Abstract
Identification of the epidermal growth factor receptor variant III (EGFRvIII) mutation in glioblastoma has become increasingly relevant in the optimization of therapy. Traditionally, determination of tumor EGFRvIII-expression has relied on tissue-based diagnostics. Here, we assess the accuracy of magnetic resonance perfusion-weighted imaging (MR-PWI) in discriminating the EGFRvIII-expressing glioblastoma subtype. We analyzed RNA from 132 primary human glioblastoma tissue samples by reverse-transcription polymerase chain reaction (RT-PCR) for the EGFRvIII and EGFR wild-type mutations and by quantitative RT-PCR for expression of vascular endothelial growth factor (VEGF). Concurrently, 3 independent observers reviewed preoperative 1.5-Tesla (T)/SE or 3.0-Tesla (T)/GE MR perfusion images to determine the maximum relative tumor blood volume (rTBV) of each of these tumors. EGFRvIII-expressing glioblastomas showed significantly higher rTBV, compared with those tumors lacking EGFRvIII expression. This association was observed in both the 1.5T/SE (P = .000) and 3.0T/GE (P = .001) cohorts. By logistic regression analysis, combining the 2 MR system cohorts, rTBV was a very strong predictor of EGFRvIII mutation (odds ratio [rTBV] = 2.70; P = .000; McFadden's ρ2 = 0.23). Furthermore, by receiver-operating characteristic curve analysis, rTBV discriminated EGFRvIII with very high accuracy (Az = 0.81). In addition, we found that VEGF upregulation was associated, although without reaching statistical significance, with EGFRvIII expression (P = .16) and with increased rTBV (F-ratio = 2.71; P = .102). These trends suggest that VEGF-mediated angiogenesis may be a potential mediator of angiogenesis to increase perfusion in EGFRvIII-expressing glioblastomas, but there are likely several other contributing factors. This study demonstrates the potential to use rTBV, a MR-PWI–derived parameter, as a noninvasive surrogate of the EGFRvIII mutation.
Keywords: EGFRvIII, glioblastoma, MR perfusion, VEGF
Glioblastoma (GBM), the most aggressive primary adult brain tumor, is associated with overall poor prognosis, despite surgical resection, chemotherapy, and radiotherapy.1 Recent investigation has highlighted the importance of defining GBM molecular subtypes for improved clinical prognostication and appropriate application of novel therapies. Traditionally, such molecular stratification has relied on genetic or histopathological analyses requiring invasive methods, including brain biopsy or surgical resection.2 Imaging-based identification of the tumor expression profile is currently receiving much attention as an alternate, noninvasive approach for the characterization of tumors.
Previous studies have proposed such a shift in the diagnostic paradigm toward using advanced imaging modalities to stratify GBMs. MR perfusion-weighted imaging (MR-PWI), one such advanced imaging technique that quantifies relative tumor blood volume (rTBV), has been shown to be useful in determining histopathological glioma grade3–5 and prognosticating treatment outcome and survival in high-grade gliomas.6–8 In a recent small cohort study, MR-PWI was correlated with histopathologic features of GBM aggressiveness.9
However, to date, MR perfusion has not been associated with any one specific GBM mutation. Epidermal growth factor receptor variant III (EGFRvIII) is a primary activating oncogene in GBMs. Identification of EGFRvIII-positive glioblastomas by MR perfusion would be clinically significant in treatment selection and determining therapeutic sensitivity. EGFRvIII, the most common variant of EGFR, expressed with an overall prevalence of 20%–30%,10 encodes an in-frame deletion of the extracellular domain of EGFR, resulting in pro-oncogenic effects.11 Studies in EGFRvIII-transfected GBM cell lines have demonstrated that EGFRvIII enhances tumorigenicity, because it leads to decreased apoptosis and increased proliferation, angiogenesis, and invasion.10 Investigations of the survival effect of EGFRvIII are mixed, with some arguing that EGFRvIII expression prognosticates lower overall survival12 and others demonstrating no significant effect.13 Additional evidence suggests that the effect of EGFRvIII on survival must be understood in the context of its complex interactions with other molecular variables.13,14
Although the effect of EGFRvIII on survival has not been clearly proven, detection of EGFRvIII would be highly clinically valuable because its expression is becoming increasingly relevant in determining GBM therapeutic sensitivity. Earlier studies showed that presence of the mutation renders GBMs resistant to chemotherapies, such as cisplatin.15 In addition, there is some evidence that coexpression of EGFRvIII along with PTEN increases GBM sensitivity to EGFR tyrosine kinase inhibitors.16–18 However, other studies have failed to demonstrate this association.19,20
The importance of identifying the EGFRvIII mutation for treatment selection has come to the forefront with the development of EGFRvIII-targeted immunotherapy21,22 and EGFRvIII-directed monoclonal antibodies.23,24 Vaccination of the EGFRvIII peptide epitope has been shown to be an efficacious immunotherapy in syngeneic murine models and in humans in 3 phase II trials (2 consecutive single institution and 1 multi-institutional).25 Moreover, detection of an immune response in humans predicts greater median overall survival.22 In addition, EGFRvIII is a target of passive immunotherapy. Monoclonal antibodies against EGFRvIII have been shown to have an antitumor effect in both in vitro and in vivo models.24 The potential of antibody-based therapies in the treatment of brain tumors is supported by active clinical trials in the pediatric and adult populations.26 As such, identification of those patients with EGFRvIII-expressing GBMs is emerging as an important clinical factor in defining the subset of patients who will respond well to novel EGFRvIII-targeted therapies.
To our knowledge, there have been no studies that demonstrate an association between expression of EGFRvIII and MR perfusion. We hypothesized that the EGFRvIII-expressing GBM subset would be associated with increased MR perfusion-derived rTBV. We surmised that expression of the EGFRvIII mutation activates a complex array of independent signals to influence a multitude of downstream factors, perturbing the local biologic environment and enhancing angiogenesis, leading to increased rTBV. As an initial step in delineating these multifaceted pathways, we include in this article a subsidiary analysis of VEGF expression in our GBM tissue cohort. Studies in GBM cell lines indicate that the EGFRvIII mutation specifically upregulates the pro-angiogenic cytokine VEGF.27 Because increased rTBV is a measure of microvasculature density and has previously been shown to reflect an underlying angiogenic mechanism,28 we further hypothesized that EGFRvIII-mediated upregulation of VEGF is one of several factors contributing to the complex mechanism manifest as increased perfusion.
To determine the usefulness of MR-PWI as a noninvasive neuroimaging surrogate of EGFRvIII expression, we analyzed the preoperative rTBV signal in a large group of 132 human GBMs and assessed the relationship of this radiographic marker to EGFRvIII mutation status. We added the expression of EGFRwt to the analysis to compare its effect on perfusion with that specifically induced by EGFRvIII and to control for its expression in the models of EGFRvIII. We also examined VEGF expression to investigate whether VEGF-mediated angiogenesis is associated with the high perfusion found in the EGFRvIII-expressing GBMs. Our preliminary data indicate a strong association between rTBV and EGFRvIII in GBMs.
