Abstract
Diffuse intrinsic pontine glioma (DIPG) is a leading cause of brain tumor–related death in children. DIPG is not surgically resectable, resulting in a paucity of tissue available for molecular studies. As such, tumor biology is poorly understood, and, currently, there are no effective treatments. In the absence of frozen tumor specimens, body fluids—such as cerebrospinal fluid (CSF), serum, and urine—can serve as more readily accessible vehicles for detecting tumor-secreted proteins. We analyzed a total of 76 specimens, including CSF, serum, urine, and normal and tumor brainstem tissue. Protein profiling of CSF from patients with DIPG was generated by mass spectrometry using an LTQ-Orbitrap-XL and database search using the Sequest algorithm. Quantitative and statistical analyses were performed with ProteoIQ and Partek Genomics Suite. A total of 528 unique proteins were identified, 71% of which are known secreted proteins. CSF proteomic analysis revealed selective upregulation of Cyclophillin A (CypA) and dimethylarginase 1 (DDAH1) in DIPG (n = 10), compared with controls (n = 4). Protein expression was further validated with Western blot analysis and immunohistochemical assays using CSF, brain tissue, serum, and urine from DIPG and control specimens. Immunohistochemical staining showed selective upregulation of secreted but not cytosolic CypA and DDAH1 in patients with DIPG. In this study, we present the first comprehensive protein profile of CSF specimens from patients with DIPG to demonstrate selective expression of tumor proteins potentially involved in brainstem gliomagenesis. Detection of secreted CypA and DDAH1 in serum and urine has potential clinical application, with implications for assessing treatment response and detecting tumor recurrence in patients with DIPG.
Keywords: brainstem glioma, CSF, CypA, DDAH1, DIPG
Brain tumors are the most common solid tumor in children. Up to 15% of pediatric brain tumors are brainstem gliomas (BSGs) of which 80% are diffuse intrinsic pontine gliomas (DIPGs).1 DIPG has a peak incidence at 6–9 years of age and carries a median survival of less than 12 months, with a 5-year survival rate less than 5%. DIPG has the highest mortality rate of all pediatric brain tumors.1–3 Current treatment consists of radiation therapy, which temporarily decreases symptoms but has no effect on overall survival. Clinical trials investigating radiation fractionation regimens and adjuvant and neoadjuvant chemotherapeutics have not led to any advancement in the treatment paradigm for DIPG, with no change in overall survival for the past 35 years.4,5
Because of its anatomic location and infiltrative nature, DIPG is not amenable to surgical resection. Diagnosis is typically made on the basis of characteristic radiographic appearance on MRI at the time of symptom onset (Fig. 1), and diagnostic biopsy is not commonly performed.6–10 As a result, there is a paucity of tissue available for study, and therefore, little is known about the molecular biology of DIPG. Rare biopsy and postmortem specimens have revealed histological characteristics typical of high-grade (WHO III or IV) glioma; however, DIPG also exhibits molecular characteristics distinct from other high-grade pediatric and adult supratentorial and BSG.11–16 Furthermore, because of the lack of response to conventional therapies that are more effective for low-grade and supratentorial gliomas, it is reasonable to conclude that DIPG may represent a biologically distinct subclass of pediatric glioma. There is currently a surge of discussion surrounding the need, use, and safety of tumor tissue biopsy from patients with DIPG.17 Until tissue acquisition from patients with DIPG becomes routine, there is a dire need for tumor biology investigation using noninvasive methodologies.
Fig. 1.
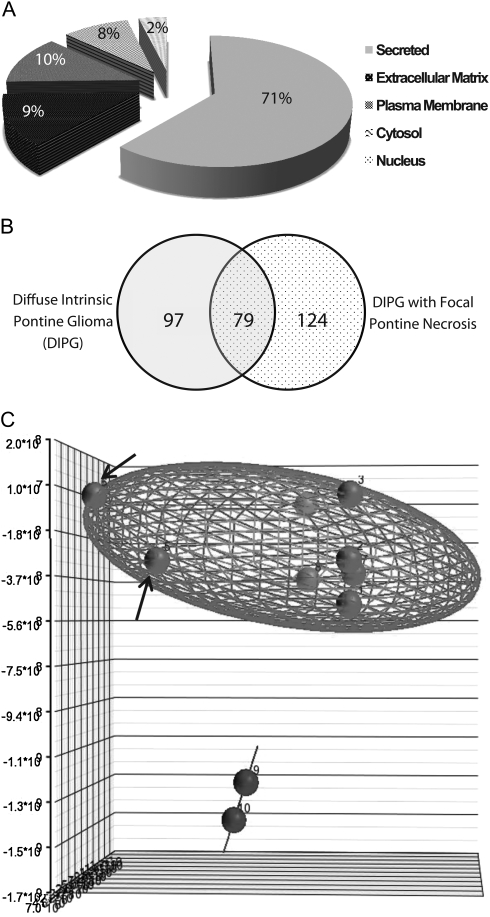
Distribution of proteins detected in analyzed cerebrospinal fluid (CSF) specimens (n = 15). (A) As expected, a large percentage (71%) of detected proteins in CSF are secreted proteins. Other proteins detected in CSF were associated with the plasma membrane (10%), extracellular matrix (9%), cytosol (8%), and nuclease (2%). (B) Overlap of proteins detected in CSF specimens from children with DIPG with homogenous radiographic appearance (n = 8) and focal pontine necrosis (n = 2). Although these 2 groups shared a large number of proteins, a subset of proteins is also uniquely expressed by each tumor type. (C) Partek Genomics Suite was used for principal component analysis (PCA) analysis. PCA showed that homogenous-appearing DIPG specimens (n = 8, red) exhibit a pattern of secreted proteins that is distinct from DIPGs with focal pontine necrosis (n = 2, blue). DIPG CSF specimens collected post-mortem (arrows) exhibit a somewhat different protein expression pattern compared to DIPG CSF specimens collected during treatment.
Cerebrospinal fluid (CSF) is a readily accessible body fluid and has been widely targeted for biomarker discovery for a variety of neurological disorders, including brain tumors.18–21 To the best of our knowledge, the only published protein analysis of pediatric BSG is our previous protein profiling of archival formalin-fixed paraffin embedded postmortem tumor specimens.22 Because of the rarity of fresh frozen DIPG tissue specimens, we tested the usefulness of CSF analysis for detection of tumor-secreted proteins and biomarkers for clinical diagnosis and treatment. Here, we present the first proteome survey of CSF from children with DIPG and report proteins that show unique upregulation in CSF from patients with BSG, compared with supratentorial glioma and controls. We further validate high expression levels of 2 potential biomarkers, cyclophillin A (CypA) and dimethylarginase-1 (DDAH1), in CSF samples from patients with DIPG and show the presence of these proteins in serum and urine samples collected from children with DIPG.
Materials and Methods
Biological Specimens
A total of 76 specimens, including CSF, serum, urine, BSG tissue, and normal brain tissue (brainstem, cerebellum, and frontal lobe), were collected in accordance with the Children's National Medical Center Institutional Review Board approvals (exemption #FWA4487 and institutional review board #4932; #3804). Patient identifiers were removed prior to evaluation, and a single sequential numerical identifier was assigned to each patient (Table 1). All brain tumor diagnoses were made radiographically by a neuroradiologist and confirmed by neuropathologic evaluation of tumor tissue specimens when available. Fifteen CSF specimens were processed for proteomic analysis: 10 specimens from patients with DIPG, 1 with glioblastoma multiforme (GBM) of the left frontal lobe, and 4 from children lacking intracranial pathology used as controls. Validation of proteomic data was performed on 48 CSF specimens, including normal controls (n = 22), pediatric low- and high-grade supratentorial glioma (n = 17), and DIPG (n = 9). CSF specimens were obtained intra-operatively or through lumbar puncture during the course of patient treatment or collected postmortem as indicated (Table 1). Tumor and surrounding normal brainstem tissue obtained postmortem from patients with DIPG (n = 10) and supratentorial glioma (n = 2) was collected for Western blot and immunohistochemical studies. Serum (n = 2) and urine specimens (n = 3) were also collected from children with DIPG during the course of treatment and compared with serum samples (n = 3) from individuals lacking intracranial pathology and urine samples (n = 3) from children with asthma.
