Abstract
Receptor protein tyrosine phosphatases (RPTPs) are involved in many cellular processes, including the regulation of adhesion, migration and cellular signaling. Many RPTPs are putative tumor suppressors because of the transcriptional and translational changes observed in their expression during tumorigenesis. Recently, RPTPs were shown to be post-translationally regulated during tumorigenesis by proteolysis in a manner similar to proteolysis of the Notch receptor. There is accumulating evidence that proteolysis of RPTPs influence their cellular function and that RPTP fragments may function as oncogenes. By exploiting what is known about RPTP ligand binding domains and crystal structures of ligand-RPTP interfaces, we describe novel molecular diagnostics that have been or can be developed to identify tumor margins and target tumor tissues.
Keywords: cell-cell interactions, cell-cell adhesion, receptor protein tyrosine phosphatase, RPTPs
INTRODUCTION
Receptor protein tyrosine phosphatases (RPTPs) are type I transmembrane proteins with intracellular tyrosine phosphatase domains linked to cell adhesion molecule (CAM)-like extracellular domains. Numerous studies demonstrate that RPTPs regulate cell-cell adhesion, cell-extracellular matrix adhesion and/or cell migration, similar to other CAMs. In the context of cancer biology this is significant, as the dissolution of stable cell-cell and increased cell-matrix adhesions have been documented as essential early steps in tumor progression. Eight RPTP subfamilies have been identified, each with distinct extracellular domain structures [1–3]. In the context of tumorigenesis, four RPTP subfamilies have been strongly implicated: the LAR Type IIa, the PTPμ Type IIb, the PTPα Type IV, and the PTPζ/β Type V subfamilies.
The extracellular domain structure of RPTPs, as with CAMs in general, determines the ligands they interact with in their environment. For example, some RPTPs bind to identical ligands, known as homophilic binding. Other RPTPs bind to different ligands, known as heterophilic binding. Depending on whether the ligands are present on other cells or in the extracellular matrix, RPTPs are able to promote either cell-cell adhesion or cell-extracellular matrix adhesion. The Type IIa, IIb, and V RPTP families mediate distinct types of cell adhesion. Type IIa RPTPs, PTPδ, PTPσ, and LAR, mediate heterophilic cell-cell adhesion with netrin-G-ligand 3 (NGL-3), heparin sulfate proteoglycans or the Laminin/nidogen complex [4–7]. However, PTPδ and specific isoforms of LAR can also mediate homophilic binding [8, 9]. Adhesion mediated by this family has been reviewed [10]. The type IIb PTPμ-like subfamily of RPTPs, including PTPμ, PTPρ, PTPκ, and PCP-2 (PTPλ), mediate homophilic cell-cell adhesion [11–16], with the exception of PCP-2 [17]. The adhesion mediated by this subfamily has been reviewed in [3, 18, 19]. Finally, the type V PTPζ/β family, including PTPζ (RPTPβ) and PTPγ, mediates heterophilic cell-matrix adhesion via the interaction with a number of different molecules, including with neural cell adhesion molecule (NCAM), the extracellular matrix protein, tenascin, and the neurite outgrowth promoting molecule, pleiotrophin [20–24].
Mis-expression and mutation of various RPTPs has been linked to tumorigenesis [25]. Recent data demonstrates that cleavage of the extracellular domain (ECD) and intracellular domain (ICD) of RPTPs regulates RPTP function and may be significant in cancer progression [26]. ECD cleavage observed for RPTPs is similar to the well-documented Notch cleavage paradigm, in which three proteases sequentially digest Notch to release biologically active protein fragments [27].
RPTP cleavage can have a number of effects. First, because they function as adhesion molecules, shedding of the RPTP’s ECD would dramatically change cell-cell and cell-matrix adhesion, likely promoting migration over adhesion. Second, release of the catalytic tyrosine phosphatase domains of cleaved RPTPs from the plasma membrane will likely impact signaling mediated by RPTPs. Evidence for this exists, as most of the RPTP ICDs translocate to the nucleus [28–31]. Finally, we suggest that the phosphatase activity of the membrane-based RPTP will differ from its nuclear-fragment, as has been described for PTPα and PTPε [32, 33]. This is likely not only due to changes in substrate availability in the different subcellular compartments, but also due to conformational changes in the tyrosine phosphatase domains that can affect their activity (see [19] for review of regulation of RPTP catalytic activity).
The best evidence exists for cleavage of type IIb RPTPs in cancer progression. Although there is evidence for the cleavage of three other RPTP subfamilies, the connection between cleavage of these RPTPs and cancer is not as clear. In general, full-length RPTPs are speculated to function as tumor suppressors [25]. We postulate that cleaved RPTP fragments function as oncogenes [26, 29]. The full length tumor suppressor versus cleaved oncogene hypothesis may explain the conflicting reports on the role of RPTPs in cancer. Cleavage of RPTPs may be preferentially triggered in cancer cells as a consequence of increased protease expression during cancer progression [34] or increased RPTP glycosylation [35]. In this review, we will summarize what is currently known about the cleavage of four RPTP subfamilies and their link to cancer progression. Finally, we will conclude with a discussion of how we can exploit the presence of cleaved RPTP fragments to develop cancer molecular diagnostic tests and molecular imaging agents.
CLEAVAGE OF RPTPs
Type IIa RPTPs
The type IIa subfamily of RPTPs are comprised exclusively of immunoglobulin (Ig) domains and fibronectin III (FNIII) repeats in their ECD. The ECD of type IIa RPTPs are proteolytically processed in the Golgi by a furin-like endo-peptidase to yield two non-covalently associated fragments [36, 37]. Members of this subfamily include LAR, PTPδ, and PTPσ (CRYPα). LAR ECD shedding was observed in high-density cell cultures [36], following PMA stimulation [38], treatment with calcium ionophores [39], and stimulation with EGF [40]. EGF stimulation increases EGFR tyrosine kinase activity and ERK1/2 activity, resulting in increased ADAM17 mediated cleavage of LAR [40]. The ECD of PTPσ is also shed in response to calcium ionophores and stimulation with the phorbol ester TPA [39]. Finally, PTPδ ECD is observed in tissue culture supernatant of COS-7 cells [41], demonstrating that all three type IIa RPTP subfamily members are cleaved and shed.
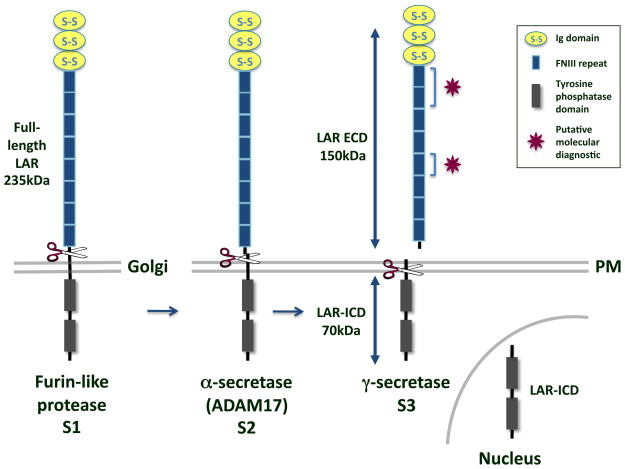
The ECDs of these RPTPs are likely cleaved by an α-secretase mechanism by an ADAM-like metalloprotease close to the plasma membrane (Figure 1). This cleavage event produces a shed fragment of the ECD and leaves the remainder of the RPTP membrane-tethered. The mechanism of LAR cleavage is similar to cleavage of Notch: the ECD is cleaved by an ADAM-like metalloprotease, likely ADAM-17 [40], to yield a 150 kD shed fragment. The remainder of the protein is then cleaved by presenilin (PS)-1 or -2 of the γ-secretase complex to yield a 70kD LAR-intracellular domain (LAR-ICD) fragment [31]. LAR-ICD is found both in the cytosol and the nucleus. The mechanisms responsible for PTPδ and PTPσ cleavage have not yet been identified, but are likely to be similar.