Materials and Methods
Population
We retrospectively reviewed records from all patients with primary GBMs (a WHO grade IV) who had tissue from surgical resection banked from January 2002 through January 2009 at the Hospital of The University of Pennsylvania (HUP). The patients did not receive any radiation or chemotherapy prior to surgical resection. We included all patients (n = 132) who had MR-PWI performed within 48 h prior to resection, using either a 1.5-Tesla/SE (n = 35) or a 3.0-Tesla/GE (n = 97) MR-system. Each patient was imaged using a single MR system. The strength of magnet used was dictated entirely by the date of the study, with earlier studies performed using the 1.5-Tesla magnet and later studies performed using the 3.0-Tesla magnet. The conversion to use of the 3.0-Tesla magnet was random with regard to the patients and is consistent with clinical practice changes in imaging patients with brain tumors at The University of Pennsylvania. The protocol was approved by the Institutional Review Board at HUP, and we obtained informed consent from each participant or each participant's guardian. An experienced neuropathologist performed the histopathologic evaluation, using the WHO classification criteria.
Magnetic Resonance (MR) Imaging
We acquired conventional MR images with either a 1.5-Tesla (Signa, GE Medical Systems) or a 3.0-Tesla (Siemens) clinical MR system. We acquired dynamic susceptibility contrast (DSC) single shot spin-echo (SE) echo-planar images using the 1.5-Tesla magnet and DSC T2* weighted gradient-echo (GE) echo-planar images using the 3.0-Tesla magnet, according to protocols previously described.29 DSC was performed 5 min after a 3 mL preloading dose of intravenous gadodiamide (Omniscan; GE Healthcare), administered according to protocols previously described.30 The preloading dose was administered to reduce the effect of contrast agent leakage on rTBV measurement.
Analysis of MR-PWI Data
We constructed blood volume maps using a standard algorithm, as previously described.31 Three independent observers drew regions of interest (ROIs) in MRIcron (http://www.sph.sc.edu/comd/rorden/mricron/install.html), based on an accepted method in the literature used to determine the region of maximum tumor perfusion.35 The observers blinded themselves to molecular subtype prior to the perfusion analysis. ROIs were drawn directly on the perfusion images, in conjunction with an overlay of the same axial level T1 gadolinium-enhanced image, to ensure accurate placement completely within the apparent margins of the neoplasm, avoiding cerebral blood vessels,5 and within T1 enhancing segments. The observers placed 15–25 voxel-sized ROIs per slice on 5 separate axial slices within regions of visually apparent maximum CBV. The mean CBV value was recorded for each axial slice and normalized to a white matter (WM) rTBV value, measured by placement of 15–25 voxels within the contralateral WM from the same axial slice. We then aggregated the 5 normalized rTBV values by taking the composite average to obtain a single rTBV value per tumor for each of the 3 observers. Because there was high interobserver correlation (the Pearson correlation coefficients varied from 0.61 to 0.74 in the 3.0T/GE cohort and from 0.71 to 0.77 in the 1.5T/SE cohort), we averaged the independent observer rTBV values to obtain a single rTBV value per tumor.
When we segmented the tumors into T1-enhancing and nonenhancing regions, we found that the areas of maximal rTBV corresponded to the gadolinium enhancing regions. Thus, tissue that was resected and banked for expression analysis was chosen, at the time of surgery, from an area of T1 enhancement, using MRI-based frameless stereotactic neuronavigation (STEALTH, Medtronic Corporation).
RT-PCR
We converted the total RNA, extracted by the TRIzol method from the tumor tissue stored in RNA later at -80°F, into cDNA fragments and subjected them to PCR analyses to assess the tumor expression of EGFRwt, EGFRvIII and GAPDH mRNAs, using the following primer sequences (written 5′ to 3′): GAPDH (forward: GTGAAGGTCGGAGTCAACGG, reverse: TGATGACAAGCTTCCCGTCTC); EGFRwt (forward-E3: ACCATCCAGGAGGTGGCTGGTTAT, EGFRvIII (forward: GAGCTCTTCGGGGAGCAG, reverse: GTGATCTGTCACCACATAATTACCTTTCT).32 PCR cycling conditions included an initial denaturing step at 94°C for 2 min, 30 cycles of denaturing at 94°C for 1 min, annealing at 55°C for 1 min, extending at 72°C for 1 min, and final extension at 72°C for 7 min.
Quantitative Real-Time PCR (RTqPCR)
We measured VEGF (isoform A) expression by RTqPCR, using a 7500 Fast Real-Time PCR System (Applied Biosystems) and primer and probe sets as previously described.33 We used 18S ribosomal RNA (Hs99999901_s1, Applied Biosystems) as an endogenous control for RNA levels. We analyzed samples in triplicate using Taqman Fast Universal Master Mix (Applied Biosystems). We performed data analysis using the 7500 Fast System SDS software version 1.4 (Applied Biosystems).
Statistical Analysis
We used statistical software SYSTAT 12 (SYSTAT) to analyze the data. According to Lilliefors Test, the rTBV data and patient age were normally distributed. The distribution of the VEGF data departed from Gaussian. Subsequently, we transformed the data using a natural logarithm. Since the transformed VEGF data was normally distributed, we used lnVEGF for the remainder of the analysis. We performed group comparisons using the 2-tailed t test.
We used multivariable logistic regression analysis to explore associations between EGFRvIII mutation and rTBV, VEGF, and EGFRwt measurements. The McFadden's ρ2 statistic was used to measure the goodness-of-fit of the model; ρ2 values of 0.20–0.40 are considered to be satisfactory. We integrated data from the 1.5T/SE and 3.0T/GE cohorts by controlling for the magnet type in the analysis, as the perfusion measurements must be standardized to account for the difference in the magnet systems. We used a linear regression to analyze associations between lnVEGF and rTBV. A subset of the population (14/132 patients) had missing VEGF data, secondary to inadequate RNA quantity to allow for further molecular analysis. We excluded these patients from all the analyses involving VEGF.
We employed receiver-operating characteristic curve (ROC) analysis to calculate the area under the curve (Az) to rank the accuracy of each parameter in the discrimination of GBM subtype. We used MedCalc for Windows, version 11.1.0 (MedCalc Software) in order to identify rTBV thresholds associated with maximum efficiency, sensitivity, specificity, and negative- and positive-predictive values (NPV and PPV). We used a Pearson correlation coefficient to assess the interobserver rTBV correlation. We used a Pearson's χ2 test to explore the dependence of occurrences of expression of EGFRwt on expression of EGFRvIII and the dependence of occurrences of expression of EGFRvIII on gender. To validate our logistic regression models, we performed 10-fold and leave-one-out cross-validation.
Results
Assessment of EGFRvIII Expression and Other Variables in GBMs
We determined the EGFRvIII status of all 132 GBM human tumor tissue samples, using reverse-transcription polymerase chain reaction (RT-PCR), a current, tissue-based, diagnostic standard. The threshold for determining presence of the mutation was determined by preliminary experiments, in which we correlated immunohistochemistry, using an EGFRvIII-specific antibody and RT-PCR in a subset of the samples. Of the 132 tumors, 30 (22.7%) were categorized as EGFRvIII positive (+) and 102 (77.3%) as EGFRvIII negative (−). The percentage EGFRvIII(+) GBMs was similar in the 2 cohorts, studied by either the 1.5T/SE (20.0%) or 3.0T/GE MR system (23.7%).