Table 1.
List of specimens used in this study.
| Patient ID | Diagnosis | Specimens Collected | Age | Sex |
|---|---|---|---|---|
| 1 | DIPG with focal pontine necrosis | CSF (S), Normal BS, Tumor | 6 y | M |
| 2 | DIPG | CSF (S) | 9 y | F |
| 3 | DIPG | CSF (V), Tumor, Normal FL | 5 y | F |
| 5 | DIPG | CSF (S) | 3 y 6 mo | M |
| 6 | DIPG | CSF (S) | 11 y | F |
| 7 | DIPG | CSF, Tumor, Normal FL | 8 y | M |
| 8 | DIPG | CSF (V) | 6 y | F |
| 9 | DIPG with focal pontine necrosis | CSF (C) | 1 y 5 mo | M |
| 10 | DIPG | Tumor, Normal Cerebellum | 8 y | M |
| 11 | DIPG | CSF (S), Normal cerebellum, Tumor | 6 y | F |
| 12 | DIPG | Tumor, Normal FL | 7 y | F |
| 13 | DIPG | Tumor | N/A | M |
| 14 | DIPG | CSF (S), Tumor | 25 y | M |
| 15 | DIPG | Tumor | N/A | N/A |
| 16 | DIPG | Tumor | 2 y | F |
| 17 | DIPG | CSF (S) | 3 y 8 mo | M |
| 18 | DIPG | Tumor | 4 y 6 mo | F |
| 19 | DIPG | Serum, Urine | 5 y 7 mo | F |
| 20 | DIPG | Serum, Urine | 4 y | M |
| 21 | Supratentorial Glioma (WHO II) | CSF (S) | 9 y 5 mo | M |
| 22 | Supratentorial Glioma (WHO I) | CSF (V) | 6 y 7 mo | F |
| 23 | Supratentorial Glioma (WHO I) | CSF (V) | 16 y 1 mo | F |
| 24 | Supratentorial Glioma (III-IV) | CSF (S) | 1 y 9 mo | M |
| 25 | Supratentorial Glioma (WHO I) | CSF | 11 y 1 mo | M |
| 26 | Supratentorial Glioma (WHO I) | CSF (S) | 1 y 4 mo | F |
| 27 | Supratentorial Glioma (WHO I) | CSF | 11 y 8 mo | M |
| 28 | Supratentorial Glioma (WHO III) | CSF (S) | 14 y 7 mo | F |
| 29 | Supratentorial Glioma (WHO III) | CSF (S) | 2 y 7 mo | F |
| 30 | Supratentorial Glioma (WHO III) | CSF (C) | 12 y 11 mo | F |
| 31 | Supratentorial Glioma (WHO IV) | CSF (S) | 9 y 6 mo | M |
| 32 | Supratentorial Glioma (WHO IV) | CSF (V) | 14 y 9 mo | F |
| 33 | Supratentorial Glioma (WHO IV) | CSF (C) | 5 y 2 mo | F |
| 34 | Supratentorial Glioma (WHO IV) | CSF (S) | 5 y 11 mo | M |
| 35 | Supratentorial Glioma (WHO III) | CSF | 10 y 2 mo | F |
| 36 | Supratentorial Glioma (III-IV) | CSF (S) | 1 y 8 mo | F |
| 37 | Supratentorial Glioma (WHO IV) | CSF (V) | 11y 11 mo | M |
| 38 | DIPG | Urine | 7 y 11 mo | M |
| 40 | Seizure | CSF (S) | 2 y | F |
| 40 | DIPG | CSF (V), Tumor, Normal FL | 1 y 2 mo | F |
| 41 | Seizure | CSF (S) | 1 y 3 mo | F |
| 42 | Fever | CSF (S) | 4 y 1 mo | M |
| 43 | ALL | CSF (S) | 14 y 10 mo | F |
| 44 | Spina bifida | CSF (V) | 6 mo | M |
| 45 | Hypdrocephalus | CSF (V) | 10 mo | F |
| 46 | Fever | CSF (S) | 11 y | M |
| 47 | Viral Meningitis | CSF (S) | 8 y | M |
| 48 | Dehydration | CSF (S) | 11 y | M |
| 49 | Negative Diagnostics | CSF (S) | N/A | N/A |
| 50 | Seizure | CSF (S) | 14 y | F |
| 51 | Negative Diagnostics | CSF (S) | 13 y | F |
| 52 | Negative Diagnostics | CSF (S) | 4 mo | F |
| 53 | Negative Diagnostics | CSF (S) | 4 y | F |
| 54 | Negative Diagnostics | CSF (S) | 14 y | M |
| 55 | Negative Diagnostics | CSF (S) | 6 y | M |
| 56 | Negative Diagnostics | CSF (S) | 15 y | M |
| 57 | Negative Diagnostics | CSF (S) | 17 y | M |
| 58 | Negative Diagnostics | CSF (S) | 1 y 6 mo | M |
| 59 | Fever | CSF (S) | 1 mo | M |
| 60 | Growth hormone deficiency | CSF (S) | 9 y | M |
| 61 | Cardiogenic shock | CSF (S) | 16 y | F |
| 70 | T-Cell ALL | Serum | 9 y | F |
| 71 | Von Willebrand | Serum | 10 y | F |
| 72 | Hives | Serum | 6 y | F |
| 80 | Normal Healthy | Urine | 8 y 4 mo | F |
| 81 | Normal Healthy | Urine | 8 y 2 mo | M |
| 82 | Normal Healthy | Urine | 6 y 5 mo | M |
Note: Specimens include frozen normal brainstem (BS), frontal lobe (FL), or DIPG tumor tissue as well as ventricular (V) or spinal (S) cerebrospinal fluid (CSF), blood serum, and urine
Specimen Processing and Protein Extraction
CSF specimens were centrifuged at 12 000 × g for 10 min at 4°C and the supernatant collected in fresh tubes. Blood collected in red-topped tubes was allowed to clot at room temperature for 30 min and then centrifuged at 1 000 × g for 15 min to isolate serum. Tissue protein extracts were prepared by homogenizing frozen tissue samples on ice using 1X SDS Laemmli sample buffer (2% SDS, 10% glycerol, 5% 2-mercaptoethanol, 0.0625 M Tris [pH, 6.8], 0.001% bromophenol blue) containing 1 mM DTT. Lysates were spun down at 14 000 × g for 30 min at 4°C and supernatants collected.
Urine specimens were passed sequentially through 100 μM, 40 μM, 5 μM, and 0.2 μM filters to remove cells and contaminating microparticles. Five milliliter aliquots of urine were added to 2 volumes of ice-cold acetone and precipitated overnight at −20°C. The sample was centrifuged at 12 000 × g for 10 min at 4°C and the supernatant removed. After air-drying, the pellet was resuspended in RIPA buffer.
The protein concentration of each CSF, serum, and urine specimen was measured using a Pierce BCA protein assay (ThermoFischer Scientific). To estimate protein concentration in tissue specimens, equal volumes of lysate were resolved by electrophoresis on 10%–20% polyacrylamide gel with a standard of known concentration. The resulting gel was stained with Coomassie Blue, and a digital image was obtained. Specimen protein concentration was determined by normalizing the total intensity of the sample band to the total band intensity of the sample with known concentration.
Proteomic Analysis of CSF
CSF samples were concentrated using 3 kDa MW Micro Bio-Spin cutoff filters (Bio-Rad). Protein concentration was determined using a Pierce BCA protein assay (ThermoFischer Scientific). Protein aliquots (110 μg) from each sample were dissolved in Laemmli sample buffer and loaded for sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) under denaturing conditions. Gels were fixed (50:10:40/methanol:acetic acid:H2O/v:v:v), stained with Biosafe Coomassie (BioRad), and washed extensively with water. Protein bands were sliced and processed for in-gel digestion, as described elsewhere.23
Resulting peptides from each band were analyzed using nano-LC (Agilent) connected to an LTQ-Orbitrap-XL instrument (ThermoFisher Scientific), operated in data-dependent mode with dynamic exclusion, in which 1 cycle of experiments consisted of a full-MS in the Orbitrap survey scan (300–2000 m/z) and 5 subsequent MS/MS scans of the most intense peaks in the LTQ. Proteins were identified using the Sequest algorithm in the Bioworks Browser (ThermoFisher Scientific) against the Uniprot database (downloaded November 2010) and indexed for fully tryptic peptides, 2 missed cleavages, and no modifications. Results were filtered on the basis of a variable threshold of Xcorr versus charge state and peptide probability with P < .05. Protein quantification was performed using ProteoIQ software (Bioinquire) with spectral count data. The following stringent filtration criteria were used: 2 or more peptides/protein, 10 or more spectra/protein, minimum peptide probability-based score with P ≤ .05, and a variable threshold of Xcorr versus charge state (Xcorr= 1.9 for z = 1, Xcorr= 2.5 for z = 2, and Xcorr= 3.5 for z = 3). Protein expression analysis was performed using Partek Genomics Suite (Partek).