Figure 1.
LAR cleavage. The type I transmembrane protein LAR RPTP is composed of 3 Ig domains and 8 FNIII repeats in its ECD and two tandem tyrosine phosphatase domains in its ICD. It is cleaved by a furin-like endoprotease in the Golgi to yield a 150kDa E-subunit and an 85kDa P-subunit that remain non-covalently attached (S1) [36, 37]. In response to phorbol esters, high cell density, or calcium ionophores, a 150kDa segment of the LAR ECD is cleaved at the plasma membrane (PM) and shed by ADAM-17/TACE (S2) [40]. The γ-secretase complex then cleaves the transmembrane domain of LAR to yield a 70kDa membrane-free fragment, LAR-ICD, which is capable of translocating to the nucleus (S3) [31]. Molecular diagnostics like the ones used to identify extracellular PTPμ fragments in GBM tissue could be designed to bind to the heterophilic [4, 6] and homophilic [9] binding sites on LAR, as indicated by the magenta stars.
The cellular consequences of LAR cleavage are not clear. One study by Haapasalo and colleagues suggests that LAR cleavage promotes stable cell-cell adhesion [31] by binding cytoplasmic β-catenin and reducing transcription of the β-catenin regulated gene, cyclin D1 [31]. Another study by Ruhe et al., however, demonstrates that LAR cleavage reduces LAR phosphatase activity and thereby promotes EGFR signaling, thus likely promoting cellular proliferation and migration [40]. The reasons for these discrepancies are likely a consequence of contextual differences, as the Haapasalo et al. study evaluated LAR cleavage in CHO cells over-expressing full-length and LAR-ICD, whereas the Ruhe et al. group examined endogenous LAR in MDA-MB-468 breast cancer cells and TACE knock-out mouse fibroblasts as well as over-expressed full-length LAR in HEK-293 cells.
LAR mRNA and protein are over-expressed in various cancer cell lines and in tissue samples, supporting its function as an oncogene, although this is not universally observed in all cancers. For example, in a microarray analysis of melanoma cell lines, LAR mRNA was either not observed or found to have reduced expression compared to normal melanocytes, suggesting that LAR functions as a tumor suppressor in these cells [42]. Antibody studies have demonstrated that LAR protein is elevated in breast cancer tissue compared to normal tissue controls [43] and in thyroid carcinoma tissue sections [44]. Neither of these studies looked at LAR cleavage products as they were published before the existence of LAR cleavage in cancer was hypothesized. However, neither of these studies excludes the possibility that part of the increased LAR expression observed may be a result of increased cleavage and/or stability of a protein fragment. Definitive demonstration of LAR cleavage in cancer can only be conducted with immunoblot analysis of potential cleavage products. Only Yang et al. [43] evaluated LAR protein on immunoblots, and most of the figures do not show lower molecular weight proteins to determine whether cleavage products are present. LAR cleavage as one mechanism of altering adhesion-based signals during tumorigenesis remains a viable hypothesis, and is supported by the existence of LAR cleavage fragments in cancer cell lines [31, 39, 40].
Full-length PTPδ and PTPσ, on the other hand, are hypothesized to function as tumor suppressors. Homozygous deletions and/or methylation of the PTPδ gene, PTPRD, has been described in a wide range of cancers, including glioblastoma multiforme (GBM), melanoma, neuroblastoma and lung carcinoma [45–49]. Addition of wild-type PTPδ to GBM cell lines harboring the PTPRD mutations suppresses cell growth [47, 49], whereas using shRNA to reduce PTPδ protein expression increases tumor size of immortalized human astrocyte cells in a mouse xenograft experiment [49]. Single nucleotide polymorphisms have been observed to occur in the PTPσ gene, PTPRS, in human cancer [50]. Additional evidence demonstrates that PTPσ expression regulates cell proliferation in A431 carcinoma cells. Induced over-expression of PTPσ in these cell lines decreased colony formation in soft agar and antagonized EGFR signaling, suggesting a potential role as a tumor suppressor [51]. Again, future studies evaluating immunoblots of cancer tissue or cell lines for cleavage fragments of type IIa RPTPs will illuminate whether there is any connection between type IIa RPTP cleavage and tumorigenesis.
Type IIb RPTPs
The ECD of the type IIb subfamily of homophilic cell-cell RPTPs are characterized by a meprin-A5 (neuropilin)-mu (MAM) domain, one Ig domain, and four FNIII repeats. The PTPμ-like RPTP intracellular domain contains a cadherin homology domain, and two tyrosine phosphatase domains, of which only the membrane proximal domain is catalytically active. ECD shedding and ICD cleavage has been observed for both PTPκ [28] and PTPμ [29], and is similar to the mechanism already described for the type IIa subfamily. Aberrant glycosylation of PTPκ has been suggested as one reason for the cleavage and shedding of PTPκ observed in tumor cells [35]. ADAM10 has been identified as the protease responsible for PTPκ-ECD cleavage [28]. Cleavage by the PS/γ-secretase complex of the membrane-tethered stump within the transmembrane domain releases an ICD fragment of the RPTP [28, 29]. Both PTPκ and PTPμ ICDs are capable of translocating to the nucleus [28, 29].
Full-length PTPμ is down-regulated in GBM [52]. Reduced PTPμ protein expression contributes to the increased migration observed in GBM cells [52]. The mechanism responsible for PTPμ down-regulation was determined to be proteolysis of the full-length protein to yield a shed ECD and membrane-released ICD [29, 53]. Notably, ECD and ICD fragments of PTPμ are present only in GBM tissue when compared to the normal tissue surrounding the tumor. It was found that PTPμ cleavage to yield PTPμ-ICD promotes GBM cell migration, growth factor independent survival and anchorage independent growth [29], suggesting that PTPμ cleavage promotes tumorigenesis. The study by Burgoyne et al. also demonstrated that phosphatase activity was necessary for some of the tumor promotion effects of PTPμ-ICD on GBM cells, as the ability of PTPμ-ICD to promote GBM cell migration was reduced when a wedge peptide that interferes with phosphatase function was used [29].
A role for PTPκ in tumor progression has also been suggested. PTPκ expression is reduced in melanoma cell lines and human tissue biopsies [54], and in Hodgkin’s lymphoma cells [55]. Over-expression of PTPκ in Hodgkin’s lymphoma cells reduces cellular proliferation and survival [55], suggesting a tumor suppressor function. Other data suggest that PTPκ may promote tumor growth. For example, the cleaved cytoplasmic domain fragment of PTPκ is catalytically active, translocates to the nucleus and promotes β-catenin-mediated transcription [28]. Furthermore, it is hypothesized that aberrant glycosylation leads to PTPκ cleavage and, ultimately, increased colon cancer cell migration, possibly by interfering with stable homophilic adhesion [35]. It is interesting that differential glycosylation may control the proteolytic cascade. Cleavage of PTPκ may, therefore, have a two-fold effect on cancer cells: loss of stable cell-cell adhesion to promote migration and increased β-catenin/Wnt signaling to promote epithelial to mesenchymal transition.