We also analyzed the EGFRwt status and found that there was a high incidence of EGFRwt expression in the overall GBM population (112/132; 84.8%), with relatively increased incidence in GBMs that concurrently expressed EGFRvIII (27/30; 90.0%), compared with the EGFRvIII(−) tumors (85/102; 83.3%). However, there was no statistically significant association between occurrences of expression of EGFRwt in relation to occurrences of expression of EGFRvIII in patients with GBMs (Pearson χ2 = 0.80; P = .371; n = 132). Gender and age of patients did not significantly vary in the EGFRvIII (+) and (−) tumor groups (Table 1).
Table 1.
Demographic data of patients with GBM stratified by presence EGFRvIII expression, MR system type, presence of the wild-type receptor (EGFRwt), and GBM location
| Variable | EGFRvIII (+) | EGFRvIII (−) | Total |
|---|---|---|---|
| Entire group, N (%) | 30 (23) | 102 (77) | 132 |
| 1.5-Tesla/SE MR-system, N (%) | 7 (20) | 28 (80) | 35 |
| 3.0-Tesla/GE MR-system, N (%) | 23 (24) | 74 (76) | 97 |
| Presence of EGFRwta, N (%) | 27 (90) | 85 (83) | 112 (85) |
| Location of GBM, N | |||
| Frontal, Parietal, Occipital | 24 | 57 | 81 |
| Temporal Insula | 6 | 44 | 50 |
| Posterior Fossa | 0 | 1 | 1 |
| Age in yearsb, median, range limits | 62, 29–83 | 61, 22–86 | 61, 22–86 |
| Genderc, N – males, females | 13, 17 | 58, 44 | 71, 61 |
| Race, N | |||
| Caucasian | 24 | 71 | 95 |
| African American | 1 | 2 | 3 |
| Asian American | 0 | 2 | 2 |
| Other/Unknown | 5 | 27 | 32 |
aPearson χ2 value (P value of χ2) = .80 (0.37).
bt test, P value = .646.
cPearson χ2 value (P value of χ2) = 1.71 (0.19).
EGFRvIII Expression in GBMs Correlates with Higher rTBV
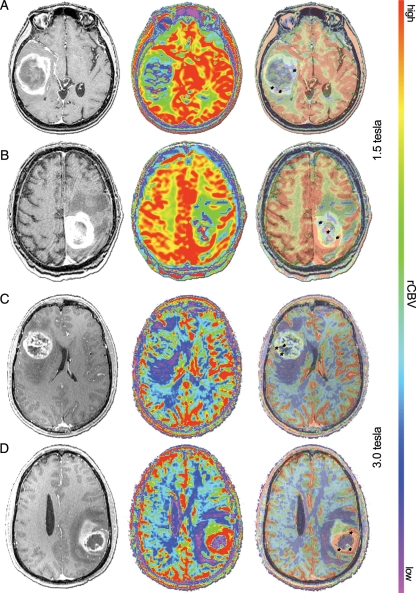
We assessed the difference in rTBV values between the EGFRvIII(+) and EGFRvIII(−) GBMs. In the 1.5T/SE cohort, mean rTBV in tumors expressing EGFRvIII (n = 7) was significantly higher than in EGFRvIII(−) tumors (EGFRvIII[+], mean = 5.23, standard deviation [SD] = 0.95; EGFRvIII[−], mean = 3.21, SD = 0.63; P = .001, t test). Similarly, in the 3.0T/GE cohort, mean rTBV in tumors expressing EGFRvIII (n = 23) was significantly higher than in EGFRvIII(−) tumors (EGFRvIII[+], mean = 7.02, SD = 1.45; EGFRvIII[−], mean = 5.60, SD = 1.32; P = 0.000, t test] (Fig. 1). The 1.5T/SE and 3.0 T/GE MR systems provide qualitatively different measures of rTBV, thus prohibiting combination of the data for t test analysis.
Fig. 1.
EGFRvIII expression correlates with higher maximum rTBV. Imaging from selected EGFRvIII (−) patients (A and C) and EGFRvIII (+) patients (B and D), with matched T1-post gadolinium (left), color 1.5-Tesla/SE MR perfusion (A and B, middle) or 3.0-Tesla/GE MR perfusion (C and D, middle), and 50% overlay (right). Black hatch marks (A–C and D, right) indicate representative voxels placed in regions of apparent maximum rTBV.
rTBV Predicts Expression of EGFRvIII
After finding significantly increased mean rTBV in the EGFRvIII-expressing tumors, we next performed logistic regression analysis to examine the potential of using the mean rTBV as a surrogate of EGFRvIII expression. In an integrated 1.5T/SE and 3.0T/GE cohort (n = 132), we found that rTBV is, in fact, an independent predictor of EGFRvIII expression [Logit(P) = 0.99(rTBV)–2.27(Magnet)–5.23, where P denotes the probability of positive EGFRvIII status] (McFadden's ρ2= 0.23, P = .000), with an odds ratio (OR) of 2.70 (95% confidence interval [CI], 1.83–3.98; P = .000). When this cohort was separated by magnet type, prediction of EGFRvIII expression remained extremely strong in both the 1.5T/SE cohort (McFadden's ρ2 = 1.00, P = .000) and the 3.0T/GE cohort (McFadden's ρ2 = 0.16, P = .000) (Table 2).
Table 2.
Multivariable logistic regression models in determination of the presence of molecular expression in GBM
| Dependent Variable | Cohort (Magnet Tesla (n)) | Regressor | P valuea | Odds Ratio (95% CI)c | Model Statistics |
|---|---|---|---|---|---|
| EGFRvIII | 1.5 (35) | rTBV Average | .000 | not in range | ρ2,b = 1.00; P = .000 |
| 3.0 (97) | rTBV Average | .000 | 2.121 (1.497–3.005) | ρ2= 0.163; P = .000 | |
| 1.5 and 3.0 (132) | rTBV Average | .000 | 2.698 (1.829–3.979) | ρ2= 0.227; P = .000 | |
| Magnet | .004 | 0.104 (0.022–0.488) | |||
| 1.5 and 3.0 (132) | rTBV Average | .000 | 2.670 (1.769–4.029) | ρ2 = 0.230; P = .000 | |
| Magnet | .005 | 8.938 (1.911–41.815) | |||
| EGFRwt | .520 | 1.667 (0.351–7.907) | |||
| 1.5 and 3.0 (118) | lnVEGF | .168 | 1.112 (0.956–1.292) | ρ2 = 0.014; P = .175 | |
| 1.5 and 3.0 (118) | rTBV Average | .000 | 2.597 (1.719–3.924) | ρ2 = 0.227; P = .000 | |
| Magnet | .016 | 12.979 (1.601–105.200) | |||
| lnVEGF | .576 | 1.084 (0.817–1.439) | |||
| EGFRwt | 1.5 and 3.0 (132) | rTBV Average | .394 | 1.164 (0.821–1.652) | ρ2 = 0.041; P = .099 |
| Magnet | .055 | 5.328 (0.962–29.500) |
aP value associated with the coefficient of regression.
bρ2 (McFadden's Rho2 statistic) values between 0.20 and 0.40 are considered very satisfactory.
cOdds Ratio box is colored gray if it is associated with a statistically significant P-value and a satisfactory McFadden's Rho2 statistic.