Protein Validation in CSF and Tumor Tissue Samples
Candidate biomarkers were validated by Western blot analysis using 48 CSF specimens, obtained from patients with DIPG (n = 10), non-BSG (n = 17), and age-matched controls (n = 22). Additional validation was performed using serum specimens from DIPG (n = 2) and healthy (n = 3) subjects and urine samples from DIPG (n = 3) and normal (n = 3) subjects as described below.
Antibodies
Rabbit polyclonal anti-CypA antibody (Cedarlane Laboratories) was used at 1:500 dilution. Goat monoclonal anti-DDAH1 antibody (Abcam) was diluted 1:300. Goat polyclonal anti-transferrin antibody (Santa Cruz Biotechnology) was diluted 1:200. Horeseradish peroxidase–labeled secondary antibodies were diluted 1:5000 (Kirkergaard and Perry Laboratories). All antibodies have been previously shown to detect the target protein at the correct molecular mass; however, depending on the tissue used, nonspecific bands were also detected at a much higher molecular weight.24–32
SDS-PAGE and Immunoblot Analysis
Serum, CSF, tissue, and urine protein concentrations were assessed using a Bradford assay and adjusted to equivalent protein concentrations with 1X SDS Laemmli sample buffer (2% SDS, 10% glycerol, 5% B-mercaptoethanol, 0.0625 M Tris [pH, 6.8], 0.001% bromophenol blue) containing 1 mM DTT. Resulting samples were subjected to electrophoresis on 4%–20% SDS-PAGE. Protein was then transferred to a nitrocellulose membrane by the electroblotting method. Membranes were incubated overnight at 4°C with blocking buffer (PBS containing 5% nonfat dry milk and 0.1% Tween 20), followed by incubation with primary antibody for an additional h at room temperature. The membranes were washed in PBST (PBS containing 0.1% Tween 20) and treated with HRP-conjugated secondary antibody for 1 h at room temperature. Protein bands were then visualized using the enhanced chemiluminescence technique.
Immunohistochemistry
Immunohistochemical analysis was performed on FFPE brain tissue sections. Paraffin was removed by incubating slides in Xylenes followed by ethanol. Slides were boiled in citrate buffer (pH, 6.0) for 15 min for antigen retrieval. After inactivating the endogenous peroxides by treating with 3% hydrogen peroxide, immunostaining was performed using an IntelliPATH FLX automatic slide processor (Biocare Medical). CypA and DDAH1 antibodies were diluted to 1:200 and 1:100, respectively. Secondary detection was performed using an IntelliPATH FLX HRP Detection Kit (Biocare Medical) for anti-rabbit and anti-goat primary antibodies.
Pathway Analysis and Data Interpretation
Pathway analysis was performed using Ingenuity Pathway Analysis software, version 1.0 (Ingenuity Systems). Accession numbers of the 528 identified CSF proteins were uploaded, and protein-protein interaction networks were generated using fold-change protein expression data. Comparison analysis of fold-change expression data was performed to reveal top molecules and pathways of interaction in BSG. Pathways of protein expression and interaction were explored using tumor specific protein sets and known pathways with biological relevance to cancer, cell-to-cell signaling, and gliomagenesis.
Results
Label-Free Proteome Profiling of CSF Specimens
CSF specimens from children with DIPG (n = 10), including 8 with homogenous radiographic appearance and 2 with regions of focal pontine necrosis, supratentorial GBM (n = 1), and normal controls (n = 4) (Table 1) were processed for proteome profiling as described in the Materials and Methods. A total of 528 unique proteins were identified (Supplementary Table S1). As expected, a large percentage (71%) of detected proteins were secreted, and 10% were identified as plasma membrane and 9% as extracellular matrix proteins (Fig. 1A). A total of 97 proteins were unique to homogenous-appearing DIPG, 124 unique to pontine gliomas with focal necrosis, and 79 shared between these 2 subtypes (Fig. 1B).
The label-free spectral counting (SC) method was used to calculate fold-change and protein expression levels across all specimens. Homogenous DIPG specimens exhibited greater protein expression dysregulation than those with necrotic features: 334 proteins showed upregulation in DIPG (fold-change 2 or more), compared with 157 proteins with more focal, atypical radiographic appearance. Seventy-three proteins (22%) were upregulated in 3 or more DIPG specimens. These include DDAH1, CypA, beta tubulin, vimentin, and members of the 14-3-3 protein family (Supplementary Table S1). Analysis of downregulated proteins (fold-change 2 or less) identified a total of 157 proteins in 3 or more DIPG specimens (Supplementary Table S1). Principle component analysis (PCA) of the relative protein expression profiles in each specimen was performed with unsupervised, nonpartitional hierarchical clustering (Partek Genomic Suite) (Fig. 1C). PCA of DIPG specimens showed that the 8 diffuse tumors clustered separately from the 2 DIPG specimens with focal necrosis. In the homogenous DIPG specimens, the 2 postmortem DIPG specimens clustered together, reflecting similarly unique protein profiles, which may be attributed to postmortem protein degradation (Fig. 1C, arrows).
Validation of CypA and DDAH as Potential Markers for DIPG
Two secreted proteins, CypA and DDAH1, found to be selectively upregulated in DIPG CSF specimens, were chosen for further validation and investigation on the basis of antibody availability, disease relevance, and CSF expression levels. Protein profiling detected insignificant SC (SC = 1) of CypA in 1 of 4 control CSF specimens tested. However, high SCs of CypA (SC more than 4) were detected in 9 of 10 CSF specimens from patients with DIPG. Similarly, DDAH1 was detected in only 1 control but in 6 of 10 DIPG specimens. Neither CypA nor DDAH1 was detected in the supratentorial glioblastoma CSF specimen tested.
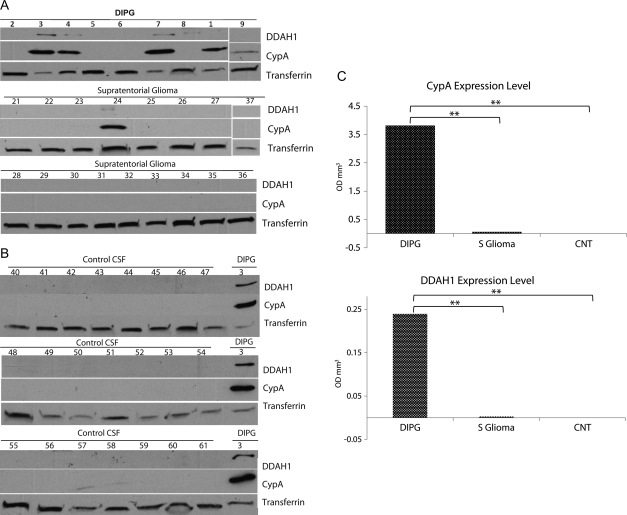
To validate our protein profiling results, Western blot analysis was performed on additional CSF specimens. CSF from patients with DIPG (n = 9), supratentorial glioma (n = 17), and controls (n = 22) were analyzed by Western blot using antibodies against CypA and DDAH1 (Fig. 2). CypA and DDAH1 expression were validated in CSF samples from 5 (55%) and 4 (44%) patients with DIPG respectively (Fig. 2A). CypA and DDAH1 co-expression was detected in only 1 low-grade (WHO-I) non-brainstem supratentorial specimen. DDAH1 was absent from all 22 of the control CSF samples tested. Low levels of CypA were detected in 2 control CSF specimens (0.09%). CypA was consistently detected at a higher level than DDAH1 in all CSF specimens tested and was selectively expressed at significantly higher intensity in high-grade (WHO-IV) specimens (Fig. 2B). Overall, CypA and DDAH1 were significantly overexpressed in DIPG, compared with supratentorial glioma and control specimens (Fig. 2C). Transferrin was used as the internal control.