Type IV RPTPs
PTPα and PTPε are two members of the type IV RPTP subfamily. These proteins have short, highly glycosylated ECDs with no known structural motifs. There is no evidence that type IV RPTPs mediate adhesion directly, but they promote integrin-mediated adhesion through the activation of src family kinases downstream of integrin receptors [56, 57]. In addition, PTPα interacts in cis with contactin1, and therefore has the potential to regulate adhesion through this interaction [58]. Unlike the mechanism proposed for the cleavage of the other RPTPs, calpain is the protease identified that cleaves PTPα and PTPε. Calpain is a calcium regulated cysteine protease, and is suggested to cleave both type IV subfamily members in the intracellular juxtamembrane domain [32]. The two fragments, PTPα-ICD (or p66α) and PTPε-ICD (p65ε), are both found in the cytosol of cells [32]. No evidence for nuclear localization of PTPα-ICD exists, while PTPε-ICD is excluded from the nucleus [33]. Notably, changes in the subcellular localization of PTPα-ICD and PTPε-ICD fragments result in a significant reduction in the ability of these RPTPs to dephosphorylate their substrates src and the voltage gated potassium channel, Kv2.1 [32]. PTPα and PTPε expression is also regulated in cancer as reviewed in [59]. While the molecular mechanism of PTPα and PTPε cleavage is distinct from that of other RPTPs, the paradigm of cleavage regulating localization and substrate availability is similar.
Type V RPTPs
PTPζ/β (Ptprz) and PTPγ are members of the type V subfamily of RPTPs that are characterized by an extracellular carbonic anhydrase-like (CA) domain, a glycosylated long unique spacer region, and a single FNIII repeat. Alternative splicing of PTPζ yields three isoforms, PTPζ-A, PTPζ-B, and PTPζ-S (also known as phosphacan) [60]. PTPζ isoforms bind to many different ligands, including the extracellular matrix protein, tenascin, the cell adhesion molecule, contactin1, the heparin binding growth factor, pleiotrophin, and the cell-cell CAMs, Nr-CAM, Ng-CAM/L1, and N-CAM [20–24]. PTPζ isoforms, therefore, have the ability to regulate several aspects of cell adhesion and migration via these interactions.
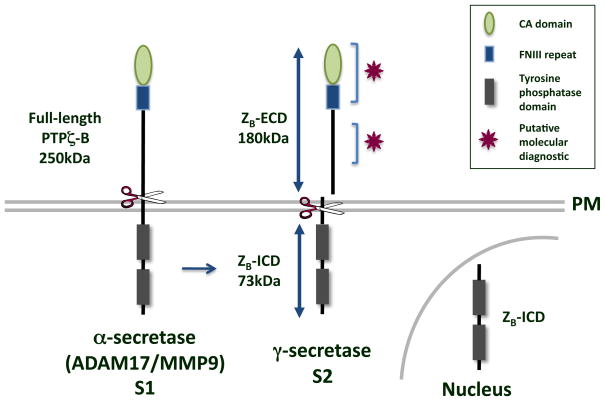
Detection of full-length protein expression of the PTPζ-A and –B isoforms was often difficult [61]. Recently, it was determined that both PTPζ-A and –B ECDs are cleaved to yield shed extracellular fragments, termed ZA-ECD and ZB-ECD, respectively [30]. Two proteases are implicated in the generation of ZA- and ZB-ECD; MMP-9 is responsible for the constitutive cleavage of the two PTPζ isoforms, while ADAM17 (TACE) is likely the protease responsible for PMA-induced cleavage (Figure 2). As has been observed for other RPTPs, the γ-secretase complex also cleaves the membrane-tethered PTPζ fragments left after MMP/ADAM cleavage into ZA- and ZB-ICDs [30]. ZB-ICD is found in the cytosol and nucleus of the cell, and thus is likely involved in as of yet unidentified novel cell signaling events, as has been observed for the other cleaved RPTP ICDs.
Figure 2.
PTPζ cleavage. The PTPζ-A and –B isoforms are cleaved in a similar manner. For simplicity’s sake, cleavage of PTPζ-B is shown. The ECD of PTPζ-B consists of a CA domain, an FNIII repeat, and a unique spacer region. S1 cleavage by either MMP-9 or ADAM-17 at the plasma membrane (PM), yields an 180kDa shed ZB-ECD [30]. S2 cleavage by γ-secretase yields the 73kDa ZB-ICD, which is found both in the cytosol and nucleus [30]. Molecular diagnostics, indicated by the magenta stars, could be developed to bind to the contactin1-binding site in the CA domain [78], the NgCAM, NCAM, NrCAM binding site in between the CA and FNIII repeat [20], the tenascin binding site in the FNIII repeat [70], or the pleiotrophin binding site in the spacer region [24].
Alteration in PTPζ expression is linked to many different cancers. Using antibodies directed to the carboxy-terminus of PTPζ, Foehr and colleagues demonstrated that PTPζ is highly expressed in colon, prostate and lung adenocarcinomas, breast carcinoma, melanoma and astrocytoma tissue [62]. In some instances, the PTPRZ gene and PTPζ mRNA are reduced in cancerous tissue compared to normal tissue [63]. The RPTPZ gene is the most differentially expressed gene when comparing grade II gliomas oligodendroglioma, oligoastrocytoma, and astrocytoma [64]. Immunohistochemistry for PTPζ strongly labeled oligodendroglioma, whereas other glioma types were not positively labeled for PTPζ. In addition, PTPζ mRNA and protein are over-expressed in various GBM cell lines compared to normal brain controls [65, 66].
Functional studies demonstrate that PTPζ isoforms regulate GBM adhesion and migration and that PTPζ may in fact act as an oncogene. Using a modified Boyden chamber assay, Muller and colleagues demonstrated that PTPζ expression was necessary for chemotaxis towards serum [65]. Furthermore, chemotaxis was augmented by pretreating the GBM cells with pleiotrophin (a PTPζ ligand) prior to addition to the Boyden chamber, which also depended upon PTPζ expression. Another study demonstrated a likely mechanism for GBM migration in vivo [66]. GBM cells themselves do not appear to migrate toward soluble pleiotrophin. Rather, they will migrate preferentially on pleiotrophin when presented as a fixed substrate, known as haptotaxis [66]. This haptotaxis is dependent on binding to PTPζ, as addition of antibodies that recognize the ECD of all three isoforms of PTPζ or use of siRNA against PTPζ, abolishes this migration [66, 67]. This same phenomenon has also been observed to occur in cortical neurons, in addition to GBM cells [68]. Proliferation of U251-MG cells was significantly reduced following decreased PTPζ expression by siRNA [67]. Finally, reducing PTPζ expression with si-RNA abolishes the tumorigenicity of U251-MG human glioblastoma cells implanted either subcutaneously or intracerebrally in nude mice [67]. These studies suggest that over-expression of PTPζ may promote tumorigenesis.
In retrospect, early studies on PTPζ suggested a function for the shed ZA-ECD and ZB-ECD fragments in cancer. Two studies, one by Sakurai et al. [69] and one by Adamsky et al. [70], suggest that either phosphacan or shed PTPζ fragments are capable of antagonizing glioma cell adhesion. PTPζ on C6 glioma cells is required for adhesion to the neural cell adhesion molecule, NgCAM/L1 [69]. After 24 hours of adhesion, however, the glioma cells spontaneously round-up and detach from the substrate. Notably, detachment from the NgCAM substrate is abolished when the conditioned media was depleted of either phosphacan or potentially ZA-ECD [69], using an antibody that we now know recognizes both forms of PTPζ [69]. Therefore, ZA-ECD may antagonize PTPζ-mediated adhesion to NgCAM. The second study demonstrated that adhesion of an astrocytoma cell line to tenascin, another PTPζ ligand, could be abolished using any Fc fusion proteins containing the FNIII repeat of PTPζ [70]. Given that both ZA- and ZB-ECD fragments contain the FNIII repeat, this again suggests a possible antagonistic function for these fragments in glioma and astrocytoma cell adhesion and the promotion of tumorigenesis.
