively lose full-length PTPµ protein by proteolysis and that the highest-grade astrocytoma, GBM, retains only PTPµ fragments. Relative levels of full-length PTPµ and PTPµ fragments in human tumors may be useful for assessing tumor grade. We also observed expression of PTPµ fragments in CD133+ glioma stem-like cells but not in CD133− cells (Fig. 1C). Since glioma stem-like cells have higher self-renewal capacity than other tumor cells and are highly resistant to chemotherapy and radiotherapy,7,10,12 we suggest that the expression of PTPµ fragments may contribute to their growth advantage in the brain.
Although U-87 MG cells are GBM derived, they expressed some level of full-length PTPµ similar to some LGAs30 when grown in 2D tissue culture at preconfluent states (Fig. 1B). However, at postconfluence in 2D tissue culture conditions, U-87 MG cells proteolyzed full-length PTPµ into fragments (Fig. 1B). Further, when U-87 MG cells were at high cell density in vivo, full-length PTPµ is completely proteolyzed into fragments similar to LN-229 glioma cells, human GBM tumors, and CD133+ glioma stem-like cells (Fig. 1A–C,32). Hence, our data suggest that glioma cell lines proteolyze PTPµ in a manner similar to GBM tumors when grown to postconfluency in vitro or at high cell density in tumor xenografts in vivo. Given that in LN-229 cells, where PTPµ is proteolyzed predominantly into PTPµ fragments, the effects of PTPµ knockdown mimic those in U87-MG cells, we hypothesize that the results we observe in our functional assays following PTPµ shRNA can be attributed to the loss of the PTPµ fragments.
We hypothesized that the PTPµ fragments present in glioma cells could promote GBM tumor growth and survival. In support of this hypothesis, we observed a decrease in the growth of glioma cells following PTPµ knockdown in tissue culture (Fig. 3A). During tumorigenesis, cells detach from the main tumor and disperse. When tumor cells disperse, they need to adapt to a new environment that may lack growth factors. Therefore, the tumor cells activate autocrine signaling, which enables them to grow. This condition can be partly mimicked in vitro by growing them in media without serum. Under these conditions, U-87 MG cells lacking PTPµ were unable to grow and survive (Figs. 3–5). Strikingly, PTPµ-depleted glioma cells failed to form tumors in intracranial xenograft tumor models (Fig. 7), demonstrating that proteolyzed PTPµ is essential for the growth and survival of GBM cells in the brain. Since PTPµ ICD fragments can translocate to the nucleus,32 they may regulate gene transcription. Given the results of our current study, we hypothesize that PTPµ ICD regulates transcription of genes whose products are involved in growth and survival signaling in GBM. For example, the Ras/mitogen-activated protein kinase and phosphatidylinositol 3-kinase/Akt signaling pathways are major downstream components of RTK signaling that regulate GBM cell growth and survival.6,14,42 Future studies will directly test the roles of the PΔE and ICD of PTPµ fragments in regulating growth and survival in glioma cells by overexpressing individual fragments in PTPµ-depleted cells.
In normal astrocytes, full-length PTPµ is upregulated at high cell density, suggesting that PTPµ could play a role in cell density–regulated phenomena like contact inhibition of growth.30 GBM is characterized by rapid cell proliferation.4,6 Contact inhibition of growth is a phenomenon of inhibition of cell growth when cells contact each other in vitro in 2D tissue culture dishes.43,44 Contact inhibition is important in normal cells to maintain tissue architecture and homeostasis.43 During tumorigenesis, cells lose contact inhibition to enable uncontrolled cell growth.43 Proteases are also upregulated during tumorigenesis.45 One of the mechanisms proposed for loss of contact inhibition during tumorigenesis is proteolysis of cell–cell adhesion receptors31 that alters the cell's ability to respond to normal extracellular cues. For example, cell surface molecules like epithelial cell adhesion molecule (Ep-CAM),46 leukocyte common antigen-related phosphatase (LAR),47,48 PTPκ,40 and PTPµ32,33 are proteolytically cleaved. Proteolytic cleavage of PTPµ results in shedding of the extracellular domain, or ectodomain, which results in cells losing contact with one another.32 This can potentially contribute to tumorigenesis by loss of contact inhibition of growth and constitutive activation of RTK signaling.