Fig. 2.
Western blot analysis of CSF specimens from patients with DIPG and supratentorial glioma using antibodies for CypA and DDAH1. (A) High levels of both CypA and DDAH1 were detected in CSF samples from DIPG. In contrast, CypA was detected only in 1 CSF specimen from a patient with supratentorial glioma. (B) DDAH1 was not detected in 22 control CSF specimens tested. Very low levels of CypA were detected in 2 control CSF samples. Transferrin was used as an internal control and a DIPG CSF specimen (patient number 3) as a positive control. (C) CypA and DDAH1 expression levels were measured by densitometry. Mean values for each group (diffuse intrinsic pontine glioma [DIPG], supratentorial glioma [S glioma], and control [CNT]) are represented by bar graphs. **P < .05.
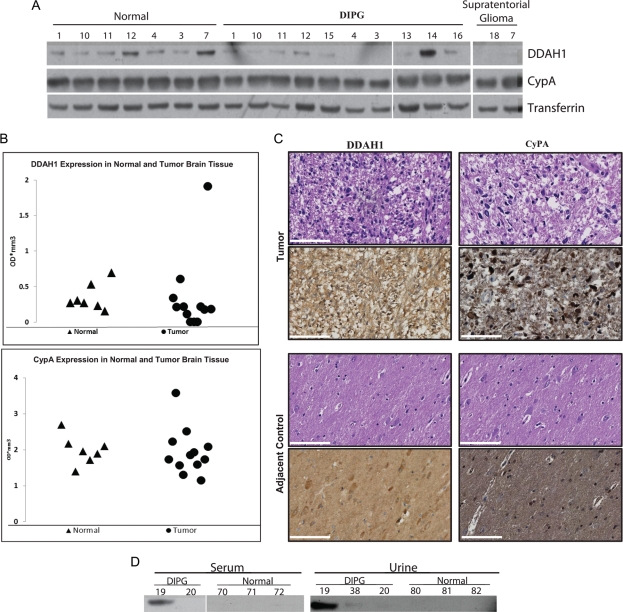
We then questioned whether cytosolic (nonsecreted) expression levels of CypA and DDAH1 differ between normal and tumor specimens. CypA and DDAH1 expression was therefore examined in DIPG tumor tissue. Total protein extracted from frozen DIPG tumor tissue (n = 10), and adjacent normal brainstem, cerebellum, or frontal lobe tissue from the same patient (n = 7) was used for Western blot analysis (Fig. 3A). CypA was detected in all DIPG tumor and normal brain tissue specimens analyzed, and DDAH1 was detected in 9 (75%) of 12 tumor and all normal brain tissue specimens (Fig. 3A). Densitometry analysis of Western blot results revealed no relative increase in cytosolic CypA or DDAH1 expression, compared with normal brain tissue from the same patient, normalized to GAPDH expression (Fig. 3B). Immunohistochemical staining of DIPG tumor tissue for CypA and DDAH1 localized expression to the cytosol and nucleus (Fig. 3C).
Fig. 3.
CypA and DDAH1 expression analysis using frozen brain tissue, serum, and urine specimens. (A) Western blot analysis showed equal expression levels of both CypA and DDAH1 in frozen diffuse intrinsic pontine glioma (DIPG) tumors (n = 12) and normal brain tissue (n = 7), indicating no alteration in cytosolic levels of these proteins. (B) Graphical representation of Western blot analysis of DIPG tumor tissue and control brain tissue based on densitometry of Western blots shown in panel A normalized to GAPDH expression levels. (C) Immunohistochemical staining of DIPG tumor tissue demonstrates cytosolic expression of CypA and DDAH1 in high-grade regions, as indicated by H&E staining. Adjacent normal regions as identified by a pathologist were used as controls. (Scale bar = 10 μm). (D) Western blot analysis of serum and urine specimens collected from patients with DIPG demonstrate expression of CypA in serum from patient 19 and urine of patients 19 and 38. Low expression level of CypA was detected in serum of one control patient (number 72) lacking intracranial pathology. DDAH1 was not detected in serum and urine of these patients (data not shown).
Detection of CypA and DDAH1 in Blood and Urine Samples from Patients With DIPG
Given the high expression levels of DDAH1 and CypA in CSF, we questioned whether we could detect these 2 potential tumor-associated proteins in serum and urine samples from patients with DIPG to explore the possibility of noninvasive clinical detection. Blood serum and urine specimens were collected from patients with DIPG and healthy individuals, and total protein was extracted. Western blot analysis of total protein extract demonstrated selective expression of CypA in blood serum (n = 1) and urine (n = 2) specimens from patients with DIPG, compared with controls (serum, n = 3; urine, n = 3) (Fig. 3D). Low expression of CypA was detected in a serum sample from 1 patient (patient 72) lacking intracranial pathology. Taken together, our results suggest that tumor-secreted CypA and DDAH1, but not the cytosolic forms, are selectively upregulated in patients with DIPG and are detectable in urine and blood. Additional studies are warranted to assess potential post translational modifications associated with secreted CypA and DDAH1 proteins.
Ingenuity Pathway Analysis
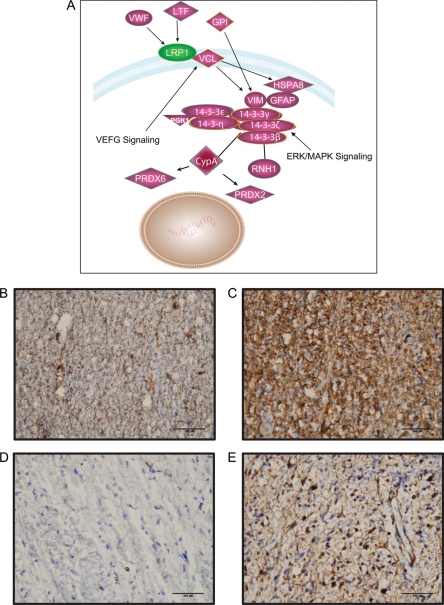
Protein expression profiles and observed fold changes were used to explore tumor-related pathways using Ingenuity Pathway Analysis software (Ingenuity Systems), which generates biologic pathways based on published molecular interactions. Top biologic functions of proteins detected in DIPG CSF included cancer tumorigenesis (74 proteins; P < .001), neurological disorder (87 proteins; P < .001), and genetic disease (130 proteins; P < .001). Functional analysis of differentially expressed proteins in DIPG specimens mapped identified proteins to canonical pathways of neoplasia, cell movement, and reaction to oxidative stress, with top molecular networks including cell-to-cell signaling, tissue development, and antigen presentation. Focused analysis of upregulated proteins (fold-change more than 2) in DIPG specimens resulted in a top network of 35 focus molecules involved in neurological disease (28 molecules; P < .001), cancer (25 molecules; P < .001), and cell movement (20 molecules; P < .001). These molecules include CYPA and DDAH1 and exhibit functional overlap with known molecular pathways implicated in gliomagenesis (Fig. 4A). To validate upregulation of proteins involved in the gliomagenesis pathway, immunohistochemical assays of DIPG tumors and adjacent normal sections were performed using antibodies against vimentin and GFAP. High expression levels of both vimentin and GFAP were detected in the tumors, compared with adjacent normal sections (Fig. 4B–E).
Fig. 4.