EXPLOITATION OF ADHESIVE PEPTIDES FOR MOLECULAR RECOGNITION OF TUMOR-DERIVED CLEAVED EXTRACELLULAR FRAGMENTS
PTPμ peptides in GBM
While the function of the shed PTPμ- and PTPκ-ECDs is not currently known, Burden-Gulley et al. demonstrated that the shed PTPμ-ECD remains in the vicinity of the GBM tumors [53]. It was hypothesized that analysis of the location of the shed PTPμ-ECD can be used to identify the tumor margin [53]. PTPμ is a homophilic binding protein that requires both its MAM and Ig domains to mediate cell-cell adhesion [15, 71–74]. The crystal structure of PTPμ’s extracellular domain has been solved [72, 73]. Based upon the surface exposed regions of the MAM and Ig domains, we devised peptides that were likely to be involved in homophilic binding [53]. Our rationale is that endogenous full-length PTPμ would be fully engaged in adhesion and unlikely to bind to an exogenous low affinity peptide. However, an extracellular fragment of PTPμ that is no longer tethered to the plasma membrane could theoretically bind an exogenous peptide that mimicked the regions involved in homophilic binding. To test this hypothesis, fluorescently tagged peptides capable of binding to PTPμ-ECD were injected intravenously in mouse xenograft tumor models. Two of the peptides designed were empirically determined to recognize the ECD of PTPμ in tumors in the animal within 2 minutes following tail vein injection [53]. These peptides were highly effective at demarcating the main tumor and its edges in both mouse flank tumors and intracranial glioma tumors [53]. Furthermore, these peptides crossed the compromised blood-brain barrier to bind intracranial tumors within 20 minutes. These data indicate that the peptides might be of clinical utility in imaging tumors due to this fast molecular recognition time frame. These data demonstrate that PTPμ-ECD shedding occurs preferentially in the tumor microenvironment and can be utilized as a novel means of identifying the tumor and its margin.
Both PTPκ and PTPρ have been implicated in cancer progression [35, 55, 75]. Shed PTPκ is found in the conditioned media of colon cancer cells [35]. Given the structural and sequence similarity between all type IIb RPTPs [73], it is likely that PTPρ and PCP-2 may also be cleaved in cancer cells. PCP-2 does not mediate homophilic cell-cell adhesion [17], so the function of its putative secreted ECD is unclear. PTPρ, like PTPμ and PTPκ, does mediate homophilic cell-cell adhesion [16, 76]. Using our knowledge of the function and structure of the type IIb RPTPs [17, 73], similar fluorescent peptides could be developed to determine whether their shed ECDs are also preferentially located in tumor microenvironments, as suggested [53].
Design of molecular diagnostics to LAR ECD fragments
A number of studies have used antibody technology to evaluate LAR protein expression in cancer cell lines and tissues [39, 40, 43, 44]. Again, these strategies do not distinguish between full-length LAR and LAR fragments. To design molecular diagnostics like the ones developed for PTPμ, we need to compare what is known about ligand binding interaction domains and surface exposed residues of LAR. Of the type IIa RPTPs, only a LAR splice variant is able to mediate homophilic binding leading to the promotion of neurite outgrowth and/or cell adhesion [8, 9]. This 11kD isoform, termed LARFN5C, encodes a short protein that contains in its ECD a novel N-terminus and only a small portion of the fifth FNIII repeat [9]. Therefore, LAR homophilic binding requires either a novel sequence contained in LARFN5C or part of its fifth FNIII repeat. Peptides designed to these regions could be used similarly to the ones we designed for PTPμ (Figure 1).
LAR binds heterophilically to the laminin-nidogen complex and to NGL-3 [4, 6]. LAR binding to laminin-nidogen occurs only with a splice variant missing 9 amino acids within the fifth FNIII repeat of LAR [6], whereas binding to NGL-3 requires the first 2 FNIII repeats [4]. It is interesting to note that two ligand interactions of LAR are dependent on changes within the fifth FNIII repeat, suggesting that this is an important part of the molecule for mediating interactions. This information combined with crystal structure analyses of the LAR ECD to identify surface exposed residues, could be used to design peptides to bind to the shed LAR ECD, as has been demonstrated for PTPμ [53].
Design of molecular diagnostics to PTPδ and PTPσ fragments
PTPδ mediates bead binding, Sf9 cell aggregation, neuronal adhesion, and promotes neurite outgrowth all in a homophilic manner [8]. The domains in the ECD of PTPδ that are responsible for homophilic binding, however, have not been characterized. PTPσ has not been shown to mediate homophilic binding. Heterophilic interactions have been documented for both PTPδ and PTPσ. PTPδ and PTPσ bind heterophilically to NGL-3 via their first two FNIII repeats [4]. PTPσ binding to unidentified heterophilic ligands within the chick visual system depends on both the Ig and FNIII repeats of PTPσ [7]. Future crystal structure analyses of both the receptors and ligands will be beneficial for the development of peptides capable of interacting with shed ECDs of these RPTPs. Based on binding experiments, the Ig and FNIII repeats of PTPδ and PTPσ are both likely to be important targets.
Design of molecular diagnostics to PTPζ fragments
Molecular diagnostic tests have been used to evaluate the expression of PTPζ in tumor tissue [62, 64, 65, 70]. Using antibodies generated to the ECD of PTPζ-B, Foehr and colleagues identified that PTPζ is over-expressed in a number of different cancer tissues [62]. Furthermore, one PTPζ antibody linked to the toxin saporin is effective at killing cancer cells in vitro and significantly decreasing tumor volume and tumor morbidity in a mouse U-87 MG subcutaneous glioma model [62]. Using Fc chimera fusion proteins of the FNIII repeat of PTPζ, Adamsky et al. demonstrated that a high level of PTPζ bound to GBM and astrocytoma cells [70]. Antibodies to the C-terminus of PTPζ have also been used to evaluate protein expression in tumors [64, 65]. These assays do not distinguish between full-length and potentially cleaved variants of PTPζ. Nevertheless, they do demonstrate the utility of tumor targeting with molecular agents that recognize RPTP extracellular fragments.
To design novel molecular diagnostic agents for PTPζ, we can exploit what is currently known about the binding domains between PTPζ and its heterophilic ligands. For example, PTPζ binds to contactin1 via its carbonic anhydrase-like (CA) domain [20, 21]. Binding to NCAM, NgCAM/L1, and NrCAM requires the unique spacer region of PTPζ found between the CA and FNIII repeat [20]. PTPζ binding to tenascin requires its FNIII repeats [70]. Finally, pleiotrophin and midkine bind to chondroitin sulfate sugars in the PTPζ spacer region [24, 77]. In theory any of these interaction domains in either the receptor or the ligand could be targeted to design heterophilic ligand-binding peptides that recognize the PTPζ –ECD (Figure 2).
The crystal structure of the carbonic anhydrase domain of PTPζ has recently been determined [78]. A crystal structure of the interface between the CA domain of the PTPζ subfamily member, PTPγ, and its ligand, contactin4 (BIG-2) was also characterized. Notably, the same domains are involved in binding between PTPζ and contactin1. Using the now characterized surface exposed residues of the PTPζ-CA domain that is necessary for the PTPζ-contactin1 binding interface, heterophilic binding peptides could be designed to identify shed PTPζ-ECDs.