Our studies suggest that GBM tumors specifically retain PTPµ fragments that contribute to growth and survival of GBM cells in the brain. A peptide inhibitor that blocks PTPµ function inhibits glioma cell migration in vitro.32 Further studies are required to determine if the peptide inhibitor can also block GBM tumor growth in vivo and could serve as a promising therapeutic strategy for the treatment of GBM.
Funding
This study was supported by the Case Center for Imaging Research, National Institutes of Health grants R01-NS051520 (S.M.B.-K.), R01-NS063971 (S.M.B.-K., J.P.B. and A.E.S.), the Peter D. Cristal Chair (A.E.S.), and the Kimble Fund (A.E.S.). This work is dedicated to Tabitha Yee-May Lou, whose endowment supports brain tumor research in the Brady-Kalnay lab.
Acknowledgments
The authors thank Catherine Doller and Scott Howell, PhD, of the Visual Sciences Research Center for providing histologic services and quantitation of data; Scott A. Becka and Jing Wang for technical assistance; and Sonya E. Craig, PhD, for assistance editing the manuscript.
Conflict of interest statement. None declared.
References
- 1.Louis DN. Molecular pathology of malignant gliomas. Annu Rev Pathol. 2006;1:97–117. doi: 10.1146/annurev.pathol.1.110304.100043. [DOI] [PubMed] [Google Scholar]
- 2.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359(5):492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 3.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK. WHO Classification of Tumours of the Central Nervous System. 4th ed. Lyon: IARC; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakada M, Nakada S, Demuth T, Tran NL, Hoelzinger DB, Berens ME. Molecular targets of glioma invasion. Cell Mol Life Sci. 2007;64(4):458–478. doi: 10.1007/s00018-007-6342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krakstad C, Chekenya M. Survival signalling and apoptosis resistance in glioblastomas: opportunities for targeted therapeutics. Mol Cancer. 2010;9:135. doi: 10.1186/1476-4598-9-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furnari FB, Fenton T, Bachoo RM, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21(21):2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 7.Huang Z, Cheng L, Guryanova OA, Wu Q, Bao S. Cancer stem cells in glioblastoma—molecular signaling and therapeutic targeting. Protein Cell. 2010;1(7):638–655. doi: 10.1007/s13238-010-0078-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamszus K, Gunther HS. Glioma stem cells as a target for treatment. Target Oncol. 2010;5(3):211–215. doi: 10.1007/s11523-010-0155-4. [DOI] [PubMed] [Google Scholar]
- 9.Ogden AT, Waziri AE, Lochhead RA, et al. Identification of A2B5+CD133-tumor-initiating cells in adult human gliomas. Neurosurgery. 2008;62(2):505–514. doi: 10.1227/01.neu.0000316019.28421.95. discussion 514–505. [DOI] [PubMed] [Google Scholar]
- 10.Singh SK, Clarke ID, Hide T, Dirks PB. Cancer stem cells in nervous system tumors. Oncogene. 2004;23(43):7267–7273. doi: 10.1038/sj.onc.1207946. [DOI] [PubMed] [Google Scholar]
- 11.Son MJ, Woolard K, Nam DH, Lee J, Fine HA. SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell. 2009;4(5):440–452. doi: 10.1016/j.stem.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 13.Gladson CL, Prayson RA, Liu WM. The pathobiology of glioma tumors. Annu Rev Pathol. 2010;5:33–50. doi: 10.1146/annurev-pathol-121808-102109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanu OO, Hughes B, Di C, et al. Glioblastoma multiforme oncogenomics and signaling pathways. Clin Med Oncol. 2009;3:39–52. doi: 10.4137/cmo.s1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan QW, Cheng CK, Nicolaides TP, et al. A dual phosphoinositide-3-kinase alpha/mTOR inhibitor cooperates with blockade of epidermal growth factor receptor in PTEN-mutant glioma. Cancer Res. 2007;67(17):7960–7965. doi: 10.1158/0008-5472.CAN-07-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goudar RK, Shi Q, Hjelmeland MD, et al. Combination therapy of inhibitors of epidermal growth factor receptor/vascular endothelial growth factor receptor 2 (AEE788) and the mammalian target of rapamycin (RAD001) offers improved glioblastoma tumor growth inhibition. Mol Cancer Ther. 2005;4(1):101–112. [PubMed] [Google Scholar]