Ingenuity Pathway Analysis (IPA) depicting dysregulated tumor proteins mapping to known pathways of glioma formation, cell migration, and response to oxidative stress. The majority of detected upregulated DIPG CSF proteins play a role in vascular endothelial growth factor (VEGF) and ERK/MAPK signaling pathways (A). The tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein (14-3-3) family (ɛ, γ, ɛ, δ, and β) is induced by VEGF and known to be involved in cell cycle mitosis. 14-3-3δ binds to Cyclophillin A (CypA), which in turn activates peroxidase 6 (PRDX6). PRDX6 is involved in oxidation-reduction regulation and protection against oxidative stress. 14-3-3β also binds to Ribonuclease inhibitor 1 (RNH1), an angiogenin regulator. The 14-3-3 complex may be upregulated in response to increased expression of extracellular proteins such as lactotransferrin (LTF), glucose-6-phosphate isomerase (GPI), and von Willebrand factor (VWF). Although low density lipoprotein receptor-related protein 1 (LRP1) is downregulated in tumor samples, its binding partner vinculin (VCL), a molecule known to be downstream of the VEGF signaling pathway, is upregulated in DIPG CSF specimens. Immunohistochemical assays validated upregulation of GFAP (B-C) and vimentin (D-E) in DIPG tumor (C and E) compared to adjacent normal sections (B and D). Although GFAP was detected at a much higher level in tumors compared to normal tissue, vimentin exhibited a more tumor-specific expression pattern. Scale bar = 200 μm.
Discussion
DIPG is a devastating pediatric brain tumor for which no effective treatment exists. Because DIPG is not amenable to surgical resection and because most patients with DIPG die at home or in hospice, historically, there has been a paucity of tumor tissue available for molecular studies. Although there is an ongoing debate within the medical community over the safety and usefulness of tumor biopsy in patients with DIPG, no robust action has been taken for biopsy acquisition and analysis. Existing analyses of rare DIPG tissue specimens underscore the premise that pediatric BSG is a biologically distinct form of glioma.15 Because of the rarity of DIPG tissue specimens for molecular analysis, there is a need for innovative methods of study. Tumor tissue protein profiling and CSF microRNA analysis using specimens from patients with glioma have been established as valuable techniques for identifying tumor-associated markers.22,33
In the case of pediatric BSG and, particularly DIPG, CSF is more readily available than tumor tissue. CSF may be in direct contact with tumor tissue, increasing the probability of identifying tumor-secreted molecules. Recent studies demonstrate the presence of a large number of quantifiable tumor-associated proteins in CSF, with the potential to identify glioma-specific molecules.18–20,34,35 Identification of specific proteins in CSF from patients with glioma is correlated with high tumor grade and poor clinical outcome and may have prognostic value.35,36 Proteins actively secreted by tumor cells could play an important role in tumorigenesis and interfere with local cellular activity in the tumor microenvironment, making CSF analysis of special interest for understanding mechanisms of gliomagenesis when access to tumor tissue is limited.37 For example, Deighton et al. (2010) reported detection of 10 differentially expressed proteins in CSF from adult patients with supratentorial glioma and demonstrated interaction between the glioma secretome and known molecular pathways of adult gliomagenesis, including TP53 and RB1.38
Recent molecular studies of pediatric glioma suggest differences in tumorigenic pathways, compared with adult disease. For example, dysregulation through PDGFRα, PTEN, and EGFR is prevalent in adult glioma.39,40 In contrast, variable EGFR amplification has been reported in children with supratentorial glioblastoma, with the majority of studies demonstrating less EGFR amplification in children than in adults.41 Of importance, rare molecular analysis of pediatric BSG tissue reveals key biological differences, compared with pediatric and adult supratentorial glioma. Sharma et al. (2010) report lack of EGFR amplification in 2 cases of BSG with PTEN deletion and a higher frequency of whole chromosomal gains (1 and 2) and segmental loss (8p, 16p, and 16q) in pediatric BSGs compared to adult and pediatric supratentorial high-grade glioma.13 Recent whole-genome profiling of pediatric DIPG has shown gains in PDGFRA in 36% of cases tested and no cases of EGFR amplification.14 Loss at 17p containing the p53 gene is reported at higher frequency in DIPGs than in supratentorial high-grade gliomas, whereas PTEN mutations are reported at higher frequency in DIPGs than in adult or pediatric supratentorial high-grade gliomas.14,15 These reports suggest that the molecular biology of pediatric BSG differs from that of adult and pediatric supratentorial glioma and, thus, may be biologically distinct.
Our data are in line with these findings and demonstrate that CSF can be studied to elucidate the molecular basis of DIPG formation. We detected 109 upregulated proteins with functional mapping to tumorigenesis in DIPG CSF, 71 of which are involved in the p53 pathway and 68 in the AKT pathway. Comparative analysis of CSF specimens stratified DIPG from normal specimens and generated a cluster pattern suggestive of distinct subtypes within DIPG samples (data not shown). However, further analysis of these subtypes may require a larger cohort of specimens and, perhaps, inclusion of protein profiles from frozen tumor specimens. Overall, although within-group heterogeneity was observed among DIPG specimens, PCA showed that DIPGs exhibit a pattern of secreted proteins that is detectable in CSF and is distinct from controls and supratentorial glioma, suggesting unique pathways of pediatric brainstem gliomagenesis (Fig. 2C). Two upregulated proteins (CypA and DDAH1) were selected for further analysis based on the availability of robust antibodies and their biological role as described below.
Cyclophillin A
Secreted CypA.
We detected and validated high expression levels of secreted CypA (peptidylprolyl isomerase A) in CSF, serum, and urine samples from patients with DIPG. In contrast, secreted CypA was detected in 1 of the 16 CSF specimens from children with supratentorial glioma (Fig. 2A) and as a faint band in 2 of the 22 control specimens tested (Fig. 2B). CypA is an 18KDa ubiquitous cytosolic chaperone protein member of the immunophilin family with peptidyl-prolyl isomerase activity.42 Elevated expression of secreted CypA and its associated membrane receptor CD147 has been demonstrated in a variety of tumor types and is linked to malignant transformation, poor cellular differentiation, and decreased survival in patients with cancer.43
The regulatory mechanisms of CypA secretion are currently unknown, but studies suggest increased CypA secretion occurs in response to oxidative stress and inflammatory mediators. Upregulated CypA expression in response to hypoxia has been shown in a variety of cancer cell types, including C6 glioma cells.44,45 Our pathway analysis of DIPG CSF also showed upregulation of proteins involved in oxidative stress (Fig. 4). The effect of secreted CypA on cellular function is mediated though transmembrane protein receptor CD147 activation, which causes phosphorylation of ERK1/2, JNK, Akt, and IkB signaling pathways to influence protein folding, gene transcription, and cell-to-cell signaling.43,46–48 CypA binding of CD147 also facilitates chemotaxis in response to stress.46,47,49,50 Secreted CypA may therefore act as a local mitogen induced by oxidative stress, either prior to or in response to malignant glial cell transformation, to influence astroglial differentiation, proliferation, and/or tumor angiogenesis. In glioblastoma multiforme, CypA secretion may be epigenetically activated during malignant transformation, with subsequent upregulation because of hypoxic conditions in regions of central tumor necrosis.44 Although central tumor necrosis is a late finding in DIPG, the cells of the developing brainstem may be more sensitive to hypoxic stress, and aberrant hypoxic stress response can lead to defective brainstem development involving VEGF signaling.51 Aberrant upregulation of secreted CypA in response to hypoxia could therefore potentially induce the rapid malignant transformation and proliferation within the neuroaxis that is seen in end-stage DIPG.
Previous research has suggested that histologically similar tumors arising in distinct anatomic regions of the brain may originate from molecularly distinct populations of progenitor cells, implying the existence of a niche microenvironment.52–54 For example, Monje et al. (2011) proposed that the ventral pontine location of DIPG suggests the presence of a local cell population with particular susceptibility to a unique signaling microenvironment favoring tumor formation.55 This difference in tumor microenvironment could therefore account for the molecular differences observed between pediatric supratentorial and BSG. In this study, we detected intracellular CypA in brainstem and supratentorial glioma tumor tissue, but secreted CypA in CSF, blood, and urine samples from patients with DIPG alone. Therefore, molecular signals in the developing brainstem microenvironment, which may not be present in the cerebral hemispheres, may have a unique effect on local cell populations within the ventral pons resulting in site-specific release of intracellular CypA from BSG cells.