CONCLUSIONS
As CAMs and tyrosine phosphatases, RPTPs have the ability to influence cell adhesion and cell signaling. Proteolysis of full-length RPTPs to release a shed ECD capable of ligand binding and a membrane freed ICD capable of dephosphorylating tyrosine residues would have a significant impact on their biological function. First, the cleavage and shedding of RPTP ECDs could reduce cell-cell or cell-matrix adhesion mediated by the intact full-length receptors. Second, the shed ECD may antagonize cell-cell or cell-matrix adhesion by “occupying” the adjacent transmembrane receptor. Third, the shed ECD may be able to activate distinct signals by occupying different receptors on other cell types. Fourth, when associated with components of the extracellular matrix, the shed ECD may form a new substrate to promote cell migration. Finally, the released ICD fragment will change from a membrane-bound enzyme to a cytosolic/nuclear-localized enzyme, with the potential to interact with novel signaling partners.
Although only a handful of RPTPs thus far have been shown to be cleaved, given the sequence similarity between subfamily members, we believe that this may be a more generalizable phenomenon. We propose that the cleavage of RPTPs is an important step in tumor progression. Techniques such as the use of fluorophore or MRI contrast-tagged homophilic or heterophilic binding peptides that are capable of interacting with shed RPTP ECDs may be extremely useful in delineating tumor margins, the tumor microenvironment and targeting tumor tissues.
The excitement around the discovery of shed ECDs in cancer is the potential for specific molecular recognition of tumors. Detection of the shed ECDs could be exploited in a number of paradigms in cancer. First, the shed ECDs could be used in non-invasive molecular imaging strategies to detect the main tumor. Second, the shed ECDs could be used to mark the invading the tumor margin and the tumor microenvironment thus enhancing surgical resection of the invasive edge of tumors. Third if the detection of shed ECDs permits tumor cell recognition, then they can also be used for molecular targeting of tumor cells to deliver therapeutics. This last theory is summarized by the exciting concept “If we can see the tumor we can destroy the tumor”.
Acknowledgments
This research was supported by the following NIH grants: RO1-NS051520, R01-NS063971, and P30-CA043703. We would like to thank the Brady-Kalnay lab for helpful discussions.
Footnotes
CONFLICT OF INTEREST
The authors have no conflicts of interest.
References
- 1.Andersen JN, Mortensen OH, Peters GH, Drake PG, Iversen LF, Olsen OH, Jansen PG, Andersen HS, Tonks NK, Moller NP. Structural and evolutionary relationships among protein tyrosine phosphatase domains. Mol Cell Biol. 2001;21(21):7117–36. doi: 10.1128/MCB.21.21.7117-7136.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brady-Kalnay SM, Tonks NK. Protein tyrosine phosphatases as adhesion receptors. Curr Opin Cell Biol. 1995;7(5):650–7. doi: 10.1016/0955-0674(95)80106-5. [DOI] [PubMed] [Google Scholar]
- 3.Brady-Kalnay SM. Protein tyrosine phosphatases. In: Beckerle M, editor. Cell Adhesion: Frontiers in Molecular Biology. Vol. 39. Oxford University Press; Oxford, UK: 2001. pp. 217–258. [Google Scholar]
- 4.Kwon SK, Woo J, Kim SY, Kim H, Kim E. Trans-synaptic adhesions between netrin-G ligand-3 (NGL-3) and receptor tyrosine phosphatases LAR, protein-tyrosine phosphatase delta (PTPdelta), and PTPsigma via specific domains regulate excitatory synapse formation. J Biol Chem. 2010;285(18):13966–78. doi: 10.1074/jbc.M109.061127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aricescu AR, McKinnell IW, Halfter W, Stoker AW. Heparan sulfate proteoglycans are ligands for receptor protein tyrosine phosphatase sigma. Mol Cell Biol. 2002;22(6):1881–92. doi: 10.1128/MCB.22.6.1881-1892.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Grady P, Thai TC, Saito H. The laminin-nidogen complex is a ligand for a specific splice isoform of the transmembrane protein tyrosine phosphatase LAR. J Cell Biol. 1998;141(7):1675–84. doi: 10.1083/jcb.141.7.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haj F, McKinnell I, Stoker A. Retinotectal ligands for the receptor tyrosine phosphatase CRYPalpha. Mol Cell Neurosci. 1999;14(3):225–40. doi: 10.1006/mcne.1999.0785. [DOI] [PubMed] [Google Scholar]
- 8.Wang J, Bixby JL. Receptor tyrosine phosphatase-delta is a homophilic, neurite-promoting cell adhesion molecular for CNS neurons. Mol Cell Neurosci. 1999;14(4–5):370–84. doi: 10.1006/mcne.1999.0789. [DOI] [PubMed] [Google Scholar]
- 9.Yang T, Bernabeu R, Xie Y, Zhang JS, Massa SM, Rempel HC, Longo FM. Leukocyte antigen-related protein tyrosine phosphatase receptor: a small ectodomain isoform functions as a homophilic ligand and promotes neurite outgrowth. J Neurosci. 2003;23(8):3353–63. doi: 10.1523/JNEUROSCI.23-08-03353.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chagnon MJ, Uetani N, Tremblay ML. Functional significance of the LAR receptor protein tyrosine phosphatase family in development and diseases. Biochem Cell Biol. 2004;82(6):664–75. doi: 10.1139/o04-120. [DOI] [PubMed] [Google Scholar]
- 11.Brady-Kalnay SM, Flint AJ, Tonks NK. Homophilic binding of PTP mu, a receptor-type protein tyrosine phosphatase, can mediate cell-cell aggregation. J Cell Biol. 1993;122(4):961–72. doi: 10.1083/jcb.122.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng J, Wu K, Armanini M, O’Rourke N, Dowbenko D, Lasky LA. A novel protein-tyrosine phosphatase related to the homotypically adhering kappa and mu receptors. J Biol Chem. 1997;272(11):7264–77. doi: 10.1074/jbc.272.11.7264. [DOI] [PubMed] [Google Scholar]
- 13.Gebbink MF, Zondag GC, Wubbolts RW, Beijersbergen RL, van Etten I, Moolenaar WH. Cell-cell adhesion mediated by a receptor-like protein tyrosine phosphatase. J Biol Chem. 1993;268(22):16101–4. [PubMed] [Google Scholar]
- 14.Sap J, Jiang YP, Friedlander D, Grumet M, Schlessinger J. Receptor tyrosine phosphatase R-PTP-kappa mediates homophilic binding. Mol Cell Biol. 1994;14(1):1–9. doi: 10.1128/mcb.14.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zondag GC, Koningstein GM, Jiang YP, Sap J, Moolenaar WH, Gebbink MF. Homophilic interactions mediated by receptor tyrosine phosphatases mu and kappa. A critical role for the novel extracellular MAM domain. J Biol Chem. 1995;270(24):14247–50. doi: 10.1074/jbc.270.24.14247. [DOI] [PubMed] [Google Scholar]