- 17.Rao RD, Mladek AC, Lamont JD, et al. Disruption of parallel and converging signaling pathways contributes to the synergistic antitumor effects of simultaneous mTOR and EGFR inhibition in GBM cells. Neoplasia. 2005;7(10):921–929. doi: 10.1593/neo.05361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doherty L, Gigas DC, Kesari S, et al. Pilot study of the combination of EGFR and mTOR inhibitors in recurrent malignant gliomas. Neurology. 2006;67(1):156–158. doi: 10.1212/01.wnl.0000223844.77636.29. [DOI] [PubMed] [Google Scholar]
- 19.Reardon DA, Quinn JA, Vredenburgh JJ, et al. Phase 1 trial of gefitinib plus sirolimus in adults with recurrent malignant glioma. Clin Cancer Res. 2006;12(3, pt 1):860–868. doi: 10.1158/1078-0432.CCR-05-2215. [DOI] [PubMed] [Google Scholar]
- 20.Kreisl TN, Lassman AB, Mischel PS, et al. A pilot study of everolimus and gefitinib in the treatment of recurrent glioblastoma (GBM) J Neurooncol. 2009;92(1):99–105. doi: 10.1007/s11060-008-9741-z. [DOI] [PubMed] [Google Scholar]
- 21.Hjelmeland AB, Lattimore KP, Fee BE, et al. The combination of novel low molecular weight inhibitors of RAF (LBT613) and target of rapamycin (RAD001) decreases glioma proliferation and invasion. Mol Cancer Ther. 2007;6(9):2449–2457. doi: 10.1158/1535-7163.MCT-07-0155. [DOI] [PubMed] [Google Scholar]
- 22.Navis AC, van den Eijnden M, Schepens JT, Hooft van Huijsduijnen R, Wesseling P, Hendriks WJ. Protein tyrosine phosphatases in glioma biology. Acta Neuropathol. 2010;119(2):157–175. doi: 10.1007/s00401-009-0614-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neel BG, Tonks NK. Protein tyrosine phosphatases in signal transduction. Curr Opin Cell Biol. 1997;9(2):193–204. doi: 10.1016/s0955-0674(97)80063-4. [DOI] [PubMed] [Google Scholar]
- 24.Östman A, Hellberg C, Böhmer FD. Protein-tyrosine phosphatases and cancer. Nat Rev Cancer. 2006;6(4):307–320. doi: 10.1038/nrc1837. [DOI] [PubMed] [Google Scholar]
- 25.Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7(11):833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 26.Julien SG, Dube N, Hardy S, Tremblay ML. Inside the human cancer tyrosine phosphatome. Nat Rev Cancer. 2011;11(1):35–49. doi: 10.1038/nrc2980. [DOI] [PubMed] [Google Scholar]
- 27.Gebbink MF, Zondag GC, Wubbolts RW, Beijersbergen RL, van Etten I, Moolenaar WH. Cell–cell adhesion mediated by a receptor-like protein tyrosine phosphatase. J Biol Chem. 1993;268(22):16101–16104. [PubMed] [Google Scholar]
- 28.Brady-Kalnay SM, Flint AJ, Tonks NK. Homophilic binding of PTP mu, a receptor-type protein tyrosine phosphatase, can mediate cell–cell aggregation. J Cell Biol. 1993;122(4):961–972. doi: 10.1083/jcb.122.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brady-Kalnay SM, Tonks NK. Identification of the homophilic binding site of the receptor protein tyrosine phosphatase PTP mu. J Biol Chem. 1994;269(45):28472–28477. [PubMed] [Google Scholar]