Furthermore, the effects of site-specific CypA secretion into the brainstem microenvironment could contribute to the distinct biology and clinical characteristics of DIPG. The young age of onset and anatomic location of DIPG, compared with supratentorial pediatric high-grade glioma, implies an interplay of signaling pathways necessary for ongoing brainstem development, such as astroglial proliferation, cytoskeletal remodeling, axonal outgrowth, myelination, and tumor cell behavior. This unique interaction in the developing brainstem could trigger unique, region-specific signaling cascades resulting in gliomagenesis. The CypA receptor CD147 has been detected in neurospheres and neural precursor cells, implying the existence of CypA-induced signaling cascade in neural development.56 Local expression of secreted CypA by glioma cells in the developing brainstem could therefore promote tumor cell proliferation and malignant transformation, resulting in the unique anatomic and histological characteristics of DIPG. Overexpression of CypA has also been shown to induce chemoresistance in cancer cells through protection from apoptosis.43 Region-specific CypA secretion by BSG cells may therefore contribute to the relative resistance to chemotherapy and radiation treatment observed in patients with DIPG, possibly explaining the lack of clinical response to therapeutics that are more effective for supratentorial adult and pediatric gliomas.
Although CypA expression was detected at high levels in CSF specimens collected after radiation, we also detected increased levels of CypA in specimens collected prior to radiation therapy, demonstrating that CypA secretion is not simply induced by cellular response to treatment. Taken together, these data also suggest that because CypA is most likely secreted by DIPG tumor cells, it may be a valuable molecule for screening for disease progression and assessing response to treatment.
Clinical Relevance of CypA
The data presented here demonstrate detection of secreted CypA in CSF, urine, and blood serum specimens from pediatric patients with DIPG. The diagnosis and treatment of DIPG is largely based on tumor appearance on MRI because tumor biopsy is rarely performed. In contrast, collection and analysis of serum or urine specimens from patients with DIPG could easily and quickly be performed. Smith et al. (2008) demonstrated detection of urinary matrix metalloproteinases correlated with the presence of a brain tumor and could serve as biomarkers indicating presence of disease, tumor recurrence, and response to treatment.57 CypA detection in the serum or urine samples from patients with DIPG could therefore potentially play a similar role in tumor diagnosis and response to treatment at varied post treatment time points in the absence of readily available tissue. The clinical course and radiographic imaging of the patients with DIPG studied here were reviewed: high levels of secreted CypA expression correlated with patients exhibiting rapid progression and regions of central tumor necrosis on MR imaging. This suggests that detection of secreted CypA could potentially facilitate subtype diagnosis, patient stratification for treatment, and the prediction of tumor response to therapy. Together, our data suggest secretion of CypA by BSG cells and emphasize the need for further investigation into its potential role in DIPG biology. Interfering with CypA function could potentially blunt astroglial proliferation and migration, inflammatory response, and tumor angiogenesis. Therefore, further studies are warranted to understand the precise role of CypA in pediatric BSG and its potential as a therapeutic target.
DDAH1
We detected selective upregulation of DDAH1 in CSF samples from patients with DIPG, compared with normal controls. DDAH1 upregulation was validated in CSF samples from 4 patients with DIPG by Western blot analysis (Fig. 2A). DDAH1 is an isoform of DDAH that is associated with neuronal nitric oxide synthase function. DDAH is a 31kD enzyme that metabolizes the asymmetric dimethlyarginine, an endogenous inhibitor of nitric oxide synthesis. As an inhibitor of asymmetric dimethlyarginine, DDAH overexpression indirectly leads to an increase in nitric oxide expression. Constitutive overexpression of DDAH1 has also been shown to increase tumor growth, vascularization, and secretion of vascular endothelial growth factor (VEGF).58–61
DDAH1 expression has been detected in a variety of tumor types, including brain tumors.59–61 In the subjects studied here, patients with DDAH1 expression in CSF exhibited an aggressive clinical course and radiographic findings consistent with rapid tumor growth and local tumor invasion, including gadolinium enhancement, central tumoral necrosis, and high perilesional signals on T2-weighted and FLAIR sequences. These data suggest that DDAH1 confers an advantage for in vivo gliomagenesis through its effects on tumor angiogenesis.
We demonstrated that differential expression of DDAH1 is detectable in CSF samples from patients with DIPG, which could have prognostic implications for patient stratification and assessing treatment outcomes. Although the mechanism of the role of DDAH1 in gliomagenesis is not entirely clear, it is thought that the angiogenic and mitotic effects of DDAH1 are influenced by local nitric oxide levels. DDAH1 also may exhibit direct protein-protein interactions to affect cell signaling and proliferation. For example, DDAH1 binds to and has overlapping RNA expression patterns with the Ras-pathway regulator neurofibromin 1 (NF1).62,63 Loss of function of tumor suppressor gene NF1 results in glioma formation in familial neurofibromatosis type 1 and is also thought to play a role in sporadic gliomagenesis. Thus, the NOS and Ras pathway could play a significant role in tumor progression and could therefore serve as a potential therapeutic target for BSG.
Conclusions
Our data demonstrate, to the best of our knowledge, the first comprehensive protein profile of CSF samples from pediatric DIPGs. Through analysis of CSF samples from 10 children with DIPG, we detected differential expression of proteins that may represent tumor-specific protein expression patterns. These data can be used to elucidate pathways of tumor formation to guide further research and development of targeted therapies for this devastating disease. High-grade pediatric BSG formation is likely to be a multifactorial process representing multiple tumor subtypes, including DIPG. Because of this inherent variability and limited access to tumor tissue, CSF analysis may be complementary to radiography and neuropathology observation to guide clinical diagnosis and management.
Supplementary Material
Supplementary material is available online at Neuro-Oncology (http://neuro-oncology.oxfordjournals.org/).
Funding
This work was supported by Isabella Kerr Molina Foundation Funds (1457), the Childhood Brain Tumor Foundation (5917), The Zickler Family Funds (0777), the Musella Foundation (1457), a Clinical and Translational Science Institutes Award (1UL1RR031988-01), the National Institutes of Health, the National Center for Research resources (UL1RR031988, 2R24HD050846-06), and core grants from the Children's National Medical Center Proteomics.
Supplementary Material
Acknowledgments
This work is dedicated to the memory of Isabella Kerr Molina. We thank Dr. Yetrib Hathout for revising the manuscript, Dr. Kristy Brown for her input into protein profiling, and Dr. Heather Gordish for her statistical analysis input.
Conflict of interest statement. None declared.