- 16.Yu J, Becka S, Zhang P, Zhang X, Brady-Kalnay SM, Wang Z. Tumor-derived extracellular mutations of PTPRT/PTPrho are defective in cell adhesion. Mol Cancer Res. 2008;6(7):1106–13. doi: 10.1158/1541-7786.MCR-07-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Becka S, Zhang P, Craig SE, Lodowski DT, Wang Z, Brady-Kalnay SM. Characterization of the adhesive properties of the type IIb subfamily receptor protein tyrosine phosphatases. Cell Commun Adhes. 2010;17(2):34–47. doi: 10.3109/15419061.2010.487957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aricescu AR, Siebold C, Jones EY. Receptor protein tyrosine phosphatase mu: measuring where to stick. Biochem Soc Trans. 2008;36(Pt 2):167–72. doi: 10.1042/BST0360167. [DOI] [PubMed] [Google Scholar]
- 19.Ensslen-Craig SE, Brady-Kalnay SM. Receptor protein tyrosine phosphatases regulate neural development and axon guidance. Dev Biol. 2004;275(1):12–22. doi: 10.1016/j.ydbio.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 20.Sakurai T, Lustig M, Nativ M, Hemperly JJ, Schlessinger J, Peles E, Grumet M. Induction of neurite outgrowth through contactin and Nr-CAM by extracellular regions of glial receptor tyrosine phosphatase beta. J Cell Biol. 1997;136(4):907–18. doi: 10.1083/jcb.136.4.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peles E, Nativ M, Campbell PL, Sakurai T, Martinez R, Lev S, Clary DO, Schilling J, Barnea G, Plowman GD, Grumet M, Schlessinger J. The carbonic anhydrase domain of receptor tyrosine phosphatase beta is a functional ligand for the axonal cell recognition molecule contactin. Cell. 1995;82(2):251–60. doi: 10.1016/0092-8674(95)90312-7. [DOI] [PubMed] [Google Scholar]
- 22.Milev P, Friedlander DR, Sakurai T, Karthikeyan L, Flad M, Margolis RK, Grumet M, Margolis RU. Interactions of the chondroitin sulfate proteoglycan phosphacan, the extracellular domain of a receptor-type protein tyrosine phosphatase, with neurons, glia, and neural cell adhesion molecules. J Cell Biol. 1994;127(6 Pt 1):1703–15. doi: 10.1083/jcb.127.6.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barnea G, Grumet M, Milev P, Silvennoinen O, Levy JB, Sap J, Schlessinger J. Receptor tyrosine phosphatase beta is expressed in the form of proteoglycan and binds to the extracellular matrix protein tenascin. J Biol Chem. 1994;269(20):14349–52. [PubMed] [Google Scholar]
- 24.Maeda N, Nishiwaki T, Shintani T, Hamanaka H, Noda M. 6B4 proteoglycan/phosphacan, an extracellular variant of receptor-like protein-tyrosine phosphatase zeta/RPTPbeta, binds pleiotrophin/heparin-binding growth-associated molecule (HB-GAM) J Biol Chem. 1996;271(35):21446–52. doi: 10.1074/jbc.271.35.21446. [DOI] [PubMed] [Google Scholar]
- 25.Ostman A, Hellberg C, Bohmer FD. Protein-tyrosine phosphatases and cancer. Nat Rev Cancer. 2006;6(4):307–20. doi: 10.1038/nrc1837. [DOI] [PubMed] [Google Scholar]
- 26.Craig SE, Brady-Kalnay SM. Cancer Cells Cut and Run. Cancer Res. 2011;71(2):303–309. doi: 10.1158/0008-5472.CAN-10-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fortini ME. Gamma-secretase-mediated proteolysis in cell-surface-receptor signalling. Nat Rev Mol Cell Biol. 2002;3(9):673–84. doi: 10.1038/nrm910. [DOI] [PubMed] [Google Scholar]
- 28.Anders L, Mertins P, Lammich S, Murgia M, Hartmann D, Saftig P, Haass C, Ullrich A. Furin-, ADAM 10-, and gamma-Secretase-Mediated Cleavage of a Receptor Tyrosine Phosphatase and Regulation of beta-Catenin’s Transcriptional Activity. Mol Cell Biol. 2006;26(10):3917–3934. doi: 10.1128/MCB.26.10.3917-3934.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burgoyne AM, Phillips-Mason PJ, Burden-Gulley SM, Robinson S, Sloan AE, Miller RH, Brady-Kalnay SM. Proteolytic cleavage of protein tyrosine phosphatase mu regulates glioblastoma cell migration. Cancer Res. 2009;69(17):6960–8. doi: 10.1158/0008-5472.CAN-09-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chow JP, Fujikawa A, Shimizu H, Suzuki R, Noda M. Metalloproteinase- and gamma-secretase-mediated cleavage of protein-tyrosine phosphatase receptor type Z. J Biol Chem. 2008;283(45):30879–89. doi: 10.1074/jbc.M802976200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haapasalo A, Kim DY, Carey BW, Turunen MK, Pettingell WH, Kovacs DM. Preseniliin/gamma-Secretase-mediated Cleavage Regulates Association of Leukocyte-Common Antigen-related (LAR) Receptor Tyrosine Phosphatase with beta-Catenin. J Biol Chem. 2007;282(12):9063–9072. doi: 10.1074/jbc.M611324200. [DOI] [PubMed] [Google Scholar]
- 32.Gil-Henn H, Volohonsky G, Elson A. Regulation of protein-tyrosine phosphatases alpha and epsilon by calpain-mediated proteolytic cleavage. J Biol Chem. 2001;276(34):31772–9. doi: 10.1074/jbc.M103395200. [DOI] [PubMed] [Google Scholar]
- 33.Gil-Henn H, Volohonsky G, Toledano-Katchalski H, Gandre S, Elson A. Generation of novel cytoplasmic forms of protein tyrosine phosphatase epsilon by proteolytic processing and translational control. Oncogene. 2000;19(38):4375–84. doi: 10.1038/sj.onc.1203790. [DOI] [PubMed] [Google Scholar]
- 34.Edwards DR, Handsley MM, Pennington CJ. The ADAM metalloproteinases. Mol Aspects Med. 2008;29(5):258–89. doi: 10.1016/j.mam.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim YS, Kang HY, Kim JY, Oh S, Kim CH, Ryu CJ, Miyoshi E, Taniguchi N, Ko JH. Identification of target proteins of N-acetylglucosaminyl transferase V in human colon cancer and implications of protein tyrosine phosphatase kappa in enhanced cancer cell migration. Proteomics. 2006;6(4):1187–91. doi: 10.1002/pmic.200500400. [DOI] [PubMed] [Google Scholar]
- 36.Streuli M, Krueger NX, Ariniello PD, Tang M, Munro JM, Blattler WA, Adler DA, Disteche CM, Saito H. Expression of the receptor-linked protein tyrosine phosphatase LAR: proteolytic cleavage and shedding of the CAM-like extracellular region. Embo J. 1992;11(3):897–907. doi: 10.1002/j.1460-2075.1992.tb05128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu Q, Lenardo T, Weinberg RA. The N-terminal and C-terminal domains of a receptor tyrosine phosphatase are associated by non-covalent linkage. Oncogene. 1992;7(6):1051–7. [PubMed] [Google Scholar]
- 38.Serra-Pages C, Saito H, Streuli M. Mutational analysis of proprotein processing, subunit association, and shedding of the LAR transmembrane protein tyrosine phosphatase. J Biol Chem. 1994;269(38):23632–41. [PubMed] [Google Scholar]