- 30.Burgoyne AM, Palomo JM, Phillips-Mason PJ, et al. PTPmu suppresses glioma cell migration and dispersal. Neuro Oncol. 2009;11(6):767–778. doi: 10.1215/15228517-2009-019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Craig SE, Brady-Kalnay SM. Cancer cells cut homophilic cell adhesion molecules and run. Cancer Res. 2011;71(2):303–309. doi: 10.1158/0008-5472.CAN-10-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burgoyne AM, Phillips-Mason PJ, Burden-Gulley SM, et al. Proteolytic cleavage of protein tyrosine phosphatase mu regulates glioblastoma cell migration. Cancer Res. 2009;69(17):6960–6968. doi: 10.1158/0008-5472.CAN-09-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burden-Gulley SM, Gates TJ, Burgoyne AM, et al. A novel molecular diagnostic of glioblastomas: detection of an extracellular fragment of protein tyrosine phosphatase mu. Neoplasia. 2010;12(4):305–316. doi: 10.1593/neo.91940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hjelmeland AB, Wu Q, Heddleston JM, et al. Acidic stress promotes a glioma stem cell phenotype. Cell Death Differ. 2011;18(5):829–840. doi: 10.1038/cdd.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tyagi M, Karn J. CBF-1 promotes transcriptional silencing during the establishment of HIV-1 latency. EMBO J. 2007;26(24):4985–4995. doi: 10.1038/sj.emboj.7601928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brady-Kalnay SM, Tonks NK. Purification and characterization of the human protein tyrosine phosphatase, PTP mu, from a baculovirus expression system. Mol Cell Biochem. 1993;127–128:131–141. doi: 10.1007/BF01076764. [DOI] [PubMed] [Google Scholar]
- 37.Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics International. 2004;11(7):36–42. [Google Scholar]
- 38.Campan M, Yoshizumi M, Seidah NG, Lee ME, Bianchi C, Haber E. Increased proteolytic processing of protein tyrosine phosphatase mu in confluent vascular endothelial cells: the role of PC5, a member of the subtilisin family. Biochemistry. 1996;35(12):3797–3802. doi: 10.1021/bi952552d. [DOI] [PubMed] [Google Scholar]
- 39.Gebbink MF, Zondag GC, Koningstein GM, Feiken E, Wubbolts RW, Moolenaar WH. Cell surface expression of receptor protein tyrosine phosphatase RPTP mu is regulated by cell-cell contact. J Cell Biol. 1995;131(1):251–260. doi: 10.1083/jcb.131.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anders L, Mertins P, Lammich S, et al. Furin-, ADAM 10-, and gamma-secretase-mediated cleavage of a receptor tyrosine phosphatase and regulation of beta-catenin's transcriptional activity. Mol Cell Biol. 2006;26(10):3917–3934. doi: 10.1128/MCB.26.10.3917-3934.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenblum ML, Knebel KD, Wheeler KT, Barker M, Wilson CB. Development of an in vitro colony formation assay for the evaluation of in vivo chemotherapy of a rat brain tumor. In Vitro Cellular & Developmental Biology. 1975;11(5):264–273. doi: 10.1007/BF02615637. [DOI] [PubMed] [Google Scholar]
- 42.Kapoor GS, O'Rourke DM. Signaling modules in glial tumors and implications for molecular therapy. In: Janigro D, editor. The Cell Cycle in the Central Nervous System. Humana Press; 2006. [Google Scholar]
- 43.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 44.Mayor R, Carmona-Fontaine C. Keeping in touch with contact inhibition of locomotion. Trends Cell Biol. 2010;20(6):319–328. doi: 10.1016/j.tcb.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mason SD, Joyce JA. Proteolytic networks in cancer. Trends Cell Biol. 2011;21(4):228–237. doi: 10.1016/j.tcb.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maetzel D, Denzel S, Mack B, et al. Nuclear signalling by tumour-associated antigen EpCAM. Nat Cell Biol. 2009;11(2):162–171. doi: 10.1038/ncb1824. [DOI] [PubMed] [Google Scholar]
- 47.Ruhe JE, Streit S, Hart S, Ullrich A. EGFR signaling leads to downregulation of PTP-LAR via TACE-mediated proteolytic processing. Cell Signal. 2006;18(9):1515–1527. doi: 10.1016/j.cellsig.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 48.Haapasalo A, Kim DY, Carey BW, Turunen MK, Pettingell WH, Kovacs DM. Presenilin/gamma-secretase-mediated cleavage regulates association of leukocyte-common antigen-related (LAR) receptor tyrosine phosphatase with beta-catenin. J Biol Chem. 2007;282(12):9063–9072. doi: 10.1074/jbc.M611324200. [DOI] [PubMed] [Google Scholar]