References
- 1.Walker DA, Punt JA, Sokal M. Clinical management of brain stem glioma. Arch Dis Child. 1999;80(6):558–564. doi: 10.1136/adc.80.6.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger MS, Edwards MS, LaMasters D, Davis RL, Wilson CB. Pediatric brain stem tumors: radiographic, pathological, and clinical correlations. Neurosurgery. 1983;12(3):298–302. doi: 10.1227/00006123-198303000-00008. [DOI] [PubMed] [Google Scholar]
- 3.Albright AL, Price RA, Guthkelch AN. Brain stem gliomas of children. A clinicopathological study. Cancer. 1983;52(12):2313–2319. doi: 10.1002/1097-0142(19831215)52:12<2313::aid-cncr2820521226>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 4.Frazier JL, Lee J, Thomale UW, Noggle JC, Cohen KJ, Jallo GI. Treatment of diffuse intrinsic brainstem gliomas: failed approaches and future strategies. J Neurosurg Pediatr. 2009;3(4):259–269. doi: 10.3171/2008.11.PEDS08281. [DOI] [PubMed] [Google Scholar]
- 5.Hargrave D, Bartels U, Bouffet E. Diffuse brainstem glioma in children: critical review of clinical trials. Lancet Oncol. 2006;7(3):241–248. doi: 10.1016/S1470-2045(06)70615-5. [DOI] [PubMed] [Google Scholar]
- 6.Barkovich AJ, Krischer J, Kun LE, et al. Brain stem gliomas: a classification system based on magnetic resonance imaging. Pediatr Neurosurg. 1990;16(2):73–83. doi: 10.1159/000120511. [DOI] [PubMed] [Google Scholar]
- 7.Epstein F. A staging system for brain stem gliomas. Cancer. 1985;56(7 Suppl):1804–1806. doi: 10.1002/1097-0142(19851001)56:7+<1804::aid-cncr2820561316>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 8.Epstein F, McCleary EL. Intrinsic brain-stem tumors of childhood: surgical indications. Journal of Neurosurgery. 1986;64(1):11–15. doi: 10.3171/jns.1986.64.1.0011. [DOI] [PubMed] [Google Scholar]
- 9.Fischbein NJ, Prados MD, Wara W, Russo C, Edwards MS, Barkovich AJ. Radiologic classification of brain stem tumors: correlation of magnetic resonance imaging appearance with clinical outcome. Pediatr Neurosurg. 1996;24(1):9–23. doi: 10.1159/000121010. [DOI] [PubMed] [Google Scholar]
- 10.Albright AL, Packer RJ, Zimmerman R, Rorke LB, Boyett J, Hammond GD. Magnetic resonance scans should replace biopsies for the diagnosis of diffuse brain stem gliomas: a report from the Children's Cancer Group. Neurosurgery. 1993;33(6):1026–1029. doi: 10.1227/00006123-199312000-00010. discussion 1029–1030. [DOI] [PubMed] [Google Scholar]
- 11.Paugh BS, Qu C, Jones C, et al. Integrated molecular genetic profiling of pediatric high-grade gliomas reveals key differences with the adult disease. J Clin Oncol. 2010;28(18):3061–3068. doi: 10.1200/JCO.2009.26.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bax DA, Mackay A, Little SE, et al. A distinct spectrum of copy number aberrations in pediatric high-grade gliomas. Clinical Cancer Research : An Official Journal of the American Association for Cancer Research. 2010;16(13):3368–3377. doi: 10.1158/1078-0432.CCR-10-0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma S, Free A, Mei Y, Peiper SC, Wang Z, Cowell JK. Distinct molecular signatures in pediatric infratentorial glioblastomas defined by aCGH. Exp Mol Pathol. 2010;89(2):169–174. doi: 10.1016/j.yexmp.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 14.Zarghooni M, Bartels U, Lee E, et al. Whole-genome profiling of pediatric diffuse intrinsic pontine gliomas highlights platelet-derived growth factor receptor alpha and poly (ADP-ribose) polymerase as potential therapeutic targets. J Clin Oncol. 2010;28(8):1337–1344. doi: 10.1200/JCO.2009.25.5463. [DOI] [PubMed] [Google Scholar]
- 15.Barrow J, Adamowicz-Brice M, Cartmill M, et al. Homozygous loss of ADAM3A revealed by genome-wide analysis of pediatric high-grade glioma and diffuse intrinsic pontine gliomas. Neuro Oncol. 2011;13(2):212–222. doi: 10.1093/neuonc/noq158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilbertson RJ, Hill DA, Hernan R, et al. ERBB1 is amplified and overexpressed in high-grade diffusely infiltrative pediatric brain stem glioma. Clin Cancer Res. 2003;9(10, pt 1):3620–3624. [PubMed] [Google Scholar]
- 17.Angelini P, Hawkins C, Laperriere N, Bouffet E, Bartels U. Post mortem examinations in diffuse intrinsic pontine glioma: challenges and chances. J Neurooncol. 2011;101(1):75–81. doi: 10.1007/s11060-010-0224-7. [DOI] [PubMed] [Google Scholar]
- 18.Rajagopal MU, Hathout Y, MacDonald TJ, et al. Proteomic profiling of cerebrospinal fluid identifies prostaglandin D2 synthase as a putative biomarker for pediatric medulloblastoma: A pediatric brain tumor consortium study. Proteomics. 2011;11(5):935–943. doi: 10.1002/pmic.201000198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohnishi M, Matsumoto T, Nagashio R, et al. Proteomics of tumor-specific proteins in cerebrospinal fluid of patients with astrocytoma: usefulness of gelsolin protein. Pathol Int. 2009;59(11):797–803. doi: 10.1111/j.1440-1827.2009.02447.x. [DOI] [PubMed] [Google Scholar]
- 20.Schuhmann MU, Zucht HD, Nassimi R, et al. Peptide screening of cerebrospinal fluid in patients with glioblastoma multiforme. Eur J Surg Oncol. 2010;36(2):201–207. doi: 10.1016/j.ejso.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 21.Leach PA, Estlin EJ, Coope DJ, Thorne JA, Kamaly-Asl ID. Diffuse brainstem gliomas in children: should we or shouldn't we biopsy? Br J Neurosurg. 2008;22(5):619–624. doi: 10.1080/02688690802366198. [DOI] [PubMed] [Google Scholar]
- 22.Nazarian J, Santi M, Hathout Y, MacDonald TJ. Protein profiling of formalin fixed paraffin embedded tissue: Identification of potential biomarkers for pediatric brainstem glioma. Proteomics - Clinical Applications. 2008;2(6):915–924. doi: 10.1002/prca.200780061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jensen ON, Wilm M, Shevchenko A, Mann M. Sample preparation methods for mass spectrometric peptide mapping directly from 2-DE gels. Methods Mol Biol. 1999;112:513–530. doi: 10.1385/1-59259-584-7:513. [DOI] [PubMed] [Google Scholar]
- 24.Berglund SR, Santana AR, Li D, Rice RH, Rocke DM, Goldberg Z. Proteomic analysis of low dose arsenic and ionizing radiation exposure on keratinocytes. Proteomics. 2009;9(7):1925–1938. doi: 10.1002/pmic.200800118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beutler E, Gelbart T, Lee P, Trevino R, Fernandez MA, Fairbanks VF. Molecular characterization of a case of atransferrinemia. Blood. 2000;96(13):4071–4074. [PubMed] [Google Scholar]
- 26.Hasegawa K, Wakino S, Tatematsu S, et al. Role of asymmetric dimethylarginine in vascular injury in transgenic mice overexpressing dimethylarginie dimethylaminohydrolase 2. Circ Res. 2007;101(2):e2–e10. doi: 10.1161/CIRCRESAHA.107.156901. [DOI] [PubMed] [Google Scholar]
- 27.Ito E, Obayashi S, Nagai A, Imamura M, Azuma H. Regulation of myometrial contractivity during pregnancy in the rat: potential role for DDAH. Mol Hum Reprod. 2009;15(8):507–512. doi: 10.1093/molehr/gap041. [DOI] [PubMed] [Google Scholar]
- 28.Richer S, Rudy D, Statkute L, Karofty K, Frankowski J. Serum iron, transferrin saturation, ferritin, and dietary data in age-related macular degeneration. Am J Ther. 2002;9(1):25–28. doi: 10.1097/00045391-200201000-00006. [DOI] [PubMed] [Google Scholar]
- 29.Siemens N, Patenge N, Otto J, Fiedler T, Kreikemeyer B. Streptococcus pyogenes M49 plasminogen/plasmin binding facilitates keratinocyte invasion via integrin-integrin-linked kinase (ILK) pathways and protects from macrophage killing. J Biol Chem. 2011;286(24):21612–21622. doi: 10.1074/jbc.M110.202671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian X, Zhao C, Zhu H, et al. Hepatitis B virus (HBV) surface antigen interacts with and promotes cyclophilin a secretion: possible link to pathogenesis of HBV infection. J Virol. 2010;84(7):3373–3381. doi: 10.1128/JVI.02555-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu M, Liu H, Fannin J, et al. Acetaminophen improves protein translational signaling in aged skeletal muscle. Rejuvenation Res. 2010;13(5):571–579. doi: 10.1089/rej.2009.1015. [DOI] [PubMed] [Google Scholar]