We explored the effect of further variables on the relationship between rTBV and EGFRvIII. The strength of the model is slightly improved by adding in an interaction factor and by controlling for patient gender (Supplementary Table S1). However, controlling for EGFRwt does not improve the model. Moreover, EGFRwt is not a significant independent regressor (P = .520) (Table 2), nor is age or gender (P = .610 and P = .158, respectively) (Supplementary material, Table S1). Of note, unlike EGFRvIII, EGFRwt expression is not predicted, with significance, by rTBV (Table 2).
rTBV Identifies EGFRvIII (+) GBMs with High Accuracy
We further validated our predictive models by applying ROC analysis to determine the accuracy of rTBV in discriminating EGFRvIII(+) GBMs. In the combined 1.5T/SE and 3.0T/GE cohorts, we found that the accuracy of rTBV in detecting the EGFRvIII mutation was very high, as determined by the area under the ROC (Az = 0.81), and was strengthened further by inclusion of an interaction factor (Az = 0.87). Similarly, the accuracy is high when analyzed in the independent MR system cohorts (Table 3) and for individual observers (Fig. 2A and B). Incorporation of EGFRwt into the analysis did not improve the accuracy of EGFRvIII discrimination (Az = 0.81).
Table 3.
ROC analysis for discrimination of EGFRvIII and EGFRwt using rTBV and/or VEGF
| Dependent Variable | Independent Variables | Cohort (MR-magnet Tesla (n)) | AUC (Az) |
|---|---|---|---|
| EGFRvIII | rTBV | 1.5 (35) | 1.00 |
| rTBV | 3.0 (97) | 0.76 | |
| rTBV, Magnet | 1.5 and 3.0 (132) | 0.81 | |
| rTBV, Magnet, EGFRwt | 1.5 and 3.0 (132) | 0.81 | |
| rTBV, Magnet, RTBVxMagnet | 1.5 and 3.0 (132) | 0.87 | |
| lnVEGF | 1.5 and 3.0 (118) | 0.59 | |
| rTBV, lnVEGF | 1.5 (33) | 1.00 | |
| rTBV, lnVEGF | 3.0 (85) | 0.75 | |
| rTBV, lnVEGF, Magnet | 1.5 and 3.0 (118) | 0.80 | |
| rTBV, lnVEGF, Magnet, rTBVxlnVEGF | 1.5 and 3.0 (118) | 0.82 | |
| EGFRwt | rTBV, Magnet | 1.5 and 3.0 (132) | 0.65 |
Fig. 2.
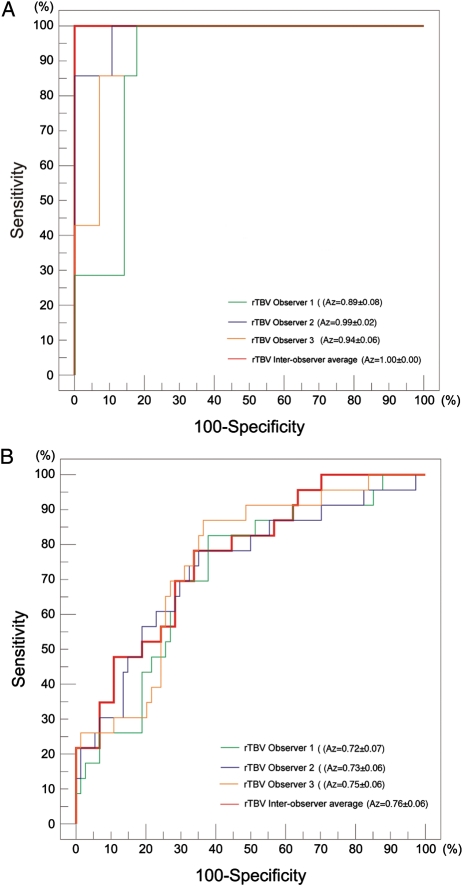
rTBV discriminates EGFRvIII expression with high accuracy, irrespective of MR-system type. ROC constructed based on rTBV values measured by 3 individual observers (green, violet, and orange lines) and based on rTBV averaged from the independent observers (red lines), using a (A) 1.5-Tesla/SE MR-system (n = 35) or (B) 3.0-Tesla/GE MR-system (n = 97). The interobserver variability in measurements, as demonstrated graphically and based on Az values, varies from 0.893 to 0.985 using the 1.5-Tesla/SE MR-system (A) and from 0.716 to 0.753 using the 3.0-Tesla/GE MR-system (B).
When using a 1.5T/SE MR system, the rTBV threshold value of 4.34, measured as the average of 3 independent observers, corresponds with maximum efficiency and is also associated with 100.0% sensitivity, specificity, positive-predictive value (PPV), and negative-predictive value (NPV) (Fig. 2A) and a 0.0% false-positive and false-negative rate.
When using a 3.0T/GE magnet, the rTBV threshold value of 5.94 corresponds with maximum efficiency and is associated with 78.3% sensitivity, 66.2% specificity, 41.9% PPV, and 90.7% NPV (Fig. 2B). This threshold is associated with a false-positive rate of 33.8%, meaning that based on rTBV classification criteria alone, a tumor would be falsely attributed a positive EGFRvIII status 33.8% of the time. In addition, the maximum efficiency threshold value of 5.94 is associated with a false-negative rate of 21.7%, meaning that, based on rTBV classification criteria alone, a tumor would be falsely attributed a negative EGFRvIII status 21.7% of the time. However, the sensitivity and specificity of the test can be changed by altering the threshold value. As such, the threshold (rTBV = 4.72) that is associated with a maximal sensitivity of 100% provides 29.7% specificity, 100% NPV, and 30.7% PPV, and the threshold (rTBV = 8.15) associated with a maximal specificity of 100% provides 21.7% sensitivity, 80.4% NPV, and 100% PPV.
We found no difference in classification accuracy in the logistic regression model with or without cross-validation. The classification accuracy and area under the curve (AUC) of the logistic regression model, without cross-validation, were 78.8% and 0.815, respectively. For 10-fold cross-validation, the classification accuracy and AUC were 78.8% and 0.764, respectively. For leave-one-out cross-validation, the classification accuracy and AUC were 78.8% and 0.773, respectively.
EGFRvIII (+) GBMs Demonstrate Enhanced VEGF Expression
To elucidate the molecular basis of elevated rTBV in the EGFRvIII-expressing subtype, we extended our study to assess VEGF expression, using quantitative RT-PCR. The expression of VEGF was enhanced, although without statistical significance, in the EGFRvIII-expressing tumors. Logarithmic values of VEGF were higher in EGFRvIII (+) tumors (mean = 3.72, SD = 2.63, range −2.35–7.38; n = 28) compared with EGFRvIII(−) GBMs (mean = 2.89, SD = 2.88, range −2.61–8.35; n = 90), with a trend toward statistical significance (P = .164, t test). In comparison, lnVEGF expression was found to be insignificantly higher in the EGFRwt(+) than in EGFRwt(−) GBMs (EGFRwt[+], mean = 3.16, SD = 2.82, n = 108; EGFRwt[−], mean = 2.32, SD = 3.05, n = 10; P = .418, t test).