- 39.Aicher B, Lerch MM, Muller T, Schilling J, Ulrich A. Cellular redistribution of protein tyrosine phosphatases LAR and PTPsigma by inducible proteolytic processing. J Cell Biol. 1997;138:681–696. doi: 10.1083/jcb.138.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruhe JE, Streit S, Hart S, Ullrich A. EGFR signaling leads to downregulation of PTP-LAR via TACE-mediated proteolytic processing. Cell Signal. 2006;18(9):1515–27. doi: 10.1016/j.cellsig.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 41.Pulido R, Krueger NX, Serra-Pages C, Saito H, Streuli M. Molecular characterization of the human transmembrane protein-tyrosine phosphatase delta. Evidence for tissue-specific expression of alternative human transmembrane protein-tyrosine phosphatase delta isoforms. J Biol Chem. 1995;270(12):6722–8. doi: 10.1074/jbc.270.12.6722. [DOI] [PubMed] [Google Scholar]
- 42.McArdle L, Rafferty MM, Satyamoorthy K, Maelandsmo GM, Dervan PA, Herlyn M, Easty DJ. Microarray analysis of phosphatase gene expression in human melanoma. Br J Dermatol. 2005;152(5):925–30. doi: 10.1111/j.1365-2133.2005.06454.x. [DOI] [PubMed] [Google Scholar]
- 43.Yang T, Zhang JS, Massa SM, Han X, Longo FM. Leukocyte common antigen-related tyrosine phosphatase receptor: increased expression and neuronal-type splicing in breast cancer cells and tissue. Mol Carcinog. 1999;25(2):139–49. [PubMed] [Google Scholar]
- 44.Konishi N, Tsujikawa K, Yamamoto H, Ishida E, Nakamura M, Shimada K, Yane K, Yamashita H, Noguchi S. Overexpression of leucocyte common antigen (LAR) P-subunit in thyroid carcinomas. Br J Cancer. 2003;88(8):1223–8. doi: 10.1038/sj.bjc.6600876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, Fulton L, Fulton RS, Zhang Q, Wendl MC, Lawrence MS, Larson DE, Chen K, Dooling DJ, Sabo A, Hawes AC, Shen H, Jhangiani SN, Lewis LR, Hall O, Zhu Y, Mathew T, Ren Y, Yao J, Scherer SE, Clerc K, Metcalf GA, Ng B, Milosavljevic A, Gonzalez-Garay ML, Osborne JR, Meyer R, Shi X, Tang Y, Koboldt DC, Lin L, Abbott R, Miner TL, Pohl C, Fewell G, Haipek C, Schmidt H, Dunford-Shore BH, Kraja A, Crosby SD, Sawyer CS, Vickery T, Sander S, Robinson J, Winckler W, Baldwin J, Chirieac LR, Dutt A, Fennell T, Hanna M, Johnson BE, Onofrio RC, Thomas RK, Tonon G, Weir BA, Zhao X, Ziaugra L, Zody MC, Giordano T, Orringer MB, Roth JA, Spitz MR, Wistuba, Ozenberger B, Good PJ, Chang AC, Beer DG, Watson MA, Ladanyi M, Broderick S, Yoshizawa A, Travis WD, Pao W, Province MA, Weinstock GM, Varmus HE, Gabriel SB, Lander ES, Gibbs RA, Meyerson M, Wilson RK. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455(7216):1069–75. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sato M, Takahashi K, Nagayama K, Arai Y, Ito N, Okada M, Minna JD, Yokota J, Kohno T. Identification of chromosome arm 9p as the most frequent target of homozygous deletions in lung cancer. Genes Chromosomes Cancer. 2005;44(4):405–14. doi: 10.1002/gcc.20253. [DOI] [PubMed] [Google Scholar]
- 47.Solomon DA, Kim JS, Cronin JC, Sibenaller Z, Ryken T, Rosenberg SA, Ressom H, Jean W, Bigner D, Yan H, Samuels Y, Waldman T. Mutational inactivation of PTPRD in glioblastoma multiforme and malignant melanoma. Cancer Res. 2008;68(24):10300–6. doi: 10.1158/0008-5472.CAN-08-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stallings RL, Nair P, Maris JM, Catchpoole D, McDermott M, O’Meara A, Breatnach F. High-resolution analysis of chromosomal breakpoints and genomic instability identifies PTPRD as a candidate tumor suppressor gene in neuroblastoma. Cancer Res. 2006;66(7):3673–80. doi: 10.1158/0008-5472.CAN-05-4154. [DOI] [PubMed] [Google Scholar]
- 49.Veeriah S, Brennan C, Meng S, Singh B, Fagin JA, Solit DB, Paty PB, Rohle D, Vivanco I, Chmielecki J, Pao W, Ladanyi M, Gerald WL, Liau L, Cloughesy TC, Mischel PS, Sander C, Taylor B, Schultz N, Major J, Heguy A, Fang F, Mellinghoff IK, Chan TA. The tyrosine phosphatase PTPRD is a tumor suppressor that is frequently inactivated and mutated in glioblastoma and other human cancers. Proc Natl Acad Sci U S A. 2009;106(23):9435–40. doi: 10.1073/pnas.0900571106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peedicayil A, Vierkant RA, Hartmann LC, Fridley BL, Fredericksen ZS, White KL, Elliott EA, Phelan CM, Tsai YY, Berchuck A, Iversen ES, Jr, Couch FJ, Peethamabaran P, Larson MC, Kalli KR, Kosel ML, Shridhar V, Rider DN, Liebow M, Cunningham JM, Schildkraut JM, Sellers TA, Goode EL. Risk of ovarian cancer and inherited variants in relapse-associated genes. PLoS One. 2010;5(1):e8884. doi: 10.1371/journal.pone.0008884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suarez Pestana E, Tenev T, Gross S, Stoyanov B, Ogata M, Bohmer FD. The transmembrane protein tyrosine phosphatase RPTPsigma modulates signaling of the epidermal growth factor receptor in A431 cells. Oncogene. 1999;18(28):4069–79. doi: 10.1038/sj.onc.1202794. [DOI] [PubMed] [Google Scholar]
- 52.Burgoyne AM, Palomo JM, Phillips-Mason PJ, Burden-Gulley SM, Major DL, Zaremba A, Robinson S, Sloan AE, Vogelbaum MA, Miller RH, Brady-Kalnay SM. PTPmu suppresses glioma cell migration and dispersal. Neuro Oncol. 2009 doi: 10.1215/15228517-2009-019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burden-Gulley SM, Gates TJ, Burgoyne AM, Cutter JL, Lodowski DT, Robinson S, Sloan AE, Miller RH, Basilion JP, Brady-Kalnay SM. A novel molecular diagnostic of glioblastomas: detection of an extracellular fragment of protein tyrosine phosphatase mu. Neoplasia. 2010;12(4):305–16. doi: 10.1593/neo.91940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McArdle L, Rafferty M, Maelandsmo GM, Bergin O, Farr CJ, Dervan PA, O’Loughlin S, Herlyn M, Easty DJ. Protein tyrosine phosphatase genes downregulated in melanoma. J Invest Dermatol. 2001;117(5):1255–60. doi: 10.1046/j.0022-202x.2001.01534.x. [DOI] [PubMed] [Google Scholar]
- 55.Flavell JR, Baumforth KR, Wood VH, Davies GL, Wei W, Reynolds GM, Morgan S, Boyce A, Kelly GL, Young LS, Murray PG. Down-regulation of the TGF-beta target gene, PTPRK, by the Epstein-Barr virus encoded EBNA1 contributes to the growth and survival of Hodgkin lymphoma cells. Blood. 2008;111(1):292–301. doi: 10.1182/blood-2006-11-059881. [DOI] [PubMed] [Google Scholar]
- 56.Harder KW, Moller NP, Peacock JW, Jirik FR. Protein-tyrosine phosphatase alpha regulates Src family kinases and alters cell-substratum adhesion. J Biol Chem. 1998;273(48):31890–900. doi: 10.1074/jbc.273.48.31890. [DOI] [PubMed] [Google Scholar]
- 57.Su J, Muranjan M, Sap J. Receptor protein tyrosine phosphatase alpha activates Src-family kinases and controls integrin-mediated responses in fibroblasts. Curr Biol. 1999;9(10):505–11. doi: 10.1016/s0960-9822(99)80234-6. [DOI] [PubMed] [Google Scholar]