- 32.Zakin MM. Regulation of transferrin gene expression. FASEB J. 1992;6(14):3253–3258. doi: 10.1096/fasebj.6.14.1426763. [DOI] [PubMed] [Google Scholar]
- 33.Baraniskin A, Kuhnhenn J, Schlegel U, et al. Identification of microRNAs in the cerebrospinal fluid as biomarker for the diagnosis of glioma. Neuro-oncology. 2012;14(1):29–33. doi: 10.1093/neuonc/nor169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roy S, Josephson SA, Fridlyand J, et al. Protein biomarker identification in the CSF of patients with CNS lymphoma. Journal of Clinical Oncology : Official Journal of the American Society of Clinical Oncology. 2008;26(1):96–105. doi: 10.1200/JCO.2007.12.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shnaper S, Desbaillets I, Brown DA, et al. Elevated levels of MIC-1/GDF15 in the cerebrospinal fluid of patients are associated with glioblastoma and worse outcome. Int J Cancer. 2009;125(11):2624–2630. doi: 10.1002/ijc.24639. [DOI] [PubMed] [Google Scholar]
- 36.Khwaja FW, Reed MS, Olson JJ, et al. Proteomic identification of biomarkers in the cerebrospinal fluid (CSF) of astrocytoma patients. J Proteome Res. 2007;6(2):559–570. doi: 10.1021/pr060240z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hathout Y, Formolo CA, Brown KJ, Nazarian J. Current Proteome Profiling Methods. In: Bouchard C, Hoffman EP, editors. Encyclopaedia of Sports Medicine Genetic and Molecular Aspects of Sport Performance. Vol XVIII. Wiley-Blackwell; 2011. pp. 46–57. [Google Scholar]
- 38.Deighton RF, McGregor R, Kemp J, McCulloch J, Whittle IR. Glioma pathophysiology: insights emerging from proteomics. Brain Pathology. 2010;20(4):691–703. doi: 10.1111/j.1750-3639.2010.00376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nicholas MK, Lukas RV, Jafri NF, Faoro L, Salgia R. Epidermal growth factor receptor - mediated signal transduction in the development and therapy of gliomas. Clin Cancer Res. 2006;12(24):7261–7270. doi: 10.1158/1078-0432.CCR-06-0874. [DOI] [PubMed] [Google Scholar]
- 40.Ohgaki H, Kleihues P. Genetic pathways to primary and secondary glioblastoma. Am J Pathol. 2007;170(5):1445–1453. doi: 10.2353/ajpath.2007.070011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suri V, Das P, Pathak P, et al. Pediatric glioblastomas: a histopathological and molecular genetic study. Neuro-oncology. 2009;11(3):274–280. doi: 10.1215/15228517-2008-092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang P, Heitman J. The cyclophilins. Genome Biol. 2005;6(7):226. doi: 10.1186/gb-2005-6-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Obchoei S, Wongkhan S, Wongkham C, Li M, Yao Q, Chen C. Cyclophilin A: potential functions and therapeutic target for human cancer. Med Sci Monit. 2009;15(11):RA221–RA232. [PubMed] [Google Scholar]
- 44.Han X, Yoon SH, Ding Y, et al. Cyclosporin A and sanglifehrin A enhance chemotherapeutic effect of cisplatin in C6 glioma cells. Oncol Rep. 2010;23(4):1053–1062. doi: 10.3892/or_00000732. [DOI] [PubMed] [Google Scholar]
- 45.Choi KJ, Piao YJ, Lim MJ, et al. Overexpressed cyclophilin A in cancer cells renders resistance to hypoxia- and cisplatin-induced cell death. Cancer Research. 2007;67(8):3654–3662. doi: 10.1158/0008-5472.CAN-06-1759. [DOI] [PubMed] [Google Scholar]
- 46.Boulos S, Meloni BP, Arthur PG, Majda B, Bojarski C, Knuckey NW. Evidence that intracellular cyclophilin A and cyclophilin A/CD147 receptor-mediated ERK1/2 signalling can protect neurons against in vitro oxidative and ischemic injury. Neurobiol Dis. 2007;25(1):54–64. doi: 10.1016/j.nbd.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 47.Jin ZG, Lungu AO, Xie L, Wang M, Wong C, Berk BC. Cyclophilin A is a proinflammatory cytokine that activates endothelial cells. Arterioscler Thromb Vasc Biol. 2004;24(7):1186–1191. doi: 10.1161/01.ATV.0000130664.51010.28. [DOI] [PubMed] [Google Scholar]
- 48.Yurchenko V, Zybarth G, O'Connor M, et al. Active site residues of cyclophilin A are crucial for its signaling activity via CD147. The Journal of Biological Chemistry. 2002;277(25):22959–22965. doi: 10.1074/jbc.M201593200. [DOI] [PubMed] [Google Scholar]
- 49.Kim JY, Kim WJ, Kim H, Suk K, Lee WH. The Stimulation of CD147 Induces MMP-9 Expression through ERK and NF-kappaB in Macrophages: Implication for Atherosclerosis. Immune Netw. 2009;9(3):90–97. doi: 10.4110/in.2009.9.3.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seko Y, Fujimura T, Taka H, Mineki R, Murayama K, Nagai R. Hypoxia followed by reoxygenation induces secretion of cyclophilin A from cultured rat cardiac myocytes. Biochemical and Biophysical Research Communications. 2004;317(1):162–168. doi: 10.1016/j.bbrc.2004.03.021. [DOI] [PubMed] [Google Scholar]
- 51.Tomita K, Izumi K, Okabe S. Roxatidine- and cimetidine-induced angiogenesis inhibition suppresses growth of colon cancer implants in syngeneic mice. J Pharmacol Sci. 2003;93(3):321–330. doi: 10.1254/jphs.93.321. [DOI] [PubMed] [Google Scholar]
- 52.Calabrese C, Poppleton H, Kocak M, et al. A perivascular niche for brain tumor stem cells. Cancer Cell. 2007;11(1):69–82. doi: 10.1016/j.ccr.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 53.Gilbertson RJ, Gutmann DH. Tumorigenesis in the brain: location, location, location. Cancer Research. 2007;67(12):5579–5582. doi: 10.1158/0008-5472.CAN-07-0760. [DOI] [PubMed] [Google Scholar]
- 54.Gilbertson RJ, Rich JN. Making a tumour's bed: glioblastoma stem cells and the vascular niche. Nat Rev Cancer. 2007;7(10):733–736. doi: 10.1038/nrc2246. [DOI] [PubMed] [Google Scholar]
- 55.Monje M, Mitra SS, Freret ME, et al. Hedgehog-responsive candidate cell of origin for diffuse intrinsic pontine glioma. Proc Natl Acad Sci USA. 2011;108(11):4453–4458. doi: 10.1073/pnas.1101657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hunt J, Cheng A, Hoyles A, Jervis E, Morshead CM. Cyclosporin A has direct effects on adult neural precursor cells. The Journal of Neuroscience : The Official Journal of the Society for Neuroscience. 2010;30(8):2888–2896. doi: 10.1523/JNEUROSCI.5991-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith ER, Zurakowski D, Saad A, Scott RM, Moses MA. Urinary biomarkers predict brain tumor presence and response to therapy. Clinical Cancer Research : An Official Journal of the American Association for Cancer Research. 2008;14(8):2378–2386. doi: 10.1158/1078-0432.CCR-07-1253. [DOI] [PubMed] [Google Scholar]
- 58.Boult JK, Walker-Samue S, Jamin Y, Leiper JM, Whitley GS, Robinson SP. Active site mutant dimethylarginine dimethylaminohydrolase 1 expression confers an intermediate tumour phenotype in C6 gliomas. J Pathol. 2011;225(3):344–352. doi: 10.1002/path.2904. [DOI] [PubMed] [Google Scholar]
- 59.Kostourou V, Robinson SP, Whitley GS, Griffiths JR. Effects of overexpression of dimethylarginine dimethylaminohydrolase on tumor angiogenesis assessed by susceptibility magnetic resonance imaging. Cancer Research. 2003;63(16):4960–4966. [PubMed] [Google Scholar]
- 60.Kostourou V, Troy H, Murray JF, et al. Overexpression of dimethylarginine dimethylaminohydrolase enhances tumor hypoxia: an insight into the relationship of hypoxia and angiogenesis in vivo. Neoplasia. 2004;6(4):401–411. doi: 10.1593/neo.04109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kostourou V, Robinson SP, Cartwright JE, Whitley GS. Dimethylarginine dimethylaminohydrolase I enhances tumour growth and angiogenesis. Br J Cancer. 2002;87(6):673–680. doi: 10.1038/sj.bjc.6600518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tokuo H, Yunoue S, Feng L, et al. Phosphorylation of neurofibromin by cAMP-dependent protein kinase is regulated via a cellular association of N(G),N(G)-dimethylarginine dimethylaminohydrolase. FEBS Lett. 2001;494(1–2):48–53. doi: 10.1016/s0014-5793(01)02309-2. [DOI] [PubMed] [Google Scholar]
- 63.Breckenridge RA, Kelly P, Nandi M, Vallance PJ, Ohun TJ, Leiper J. A role for Dimethylarginine Dimethylaminohydrolase 1 (DDAH1) in mammalian development. Int J Dev Biol. 2009;54(1):215–220. doi: 10.1387/ijdb.072356rb. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.