EGFRvIII Expression is Associated With a Trend of VEGF Upregulation
Logistic regression analysis showed that lnVEGF expression predicts EGFRvIII status, with a trend toward statistical significance (OR = 1.11; 95% CI, 0.96–1.29; P = .168; McFadden's ρ2 = 0.01, P = .175). However, the combination of lnVEGF expression with rTBV does not improve prediction of EGFRvIII status, compared with rTBV alone (See Table 2 and Supplementary Table S1 for a breakdown by MR magnet system). Similarly, by ROC analysis, lnVEGF can discriminate the EGFRvIII mutation with moderate accuracy (Az = 0.59), but adding lnVEGF to rTBV does not improve accuracy (Az = 0.80), compared with rTBV alone.
VEGF Expression is Associated with Higher rTBV
After observing the significant association between rTBV and EGFRvIII and the correlation trending toward significance between lnVEGF and EGFRvIII, we explored an association between lnVEGF and rTBV. Although we found that rTBV is not significantly associated with lnVEGF (F-ratio = 2.71, P = .102), the analysis did show a trend toward statistical significance, controlling in the regression model for the statistically significant effect of the magnet type (F-ratio = 34.78, for the model, P = .000).
Discussion
The present study demonstrates a novel use for MR perfusion in the differentiation of an important GBM molecular subtype. Previous studies showed that rTBV correlates with glioma grade.3 Thus, it is intuitive that EGFRvIII-expressing GBMs, which have been shown to be more tumorigenic in biologic systems, would be associated with relatively higher rTBV. Accordingly, we show that the expression of EGFRvIII in human glioblastoma correlates with increased rTBV. In a cohort of 132 human primary GBMs, we found that the EGFRvIII-expressing subset exhibited statistically significant increased rTBV. This association, initially observed using a 1.5-Tesla(T)/SE MR-system, was subsequently corroborated in an expanded cohort and analyzed by a 3.0-Tesla(T)/GE MR system. The conversion to use of the 3.0T/GE magnet was random with regard to the patients and is consistent with clinical practice changes in imaging brain tumors in patients at The University of Pennsylvania.
Moreover, we constructed logistic models from our data, combining the 1.5T/SE and 3.0T/GE cohorts, and we observed that rTBV can discriminate the EGFRvIII-expressing subtype with efficient classificatory power and high predictive capacity. Of note, the predictive power was strong in each of the 1.5T/SE and 3.0-T/GE cohorts, when assessed independently. The advantage of combining the 2 magnet-type cohorts is that it allows for expansion of the overall cohort size.
For the purpose of using threshold values of rTBV in a clinical setting, it is necessary to separate the 1.5T/SE and 3.0T/GE MR systems. We determined a threshold value for the 1.5T/SE MR-system (rTBV = 4.34) that provides extremely high sensitivity and specificity. For the 3.0T/GE MR-system, the maximum efficiency threshold (rTBV = 5.94) is also associated with high, although slightly lower, sensitivity and specificity, comparable to widely used standard diagnostic tests. Because our study focuses on an imaging biomarker and because of the novelty of such molecular imaging work, there are few data available on accepted diagnostic test statistical parameters. That being said, further modeling in additional patient cohorts will hopefully allow us to improve on the false-negative rate (21.7%) observed to date in the 3.0T/GE cohort.
The association between rTBV and EGFRvIII is demonstrated regardless of whether the 1.5T/SE or 3.0T/GE MR system was used. This improves the generalizability of our results and has practical implications, because although 3.0-Tesla scanners have been increasingly introduced into clinical centers, the older 1.5-T magnets have not been entirely phased out. Thus, both magnet types are still in use and are likely to be for some time in the future. Of note, in the 1.5T/SE cohort, rTBV demonstrated very high accuracy in the identification of the EGFRvIII mutation, superior to that found using measurements obtained with the 3.0T/GE magnet. This disparity was likely attributable not only to the difference in magnetic field strength but also to the difference in pulse sequence between the 2 series. The 3.0-Tesla perfusion data are collected using gradient echo single shot echo planar imaging, whereas the 1.5-Tesla perfusion data are obtained using spin echo single shot echo planar imaging. It is well-known that spin echo functional images have greater specificity to microvascular perfusion than do gradient echo images, which provide both microvascular and macrovascular volume measurements.34 This difference in pulse sequence between the 1.5-Tesla and 3.0-Tesla cohorts could account for the variance in the strength of the association between rTBV and EGFRvIII in the 2 cohorts, because EGFRvIII may effect more predominately microvascular downstream events. In addition, the cohort studied by the 1.5-Tesla/SE MR system (n = 35) was smaller than that studied using the 3.0-Tesla/GE magnet (n = 97). Thus, the more powerful correlation demonstrated in the 1.5-Tesla/SE cohort may be attributable to the small sample size. However there is a very strong association between rTBV and EGFRvIII in the expanded 3.0-Tesla cohort. We obtained overall higher rTBV values for the 3.0T/GE cohort compared with the 1.5T/SE cohort. This difference in rTBV magnitude is related to 2 factors, including the difference in pulse sequence readout EPI (with GE producing a higher value compared to SE) and the difference in field strength (with the 3.0 Tesla magnet producing a higher strength field compared with the 1.5 Tesla magnet).
We measured maximum rTBV values by using a method that has been accepted in the literature.35 This method requires the observer to choose areas of maximal perfusion on the basis of the apparent brightness of the image. Although this approach likely compromises the accuracy of identifying the maximal value, given the subjective visual factor involved, it lends itself to being used in a clinical setting, where a more complex method would be too labor-intensive. We believe that we could optimize the classificatory performance of rTBV by assessing EGFRvIII status according to a continuous (RTqPCR) rather than a binary quantification scheme.
Although rTBV was a strong predictor of EGFRvIII status, it failed to predict expression of EGFRwt (Tables 2 and 3). This supports our hypothesis that the EGFRvIII mutation modifies the EGFR signal, in both intensity36 and quality10,36 with specific biologic effects reflected in perfusion imaging. In addition, we found no correlation between EGFRwt and EGFRvIII, consistent with the finding that controlling for EGFRwt does not improve the predictive model of EGFRvIII. Although patient age had no effect on the performance of the model, controlling for gender did slightly improve its predictive capacity (Supplementary Table S1).
Of note, the performance of our logistic regression model was stable under cross-validation. However, a limitation of this study is that the models were not initially obtained in a training set. Thus, the findings of this study need replicating in a truly independent data set.
Noninvasive imaging-based diagnostics have attracted much attention in recent years as a means to characterize tumor behavior and to monitor response to therapy.37 A small number of previous studies have also explored the ability to use imaging to detect molecular profiles. Others have explored the diversity, both inter- and intratumoral, in the basic anatomic MRI features of GBMs and have shown that this diversity reflects underlying variation in gene expression.9,38–40 However, these prior studies have been limited to anatomic MRI features, such as contrast enhancement. Here, we contribute to the new paradigm of imaging-based molecular diagnostics by moving beyond basic MRI features and showing the potential of using a physiologic MRI modality, MR perfusion, to predict a specific tumor mutation. Moreover, we demonstrate this finding in a large human cohort.