- 58.Zeng L, D’Alessandri L, Kalousek MB, Vaughan L, Pallen CJ. Protein tyrosine phosphatase alpha (PTPalpha) and contactin form a novel neuronal receptor complex linked to the intracellular tyrosine kinase fyn. J Cell Biol. 1999;147(4):707–14. doi: 10.1083/jcb.147.4.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pallen CJ. Protein tyrosine phosphatase alpha (PTPalpha): a Src family kinase activator and mediator of multiple biological effects. Curr Top Med Chem. 2003;3(7):821–35. doi: 10.2174/1568026033452320. [DOI] [PubMed] [Google Scholar]
- 60.Peles E, Schlessinger J, Grumet M. Multi-ligand interactions with receptor-like protein tyrosine phosphatase beta: implications for intercellular signaling. Trends Biochem Sci. 1998;23(4):121–4. doi: 10.1016/s0968-0004(98)01195-5. [DOI] [PubMed] [Google Scholar]
- 61.Levy JB, Canoll PD, Silvennoinen O, Barnea G, Morse B, Honegger AM, Huang JT, Cannizzaro LA, Park SH, Druck T, et al. The cloning of a receptor-type protein tyrosine phosphatase expressed in the central nervous system. J Biol Chem. 1993;268(14):10573–81. [PubMed] [Google Scholar]
- 62.Foehr ED, Lorente G, Kuo J, Ram R, Nikolich K, Urfer R. Targeting of the receptor protein tyrosine phosphatase beta with a monoclonal antibody delays tumor growth in a glioblastoma model. Cancer Res. 2006;66(4):2271–8. doi: 10.1158/0008-5472.CAN-05-1221. [DOI] [PubMed] [Google Scholar]
- 63.Yamakawa T, Kurosawa N, Kadomatsu K, Matsui T, Itoh K, Maeda N, Noda M, Muramatsu T. Levels of expression of pleiotrophin and protein tyrosine phosphatase zeta are decreased in human colorectal cancers. Cancer Lett. 1999;135(1):91–6. doi: 10.1016/s0304-3835(98)00275-4. [DOI] [PubMed] [Google Scholar]
- 64.Hagerstrand D, Smits A, Eriksson A, Sigurdardottir S, Olofsson T, Hartman M, Nister M, Kalimo H, Ostman A. Gene expression analyses of grade II gliomas and identification of rPTPbeta/zeta as a candidate oligodendroglioma marker. Neuro Oncol. 2008;10(1):2–9. doi: 10.1215/15228517-2007-041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Muller S, Kunkel P, Lamszus K, Ulbricht U, Lorente GA, Nelson AM, von Schack D, Chin DJ, Lohr SC, Westphal M, Melcher T. A role for receptor tyrosine phosphatase zeta in glioma cell migration. Oncogene. 2003;22(43):6661–8. doi: 10.1038/sj.onc.1206763. [DOI] [PubMed] [Google Scholar]
- 66.Ulbricht U, Brockmann MA, Aigner A, Eckerich C, Muller S, Fillbrandt R, Westphal M, Lamszus K. Expression and function of the receptor protein tyrosine phosphatase zeta and its ligand pleiotrophin in human astrocytomas. J Neuropathol Exp Neurol. 2003;62(12):1265–75. doi: 10.1093/jnen/62.12.1265. [DOI] [PubMed] [Google Scholar]
- 67.Ulbricht U, Eckerich C, Fillbrandt R, Westphal M, Lamszus K. RNA interference targeting protein tyrosine phosphatase zeta/receptor-type protein tyrosine phosphatase beta suppresses glioblastoma growth in vitro and in vivo. J Neurochem. 2006;98(5):1497–506. doi: 10.1111/j.1471-4159.2006.04022.x. [DOI] [PubMed] [Google Scholar]
- 68.Maeda N, Noda M. Involvement of receptor-like protein tyrosine phosphatase zeta/RPTPbeta and its ligand pleiotrophin/heparin-binding growth-associated molecule (HB-GAM) in neuronal migration. J Cell Biol. 1998;142(1):203–16. doi: 10.1083/jcb.142.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sakurai T, Friedlander DR, Grumet M. Expression of polypeptide variants of receptor-type protein tyrosine phosphatase beta: the secreted form, phosphacan, increases dramatically during embryonic development and modulates glial cell behavior in vitro. J Neurosci Res. 1996;43(6):694–706. doi: 10.1002/(SICI)1097-4547(19960315)43:6<694::AID-JNR6>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 70.Adamsky K, Schilling J, Garwood J, Faissner A, Peles E. Glial tumor cell adhesion is mediated by binding of the FNIII domain of receptor protein tyrosine phosphatase beta (RPTPbeta) to tenascin C. Oncogene. 2001;20(5):609–18. doi: 10.1038/sj.onc.1204119. [DOI] [PubMed] [Google Scholar]
- 71.Brady-Kalnay SM, Tonks NK. Identification of the homophilic binding site of the receptor protein tyrosine phosphatase PTP mu. J Biol Chem. 1994;269(45):28472–7. [PubMed] [Google Scholar]
- 72.Aricescu AR, Hon WC, Siebold C, Lu W, van der Merwe PA, Jones EY. Molecular analysis of receptor protein tyrosine phosphatase mu-mediated cell adhesion. Embo J. 2006;25(4):701–12. doi: 10.1038/sj.emboj.7600974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aricescu AR, Siebold C, Choudhuri K, Chang VT, Lu W, Davis SJ, van der Merwe PA, Jones EY. Structure of a tyrosine phosphatase adhesive interaction reveals a spacer-clamp mechanism. Science. 2007;317(5842):1217–20. doi: 10.1126/science.1144646. [DOI] [PubMed] [Google Scholar]
- 74.Cismasiu VB, Denes SA, Reilander H, Michel H, Szedlacsek SE. The MAM (meprin/A5-protein/PTPmu) domain is a homophilic binding site promoting the lateral dimerization of receptor-like protein-tyrosine phosphatase mu. J Biol Chem. 2004;279(26):26922–31. doi: 10.1074/jbc.M313115200. [DOI] [PubMed] [Google Scholar]
- 75.Wang Z, Shen D, Parsons DW, Bardelli A, Sager J, Szabo S, Ptak J, Silliman N, Peters BA, van der Heijden MS, Parmigiani G, Yan H, Wang TL, Riggins G, Powell SM, Willson JK, Markowitz S, Kinzler KW, Vogelstein B, Velculescu VE. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science. 2004;304(5674):1164–6. doi: 10.1126/science.1096096. [DOI] [PubMed] [Google Scholar]
- 76.Zhang P, Becka S, Craig SE, Lodowski DT, Brady-Kalnay SM, Wang Z. Cancer-derived mutations in the fibronectin III repeats of PTPRT/PTPrho inhibit cell-cell aggregation. Cell Commun Adhes. 2009;16(5–6):146–53. doi: 10.3109/15419061003653771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maeda N, Ichihara-Tanaka K, Kimura T, Kadomatsu K, Muramatsu T, Noda M. A receptor-like protein-tyrosine phosphatase PTPzeta/RPTPbeta binds a heparin-binding growth factor midkine. Involvement of arginine 78 of midkine in the high affinity binding to PTPzeta. J Biol Chem. 1999;274(18):12474–9. doi: 10.1074/jbc.274.18.12474. [DOI] [PubMed] [Google Scholar]
- 78.Bouyain S, Watkins DJ. The protein tyrosine phosphatases PTPRZ and PTPRG bind to distinct members of the contactin family of neural recognition molecules. Proc Natl Acad Sci U S A. 2010;107(6):2443–8. doi: 10.1073/pnas.0911235107. [DOI] [PMC free article] [PubMed] [Google Scholar]