Because of the emergence of promising therapies directed specifically at EGFRvIII, including EGFRvIII peptide vaccination and EGFRvIII-directed monoclonal antibodies,24,25 it is becoming increasingly important to be able to identify the EGFRvIII mutation. Imaging-based identification of EGFRvIII could theoretically offer several advantages over tissue diagnosis. It would provide preoperative insight into molecular composition of the tumor, which might guide the preoperative use of EGFRvIII-directed therapies to shrink the tumor. Such techniques are not commonly used but may be in the future. Moreover, standard tissue-based methods are often associated with sampling errors due to improper processing of tumor tissues and tumor heterogeneity (EGFRvIII variants are not necessarily found uniformly throughout the tumor), resulting in some cases with false assignation of molecular profile. MR-PWI might avoid such sampling errors because it provides an in vivo scan, thus providing a greater scope of view to visualize the entire tumor and its surrounding brain tissue. Thus, MR-PWI, which is widely available and can easily be added to the conventional MRI at a low cost,41 might be used to verify tissue-based diagnosis or even eventually to replace it in selected cases. Imaging-based diagnosis might provide further use postoperatively to assess whether the residual tumor has high expression of EGFRvIII and is therefore susceptible to EGFRvIII-directed therapies. Moreover, such molecular profiling would be important in monitoring treatment response and in the characterization of recurrent tumors.
We attempted to elucidate the biologic basis of the association between the EGFRvIII mutation and rTBV enhancement by exploring differential expression of VEGF in the GBM cohort. A previous study performed by our group showed a strong correlation between the EGFRvIII mutation and VEGF expression in GBM cell lines.27 Here, we extended this analysis and assessed this correlation using actual human GBM tissue. We found that EGFRvIII expression is associated with an upregulation of VEGF, but this upregulation was not statistically significant. This is consistent with the fact that VEGF expression is mediated by several other upstream receptors.42 Of note, unlike EGFRvIII, the EGFRwt subgroup was not associated with enhanced VEGF expression. This suggests that the EGFRvIII mutation may modify the EGFRwt receptor with effects on the pro-angiogenic cytokine.
We found that VEGF and rTBV are associated, but this association does not reach statistical significance. It is unlikely that the lack of significance, in contrast to previous smaller studies,28 reflects an inadequacy of sample size, because our cohort was quite large. Instead, it may in part reflect an inherent technical limitation in measuring rTBV. It is well-known that VEGF leads to the formation of leaky vessels and vasogenic edema, which leads to an increase in contrast extravasation and an artificial underestimation of perfusion,43 a phenomenon that is particularly seen with high-grade gliomas and their disruption of the blood-brain barrier. We account for this phenomenon by preloading with contrast prior to PWI acquisition. However, this method is not perfect. Future studies require the use of contrast agents with greater blood-pooling properties and/or further correcting for extravasation.44 Our regression model of EGFRvIII was not improved when VEGF was added to rTBV, perhaps reflecting the strong dominance of rTBV in the prediction EGFRvIII. Alternatively, it might suggest the redundancy of rTBV and VEGF due to the presence of an underlying association.
These statistically insignificant trends suggest potential hypotheses to explain the molecular basis of the correlation between EGFRvIII-expression and increased perfusion: (1) VEGF may be involved in, although does not fully account for, the finding of increased tumor perfusion; (2) EGFRvIII represents one of several upstream modulators of VEGF expression; and (3) EGFRvIII may lead to enhancement of rTBV by angiogenic pathways, including, but not limited to, VEGF-mediated angiogenesis, and, likely, by non-angiogenic recruitment of blood vessels and other mechanisms not yet defined. Of note, we did not aim or expect to reduce changes in either angiogenesis or perfusion to a single angiogenic factor or variable. Our findings are in line with the paradigm of EGFRvIII affecting a multitude of downstream factors and of VEGF representing one of several molecules involved in a complex array of independent signals leading to angiogenesis and, in turn, perfusion. Moreover, our data are consistent with findings by Jain et al. that point to the multistep nature of angiogenesis and suggest that cerebral blood volume is a measure of a more mature stage of vascular genesis as compared with VEGF, which is involved in earlier stages of angiogenesis, and more closely correlated to leaky vessels.45 The VEGF data presented in this study sets the stage for further analysis of this mechanism. Follow-up work will measure other angiogenic markers and microvascular density to precisely delineate the presumed microvascular mechanisms contributing to upregulation of rTBV in primary GBMs. Of note, the statistical analyses presented in this article demonstrate associations among variables. We are suggesting potential biologic mechanisms that would be consistent with the associations that we have demonstrated.
Because EGFR amplification has previously been associated with conventional T1- and T2-weighted MR measurements,46 we are currently exploring the contribution of EGFR amplification to MR perfusion. In addition, ongoing work will look at the association between rTBV and clinical information, including patient survival, and other important GBM mutations, such as loss of PTEN. Finally, an interesting question would be whether recurrent GBMs that express EGFRvIII demonstrate an increase in rTBV compared with primary tumors in which EGFRvIII was not expressed. This is the subject of an additional study.
The tumor tissue used for analysis of molecular markers was resected from T1-enhancing regions. Because we found that the T1-enhancing segments corresponded to the areas of maximal MR perfusion values (as discussed in the Materials and Methods section), we studied molecular and imaging characteristics from matched tumor regions. Further studies will aim to rule out the possibility of mismatch between secreted protein and transcript levels.
Collectively, our data support a statistically significant correlation between rTBV and EGFRvIII mutation in GBMs and a weaker correlation of these variables to VEGF. Demonstration of these associations establishes the groundwork for further exploration of using MR perfusion as a noninvasive imaging surrogate of GBM EGFRvIII status.
Supplementary Material
Funding
This work was supported by the National Institutes of Health (R01-CA090586 to D.M.O., R01-2R56CA0905896-06A1 to D.M.O.), Terri Ann for a Cure, and the For Pete's Sake Fund.
Supplementary Material
Acknowledgments
These results were previously presented in part at a plenary session at the 2009 American Association for Cancer Research annual meeting and at the 2009 Society for Neuro-Oncology annual meeting.
Conflict of interest statement. None declared.
References
- 1.Krex D, Klink B, Hartmann C, et al. Long-term survival with glioblastoma multiforme. Brain. 2007;130(pt 10):2596–2606. doi: 10.1093/brain/awm204. doi:10.1093/brain/awm204. [DOI] [PubMed] [Google Scholar]
- 2.Bernays RL, Kollias SS, Khan N, Brandner S, Meier S, Yonekawa Y. Histological yield, complications, and technological considerations in 114 consecutive frameless stereotactic biopsy procedures aided by open intraoperative magnetic resonance imaging. J Neurosurg. 2002;97(2):354–362. doi: 10.3171/jns.2002.97.2.0354. doi:10.3171/jns.2002.97.2.0354. [DOI] [PubMed] [Google Scholar]
- 3.Law M, Yang S, Wang H, et al. Glioma grading: sensitivity, specificity, and predictive values of perfusion MR imaging and proton MR spectroscopic imaging compared with conventional MR imaging. AJNR Am J Neuroradiol. 2003;24(10):1989–1998. [PMC free article] [PubMed] [Google Scholar]
- 4.Arvinda HR, Kesavadas C, Sarma PS, et al. Glioma grading: sensitivity, specificity, positive and negative predictive values of diffusion and perfusion imaging. J Neurooncol. 2009;94(1):87–96. doi: 10.1007/s11060-009-9807-6. doi:10.1007/s11060-009-9807-6. [DOI] [PubMed] [Google Scholar]
- 5.Gasparetto EL, Pawlak MA, Patel SH, et al. Posttreatment recurrence of malignant brain neoplasm: accuracy of relative cerebral blood volume fraction in discriminating low from high malignant histologic volume fraction. Radiology. 2009;250(3):887–896. doi: 10.1148/radiol.2502071444. doi:10.1148/radiol.2502071444. [DOI] [PubMed] [Google Scholar]
- 6.Hirai T, Murakami R, Nakamura H, et al. Prognostic value of perfusion MR imaging of high-grade astrocytomas: long-term follow-up study. AJNR Am J Neuroradiol. 2008;29(8):1505–1510. doi: 10.3174/ajnr.A1121. doi:10.3174/ajnr.A1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saraswathy S, Crawford FW, Lamborn KR, et al. Evaluation of MR markers that predict survival in patients with newly diagnosed GBM prior to adjuvant therapy. J Neurooncol. 2009;91(1):69–81. doi: 10.1007/s11060-008-9685-3. doi:10.1007/s11060-008-9685-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galban CJ, Chenevert TL, Meyer CR, et al. The parametric response map is an imaging biomarker for early cancer treatment outcome. Nat Med. 2009;15(5):572–576. doi: 10.1038/nm.1919. doi:10.1038/nm.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barajas RF, Jr, Hodgson JG, Chang JS, et al. Glioblastoma multiforme regional genetic and cellular expression patterns: influence on anatomic and physiologic MR imaging. Radiology. 2010;254(2):564–576. doi: 10.1148/radiol.09090663. doi:10.1148/radiol.09090663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gan HK, Kaye AH, Luwor RB. The EGFRvIII variant in glioblastoma multiforme. J Clin Neurosci. 2009;16(6):748–754. doi: 10.1016/j.jocn.2008.12.005. doi:10.1016/j.jocn.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 11.Zhan Y, O'Rourke DM. SHP-2-dependent mitogen-activated protein kinase activation regulates EGFRvIII but not wild-type epidermal growth factor receptor phosphorylation and glioblastoma cell survival. Cancer Res. 2004;64(22):8292–8298. doi: 10.1158/0008-5472.CAN-03-3143. doi:10.1158/0008-5472.CAN-03-3143. [DOI] [PubMed] [Google Scholar]
- 12.Feldkamp MM, Lala P, Lau N, Roncari L, Guha A. Expression of activated epidermal growth factor receptors, Ras-guanosine triphosphate, and mitogen-activated protein kinase in human glioblastoma multiforme specimens. Neurosurgery. 1999;45(6):1442–1453. doi: 10.1097/00006123-199912000-00034. doi:10.1097/00006123-199912000-00034. [DOI] [PubMed] [Google Scholar]
- 13.Pelloski CE, Ballman KV, Furth AF, et al. Epidermal growth factor receptor variant III status defines clinically distinct subtypes of glioblastoma. J Clin Oncol. 2007;25(16):2288–2294. doi: 10.1200/JCO.2006.08.0705. doi:10.1200/JCO.2006.08.0705. [DOI] [PubMed] [Google Scholar]
- 14.Shinojima N, Tada K, Shiraishi S, et al. Prognostic value of epidermal growth factor receptor in patients with glioblastoma multiforme. Cancer Res. 2003;63(20):6962–6970. [PubMed] [Google Scholar]
- 15.Nagane M, Levitzki A, Gazit A, Cavenee WK, Huang HJ. Drug resistance of human glioblastoma cells conferred by a tumor-specific mutant epidermal growth factor receptor through modulation of Bcl-XL and caspase-3-like proteases. Proc Natl Acad Sci USA. 1998;95(10):5724–5729. doi: 10.1073/pnas.95.10.5724. doi:10.1073/pnas.95.10.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mellinghoff IK, Wang MY, Vivanco I, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353(19):2012–2024. doi: 10.1056/NEJMoa051918. doi:10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 17.Voelzke WR, Petty WJ, Lesser GJ. Targeting the epidermal growth factor receptor in high-grade astrocytomas. Curr Treat Options Oncol. 2008;9(1):23–31. doi: 10.1007/s11864-008-0053-5. doi:10.1007/s11864-008-0053-5. [DOI] [PubMed] [Google Scholar]
- 18.Raizer JJ. HER1/EGFR tyrosine kinase inhibitors for the treatment of glioblastoma multiforme. J Neurooncol. 2005;74(1):77–86. doi: 10.1007/s11060-005-0603-7. doi:10.1007/s11060-005-0603-7. [DOI] [PubMed] [Google Scholar]
- 19.van den Bent MJ, Brandes AA, Rampling R, et al. Randomized phase II trial of erlotinib versus temozolomide or carmustine in recurrent glioblastoma: EORTC brain tumor group study 26034. J Clin Oncol. 2009;27(8):1268–1274. doi: 10.1200/JCO.2008.17.5984. doi:10.1200/JCO.2008.17.5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown PD, Krishnan S, Sarkaria JN, et al. Phase I/II trial of erlotinib and temozolomide with radiation therapy in the treatment of newly diagnosed glioblastoma multiforme: North Central Cancer Treatment Group Study N0177. J Clin Oncol. 2008;26(34):5603–5609. doi: 10.1200/JCO.2008.18.0612. doi:10.1200/JCO.2008.18.0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li G, Mitra S, Wong AJ. The epidermal growth factor variant III peptide vaccine for treatment of malignant gliomas. Neurosurg Clin N Am. 2010;21(1):87–93. doi: 10.1016/j.nec.2009.08.004. doi:10.1016/j.nec.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 22.Choi BD, Archer GE, Mitchell DA, et al. EGFRvIII-targeted vaccination therapy of malignant glioma. Brain Pathol. 2009;19(4):713–723. doi: 10.1111/j.1750-3639.2009.00318.x. doi:10.1111/j.1750-3639.2009.00318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang W, Barth RF, Wu G, et al. Molecular targeting and treatment of EGFRvIII-positive gliomas using boronated monoclonal antibody L8A4. Clin Cancer Res. 2006;12(12):3792–3802. doi: 10.1158/1078-0432.CCR-06-0141. doi:10.1158/1078-0432.CCR-06-0141. [DOI] [PubMed] [Google Scholar]
- 24.Hadjipanayis CG, Machaidze R, Kaluzova M, et al. EGFRvIII Antibody-Conjugated Iron Oxide Nanoparticles for Magnetic Resonance Imaging-Guided Convection-Enhanced Delivery and Targeted Therapy of Glioblastoma. Cancer Res. 2010;70(15):6303–6312. doi: 10.1158/0008-5472.CAN-10-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sampson JH, Archer GE, Mitchell DA, Heimberger AB, Bigner DD. Tumor-specific immunotherapy targeting the EGFRvIII mutation in patients with malignant glioma. Semin Immunol. 2008;20(5):267–275. doi: 10.1016/j.smim.2008.04.001. doi:10.1016/j.smim.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fouladi M, Stewart CF, Blaney SM, et al. Phase I trial of lapatinib in children with refractory CNS malignancies: a Pediatric Brain Tumor Consortium study. J Clin Oncol. 2010;28(27):4221–4227. doi: 10.1200/JCO.2010.28.4687. doi:10.1200/JCO.2010.28.4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maity A, Pore N, Lee J, Solomon D, O'Rourke DM. Epidermal growth factor receptor transcriptionally up-regulates vascular endothelial growth factor expression in human glioblastoma cells via a pathway involving phosphatidylinositol 3′-kinase and distinct from that induced by hypoxia. Cancer Res. 2000;60(20):5879–5886. [PubMed] [Google Scholar]
- 28.Maia AC, Jr, Malheiros SM, da Rocha AJ, et al. MR cerebral blood volume maps correlated with vascular endothelial growth factor expression and tumor grade in nonenhancing gliomas. AJNR Am J Neuroradiol. 2005;26(4):777–783. [PMC free article] [PubMed] [Google Scholar]
- 29.Kapoor GS, Gocke TA, Chawla S, et al. Magnetic resonance perfusion-weighted imaging defines angiogenic subtypes of oligodendroglioma according to 1p19q and EGFR status. J Neurooncol. 2009;92(3):373–386. doi: 10.1007/s11060-009-9880-x. doi:10.1007/s11060-009-9880-x. [DOI] [PubMed] [Google Scholar]
- 30.Wang S, Kim S, Chawla S, et al. Differentiation between glioblastomas, solitary brain metastases, and primary cerebral lymphomas using diffusion tensor and dynamic susceptibility contrast-enhanced MR imaging. AJNR Am J Neuroradiol. 2011;32(3):507–514. doi: 10.3174/ajnr.A2333. doi:10.3174/ajnr.A2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cha S, Knopp EA, Johnson G, Wetzel SG, Litt AW, Zagzag D. Intracranial mass lesions: dynamic contrast-enhanced susceptibility-weighted echo-planar perfusion MR imaging. Radiology. 2002;223(1):11–29. doi: 10.1148/radiol.2231010594. doi:10.1148/radiol.2231010594. [DOI] [PubMed] [Google Scholar]
- 32.Yoshimoto K, Dang J, Zhu S, et al. Development of a real-time RT-PCR assay for detecting EGFRvIII in glioblastoma samples. Clin Cancer Res. 2008;14(2):488–493. doi: 10.1158/1078-0432.CCR-07-1966. doi:10.1158/1078-0432.CCR-07-1966. [DOI] [PubMed] [Google Scholar]
- 33.Yuan A, Yu CJ, Luh KT, et al. Quantification of VEGF mRNA expression in non-small cell lung cancer using a real-time quantitative reverse transcription-PCR assay and a comparison with quantitative competitive reverse transcription-PCR. Lab Invest. 2000;80(11):1671–1680. doi: 10.1038/labinvest.3780177. doi:10.1038/labinvest.3780177. [DOI] [PubMed] [Google Scholar]
- 34.Boxerman JL, Hamberg LM, Rosen BR, Weisskoff RM. MR contrast due to intravascular magnetic susceptibility perturbations. Magn Reson Med. 1995;34(4):555–566. doi: 10.1002/mrm.1910340412. doi:10.1002/mrm.1910340412. [DOI] [PubMed] [Google Scholar]
- 35.Wetzel SG, Cha S, Johnson G, et al. Relative cerebral blood volume measurements in intracranial mass lesions: interobserver and intraobserver reproducibility study. Radiology. 2002;224(3):797–803. doi: 10.1148/radiol.2243011014. doi:10.1148/radiol.2243011014. [DOI] [PubMed] [Google Scholar]
- 36.Hatanpaa KJ, Burma S, Zhao D, Habib AA. Epidermal growth factor receptor in glioma: signal transduction, neuropathology, imaging, and radioresistance. Neoplasia. 2010;12(9):675–684. doi: 10.1593/neo.10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harry VN, Semple SI, Parkin DE, Gilbert FJ. Use of new imaging techniques to predict tumour response to therapy. Lancet Oncol. 2010;11(1):92–102. doi: 10.1016/S1470-2045(09)70190-1. doi:10.1016/S1470-2045(09)70190-1. [DOI] [PubMed] [Google Scholar]
- 38.Diehn M, Nardini C, Wang DS, et al. Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proc Natl Acad Sci USA. 2008;105(13):5213–5218. doi: 10.1073/pnas.0801279105. doi:10.1073/pnas.0801279105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pope WB, Chen JH, Dong J, et al. Relationship between gene expression and enhancement in glioblastoma multiforme: exploratory DNA microarray analysis. Radiology. 2008;249(1):268–277. doi: 10.1148/radiol.2491072000. doi:10.1148/radiol.2491072000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Meter T, Dumur C, Hafez N, Garrett C, Fillmore H, Broaddus WC. Microarray analysis of MRI-defined tissue samples in glioblastoma reveals differences in regional expression of therapeutic targets. Diagn Mol Pathol. 2006;15(4):195–205. doi: 10.1097/01.pdm.0000213464.06387.36. doi:10.1097/01.pdm.0000213464.06387.36. [DOI] [PubMed] [Google Scholar]
- 41.Wetzel SG, Cha S, Law M, et al. Preoperative assessment of intracranial tumors with perfusion MR and a volumetric interpolated examination: a comparative study with DSA. AJNR Am J Neuroradiol. 2002;23(10):1767–1774. [PMC free article] [PubMed] [Google Scholar]
- 42.Ferrara N. Vascular endothelial growth factor: basic science and clinical progress. Endocr Rev. 2004;25(4):581–611. doi: 10.1210/er.2003-0027. doi:10.1210/er.2003-0027. [DOI] [PubMed] [Google Scholar]
- 43.Jain RK, di Tomaso E, Duda DG, Loeffler JS, Sorensen AG, Batchelor TT. Angiogenesis in brain tumours. Nat Rev Neurosci. 2007;8(8):610–622. doi: 10.1038/nrn2175. doi:10.1038/nrn2175. [DOI] [PubMed] [Google Scholar]
- 44.Boxerman JL, Schmainda KM, Weisskoff RM. Relative cerebral blood volume maps corrected for contrast agent extravasation significantly correlate with glioma tumor grade, whereas uncorrected maps do not. AJNR Am J Neuroradiol. 2006;27(4):859–867. [PMC free article] [PubMed] [Google Scholar]
- 45.Jain R, Gutierrez J, Narang J, et al. In vivo correlation of tumor blood volume and permeability with histologic and molecular angiogenic markers in gliomas. AJNR Am J Neuroradiol. 2011;32(2):388–394. doi: 10.3174/ajnr.A2280. doi:10.3174/ajnr.A2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aghi M, Gaviani P, Henson JW, Batchelor TT, Louis DN, Barker FG., 2nd Magnetic resonance imaging characteristics predict epidermal growth factor receptor amplification status in glioblastoma. Clin Cancer Res. 2005;11(24, pt 1):8600–8605. doi: 10.1158/1078-0432.CCR-05-0713. doi:10.1158/1078-0432.CCR-05-0713. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.